Abstract
Smoking tobacco is a known risk factor for the development of colorectal cancer, and for mortality associated with the disease. Smoking has been reported to be associated with changes in DNA methylation in blood and in lung tumour tissues, although there has been scant investigation of how epigenetic factors may be implicated in the increased risk of developing colorectal cancer. To identify epigenetic changes associated with smoking behaviours, we performed epigenome-wide analysis of DNA methylation in colorectal tumours from 36 never smokers, 47 former smokers and 13 active smokers, and adjacent mucosa from 49 never smokers, 64 former smokers and 18 active smokers. Our analyses identified 15 CpG sites within the APC 1A promoter that were significantly hypermethylated and 14 CpG loci within the NFATC1 gene body that were significantly hypomethylated (pLIS<1×10−5) in tumours of active smokers. The APC 1A promoter was hypermethylated in 7 of 36 tumours from never smokers (19%), 12 of 47 tumours from former smokers (26%), and 8 of 13 tumours from active smokers (62%). Promoter hypermethylation was positively associated with duration of smoking (Spearman rank correlation, ρ=0.26, p=0.03) and was confined to tumours, with hypermethylation never being observed in adjacent mucosa. Further analysis of adjacent mucosa revealed significant hypomethylation of four loci associated with the TNXB gene in tissue from active smokers. Our findings provide exploratory evidence for hypermethylation of the key tumour suppressor gene APC being implicated in smoking-associated colorectal carcinogenesis. Further work is required to establish the validity of our observations in independent cohorts.
Keywords: Smoking, Tobacco, Colorectal cancer, Epigenetics, DNA methylation, APC
Introduction
Smoking tobacco is a risk factor for many forms of cancer, including colorectal cancer (CRC). Ever-smokers, which includes both current and former smokers, have an 18% increase in the risk of developing the disease relative to individuals who have never smoked [1], and the risk is greatest for the development of tumours in the rectum. In addition to increased incidence, active smokers have a 23% greater risk of CRC-related mortality [2] and patients who are former smokers still display increased risk of all-cause mortality [3]. The duration and intensity of smoking are known to modify risk, with individuals who have smoked for ≥30 years and those with ≥20 pack-years of smoking each displaying a 40% increase in risk of CRC-related mortality [3]. However, the mechanisms by which smoking tobacco increases CRC risk have not been elucidated. It has been hypothesised that the carcinogenic products of cigarette smoke may reach the colorectum through the blood, and are implicated in the early initiation of cancer, as opposed to furthering the development of existing adenomas [4].
Smoking is associated with alterations in DNA methylation, an epigenetic modifier of gene expression, in healthy individuals. Such epigenetic events display tissue-specificity [5] and differ by ethnicity [6,7], and can serve as markers of long-term exposure to tobacco smoke [8]. Several studies examining the blood of smokers have reported differential methylation of loci within the aryl hydrocarbon receptor repressor (AHRR) gene [6,9], a putative tumour-suppressor which mediates the detoxification of products in cigarette smoke, and the coagulation factor II (thrombin) receptor-like 3 (F2RL3) gene [6,8–10], implicated in blood clotting. Associations have been identified between smoking-related changes in DNA methylation of AHRR, F2RL3 and LINE1 elements measured in blood and the risk of cancer [11] and mortality from the disease [12].
Further to these observations in healthy individuals there is evidence that smoking is associated with epigenetic changes in tumour tissue. Epigenome-wide association studies have identified distinct methylation profiles in lung tumours from smokers and non-smokers [13], and candidate-gene approaches have identified smoking-related changes in the methylation of cyclin-dependent kinase inhibitor 2A (CDKN2A/p16) and runt-related transcription factor 3 (RUNX3) in bladder tumours [14,15] and CDKN2A/p16 and O-6-methylguanine-DNA methyltransferase (MGMT) in lung tumours [16]. Smoking-related epigenetic events may occur early in carcinogenesis, as demonstrated by their observation in stage I non-small cell lung cancers [17]. However, the evidence for smoking-associated epigenetic dysregulation in CRC is currently limited. Smoking has been reported to be associated with microsatellite instability and positive CpG island methylator phenotype (CIMP) status [18], but there has otherwise been scant research into DNA methylation in colorectal tumours classified by smoking status.
In this study, we investigated whether epigenetic factors may be implicated in the increased risk of CRC among tobacco smokers by analysing epigenetic patterns in colorectal tumours and neighbouring mucosa in relation to smoking behaviours. We utilised the Illumina HumanMethylation450 microarray platform to analyse DNA methylation in samples taken from a total of 137 colorectal cancer patients, 51 of whom had never smoked (‘never smokers’), 68 who had been smokers but had ceased at least two years prior to cancer diagnosis (‘former smokers’), and 18 who smoked at the point of diagnosis (‘active smokers’). We report that promoter 1A of the APC gene, commonly inactivated in CRC, is hypermethylated in the tumours of active smokers. Methylation of this region is associated with duration of smoking, and hypermethylation (β>0.2) was never observed in adjacent mucosa. Our results suggest that the increased risk of CRC development among smokers may progress through epigenetic inactivation of the key tumour suppressor gene APC.
Material and Methods
The ColoCare Study
The ColoCare consortium is a multicentre initiative of interdisciplinary research on outcomes associated with colorectal cancer, with sites at the Fred Hutchison Cancer Research Center (Seattle, USA), Moffit Cancer Center (Tampa, USA), and from 2010 at the German Cancer Research Center (Heidelberg, Germany). This study exclusively focussed upon patients recruited in Heidelberg. ColoCare has been approved by the ethics committee of the University of Heidelberg medical faculty. Patients were enrolled to this prospective cohort at the point of diagnosis, having given informed consent, with biospecimens and data collected at regularly scheduled intervals of 3, 6, 12, 24 and 36 months post-surgery. Medical factors were abstracted from patients’ charts and records from the University Hospital of Heidelberg. Data on dietary habits, exercise and physical activity, smoking habits, medication, socio-demographic information, and quality of life were collected via questionnaires. To date, 500 patients have been recruited at the Heidelberg site.
Tissue samples
Tissue samples were collected from patients undergoing surgery at the University Hospital of Heidelberg, and were reviewed by pathologists to ensure their quality and origin. Tumour samples were collected from 36 patients who had never smoked, 47 who were former smokers, and 13 who were active smokers at the point of diagnosis. Mucosa was taken from adjacent to tumours from 49 never smokers, 64 former smokers and 18 active smokers. A summary of patient characteristics is provided in Table 1.
Table 1.
Clinical and demographic characteristics of the patients.
| Adjacent mucosa | Tumour | ||||||
|---|---|---|---|---|---|---|---|
| Never | Former | Active | Never | Former | Active | ||
| Patients | n | 49 | 64 | 18 | 36 | 47 | 13 |
| Age | Mean | 63.7 | 65.5 | 56.6 | 63.9 | 65.1 | 59.6 |
| SD | 11.8 | 10.9 | 12.5 | 11.5 | 10.3 | 9.8 | |
| Range | 34 – 82 | 38 – 89 | 22 – 79 | 41 – 82 | 38 – 89 | 35 – 79 | |
| Gender | Male | 24 | 48 | 10 | 21 | 37 | 6 |
| Female | 25 | 16 | 8 | 15 | 10 | 7 | |
| Stage | I | 8 | 8 | 4 | 3 | 5 | 1 |
| II | 15 | 24 | 7 | 12 | 17 | 7 | |
| III | 13 | 18 | 5 | 12 | 14 | 4 | |
| IV | 11 | 14 | 2 | 9 | 11 | 1 | |
| Pack-years | Mean (years) | - | 11.5 | 18.7 | - | 12.6 | 16.4 |
| 0 – 9 (n) | - | 31 | 5 | - | 22 | 4 | |
| 10 – 19 (n) | - | 15 | 4 | - | 11 | 3 | |
| ≥20 (n) | - | 11 | 6 | - | 11 | 4 | |
| Duration | Mean (years) | - | 18.8 | 31.8 | - | 19.6 | 37.6 |
| 0 – 9 (n) | - | 18 | 2 | - | 11 | 0 | |
| 10 – 19 (n) | - | 17 | 3 | - | 11 | 2 | |
| 20 – 29 (n) | - | 13 | 2 | - | 8 | 0 | |
| ≥30 (n) | - | 15 | 17 | - | 11 | 9 | |
Data are provided regarding the age (mean, standard deviation, and range), gender, tumour stage and pack-years of smoking for the patients according to smoking status at the point of cancer diagnosis.
DNA isolation
DNA was extracted from fresh-frozen tissue using the QIAamp AllPrep DNA/RNA mini kit (Qiagen) according to the manufacturer’s instructions.
Illumina Infinium HumanMethylation450 BeadChip microarrays
DNA microarrays were performed at the Genomics and Proteomics Core Facility at the German Cancer Research Center (Heidelberg, Germany). One μg of genomic DNA was bisulphite-converted using the EZ DNA Methylation kit (Zymo Research) according to the manufacturer’s instructions. The microarray assays were then performed according to the Illumina Infinium HD Methylation protocol.
Microarray data analysis
Microarray data was pre-processed using the Illumina Genome Studio software program before analysis using the R minfi package. Background correction and dye-bias normalisation were performed using noob [19], and functional normalisation was performed to remove batch effects and inner technical variability and adjust for Type I/II probe fluorescence effect, as described elsewhere [20]. Prior to background correction and normalisation, probes with detection p values >0.01 in 10% of samples (n=662) or bead counts less than three in 10% of samples (n=162) were removed. Probes with SNPs within 10 bp of the target CpG with minor allele frequencies of >0.01 (n=19,099) and mapping to the X and Y chromosomes (n=11,150) were removed. Subsequently, a total of 456,144 probes were taken forward for analysis.
Loci that were differentially methylated by smoking behaviours were identified by fitting a linear least-squares regression model across the conditions followed by computing moderated t-statistics for every CpG site, as described in the limma pipeline [21]. Due to the non-independent structure of the univariate t-statistics, we used a non-homogenous hidden Markov model (NHMM) to incorporate the dependence coming from the chromosomal positions of CpGs in the test statistics, as proposed and described elsewhere [22]. In brief, t-statistics were z-score transformed and distances (base pairs) between CpGs calculated and used as dependence structure in the NHMM. The NHMM parameters were estimated by expectation maximisation with randomised initial values. To avoid local maxima in the maximisation algorithm we used 30 initialisations and chose the initialisation with the smallest Bayesian information criteria (BIC). This provides a reproducible local index of significance (LIS), as previously defined [23], and can be interpreted as dependence corrected p value (pLIS). For computational efficiency we performed the analysis by chromosome and pooled the results afterwards, with significance defined as pLIS<1×10−5. The pLIS scores were computed using the R package NHMMfdr. Comparisons were made between never smokers and active smokers and between never smokers and former smokers in tumour and adjacent mucosa tissues. To identify loci that are differentially methylated between tumours and adjacent mucosa in a smoking-specific manner, we compared the differences in active smokers of tumour and mucosa with the differences among never smokers of tumour and mucosa. All analyses were adjusted for age and sex in the linear regression model.
The methylation microarray dataset is available from the NCBI Gene Expression Omnibus repository (accession number: GSE101764).
Identification of probe-associated SNPs
To account for false positives stemming from genetic variation, we used the UCSC Genome Browser and NCBI dbSNP databases [24,25] to identify single nucleotide polymorphisms (SNPs) within the 50-mer probes of the microarray for sites identified as significantly differentially methylated by smoking behaviours. The unconverted DNA sequences (‘SourceSeq’) for each significantly-different probe in tumour tissue and adjacent mucosa were extracted from the GenomeStudio output file and were used to perform a BLAT search using the UCSC Genome Browser [24]. The minor allele frequencies for all SNPs located within the probe sequences were identified using the UCSC Genome Browser and the NCBI dbSNP database [24,25]. Data from across all ethnicities or, where available, European populations was recorded, using estimates from studies with the largest sample sizes.
Statistical analyses
Associations between DNA methylation and smoking habits were calculated using data on pack-years and duration of smoking for each patient, and time since cessation among former smokers. Detailed data on smoking habits was available for 87 patients from whom tumour tissue was taken and 115 patients providing adjacent mucosa. Associations between DNA methylation (beta values) and intensity (pack-years) and duration (years) of smoking were identified using Spearman’s rank correlation coefficient, as were associations with time since cessation of smoking (years). Associations between tumour location and APC promoter 1A hypermethylation were calculated using Fisher’s exact test. Statistical significance was defined as p<0.05.
Results
Characteristics of the patients
Details of the CRC patients from whom samples of colorectal tumours and adjacent mucosa were obtained are provided in Table 1. Tumour tissue was obtained from 36 never smokers, 47 former smokers and 13 active smokers, while adjacent mucosa was taken from 49 patients who were never smokers, 64 who were former smokers and 18 active smokers. Matched pairs of tumour and adjacent mucosa tissue were available for 89 of the patients (33 never smokers, 43 former smokers and 13 active smokers). The mean level of smoking was 18.7 pack-years among active smokers and 12.7 pack-years among former smokers. The mean duration of smoking was 37.6 years among active smokers and 19.6 years among former smokers.
The APC promoter 1A is hypermethylated in the tumours of active smokers
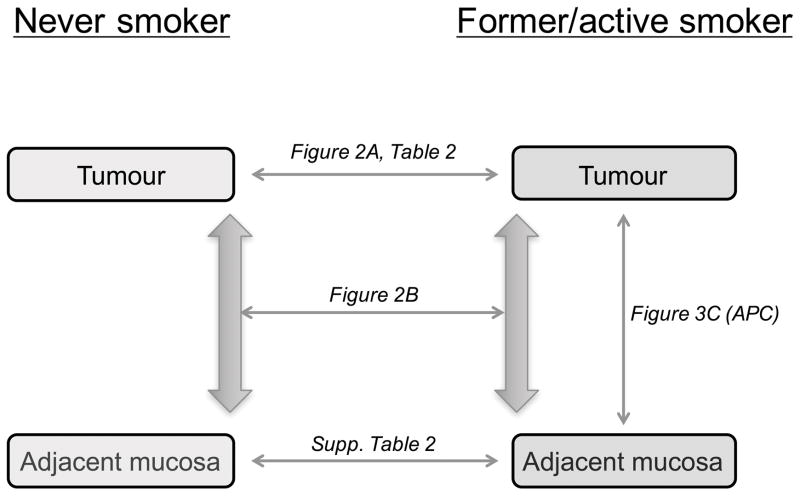
Epigenome-wide analysis of DNA methylation in 96 colorectal tumours and 131 samples of adjacent mucosa was performed using the Illumina Infinium HumanMethylation450 BeadChip microarray platform at the German Cancer Research Center Genomics and Proteomics Core Facility (Heidelberg, Germany). An overview of performed analyses with the different comparisons is shown in Figure 1.
Figure 1.
Overview of analyses by smoking behaviours in tumours and adjacent mucosa. Differentially methylated sites between smokers and never smokers were identified in tumour tissue and in adjacent mucosa. Further analyses were performed to identify sites displaying smoking-specific differential methylation between tumours and adjacent mucosa.
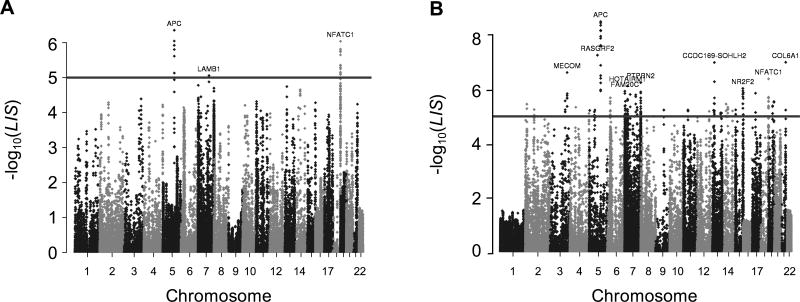
We identified 21 CpG sites where methylation was significantly different between tumours from patients who had never smoked and those who were active smokers at the point of diagnosis (Figure 2A). These mapped to 14 loci within the NFATC1 gene, 6 within the APC gene, and 1 within LAMB1 (Table 2). The 14 loci that mapped to the NFATC1 gene were distributed throughout the gene body and predominantly located in CpG islands. In contrast, each of the six loci associated with APC corresponded to the 1A promoter region and were within a span of 83 bp. Median beta values at each of the six CpG sites were 0.41–0.53 higher in active smokers in comparison to never smokers. No CpG sites were differentially methylated between tumours from former smokers and never smokers.
Figure 2.
Manhattan plots showing differentially methylated sites between never and active smokers. Results of the analyses between tumours from never and active smokers (A) and differential methylation between tumours and adjacent mucosa unique to active smokers (B). Gene symbols of the genes associated with the most significantly different sites are provided. The threshold (line) represents statistical significance (pLIS<1×10−5)
Table 2.
CpG sites with differential methylation by smoking status in tumours
| Probe ID | Chromosomal location | Gene | Gene region | Island status | Mean β-value | LIS p value | |
|---|---|---|---|---|---|---|---|
| Never | Active | ||||||
| cg08571859 | chr5:112073350 | APC | TSS1500 | Open sea | 0.11 | 0.36 | 7.4 × 10−6 |
| cg14511739 | chr5:112073373 | APC | TSS200 | Open sea | 0.11 | 0.39 | 1.2 × 10−6 |
| cg22035501 | chr5:112073426 | APC | TSS200 | Open sea | 0.12 | 0.42 | 4.4 × 10−7 |
| cg11613015 | chr5:112073433 | APC | TSS200 | Open sea | 0.10 | 0.34 | 9.0 × 10−7 |
| cg14479889 | chr5:112073426 | APC | TSS200 | Open sea | 0.12 | 0.38 | 1.6 × 10−6 |
| cg16970232 | chr5:112073433 | APC | TSS200 | Open sea | 0.13 | 0.40 | 2.4 × 10−6 |
| cg04744624 | chr7: 107641770 | LAMB1 | Body | N_Shore | 0.23 | 0.41 | 8.8 × 10−6 |
| cg15138382 | chr18: 77186504 | NFATC1 | Body/5′ UTR | Island | 0.89 | 0.75 | 1.8 × 10−6 |
| cg05302701 | chr18: 77196320 | NFATC1 | Body/5′ UTR | Island | 0.81 | 0.68 | 4.4 × 10−6 |
| cg18092363 | chr18: 77202678 | NFATC1 | Body/5′ UTR | Island | 0.94 | 0.85 | 7.9 × 10−6 |
| cg26550337 | chr18: 77203542 | NFATC1 | Body/5′ UTR | Island | 0.81 | 0.70 | 1.5 × 10−6 |
| cg26100137 | chr18: 77203667 | NFATC1 | Body/5′ UTR | Island | 0.97 | 0.90 | 2.3 × 10−6 |
| cg22279865 | chr18: 77204561 | NFATC1 | Body/5′ UTR | S_Shore | 0.93 | 0.87 | 4.5 × 10−6 |
| cg00445548 | chr18: 77207209 | NFATC1 | Body/5′ UTR | Island | 0.93 | 0.82 | 7.1 × 10−6 |
| cg02675550 | chr18: 77208807 | NFATC1 | Body/5′ UTR | Island | 0.86 | 0.75 | 5.6 × 10−6 |
| cg21242663 | chr18: 77208881 | NFATC1 | Body | Island | 0.90 | 0.81 | 8.1 × 10−6 |
| cg25595641 | chr18: 77208991 | NFATC1 | Body | Island | 0.94 | 0.86 | 1.8 × 10−6 |
| cg21806238 | chr18: 77210990 | NFATC1 | Body | Island | 0.92 | 0.84 | 5.1 × 10−6 |
| cg16253249 | chr18: 77211212 | NFATC1 | Body | Island | 0.81 | 0.74 | 1.8 × 10−6 |
| cg03239925 | chr18: 77230795 | NFATC1 | Body | Island | 0.74 | 0.63 | 7.0 × 10−6 |
| cg22324981 | chr18: 77283493 | NFATC1 | Body | N_Shore | 0.80 | 0.58 | 9.2 × 10−7 |
Loci with significantly different methylation between tumours from never smokers and active smokers are listed, including Illumina annotation data. Median beta values are provided, along with pLIS values.
Smoking-specific differential methylation between tumours and adjacent mucosa
We performed further analysis to identify genes that may be implicated in smoking-associated carcinogenesis by identifying loci that are differentially methylated between tumours and adjacent mucosa among active smokers but not never smokers. We identified 148 loci that were significantly differentially methylated between these conditions (Figure 2B, supplementary material, Table S1). This included all six of the loci previously identified within the APC 1A promoter and 9 of the 14 sites previously identified within the NFATC1 gene body. The nine sites with greatest statistical significance all mapped to the APC 1A promoter, and a further six significantly differentially methylated sites were also identified within this region. The average beta values in tumours and adjacent mucosa from active smokers differed by >0.24 at each of the 15 sites of the APC 1A promoter, while differing by <0.10 in the same tissues from never smokers. Other genes prominently identified by this analysis included receptor-type tyrosine-protein phosphatase N2 (PTPRN2) and sidekick cell adhesion molecule 1 (SDK1).
APC promoter 1A methylation and tumour pathology
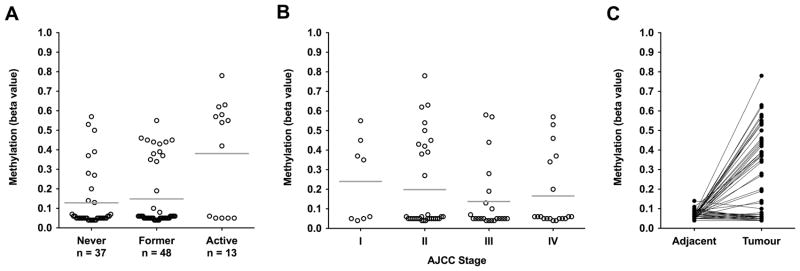
Our epigenome-wide analysis identified the APC promoter 1A as the leading target for smoking-associated methylation changes. This was confirmed by cross-validation analysis, which identified this region as the most predictive to distinguish between tumours from never and active smokers (supplementary material, Figure S1). We sought to further characterise methylation of this region by tumour pathology and smoking behaviours. Expanded analysis across the 15 significantly differentially methylated loci mapping to the APC 1A promoter revealed distinct hypermethylation in some patients (Figure 3A). Defining hypermethylation as mean beta values of >0.2, in accordance with our observed values across all tumours, the APC 1A promoter was hypermethylated in 7 of 36 tumours from never smokers (19%), 12 of 47 tumours from former smokers (26%), and 8 of 13 tumours from active smokers (62%). Across all smoking behaviours, hypermethylation was observed at all AJCC stages, including 4 of 8 stage I tumours (Figure 3B), and was more common in tumours located in the rectum (14 of 38 tumours, 37%) and distal colon (8 of 25, 32%) than in the proximal colon (2 of 14, 14%), but not significantly so (Fisher’s exact test, p=0.18 and p=0.28 respectively). We identified no associations between methylation at the six differentially methylated loci within the APC 1A promoter and alcohol consumption (grams/day) or BMI (both p > 0.05). Hypermethylation of the 1A promoter was significantly more frequent among women (Fisher’s exact test, p=0.02) and was associated with younger age (Spearman rank correlation, ρ=−0.28, p=0.01).
Figure 3.
Methylation of the APC promoter 1A in tumours and matched adjacent mucosa. Mean methylation levels (beta values) for each patient were calculated across the 15 CpG sites mapping to the 1A promoter that were identified as differentially methylated by smoking status (Figure 2). (A) promoter methylation in tumours by patient smoking status. Mean values by smoking status are indicated by horizontal lines. (B) promoter methylation in tumours by AJCC stage in all patients. Mean values by stage are indicated by horizontal lines. (C) promoter methylation in matched samples of tumours and adjacent mucosa from 89 patients (33 never smokers, 43 former smokers, and 13 active smokers). Lines indicate matched samples from the same patient.
Methylation of the APC 1A promoter is associated with duration of smoking
To explore the relation between the intensity and duration of smoking with methylation of the APC 1A promoter, we utilised data for the 72 former and active smokers in this study regarding intensity (pack-years) and smoking duration (length of time for which the patient smoked). Additionally, for the 47 former smokers, the relation with the length of time between cessation of smoking and cancer diagnosis was also assessed. Greater duration of smoking was significantly and positively associated with increased methylation at cg14479889 (ρ=0.27, p=0.03) and trended towards significance at each of the other five differentially methylated loci (ρ>0.19, p<0.09) (Table 3). Most notably, the average methylation (beta values) across the 15 differentially methylated loci mapped to this promoter region was significantly and positively associated with duration of smoking (ρ=0.26, p=0.03). No significant associations were observed with pack-years of smoking (p>0.29) or time between cessation of smoking and cancer diagnosis among former smokers (p>0.16).
Table 3.
Associations between DNA methylation and smoking intensity and duration in tumours
| Probe ID | Chromosomal location | Gene | Pack-years | Duration | Time since cessation | |||
|---|---|---|---|---|---|---|---|---|
| ρ | p | ρ | p | ρ | p | |||
| cg08571859 | chr5:112073350 | APC | −0.07 | 0.31 | 0.22 | 0.06 | −0.05 | 0.41 |
| cg14511739 | chr5:112073373 | APC | −0.01 | 0.47 | 0.19 | 0.09 | −0.05 | 0.41 |
| cg22035501 | chr5:112073426 | APC | −0.08 | 0.29 | 0.19 | 0.09 | −0.11 | 0.31 |
| cg11613015 | chr5:112073433 | APC | −0.02 | 0.44 | 0.20 | 0.08 | −0.14 | 0.27 |
| cg14479889 | chr5:112073426 | APC | −0.05 | 0.37 | 0.27 | 0.03 | −0.21 | 0.17 |
| cg16970232 | chr5:112073433 | APC | −0.01 | 0.48 | 0.21 | 0.07 | −0.10 | 0.32 |
| Promoter 1A | chr5:112,072,710 - 112,073,585 | APC | 0.02 | 0.44 | 0.26 | 0.03 | −0.20 | 0.19 |
Spearman’s rank correlation coefficients were calculated for each of the significantly different loci in tumour tissue, using data from former (n=47) and active (n=13) smokers. Correlations were calculated between methylation (beta values) and the pack-years of smoking or duration (years) of smoking. Additionally, for former smokers, correlations between methylation and time since cessation were calculated. ρ and p values are provided, with significant values highlighted in bold.
The APC promoter 1A is not hypermethylated in the mucosa adjacent to tumours
We examined APC promoter 1A methylation in mucosa adjacent to tumours, to determine whether hypermethylation of this region exists as a field defect. Matched tumour and adjacent mucosal tissue were available for 24 of the 27 patients with tumoural hypermethylation of the 1A promoter (irrespective of smoking status). No promoter hypermethylation was observed in the adjacent mucosa from any of the 24 patients (Figure 3C, average beta < 0.11), or individually at any of the six differentially methylated loci (β<0.13) (supplementary material, Figure S2).
TNXB is differentially methylated in the adjacent mucosa of smokers
To gain insight into how smoking may act upon the colon, such as through carcinogenic compounds from cigarette smoke carried in the blood or chronic inflammation, we performed epigenome-wide analyses of DNA methylation in adjacent mucosa by smoking behaviours. We identified four sites within a 500 bp region that map to the tenascin XB (TNXB) gene body that were significantly hypomethylated in mucosa from active smokers (supplementary material, Table S2). No differentially methylated loci were observed between former and never smokers.
Discussion
In this study, we investigated how epigenetic factors may be implicated in conferring the increased risk of colorectal cancer among smokers by performing epigenome-wide analysis of DNA methylation in samples of tumours and adjacent mucosa by smoking behaviours. We report that smoking at the time of diagnosis is significantly associated with hypermethylation of the 1A promoter of APC, a key tumour suppressor gene that has been extensively studied with regard to colorectal cancer. Hypermethylation was unique to tumour tissue and was associated with the duration for which the patient has smoked. We observed that hypermethylation of this promoter was more common in the rectum and distal colon, in concordance with evidence that the association between smoking and CRC risk is greatest for developing tumours in the rectum [1,26]. Our findings may implicate the epigenetic silencing of APC in smoking-associated colorectal carcinogenesis. However, due to the relatively small number of patients who were active smokers at diagnosis, our results should be considered exploratory at this stage. We have been unable to validate our observations in an independent cohort due to the absence of publicly-available datasets incorporating smoking history, and insufficient numbers of active smokers at diagnosis within other studies. Further work in external cohorts is required to examine the validity of our observations.
APC is a tumour suppressor gene and regulator of the Wnt signalling pathway, which acts via regulation of β-catenin degradation and localisation. Loss of APC function has been proposed as a key early event in the development of sporadic colorectal cancer [27], with inactivation frequently occurring through mutations, especially in the mutation cluster region [28], and promoter methylation [29]. Expression of the 1A mRNA isoform of APC is regulated in part through methylation of promoter 1A (chr5:112,072,710–112,073,585) [30], and this region is aberrantly methylated in colorectal, breast and lung tumours, resulting in transcriptional silencing and increased activation of the Wnt signalling pathway [29,31]. We observed significantly greater methylation of this region in tumours from patients who were active smokers at the point of diagnosis, thereby linking smoking behaviours to silencing of this key tumour suppressor gene. It has been reported elsewhere that smoking is associated with mutations in TP53 and BRAF but not APC [32], which together with our study may suggest that inactivation of this gene more commonly occurs through epigenetic dysregulation in smoking-associated CRC than through genetic changes. Median promoter methylation levels (beta values) were approximately 0.5 higher in active smokers (Figure 3), consistent with monoallelic methylation of the promoter. Although evidence from the mouse model suggests that inactivation of both alleles is required for tumourigenesis [33], monoallelic methylation of the APC promoter 1A is a frequent event in human colorectal tumours [31,34] and cancer cell lines [35], and has been reported in gastric tumours [36].
Interestingly, hypermethylation of the APC promoter 1A was never present in mucosa adjacent to the tumours. Methylation at each interrogated CpG site within promoter 1A was very highly conserved in adjacent mucosa, while in direct contrast there was substantial variation in promoter methylation between tumour samples (Figure 3C, supplementary material, Figure S2). Cancer is associated with significantly greater variability in DNA methylation than is found in healthy tissue, and this loss of stability and increased stochastic variation may facilitate malignant cells to adapt to changes in their microenvironments [37,38]. Genetic and epigenetic alterations implicated in carcinogenesis are sometimes present in the surrounding tissue as field defects [39,40], and increased variation in DNA methylation has been observed in cytologically-normal cells from individuals later diagnosed with cervical cancer [41]. However, we observed that methylation of the APC promoter 1A was still highly conserved in adjacent mucosa, in line with studies reporting an absence of APC hypermethylation in colonic mucosa [31]. Mutations in APC are sufficient to induce polyp formation in mice [42,43] and humans [44], and we therefore speculate that this absence of APC hypermethylation in adjacent mucosa may be due to the key role for loss of APC in driving carcinogenesis. Indeed, we observed hypermethylation of the 1A promoter in half of stage I tumours (Figure 3B). This hypothesis is further supported by evidence of APC promoter methylation being an early event in colorectal carcinogenesis that is detectable in small (<15 mm) adenomas [31].
We observed a significant association between promoter 1A hypermethylation and duration of smoking (Table 3), but further work is required to expand upon the relation between the intensity and duration of exposure and epigenetic events in CRC. Indeed, promoter hypermethylation was not observed in tumours from any of the 8 former smokers who had smoked for >35 years, and our epigenome-wide analysis did not identify any differentially methylated sites between never smokers and former smokers in tumours or adjacent mucosa. The cessation of smoking is known to reduce the risk of CIMP-high colorectal cancer and patients who quit >10 years prior to diagnosis display similar risk of CIMP-high tumours to never smokers [18]. Furthermore, it is known that methylation of the AHRR and F2RL3 genes returns to normal levels with increasing time since cessation [9]. Therefore, as only 9 of the 40 former smokers in this study for whom there is relevant data ceased smoking <10 years prior to diagnosis, we speculate that the time since cessation may also be a significant factor in the risk of hypermethylation of APC promoter 1A.
Our epigenome-wide analysis also identified the NFATC1 gene body as being hypomethylated in tumours from smokers (Figure 2). This gene encodes a transcription factor implicated in T cell activation. Epigenetic dysregulation of this gene has been observed in hepatocellular carcinoma [45] and lymphomas [46] while hypomethylation has been reported in healthy individuals with lower socioeconomic status [47]. Our study is the first to report hypomethylation of NFATC1 in colorectal tumours. Overexpression of the gene is associated with worse prognosis in stage II and III colorectal cancer patients, which may occur through the promotion of cell migration and metastasis [48,49]. As the ColoCare Study began to recruit patients in October 2010, we are currently unable to determine whether NFATC1 methylation is associated with patient prognosis in this cohort. We will be able to address this question in time as further data regarding patient outcomes is collected.
Our data suggests that smoking is not associated with the accumulation of widespread epigenetic defects in the adjacent mucosa. Methylation of the APC promoter 1A occurs independently of other epigenetic events in CRC [31], and we identified only one gene, TNXB, as differentially methylated in the adjacent mucosa of active smokers (Table 3). This may be considered to be in contrast to the findings of Paun et al [50], who reported disruption of normal gene methylation profiles in the normal rectal mucosa of smokers. We speculate that this may be the product of our analyses identifying genes implicated in malignant transformation due to our comparison of tumours and adjacent mucosa, while Paun et al examined rectal mucosa prior to the advent of tumour formation. To our knowledge, ours is the first study to observe differential methylation of TNXB by smoking behaviours. Further work is required to investigate how this extracellular matrix glycoprotein could be implicated in smoking-associated carcinogenesis.
Further to the inability to confirm our findings in an independent cohort, the comparatively low number of patients who actively smoked at the point of diagnosis is a limitation of this study, and one which could inhibit the identification of associations between smoking and methylation. We therefore incorporated the chromosomal position into test statistics by means of a NHMM, which also served to reduce the probability of secluded differentially methylated CpGs and hence most likely false positives. A particular strength of this study is the analysis of both tumour tissue and adjacent mucosa, which has enabled us to gain greater insight by identifying epigenetic events associated with smoking that are uniquely found in tumour tissue (hypermethylation of the APC promoter 1A) and to establish an absence of field defects associated with smoking in the neighbouring mucosa.
In conclusion, we report exploratory evidence for hypermethylation of the APC promoter 1A being implicated in the development of colorectal tumours among smokers. Methylation of this region was significantly associated with smoking at the point of diagnosis and with the duration of time for which the patient smoked, and hypermethylation was confined to tumours. Further work is required to validate our observations in independent cohorts, and to identify implications for patient prognosis.
Supplementary Material
Prediction performances of the top-six methylation features
Methylation of differentiated methylated sites within the APC promoter 1A in matched tumours and adjacent mucosa
CpG sites with smoking-specific differential methylation between tumours and adjacent mucosa
CpG sites with differential methylation by smoking status in adjacent mucosa
Acknowledgments
This study was funded by the German Consortium for Translational Cancer Research (DKTK). The authors would like to thank all ColoCare study participants and the entire ColoCare study team in Heidelberg, especially Dr Werner Diehl for data acquisition and documentation, and Judith Kammer, Susanne Jakob and Torsten Koelsch for patient recruitment and tissue collection. We are grateful to Dr. Melanie Bewerunge-Hudler and the Genomics and Proteomics Core Facility at the German Cancer Research Center (Heidelberg, Germany) for running the Illumina Infinium HumanMethylation450 BeadChip microarrays. The ColoCare Study and Consortium has been designed and first implemented at the Fred Hutchinson Cancer Research Center, Seattle, USA (PIs: Ulrich/Grady) and protocols have been used with permission in Heidelberg, Germany (PI: Ulrich). The ColoCare Study site in Heidelberg was funded by the Matthias Lackas Foundation, the German Consortium for Translational Cancer Research (DKTK), the Division of Preventive Oncology at the German Cancer Research Center, and the National Institutes of Health (NIH R01 189184 and NIH U01 CA206110). Hagen Klett and Melanie Boerries were additionally funded by the German Ministry of Education and Research (BMBF) within the e:Med consortium “DeCaRe-Delineating Cardiac Regeneration”. Hauke Busch acknowledges support from the DFG Cluster of Excellence EXC-306 ‘Inflammation at Interfaces’.
Footnotes
Author contributions statement
CU conceived the cohort study. TB, RT, NH, CU and KM conceived the investigation into smoking. JB, LZ, MS, AU, PS and EH organised and performed the sample collection. RT, BG, DS, SS, CAM, PSK and H Brenner were involved in data collection and organisation. TB, HK, RT, H Busch and MB analysed the DNA methylation data. TB, HK, RT, CU and KM performed data interpretation. TB wrote the manuscript, with figures generated by TB and HK. All authors were involved in writing and had final approval of the submitted manuscript.
The authors report no conflicts of interest.
References
- 1.Botteri E, Iodice S, Bagnardi V, et al. Smoking and colorectal cancer: a meta-analysis. JAMA. 2008;300:2765–2778. doi: 10.1001/jama.2008.839. [DOI] [PubMed] [Google Scholar]
- 2.Parajuli R, Bjerkaas E, Tverdal A, et al. Cigarette smoking and colorectal cancer mortality among 602,242 Norwegian males and females. Clin Epidemiol. 2014;6:137–145. doi: 10.2147/CLEP.S58722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walter V, Jansen L, Hoffmeister M, et al. Smoking and survival of colorectal cancer patients: systematic review and meta-analysis. Ann Oncol. 2014;25:1517–1525. doi: 10.1093/annonc/mdu040. [DOI] [PubMed] [Google Scholar]
- 4.Giovannucci E, Martínez ME. Tobacco, colorectal cancer, and adenomas: a review of the evidence. J Natl Cancer Inst. 1996;88:1717–1730. doi: 10.1093/jnci/88.23.1717. [DOI] [PubMed] [Google Scholar]
- 5.Novakovic B, Ryan J, Pereira N, et al. Postnatal stability, tissue, and time specific effects of AHRR methylation change in response to maternal smoking in pregnancy. Epigenetics. 2014;9:377–386. doi: 10.4161/epi.27248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott HR, Tillin T, McArdle WL, et al. Differences in smoking associated DNA methylation patterns in South Asians and Europeans. Clin Epigenetics. 2014;6:4. doi: 10.1186/1868-7083-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leng S, Liu Y, Thomas CL, et al. Native American ancestry affects the risk for gene methylation in the lungs of Hispanic smokers from New Mexico. Am J Respir Crit Care Med. 2013;188:1110–1116. doi: 10.1164/rccm.201305-0925OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Yang R, Burwinkel B, et al. F2RL3 methylation as a biomarker of current and lifetime smoking exposures. Environ Health Perspect. 2014;122:131–137. doi: 10.1289/ehp.1306937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shenker NS, Polidoro S, van Veldhoven K, et al. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum Mol Genet. 2013;22:843–851. doi: 10.1093/hmg/dds488. [DOI] [PubMed] [Google Scholar]
- 10.Breitling LP, Yang R, Korn B, et al. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am J Hum Genet. 2011;88:450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andreotti G, Karami S, Pfeiffer RM, et al. LINE1 methylation levels associated with increased bladder cancer risk in pre-diagnostic blood DNA among US (PLCO) and European (ATBC) cohort study participants. Epigenetics. 2014;9:404–415. doi: 10.4161/epi.27386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Yang R, Burwinkel B, et al. F2RL3 methylation in blood DNA is a strong predictor of mortality. Int J Epidemiol. 2014;43:1215–1225. doi: 10.1093/ije/dyu006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan Q, Wang G, Huang J, et al. Epigenomic analysis of lung adenocarcinoma reveals novel DNA methylation patterns associated with smoking. Onco Targets Ther. 2013;6:1471–1479. doi: 10.2147/OTT.S51041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marsit CJ, Karagas MR, Schned A, et al. Carcinogen exposure and epigenetic silencing in bladder cancer. Ann N Y Acad Sci. 2006;1076:810–821. doi: 10.1196/annals.1371.031. [DOI] [PubMed] [Google Scholar]
- 15.Wolff EM, Liang G, Cortez CC, et al. RUNX3 methylation reveals that bladder tumors are older in patients with a history of smoking. Cancer Res. 2008;68:6208–6214. doi: 10.1158/0008-5472.CAN-07-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Lan Q, Siegfried JM, et al. Aberrant promoter methylation of p16 and MGMT genes in lung tumors from smoking and never-smoking lung cancer patients. Neoplasia. 2006;8:46–51. doi: 10.1593/neo.05586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lokk K, Vooder T, Kolde R, et al. Methylation markers of early-stage non-small cell lung cancer. PLoS One. 2012;7:e39813. doi: 10.1371/journal.pone.0039813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishihara R, Morikawa T, Kuchiba A, et al. A prospective study of duration of smoking cessation and colorectal cancer risk by epigenetics-related tumor classification. Am J Epidemiol. 2013;178:84–100. doi: 10.1093/aje/kws431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Triche TJ, Weisenberger DJ, Van Den Berg D, et al. Low-level processing of Illumina Infinium DNA methylation beadarrays. Nucleic Acids Res. 2013;41:e90. doi: 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fortin JP, Labbe A, Lemire M, et al. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014;15:503. doi: 10.1186/s13059-014-0503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 22.Kuan PF, Chiang DY. Integrating prior knowledge in multiple testing under dependence with applications to detecting differential DNA methylation. Biometrics. 2012;68:774–783. doi: 10.1111/j.1541-0420.2011.01730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun W, Cai T. Large-scale multiple testing under dependence. J R Stat Soc B. 2009;71:393–424. [Google Scholar]
- 24.Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sherry ST, Ward M, Sirotkin K. DbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–679. [PubMed] [Google Scholar]
- 26.Cheng J, Chen Y, Wang X, et al. Meta-analysis of prospective cohort studies of cigarette smoking and the incidence of colon and rectal cancers. Eur J Cancer Prev. 2015;24:6–15. doi: 10.1097/CEJ.0000000000000011. [DOI] [PubMed] [Google Scholar]
- 27.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 28.Miyoshi Y, Nagase H, Ando H, et al. Somatic mutations of the APC gene in colorectal tumors: mutation cluster region in the APC gene. Hum Mol Genet. 1992;1:229–233. doi: 10.1093/hmg/1.4.229. [DOI] [PubMed] [Google Scholar]
- 29.Hiltunen MO, Alhonen L, Koistinaho J, et al. Hypermethylation of the APC (adenomatous polyposis coli) gene promoter region in human colorectal carcinoma. Int J Cancer. 1997;70:644–648. doi: 10.1002/(sici)1097-0215(19970317)70:6<644::aid-ijc3>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 30.Segditsas S, Sieber OM, Rowan A, et al. Promoter hypermethylation leads to decreased APC mRNA expression in familial polyposis and sporadic colorectal tumours, but does not substitute for truncating mutations. Exp Mol Pathol. 2008;85:201–206. doi: 10.1016/j.yexmp.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 31.Esteller M, Sparks A, Toyota M, et al. Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res. 2000;60:4366–4371. [PubMed] [Google Scholar]
- 32.Chen K, Xia G, Zhang C, et al. Correlation between smoking history and molecular pathways in sporadic colorectal cancer: a meta-analysis. Int J Clin Exp Med. 2015;8:3241–3257. [PMC free article] [PubMed] [Google Scholar]
- 33.Samuel MS, Suzuki H, Buchert M, et al. Elevated Dnmt3a activity promotes polyposis in Apc(min) mice by relaxing extracellular restraints on Wnt signaling. Gastroenterology. 2009;137:902–13. 913.e1–11. doi: 10.1053/j.gastro.2009.05.042. [DOI] [PubMed] [Google Scholar]
- 34.Arnold CN, Goel A, Niedzwiecki D, et al. APC promoter hypermethylation contributes to the loss of APC expression in colorectal cancers with allelic loss on 5q. Cancer Biol Ther. 2004;3:960–964. doi: 10.4161/cbt.3.10.1113. [DOI] [PubMed] [Google Scholar]
- 35.Lind GE, Thorstensen L, Løvig T, et al. A CpG island hypermethylation profile of primary colorectal carcinomas and colon cancer cell lines. Mol Cancer. 2004;3:28. doi: 10.1186/1476-4598-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clément G, Bosman FT, Fontolliet C, et al. Monoallelic methylation of the APC promoter is altered in normal gastric mucosa associated with neoplastic lesions. Cancer Res. 2004;64:6867–6873. doi: 10.1158/0008-5472.CAN-03-2503. [DOI] [PubMed] [Google Scholar]
- 37.Barrow TM, Barault L, Ellsworth RE, et al. Aberrant methylation of imprinted genes is associated with negative hormone receptor status in invasive breast cancer. Int J Cancer. 2015;137:537–547. doi: 10.1002/ijc.29419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansen KD, Timp W, Bravo HC, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ellsworth DL, Ellsworth RE, Love B, et al. Genomic patterns of allelic imbalance in disease free tissue adjacent to primary breast carcinomas. Breast Cancer Res Treat. 2004;88:131–139. doi: 10.1007/s10549-004-1424-7. [DOI] [PubMed] [Google Scholar]
- 40.Shen L, Kondo Y, Rosner GL, et al. MGMT promoter methylation and field defect in sporadic colorectal cancer. J Natl Cancer Inst. 2005;97:1330–1338. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]
- 41.Teschendorff AE, Jones A, Fiegl H, et al. Epigenetic variability in cells of normal cytology is associated with the risk of future morphological transformation. Genome Med. 2012;4:24. doi: 10.1186/gm323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oshima M, Oshima H, Kitagawa K, et al. Loss of Apc heterozygosity and abnormal tissue building in nascent intestinal polyps in mice carrying a truncated Apc gene. Proc Natl Acad Sci U S A. 1995;92:4482–4486. doi: 10.1073/pnas.92.10.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shibata H, Toyama K, Shioya H, et al. Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science. 1997;278:120–123. doi: 10.1126/science.278.5335.120. [DOI] [PubMed] [Google Scholar]
- 44.Lamlum H, Papadopoulou A, Ilyas M, et al. APC mutations are sufficient for the growth of early colorectal adenomas. Proc Natl Acad Sci U S A. 2000;97:2225–2228. doi: 10.1073/pnas.040564697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song MA, Tiirikainen M, Kwee S, et al. Elucidating the landscape of aberrant DNA methylation in hepatocellular carcinoma. PLoS One. 2013;8:e55761. doi: 10.1371/journal.pone.0055761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akimzhanov A, Krenacs L, Schlegel T, et al. Epigenetic changes and suppression of the nuclear factor of activated T cell 1 (NFATC1) promoter in human lymphomas with defects in immunoreceptor signaling. Am J Pathol. 2008;172:215–224. doi: 10.2353/ajpath.2008.070294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stringhini S, Polidoro S, Sacerdote C, et al. Life-course socioeconomic status and DNA methylation of genes regulating inflammation. Int J Epidemiol. 2015;44:1320–1330. doi: 10.1093/ije/dyv060. [DOI] [PubMed] [Google Scholar]
- 48.Jauliac S, López-Rodriguez C, Shaw LM, et al. The role of NFAT transcription factors in integrin-mediated carcinoma invasion. Nat Cell Biol. 2002;4:540–544. doi: 10.1038/ncb816. [DOI] [PubMed] [Google Scholar]
- 49.Tripathi MK, Deane NG, Zhu J, et al. Nuclear factor of activated T-cell activity is associated with metastatic capacity in colon cancer. Cancer Res. 2014;74:6947–6957. doi: 10.1158/0008-5472.CAN-14-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paun BC, Kukuruga D, Jin Z, et al. Relation between normal rectal methylation, smoking status, and the presence or absence of colorectal adenomas. Cancer. 2010;116:4495–4501. doi: 10.1002/cncr.25348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Prediction performances of the top-six methylation features
Methylation of differentiated methylated sites within the APC promoter 1A in matched tumours and adjacent mucosa
CpG sites with smoking-specific differential methylation between tumours and adjacent mucosa
CpG sites with differential methylation by smoking status in adjacent mucosa