Abstract
Marine organisms adapt to complex temporal environments that include daily, tidal, semi-lunar, lunar and seasonal cycles. However, our understanding of marine biological rhythms and their underlying molecular basis is mainly confined to a few model organisms in rather simplistic laboratory settings. Here, we use new empirical data and recent examples of marine biorhythms to highlight how field ecologists and laboratory chronobiologists can complement each other's efforts. First, with continuous tracking of intertidal shorebirds in the field, we reveal individual differences in tidal and circadian foraging rhythms. Second, we demonstrate that shorebird species that spend 8–10 months in tidal environments rarely maintain such tidal or circadian rhythms during breeding, likely because of other, more pertinent, temporally structured, local ecological pressures such as predation or social environment. Finally, we use examples of initial findings from invertebrates (arthropods and polychaete worms) that are being developed as model species to study the molecular bases of lunar-related rhythms. These examples indicate that canonical circadian clock genes (i.e. the homologous clock genes identified in many higher organisms) may not be involved in lunar/tidal phenotypes. Together, our results and the examples we describe emphasize that linking field and laboratory studies is likely to generate a better ecological appreciation of lunar-related rhythms in the wild.
This article is part of the themed issue ‘Wild clocks: integrating chronobiology and ecology to understand timekeeping in free-living animals’.
Keywords: circadian, tidal, lunar, shorebirds, invertebrates, molecular
1. Introduction
As the Earth rotates around its axis every 24 h, it generates relentless rhythms of light and dark, heat and cold. In addition, the tilt of the Earth's axis produces the annual seasonal rhythms that so dramatically modulate the light and dark cycles as we move towards the polar extremes [1,2]. The rotation of the Earth and the gravitational pull of the Sun and the Moon deform the mass of the oceans, producing the rise and fall of sea levels every 12.4 h. When the Earth, Moon and Sun are in alignment during new and full moon every 15 days, the gravitational pull on the Earth's oceans is at its maximum, producing the high-amplitude spring tides (figure 1a). When the Sun and Moon are at right angles when viewed from the Earth (Moon's first or third quarter), the gravitational pull on the oceans is reduced, generating the low-amplitude neap tides (figure 1a). Furthermore, when the Moon orbits off the equatorial plane the tide is higher at night than during the day, a phenomenon termed ‘diurnal inequality’ ([3] and figure 1b). Finally, there is the waxing and waning of the Moon itself with its 14.8 day semi-lunar and 29.6 day lunar cycles.
Figure 1.
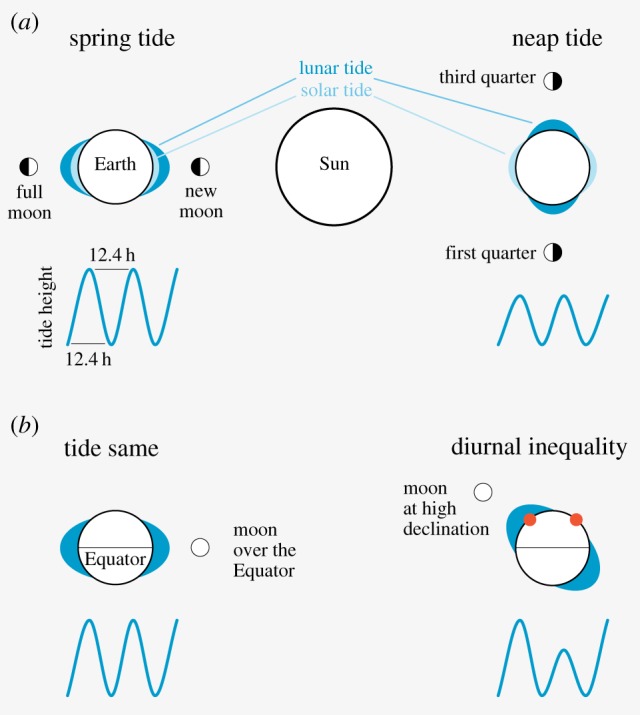
Variation in high-tide levels. (a) When the Sun, Moon and Earth are in alignment during new or full moon (i.e. twice a month) the gravitational pull on the oceans is strongest, producing the high-amplitude spring tides, i.e. lunar tide (dark blue) and sun tide (light blue) combine. In contrast, when the Moon is in its first or third quarter the gravitational pull on the oceans is reduced, leading to the low amplitude neap tides. (b) If the Moon orbits directly over the Equator, the day and night tides are similar, whereas when the Moon orbits at high declination the night tides are higher than the day tides (diurnal inequality; indicated by red dots).
For hundreds of millions of years these geophysical cycles have shaped the behaviour and physiology of organisms. Not surprisingly, nearly all terrestrial and marine species (including some bacteria) show circadian phenotypes [4]. In addition, organisms living in intertidal zones also show tidal, semi-lunar and lunar cycles [5]. However, marine biorhythms are rarely studied in higher vertebrates [6]. Also, whereas genetic studies of circadian rhythms have a 45-year history, particularly in the model organisms of mouse and Drosophila, until recently a similar approach to studying rhythms in intertidal (non-model) organisms was not feasible. However, in the past few years, the advent of genomic technologies that are applicable to any species has initiated the mechanistic study of tidal and lunar cycles of behaviour and physiology [7].
Here, our aims are threefold. We first address the scarcity of data on intertidal higher vertebrates by investigating the interactions between tidal and daily cycles in the foraging movements and incubation rhythms of shorebirds. We then discuss some fresh studies that have illuminated the role of circadian clock genes in the intertidal behaviour and physiology of arthropods and worms. Finally, we use our findings and the reported examples to highlight how collaborations between field ecologists and chronobiologists may uncover fundamental adaptive principles about biorhythms in the wild.
2. Tidal rhythms in shorebirds
Substantial numbers of shorebird species live and feed, at least for part of the year, in tidal habitats [8,9]. Some of these tidal populations are sedentary in tidal environments, and face day–night fluctuations of illumination throughout the year (e.g. several species of oystercatcher, Haematopus; [10]). Other populations are migratory and live in the coastal nonbreeding areas during 8–10 months of the year, where they cope with a combination of tidal and day–night environmental rhythms (e.g. bar-tailed godwit, Limosa lapponica; sanderling, Calidris alba; and red knot, Calidris canutus), and breed in Arctic non-tidal environments for two months of the year, where day–night environmental rhythms are damped [8,9]. Shorebirds manage the interplay between circadian and tidal environmental, but how they schedule their behaviour to the interacting environmental rhythms is unclear [11]. Indeed, the behavioural rhythms of shorebirds under such circumstances are relatively unexplored (but see [12,13]).
To anticipate tidal foraging opportunities, it is assumed that these species have activity patterns with a period length resembling the tidal period. We might expect shorebirds that use tides throughout the whole year to exhibit incubation rhythms with tidal periods [14] more readily than shorebirds that only use tides away from their breeding grounds. Nevertheless, as changing to a different rhythm may be costly [15], the tidal activity patterns could carry over to incubation even for shorebirds that are tidal only when away from their breeding grounds.
The aims of our shorebird study are twofold. We used novel automated-tracking technology [16] to first describe the foraging rhythms of red knots at Banc d'Arguin, their coastal Mauritanian wintering ground—an environment with both tidal rhythms and strong diel fluctuations in light intensity (see [17]. Second, we analyse data from a recent comparative study on shorebirds that incubate biparentally [14,18], to reveal whether shorebirds with tidal life-histories keep tidal rhythms also during incubation [14].
(a). The tidal rhythm of red knots
Red knots, C. canutus, are long-distance migratory shorebirds that breed in the High Arctic and live in coastal intertidal environments during the rest of the year [19,20], where they almost exclusively eat hard-shelled molluscs ingested whole and crushed in their large muscular gizzards [21]. When the tide goes out and the intertidal mudflats become available they take the opportunity to feed, being forced to retreat to shoreline high-tide roost during the high-water periods [22]. However, the individual variation in foraging rhythm of knots (and of any other intertidal bird) is unknown.
We found that the distance of red knots to their roosting site followed the tidal as well as the day–night rhythm (tidal = 88% of individuals, daily = 57%, both rhythms = 52%; N = 42 individuals with more than 50 h of observation; median [range] = 19 [2–34] days of observation per individual; for methods see Supplementary Information [16]). At high tide, the birds were generally close to the roost and as the tide retreated, birds moved away from it (figure 2a). How far the birds moved was modulated by time of day, but in a bird-specific manner (figure 2b). For example, one bird usually roamed between 400 and 600 m from its roost when the low tide occurred during the day (figure 3a, light blue), but often went to mudflats further than 1 km from its roost when the low tide occurred at night (figure 3a, dark blue). In this particular bird it seems that an approximately 15 day semi-lunar pattern also emerges where the distance travelled at night is greater and is particularly consolidated when the low tide is at its lowest ebb.
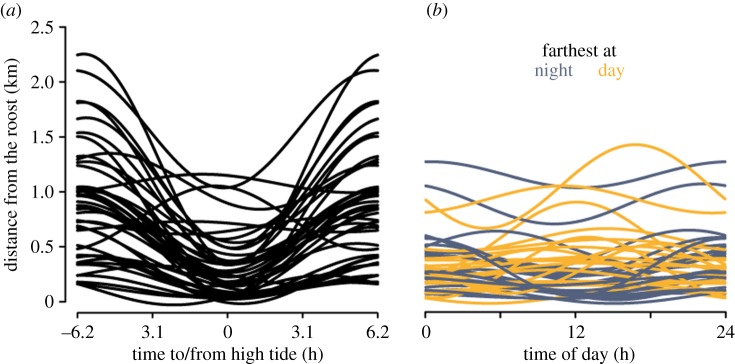
Figure 2.
Distance of redknots to their to the closest roost relative to high tide (a) and time of day (b). Each line depicts the model prediction for a single individual (N = 42 individuals; see [16] for details.
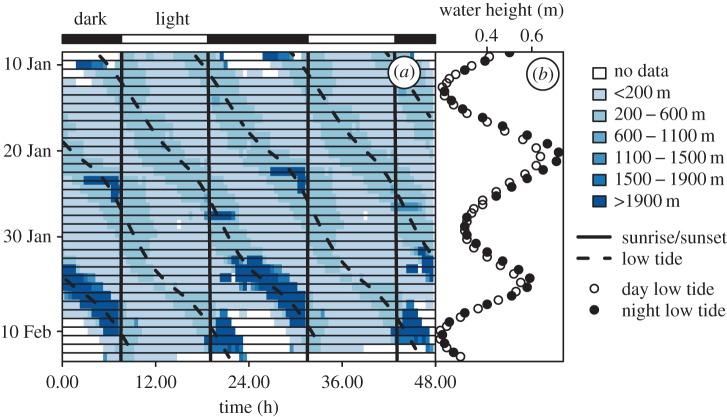
Figure 3.
The distance of a radio-tagged red knot to its roost. (a) The distance to the main roost (the darker the blue, the farther the knot travelled). Sunrise and sunset are given by the solid vertical lines and the day and night are indicated above the actogram. The low tide times are given by the dashed lines. For actograms see [16]. (b) Differences in low-tide water height between day (open circles) and night (filled circles) and during the neap–spring lunar cycle.
The reported tidal rhythms (figure 2a) reflect red knots' feeding on molluscs that are only available during low tide. However, why red knots varied so much in how far they travelled during the night and during the day remains unclear. Such daily rhythms (superimposed on the tidal rhythm) can be partly a consequence of the slightly higher tide during the night (figure 3b), reducing the maximal extent of the available foraging area. However, why some individuals foraged further from the roost during the night is unclear and unlikely a consequence of dynamics in searching efficiency or food availability. That is, red knots forage by touch rather than by sight [23] and the burying depths of their main prey are not expected to differ between day and night. An alternative explanation for the individual differences may be individual experience with predators. During the day, red knots are predated mainly by large falcons [24,25], and during the night by owls [26–28] . Thus, depending on the local distributions of these two kinds of predators and individual experiences with these predators, the red knot's perceived ‘landscape of fear’ [29], and hence its movement choices, may differ between individuals and between day and night, something worthy of future investigations.
The individuality of red knot tidal movements and hence the investigation of among-individual variation in behavioural rhythms in the wild contrast starkly with laboratory studies where individual subjects, for methodological reasons, are often chosen to be as similar as possible. Although foraging rhythms of red knots appear related to both tidal and daily environmental fluctuations, quantitative studies from different locations are required to validate the generality of these behavioural rhythms, as well as to explore (albeit in a correlative manner) the hypotheses about possible ecological causes of such biorhythms. Also, to demonstrate whether individuals will free-run with circatidal or circadian rhythm or with both of these rhythms, and hence to demonstrate whether these rhythms are truly endogenous, we would need to keep red knots under constant conditions. Such observations will also reveal whether the among-individual differences are endogenous.
(b). Do tidal shorebirds maintain a tidal incubation rhythm?
In a recent study of 32 species of shorebirds with biparental care, only in 5% of 584 nests did the shorebird pairs display an incubation period length that might have been entrained by the tide [14]. This is surprising, given that half of the studied species live in intertidal habitats away from their breeding grounds [14]. Interestingly, from populations known to forage on intertidal habitats at their breeding grounds (N = 10), pairs in only 3 out of 74 nests displayed a period length entrained by the tide. In contrast, incubation rhythms with periods that do not follow the 24 h light–dark cycle were more common and the deviations from 24 h increased in shorebirds breeding at high latitudes.
Although these findings support the existence of a latitudinal cline in incubation rhythms, a substantial number of rhythms defied the 24 h day even at low and mid latitudes. These results might reflect an underestimation of tidal and circadian patterns in incubating shorebirds because the method used depicted only the dominant period of the incubation rhythm, yet other less-dominant periodicities were rare [14]. Importantly, the study suggests that other factors (such as risk of predation and synchronization of the clock between the two parents) might be much more important than any geophysically imposed variable, hence the extremely variable and generally non-daily/tidal rhythmicity in incubation [14].
In summary, these findings suggest that tidal life-history seems to play, at best, a negligible role in determining incubation rhythms, even in shorebirds that forage with the tide during breeding. They corroborate the observations on pre-incubation activities of shorebirds on their Arctic breeding grounds; birds were active around the clock without significant tidal periodicity [30]. Chronobiologists might ask whether these variable cycles of incubation mask an otherwise endogenous circatidal rhythm. Unfortunately, to study any such tidal cycle, birds would have to be removed from the entraining stimuli, conspecifics and any potential predators and placed in free-running constant conditions for several days, something that is impractical during breeding.
3. Molecular studies of tidal rhythms
The work described above suggests that tidal and circadian rhythms in foraging shorebirds reflect adjustments to the complex temporal environment in which they live. However, other factors beyond circadian day–night or tidal rhythms, such as predation or behaviour of conspecifics (which themselves may have clock-like features), may outweigh the entrainment of behaviour imposed by these geophysical variables [14]. Still, circadian rhythms are identified in nearly all higher organisms and, for example, migratory birds use the clock for navigation and to compensate for the movement of the sun [31]. Consequently, given the ubiquity of biological rhythmicity, considerable effort has been expended over five decades to identify the genetic and molecular bases for these behavioural rhythms. The discovery of the molecular basis of the circadian clock was a defining moment in the study of gene regulation of complex phenotypes [32].
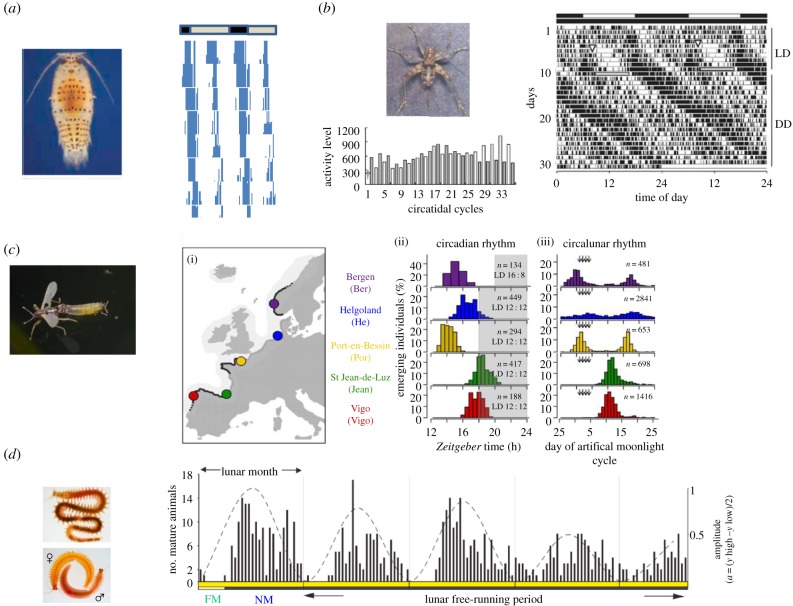
Despite insects and crustaceans having long been studied for lunar-related rhythms at the behavioural level [6], we have been missing a genetically tractable model species from intertidal habitats. Here, we introduce four organisms (figure 4) where molecular interventions were recently used to illuminate the molecular bases of lunar-related rhythms. Specifically, we highlight the finding of tidal activity rhythms in the marine isopod Eurydice pulchra and the mangrove cricket, Apteronemobius asahinai, semi-lunar emergence rhythms of the marine midge, Clunio marinus, and the lunar reproductive cycles of the bristle worm Platynereis dumerilii.
Figure 4.
Examples of lunar-related rhythms of invertebrates. (a) Eurydice adult with chromatophores (black dots on the dorsal surface of cuticle) and swimming activity of a single individual over 9 days in constant darkness). The animal was taken during a spring tide from Bangor, Wales, UK and placed immediately in constant darkness (DD). The approximate natural light (grey)/dark (black) cycle on the day the animal was harvested is shown as a bar above the actogram and each day's activity is double plotted on a horizontal 48 h scale so that so that each row represents two consecutive days. Note that the movement to the right on every successive day reveals a tidal period longer than 12 h and the night-time activity is greater than that of the daytime (diurnal inequality). Adapted from [30]. (b) Mangrove cricket and an actogram for single individual placed in 12 L: 12 D for 8 days then allowed to free run in DD, during which there is more intense activity in the dark phase compared with the light phase (see the histogram) which drifts towards the right reflecting the predominantly 24.8–25.5 h rhythm which is about twice the tidal period of approximately 12.4 h. The histogram shows the night-time burst of activity (filled columns) being greater than the daytime burst (unfilled columns) for a few cycles but as this is modulated by the circadian clock, it drifts out of phase with the tidal cycle; so after many cycles, the daytime tidal episode is greater than the night-time (adapted from [33]). The cricket image is taken from http://mangrove.nus.edu.sg/guidebooks/text/2010.htm. (c) Midge C. marinus and five natural populations (i) with different phases of emergence (ii) and semi-lunar or lunar frequency during day of emergence (iii). Image is taken from https://www.flickr.com/photos/davidh-j/6270311922 and figure was adapted from [34]. (d) Premature adult, and adult male and female Platynereis dumerilii. Lunar maturation cycle of single individual over several months. FM, full moon simulated by dim light. NM, new moon. Lunar month in days plotted as horizontal yellow bar. Adapted from [35].
(a). Circadian and circatidal rhythms in a marine isopod and a mangrove cricket
Eurydice pulchra is a marine isopod that lives in the intertidal zone around northern European coasts (figure 4a). As the tide comes in, Eurydice swims out of its sandy burrow and forages. As the tide goes out, Eurydice buries itself back into the sand so it is not dragged out to sea [33,36]. In constant darkness, Eurydice exhibits an endogenous circatidal swimming rhythm of 12.4 h (figure 4a) which can be reset by vibration stimuli, and is temperature compensated, thereby showing all the hallmarks of a true clock [36]. Interestingly the swimming pattern usually shows the diurnal inequality phenomenon at temperate latitudes (figure 1b), so nocturnal high-tide swimming is considerable greater than daytime swimming (figure 4a). This modulation in swimming is regulated by the circadian clock because under bright light it is disrupted, whereas the tidal 12.4 h swimming period is unaffected, suggesting an independence of circadian and tidal oscillators [36].
Moreover, Eurydice is called the ‘speckled sea louse’ because it carries pigmented spots, chromatophores that expand during the day and contract at night (figure 4a) [33,36]. This 24 h cycle is likely regulated by a circadian clock because the 24 h cycle persists under constant darkness, can be reset by light and is disrupted by constant bright light [33,36]). Indeed, knockdown of Eurydice's period gene, whose Drosophila orthologue plays a central role in the molecular clock machinery of Drosophila melanogaster, has a similar effect to constant light, with circadian cycles in chromatophore dispersion and in Eurydice timeless mRNA disrupted. Yet the very same canonical clock gene misregulation has little effect on the circatidal swimming periodicity of 12.4 h [36].
Although these results invoke separate circatidal and circadian oscillators, pharmacological inhibitors of Eurydice’s casein kinase 1ɛ (CK1ɛ), which phosphorylates PER protein in D. melanogaster and hence could also inhibit similar post-translational modification of Eurydice's PER protein, lengthened both tidal swimming and the circadian chromatophore cycle [36]. This might suggest that the two oscillators share a common pathway. However CK1ɛ has many targets, so the inhibitor might render CK1ɛ less able to phosphorylate a tidally relevant protein that we have yet to identify. It is unlikely that any effect of the inhibitor on Eurydice’s PER protein phosphorylation is mediating tidal lengthening because direct disruption of Eurydice’s period gene mRNA through RNA interference had no effect on this phenotype [36].
The circadian day–night modulation of the tidal swimming rhythms in Eurydice is also observed in the locomotor activity of the mangrove cricket [37] (figure 4b). However, the periodicity of the cricket's locomotor activity pattern is circatidal and approximately 12.4 h. Elegant genetic studies have used RNAi-mediated knockdown of the canonical clock genes in this species, period and Clock (in insects and mammals CLOCK protein is one of a pair of molecules that activate period and timeless gene transcription). The knock-down left 12.4 h tidal rhythms intact, but disrupted the circadian modulation of alternate bouts of locomotor activity [38,39]. As in Eurydice, these gene knockdowns suggest that the two molecular oscillators underlying circadian and tidal rhythms are largely independent of each other. Moreover, surgical ablation of the optic lobes (likely location of the circadian oscillator) disrupted the circadian locomotor pattern, but as with the gene knockdown, the tidal rhythm remained intact [34]). Consequently, molecular mechanisms of the two oscillators not only may be independent, but also may reside in different groups of neurons.
(b). Circadian and semi-lunar emergence of the marine midge
Perhaps the best-known example of a moon-related phenotype in insects is the semi-lunar emergence rhythms in the marine midge, C. marinus (figure 4c), first studied by Neumann and collaborators 50 years ago (e.g. [40]). During full and new moon, millions of males and females of the midge emerge from the sea as low tide exposes the habitats where they have developed from eggs to pupae (figure 4c). These adults mate and live for a few hours, so it is critical that they emerge synchronously during those few hours of low tide. The timing of the lowest tide can be predicted from the lunar calendar, but these critical few hours during the day vary from location to location [40]). Thus, the emergence of the marine midge has to rely on two clocks, one circa-semi-lunar or circalunar, and the other circadian.
A recent and spectacular molecular genetic study used populations of midges living in different European locations (figure 4c), in combination with the fully referenced draft genome of the midge generated de novo [7], to identify the genetic bases of semi-lunar or lunar and circadian rhythms. First, the local circadian adaptations mapped to the gene encoding calcium/calmodulin-dependent kinase II.1 (CaMKII) [7]. Importantly, mutations in the homologous gene can disrupt circadian timing in the mouse [41] and D. melanogaster [35,42]. Secondly and more importantly for lunar-related phenotypes, the genetic mapping experiment localized a chromosomal region responsible for the population differences in semi-lunar versus lunar emergence timing [7]. Lack of canonical clock circadian genes mapping to this region implies that a novel timing gene (or genes) contributes to the lunar phenotype.
(c). Circadian activity and lunar reproductive cycles of the bristle worm
Finally, the bristle worm P. dumerilii (figure 4d) spawns in a monthly rhythm, in which the number of worms that are sexually mature peaks around the time of new moon and troughs at full moon (figure 4d) [43,44]. This monthly rhythm appears to be driven by exposure to moonlight during full moon because the monthly cycle of reproductive maturity can be entrained in the laboratory by nocturnal dim light lasting for eight consecutive nights during the month (figure 4d). Also, the monthly maturity rhythm will free-run for several months under constant darkness, but not under constant light or in constant darkness without previous moonlight exposure, suggesting a true circalunar cycle [43]. In addition, the worms show circadian locomotor rhythms particularly in light–dark cycles. The strength of this rhythm is modulated by the phases of the moon, suggesting a crosstalk between the two oscillators [43].
When the worms were treated with the same CK1ɛ/δ kinase inhibitor used in Eurydice, circadian locomotor behaviour and circadian gene expression of canonical clock genes were severely disrupted, but the circalunar maturity rhythm was essentially unaffected. The authors' conclusions resonated with those from Eurydice and the mangrove cricket, in that the circadian oscillators appeared to be molecularly independent from the circalunar clocks [43]. The only possible inconsistency between the discussed studies concerns tidal and lunar periodicity. The CK1ɛ inhibitor influenced the tidal periodicity in Eurydice, but not the lunar cycle in bristle worm. Likely, there are important differences in the mechanisms that generate 12.4 h tidal and 29 day lunar rhythms even though they are clearly geophysically and astronomically related. However, the maturity rhythm of the bristle worm was monitored only for two months after the inhibition. Thus, a period difference between the inhibited and control animals might have gone undetected. It would require several more months of expensive drug exposure and several cycles of monitoring of the maturity rhythm to state definitively that there was no effect on the period of the free-running maturation cycle.
The above examples used molecular manipulations in vivo allied to the analysis of behavioural and molecular phenotypes in non-model invertebrates. Such analyses are much more difficult to perform compared with model organisms like D. melanogaster or the mouse but they have led to an understanding of what does NOT constitute the tidal oscillator. From three independent studies in Eurydice, mangrove crickets and the bristle worm, the consensus of opinion suggests that lunar-related rhythms may not be generated by the canonical circadian clock genes. Some caution should still be reserved in accepting this conclusion, particularly concerning the CK1ε inhibitor, which dramatically affects the period of Eurydice’s tidal swimming. In addition, if the tidal oscillator in the mangrove cricket is more robust than the circadian oscillator that modulates its tidal locomotor episodes, then RNAi-mediated knockdown may not knock-down period or Clock genes far enough to affect the tidal oscillator. Unfortunately, both organisms are difficult to rear in the laboratory so the use of gene editing tools to create null-mutants is unlikely in the near future.
4. General conclusion and outlook
We have documented the crosstalk between the tidal and circadian rhythms in the distance that a red knot moved from its roost during foraging (figure 3). This is reminiscent of the circadian modulation of tidal behaviour observed in both Eurydice and the mangrove cricket. Thus, we suspect that in all these organisms the brain centres dedicated to expressing tidal and circadian phenotypes will be anatomically connected and, therefore, signalling reciprocally to each other.
The next challenge is to find which genes encode tidal/lunar time in the above-described invertebrates. Once invertebrate lunar/tidal genes are identified, homology should allow the isolation of similar genes in vertebrates like red knots. We might predict that the tidal genes that generate the approximately 12.4 h behavioural cycles might also encode cycling mRNAs by analogy with their circadian counterparts. Might these (as yet unidentified) putatively 12 h tidally cycling mRNAs show among-individual fluctuations to account for the variation in tidal rhythms observed in red knots? Could these mRNAs still be cycling in the biparental incubating species but their output is suppressed? Would any future identification of a tidally cycling mRNA in a tidal vertebrate suggest a co-option of a previously 12 h cycling mRNA in a terrestrial circadian species [45,46] that was re-used to generate tidal phenotypes when the species moved to an intertidal environment?
Whatever the identity of these tidal or lunar genes, the conservation of circadian genes in invertebrates and vertebrates might suggest that the same will be true also for tidal and lunar genes [47]. Tidal genes will initially be identified in invertebrates, but homology with vertebrate genes will be expected to open up interesting possibilities for mechanistic studies of the clock in intertidal birds. For example, using in situ hybridization will identify the brain regions that have tidally cycling molecules and comparing these regions with those areas that show circadian cycling molecules will detect both oscillators.
In addition, we must not forget the obvious, that behavioural ecology scenarios are far more complex than those we play out in the confines of the laboratory. As we have learned with shorebirds, individuals vary in their foraging rhythms, and behavioural rhythms during incubation are very loosely coupled to the major environmental cycles [14]. Consequently, the modulation of molecular rhythms by other selection pressures will provide a novel background against which to study biological rhythmicity within an ecologically realistic framework. Indeed, when rodents or flies are placed in semi-natural environments and their circadian rhythms monitored, quite startling results can be observed that could not have been predicted from laboratory studies and which question some of the assumptions made about the adaptive value of the circadian clock [48–50, but see also 51]. As with the incubation study of biparental shorebirds [14], when realistic scenarios are used to study biological rhythms, the results do not meet expectations. We, therefore, encourage behavioural ecologists and chronobiologists to seek collaborations, particularly as the long-term spatial and temporal monitoring of individuals in the field becomes feasible [52] and the new post-genomic age allows molecular study of organisms other than laboratory flies or mice. We anticipate that a fertile hybrid area of research will evolve, perhaps slowly at first, but with a real potential to significantly illuminate our understanding of the functional and adaptive roles of biological rhythms.
Acknowledgements
C.P.K. thanks the BBSRC for funding the Eurydice study and Tobias Kaiser for generously providing him with a final draft of his study on C. marinus while it was still in press. M.B. thanks everyone who made the incubation study possible, M. Valcu for statistical advice, Borce and Maje for patience, the Open Science Framework for the free data and script deposition, and B. Kempenaers (MPIO) for funding and support. M.B. did part of this work as a PhD student in the International Max Planck Research School for Organismal Biology.
Ethics
Research using animals shown in the case studies adhered to local guidelines and appropriate ethical approval and licenses were obtained.
Data accessibility
Information on (methods and actograms) data and results for the red knot foraging and additional incubation results are freely available at https://osf.io/xby9t/ [16]. Data for the shorebird incubation results were retrieved from Open Science Framework, https://osf.io/wxufm/, which also contains further supporting information from the original study [14].
Authors' contributions
C.P.K., M.B., T.P. conceived the paper. T.O., A.I.B. and T.P. are responsible for the red knot data, which T.O. with M.B. analysed. M.B. is responsible for the incubation case study and figure 1 and C.P.K. for reviewing the molecular case studies in invertebrates. C.P.K. also drafted the general introduction and discussion, and M.B. drafted the shorebird introduction and discussion. C.P.K. and M.B. wrote the final version of the paper with input from all authors.
Competing interests
We have no competing interests.
Funding
C.P.K. is funded by the BBSRC (grant no. BB/K009702/1), M.B. is funded by the EU Marie Curie individual fellowship (grant no. 658425, ‘Social Jet Lag’ project), Czech University of Life Sciences and Max Planck Society. The research on red knots in Mauritania was funded by an NWO-VIDI grant awarded to Jan A. van Gils (grant no. 864.09.002) and an NWO-TOP grant (‘Shorebirds in space’, grant no. 854.11.004) awarded to T.P.
References
- 1.Alerstam T. 1990. Bird migration. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 2.Hut RA, Paolucci S, Dor R, Kyriacou CP, Daan S. 2013. Latitudinal clines: an evolutionary view on biological rhythms. Proc. R. Soc. B 280, 20130433 ( 10.1098/rspb.2013.0433) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de la Iglesia HO, Johnson CH. 2013. Biological clocks: riding the tides. Curr. Biol. 23, R921–R923. ( 10.1016/j.cub.2013.09.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunlap J. 2004. Chronobiology. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 5.Naylor E. 2010. Chronobiology of marine organisms. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 6.Palmer JD. 2000. The clocks controlling the tide-associated rhythms of intertidal animals. Bioessays 22, 32–37. ( 10.1002/(SICI)1521-1878(200001)22:1%3C32::AID-BIES7%3E3.0.CO;2-U) [DOI] [PubMed] [Google Scholar]
- 7.Kaiser TS, et al. 2016. The genomic basis of circadian and circalunar timing adaptations in a midge. Nature 540, 69–73. ( 10.1038/nature20151) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.del Hoyo J, Elliott A, Sargatal J. 1996. Handbook of the birds of the world. Barcelona: Lynx Edicions. [Google Scholar]
- 9.van de Kam J, Ens B, Piersma T, Zwart L. 2004. Shorebirds: an illustrated behavioural ecology. Zeist, The Netherlands: KNNV Publishing. [Google Scholar]
- 10.Ens BJ, Underhill LG. 2014. Synthesis of oystercatcher conservation assessments: general lessons and recommendations. Int. Wader Stud. 20, 5–22. [Google Scholar]
- 11.Helm B, Gwinner E, Koolhaas A, Battley P, Schwalb I, Dekinga A, Piersma T. 2012. Avian migration: temporal multitasking and a case study of melatonin cycles in waders. In The neurobiology of circadian timing (eds A Kalsbeek, M Merrow, T Roenneberg, RG Foster), pp. 457–479. Amsterdam: Elsevier. [DOI] [PubMed] [Google Scholar]
- 12.Daan S, Koene P. 1981. On the timing of foraging flights by oystercatchers, Haematopus ostralegus, on tidal mudflats. Neth. J. Sea Res. 15, 1–22. ( 10.1016/0077-7579(81)90002-8) [DOI] [Google Scholar]
- 13.Hötker H. 1995. [Activity rhythms of shelducks (Tadorna tadorna) and waders (Charadrii) on the North Sea coast]. J. Ornithol. 136, 105–126 (in German with English abstract) ( 10.1007/BF01651234) [DOI] [Google Scholar]
- 14.Bulla M, et al. 2016. Defying the 24-h day: unexpected diversity in socially synchronized rhythms of shorebirds Nature 540, 109–113. ( 10.1038/nature20563) [DOI] [PubMed] [Google Scholar]
- 15.Foster RG, Wulff K. 2005. The rhythm of rest and excess. Nat. Rev. Neurosci. 6, 407–414. ( 10.1038/nrn1670) [DOI] [PubMed] [Google Scholar]
- 16.Bulla M, Oudman T, Bijleveld AI. 2017. Supporting information for ‘Marine biorhythms: bridging chronobiology and ecology’ Open Sci. Framework. ( 10.17605/OSF.IO/XBY9T) [DOI] [PMC free article] [PubMed]
- 17.Zwarts L, Blomert A, Hupkes R. 1990. Increase of feeding time in waders preparing for spring migration from the Banc d'Arguin, Mauritania. Ardea 78, 237–256. [Google Scholar]
- 18.Bulla M, et al. 2016. Supplementary data 3 – study sites: location, population wing length, monitoring method, tide. Version 11. figshare ( 10.6084/m9.figshare.1536260.v11) [DOI] [Google Scholar]
- 19.Piersma T. 2007. Using the power of comparison to explain habitat use and migration strategies of shorebirds worldwide. J. Ornithol. 148(Suppl. 1), S45–S59. ( 10.1007/s10336-007-0240-3) [DOI] [Google Scholar]
- 20.Buehler DM, Piersma T. 2008. Travelling on a budget: predictions and ecological evidence for bottlenecks in the annual cycle of long-distance migrants. Phil. Trans. R. Soc. B 363, 247–266. ( 10.1098/rstb.2007.2138) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Gils JA, Piersma T, Dekinga A, Battley PF. 2006. Modelling phenotypic flexibility: an optimality analysis of gizzard size in red knots (Calidris canutus). Ardea 94, 409–420. [Google Scholar]
- 22.Piersma T, Hoekstra R, Dekinga A, Koolhaas A, Wolf P, Battley PF, Wiersma P. 1993. Scale and intensity of intertidal habitat use by knots Calidris canutus in the western Wadden Sea in relation to food, friends and foes. Neth. J. Sea Res. 31, 331–357. ( 10.1016/0077-7579(93)90052-T) [DOI] [Google Scholar]
- 23.Piersma T, van Aelst R, Kurk K, Berkhoudt H, Maas LRM. 1998. A new pressure sensory mechanism for prey detection in birds: the use of principles of seabed dynamics? Proc. R. Soc. Lond. B 265, 1377–1383. ( 10.1098/rspb.1998.0445) [DOI] [Google Scholar]
- 24.Bijlsma RG. 1990. Predation by large falcons on wintering waders on the Banc d'Arguin, Mauritania. Ardea 78, 82. [Google Scholar]
- 25.van den Hout PJ, van Gils JA, Robin F, van der Geest M, Dekinga A, Piersma T. 2014. Interference from adults forces young red knots to forage for longer and in dangerous places. Anim. Behav. 88, 137–146. ( 10.1016/j.anbehav.2013.11.020) [DOI] [Google Scholar]
- 26.Sitters HP, Gonzalez PM, Piersma T, Baker AJ, Price DJ. 2001. Day and night feeding habitat of Red Knots in Patagonia: profitability versus safety? J. Field Ornithol. 72, 86–95. ( 10.1648/0273-8570-72.1.86) [DOI] [Google Scholar]
- 27.Rogers DI, Battley PF, Piersma T, Van Gils JA, Rogers KG. 2006. High-tide habitat choice: insights from modelling roost selection by shorebirds around a tropical bay. Anim. Behav. 72, 563–575. ( 10.1016/j.anbehav.2005.10.029) [DOI] [Google Scholar]
- 28.Piersma T, Gill REJ, de Goeij P, Dekinga A, Shepherd ML, Ruthrauff D, Tibbitts L. 2006. Shorebird avoidance of nearshore feeding and roosting areas at night correlates with presence of a nocturnal avian predator. Wader Study Group Bull. 109, 73–76. [Google Scholar]
- 29.Laundré JW, Hernández L, Ripple WJ. 2010. The landscape of fear: ecological implications of being afraid. Open Ecol. J. 3, 1–7. ( 10.2174/1874213001003030001) [DOI] [Google Scholar]
- 30.Steiger SS, Valcu M, Spoelstra K, Helm B, Wikelski M, Kempenaers B. 2013. When the sun never sets: diverse activity rhythms under continuous daylight in free-living arctic-breeding birds. Proc. R. Soc. B 280, 20131016 ( 10.1098/rspb.2013.1016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beason RC, Wiltschko W. 2015. Cues indicating location in pigeon navigation. J. Comp. Physiol. A Neuroethol Sens. Neural Behav. Physiol. 201, 961–967. ( 10.1007/s00359-015-1027-2) [DOI] [PubMed] [Google Scholar]
- 32.Ozkaya O, Rosato E. 2012. The circadian clock of the fly: a neurogenetics journey through time. Adv. Genet. 77, 79–123. ( 10.1016/B978-0-12-387687-4.00004-0) [DOI] [PubMed] [Google Scholar]
- 33.Wilcockson D, Zhang L. 2008. Circatidal clocks. Curr. Biol. 18, R753–R755. ( 10.1016/j.cub.2008.06.041) [DOI] [PubMed] [Google Scholar]
- 34.Takekata H, Numata H, Shiga S. 2014. The circatidal rhythm persists without the optic lobe in the mangrove cricket Apteronemobius asahinai. J. Biol. Rhythms 29, 28–37. ( 10.1177/0748730413516309) [DOI] [PubMed] [Google Scholar]
- 35.Weber F, Hung HC, Maurer C, Kay SA. 2006. Second messenger and Ras/MAPK signalling pathways regulate CLOCK/CYCLE-dependent transcription. J. Neurochem. 98, 248–257. ( 10.1111/j.1471-4159.2006.03865.x) [DOI] [PubMed] [Google Scholar]
- 36.Zhang L, Hastings MH, Green EW, Tauber E, Sladek M, Webster SG, Kyriacou CP, Wilcockson DC. 2013. Dissociation of circadian and circatidal timekeeping in the marine crustacean Eurydice pulchra. Curr. Biol. 23, 1863–1873. ( 10.1016/j.cub.2013.08.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Satoh A, Yoshioka E, Numata H. 2008. Circatidal activity rhythm in the mangrove cricket Apteronemobius asahinai. Biol. Lett. 4, 233–236. ( 10.1098/rsbl.2008.0036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takekata H, Matsuura Y, Goto SG, Satoh A, Numata H. 2012. RNAi of the circadian clock gene period disrupts the circadian rhythm but not the circatidal rhythm in the mangrove cricket. Biol. Lett. 8, 488–491. ( 10.1098/rsbl.2012.0079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takekata H, Numata H, Shiga S, Goto SG. 2014. Silencing the circadian clock gene Clock using RNAi reveals dissociation of the circatidal clock from the circadian clock in the mangrove cricket. J. Insect Physiol. 68, 16–22. ( 10.1016/j.jinsphys.2014.06.012) [DOI] [PubMed] [Google Scholar]
- 40.Neumann D, Heimbach F. 1984. Time cues for semilunar reproduction rhythms in European populations of Clunio marinus. II The influence of tidal temperature cycles. Biol. Bull. 166, 509–524. ( 10.2307/1541158) [DOI] [Google Scholar]
- 41.Kon N, et al. 2014. CaMKII is essential for the cellular clock and coupling between morning and evening behavioral rhythms. Genes Dev. 28, 1101–1110. ( 10.1101/gad.237511.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harrisingh MC, Wu Y, Lnenicka GA, Nitabach MN. 2007. Intracellular Ca2+ regulates free-running circadian clock oscillation in vivo. J. Neurosci. 27, 12 489–12 499. ( 10.1523/JNEUROSCI.3680-07.2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zantke J, Ishikawa-Fujiwara T, Arboleda E, Lohs C, Schipany K, Hallay N, Straw AD, Todo T, Tessmar-Raible K. 2013. Circadian and circalunar clock interactions in a marine annelid. Cell. Rep. 5, 99–113. ( 10.1016/j.celrep.2013.08.031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raible F, Tessmar-Raible K. 2014. Platynereis dumerilii. Curr. Biol. 24, R676–R677. ( 10.1016/j.cub.2014.06.032) [DOI] [PubMed] [Google Scholar]
- 45.Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. 2002. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr. Biol. 12, 540–550. ( 10.1016/S0960-9822(02)00759-5) [DOI] [PubMed] [Google Scholar]
- 46.Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB. 2009. Harmonics of circadian gene transcription in mammals. PLoS Genet. 5, e1000442 ( 10.1371/journal.pgen.1000442) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clayton JD, Kyriacou CP, Reppert SM. 2001. Keeping time with the human genome. Nature 409, 829–831. ( 10.1038/35057006) [DOI] [PubMed] [Google Scholar]
- 48.Gattermann R, et al. 2008. Golden hamsters are nocturnal in captivity but diurnal in nature. Biol. Lett. 4, 253–255. ( 10.1098/rsbl.2008.0066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daan S, et al. 2011. Lab mice in the field: unorthodox daily activity and effects of a dysfunctional circadian clock allele. J. Biol. Rhythms 26, 118–129. ( 10.1177/0748730410397645) [DOI] [PubMed] [Google Scholar]
- 50.Vanin S, Bhutani S, Montelli S, Menegazzi P, Green EW, Pegoraro M, Sandrelli F, Costa R, Kyriacou CP. 2012. Unexpected features of Drosophila circadian behavioural rhythms under natural conditions. Nature 484, 371–375. ( 10.1038/nature10991) [DOI] [PubMed] [Google Scholar]
- 51.Spoelstra K, Wikelski M, Daan S, Loudon AS, Hau M. 2016. Natural selection against a circadian clock gene mutation in mice. Proc. Natl Acad. Sci. USA 113, 686–691. ( 10.1073/pnas.1516442113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dominoni D, Åkesson S, Klaassen R, Spoelstra K, Bulla M. 2017. Methods in field chronobiology. Phil. Trans. R. Soc. B 372, 20160247 ( 10.1098/rstb.2016.0247) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Bulla M, Oudman T, Bijleveld AI. 2017. Supporting information for ‘Marine biorhythms: bridging chronobiology and ecology’ Open Sci. Framework. ( 10.17605/OSF.IO/XBY9T) [DOI] [PMC free article] [PubMed]
Data Availability Statement
Information on (methods and actograms) data and results for the red knot foraging and additional incubation results are freely available at https://osf.io/xby9t/ [16]. Data for the shorebird incubation results were retrieved from Open Science Framework, https://osf.io/wxufm/, which also contains further supporting information from the original study [14].