Abstract
The theory of species coexistence is a key concept in ecology that has received much attention. The role of rapid evolution for determining species coexistence is still poorly understood although evolutionary change on ecological time-scales has the potential to change almost any ecological process. The influence of evolution on coexistence can be especially pronounced in microbial communities where organisms often have large population sizes and short generation times. Previous work on coexistence has assumed that traits involved in resource use and species interactions are constant or change very slowly in terms of ecological time-scales. However, recent work suggests that these traits can evolve rapidly. Nevertheless, the importance of rapid evolution to coexistence has not been tested experimentally. Here, we show how rapid evolution alters the frequency of two bacterial competitors over time when grown together with specialist consumers (bacteriophages), a generalist consumer (protozoan) and all in combination. We find that consumers facilitate coexistence in a manner consistent with classic ecological theory. However, through disentangling the relative contributions of ecology (changes in consumer abundance) and evolution (changes in traits mediating species interactions) on the frequency of the two competitors over time, we find differences between the consumer types and combinations. Overall, our results indicate that the influence of evolution on species coexistence strongly depends on the traits and species interactions considered.
Keywords: community dynamics, experimental evolution, eco-evolutionary dynamics, interaction, Pseudomonas fluorescens SBW25, Tetrahymena thermophila
1. Introduction
The diversity observed in microbial communities living in seemingly simple environments is overwhelming. One of the fundamental questions in (microbial) community ecology is how a large number of species can coexist while sharing only one or few resources [1–3]. Several conditions have been observed to slow down or prevent competitive exclusion, e.g. differences in resource use, trade-offs, spatial and temporal separation of competitors and nonlinear species interactions [4]. Furthermore, individual traits of species that determine fitness differences, niche overlap and species interactions with density or frequency dependency are important factors in regulating the coexistence of competing species. Generally, equalizing fitness among species and decreasing their niche overlap (stabilizing effect) facilitate the coexistence of competitors.
Consumers such as predators, parasites or herbivores can have significant effects on the outcome of competition and thus the maintenance of species diversity [4,5]. In microbial communities, lysis caused by bacteriophages (i.e. phages) and predation by protists represent major causes of mortality, and these interactions can be very important for the coexistence of bacteria [6]. They can, for example, result in apparent competition or create new niches where each species has its own specialist consumer (e.g. [1,7–9]). In particular, specialist and generalist consumers can have qualitatively different effects on coexistence of resource (prey) populations [10]. When specialist consumers dominate food webs, prey species are mainly connected through shared basal resources (e.g. nutrients). With generalists as consumers, prey species are also connected via the shared consumers and coexistence of prey species is possible through niche differentiation into grazing resistance (i.e. reduced vulnerability to predation [11]) and competitive ability [12,13].
Classic ecological theory on coexistence has focused on interspecific differences in traits. However, more recent work has shown that traits and species interactions within food webs are not rigid and that evolutionary change—de novo mutations and changes in genotype frequencies from standing genetic variation over time—can occur at the same temporal scale as ecological processes, as recently reviewed in [14]. Weakly defended but highly competitive prey genotypes can, for example, coexist with highly defended but weakly competitive prey genotypes in the presence of a predator where the frequency of the two prey genotypes fluctuates over time [9,15–20]. Besides trait polymorphisms within a population, consumer–resource interactions can lead to rapid de novo evolution and trait divergence between prey species [21,22], as well as increasing intraspecific trait variation in prey populations [15,16,20]. Similarly, host–parasite interactions often give rise to rapid coevolution of bacteria and their parasitic phages (e.g. [23–25]), which has also been observed with other microbes [26]. Differences in consumers and their interactions with the resource population can lead to different evolutionary changes in the traits of resource populations. Specialized phages typically lead to phage-resistant mutants rapidly overtaking bacterial populations [24,27], which has been shown to affect species coexistence [28]. Defence traits against grazing by a generalist consumer, such as protozoa, typically evolve at a slower pace and provide only partial protection [22].
Trait evolution in response to the presence of predators and phage also depends on community structure leading to diffuse coevolution [29]. Increasing community complexity at the competitor or consumer level has been commonly observed to slow down evolutionary change, because population sizes are reduced, lowering the supply of adaptive mutations and/or encounter rates [30–32]. Nevertheless, little is known about how consumer community composition affects the rapid evolution and coexistence of competing species. On the basis of previous work on consumer-mediated coexistence and the fact that rapid evolution can alter species interactions, we hypothesize that the effect of evolutionary change on the coexistence of competitors depends on the consumer community present.
In order to test how adaptive evolution in consumer resource communities can alter coexistence over time, we used an experimental evolution approach to manipulate the community structure in simple laboratory microcosms. Specifically, we ask how changes in frequencies of two competing bacterial populations change with food web structure and whether and how rapid evolutionary changes in traits mediating species interactions alter these frequencies over time in relation to population size changes. For this purpose, we tracked the population and evolutionary dynamics of two bacterial species, Pseudomonas fluorescens and Escherichia coli when (i) growing without a consumer (treatment hereafter: ‘Bacteria’), (ii) with a generalist consumer (one ciliate species, Tetrahymena thermophila; ‘Ciliate’), (iii) with specialized consumers (specific phages for both bacteria: T4 for E. coli and ϕ2 for P. fluorescens; ‘Phage’) and (iv) with all three consumers (All). We used the same isogenic lines of the bacteria to inoculate the experiments to minimize the standing genetic variation in the beginning of the experiment, simplifying the experimental set-up. We assessed the ecological dynamics by measuring changes in population size for 60 days and evolutionary dynamics by testing whether the bacteria evolved resistance against the phage and defence against the ciliate (measured as prey defence level, D) as well as bacterial resource use (measured as growth in used medium of the other bacterial species).
With this experimental set-up, we could test the combined effects of specialist and generalist consumers on trait evolution of two competing bacteria and whether and how these evolutionary changes and changes in consumer densities (ecology) alter coexistence of the two competitors over time, here measured as changes in the frequency of the two bacterial species. Ideally, one would contrast experiments with and without evolution to test for the role of evolution for coexistence. However, as evolutionary adaptation in bacterial populations in experimental studies lasting for hundreds of generations is common [33], evolution cannot be suppressed without extensive experimental manipulation and temporal interruption of species interactions. To disentangle the roles of ecology and evolution in coexistence of the two bacteria over time, we used the Geber method [14,34,35] to decompose the effects of rapid adaptation on coexistence a posteriori. The method is based on the notion that the frequency of Pseudomonas fluorescens depends on densities of the bacteria and consumers (i.e. ecology) and how well adapted the bacteria are against these negative interactions (i.e. evolution via heritable defence traits). We assume here that all measured trait changes are heritable rather than being plastic. The relative importance of these changes can be partitioned with a two-way ANOVA.
We found that manipulating consumer community structure affected the frequencies of the two bacteria over time and that predation by the generalist was the primary force facilitating coexistence within and across populations. Moreover, the highest frequency of P. fluorescens was observed in communities with both the generalist and the specialists present. Our results show that the relative contributions of evolution (temporal changes in the resistance and/or defence traits) and ecology (population sizes of the consumers) to changes in the frequency of the two bacteria over time were strongly dependent on community structure and time. Interestingly, the contribution of evolution in the frequency changes of the two bacteria was negligible when phages were present.
2. Material and methods
(a). Model system
As host/prey, we used two bacterial species: P. fluorescens SBW25 [36] and E. coli ATCC 11303. As a specialist consumer species, we used two lytic bacteriophages: ϕ2 [24] specific to P. fluorescens and T4 ATCC 11303-B4 specific to E. coli. As a generalist consumer, capable of consuming both bacterial species, we used the ciliated protozoan T. thermophila 1630/1U (CCAP). Generally, the ancestral E. coli grows faster and reaches higher densities when cultured alone (electronic supplementary material, figure S1), and is more limited by predation at high bacterial densities than P. fluorescens (electronic supplementary material, figure S2). Prior to the experiments, all bacterial and phage stocks were kept at –80°C and ciliate stocks were cultured axenically in proteose peptone yeast extract (PPY) medium containing 20 g of proteose peptone and 2.5 g of yeast extract in 1 l of deionized water. All treatments were started from clonal cultures of bacteria and phages to achieve minimal initial genetic variability in populations.
(b). Microcosm experiment and manipulating community structure
Experiments were conducted in standard 25 ml glass vials (a microcosm type previously used, e.g. in [21,24,33,37–39]) with 6 ml medium containing M9 salts and King's B (KB) nutrients at 1% concentration (1% KB: 0.2 g l−1 Peptone number 3 and 0.1 ml l−1 glycerol). All treatments were replicated three times. Every 48 h, 1% of each culture (approx. (0.4–1.6) × 105 cells) was transferred to a new vial containing fresh culture medium. Microcosms were kept at 28 ± 0.1°C and shaken constantly (50 r.p.m.). We manipulated the community structure by having (i) bacteria only (in all treatments both species present), (ii) bacteria and generalist ciliate consumer, (iii) bacteria and both of their specific phage consumers and (iv) bacteria and all three consumers. The duration of the experiment was 60 days, representing approximately 340 bacterial and 190 ciliate generations.
(c). Community dynamics
During each transfer, a 0.5 ml subsample from each vial was frozen with 0.5 ml of 80% glycerol and kept at –80°C for later analysis (ciliates do not survive freezing under these conditions). Total bacterial biomass was estimated as optical density (OD) at 600 nm (UV-1800 spectrophotometer, Shimadzu, Japan). We estimated the relative frequency of the two bacterial species with selective agar plating of dilution series. For P. fluorescens, we plated samples on agar containing CFC selective supplement (CFC supplement: 10 mg of cetrimide and fucidin and 50 mg cephalosporin in 1 l of PPY agar). For E. coli, we used Tryptone Bile X-Glucuronide agar. We cultured all samples for 48 h: P. fluorescens at 28°C and E. coli at 37°C to obtain optimal growth for the target species. With these selective media and culture conditions, we were able to clearly distinguish and enumerate both bacterial species from mixed samples. At this point, we also isolated 16 individual colonies (clones) from both species that were stored at –80°C for later analysis. Phage densities were enumerated with plaque assay in which the two phage species were distinguished by plating mixed phage samples to agar plates to two separate agar plates, each containing one host species. Tetrahymena thermophila cell densities were enumerated directly from live 2.5 µl subsamples using a compound microscope (Zeiss Axioskop 2 plus, Oberkochen, Germany).
(d). Evolutionary changes in predator defence and phage resistance
Evolution of the prey defence trait D against predator grazing was quantified with a simple, ecologically relevant bioassay, as described in detail in [21]. Briefly, after thawing cryopreserved bacterial clones, we grew samples in liquid culture (1% KB) for 24 h, corresponding to approximately 10 bacterial generations, so that the subsequently measured phenotypic differences resulted from evolutionary change rather than an induced defence mechanism. We then mixed all 16 clones in equal proportions, added 100 µl of the mixture into 2 ml of fresh culture medium, and added 2100 ciliates from the stock: that is, we used naive predators as a standard for consumer feeding on genetically differentiated prey. Predator numbers were counted after 48 h, and differences in predator densities compared with predators grown on naive prey were taken as an estimate for the prey defence level D. Prey defence trait values were calculated as relative fitness by 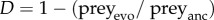 , where preyevo is the predator density after feeding on evolved prey, and preyanc is the predator density after feeding on ancestral prey. Ciliate coevolution was measured by comparing ancestor ciliates and populations isolated at the end of the experiment. These were fed with sympatric bacterial populations evolved with ciliates in the experiment. However, we did not observe any ciliate coevolution in our experiment (detailed methods and data in the electronic supplementary material).
, where preyevo is the predator density after feeding on evolved prey, and preyanc is the predator density after feeding on ancestral prey. Ciliate coevolution was measured by comparing ancestor ciliates and populations isolated at the end of the experiment. These were fed with sympatric bacterial populations evolved with ciliates in the experiment. However, we did not observe any ciliate coevolution in our experiment (detailed methods and data in the electronic supplementary material).
We also estimated the bacterial phage resistance of 16 clones from each population using a modified version of a streak test commonly used in bacterial phage studies (e.g. [23,24,37]). We first cultured each clone overnight in a liquid medium (PPY medium in 96-well plates, each well containing one clone) and then used a multichannel pipette to inoculate 10 µl samples into a Petri dish containing PPY agar and a thin layer of soft agar with a high concentration of the coevolved phage isolated from the same time point as the host clones (cf. [24]). The ability to form visible colonies (indicating resistance) was evaluated for each clone after 24 h.
(e). Data analyses
We used generalized estimating equation models (geeGLMs) to compare the frequency of P. fluorescens, bacteria densities, ciliate densities, D for E. coli and P. fluorescens, phage ϕ2 and phage T4 numbers, as well as average resistance against the phages as data were distributed non-normally and to account for the temporal autocorrelation of the time series. We used the function geeglm from R package geepack [40–42] with the family Gamma (for the comparison of the frequency of P. fluorescens and phage resistance, we added 0.01 to all data in order to have only positive values; we added 1 to all phage number values in order to have only positive values). We used Kruskal–Wallis rank sum tests to test for differences in the coefficient of variation of defence level as CV data were non-normal. We used, for each bacteria species, Kruskal–Wallis rank sum tests to test for differences between resource use at the start and end between treatments (non-normal data). All analyses were performed in R [43].
(f). Eco–evo contribution
We tested how the community composition affected the contribution of ecology and evolution in the change in the frequency of the bacteria using the Geber method [14] described in [34]. More details and an example are shown in the electronic supplementary material. Specifically, we asked how the change in the frequency of P. fluorescens is explained by ciliate and E. coli densities (i.e. ecology) and defence traits (i.e. evolution) or phage densities (i.e. ecology) and resistance trait (i.e. evolution) over time. We calculated the contributions of ecology and evolution for all time points for which we had density and P. fluorescens defence and/or resistance data and used the data specific to these time points. Since we use species frequencies, the results are qualitatively identical to the results that would be obtained using E. coli as a focal competitor. As we found no evidence for evolutionary change in the predator and resource use of the bacteria (see above and electronic supplementary material), we did not include these evolutionary processes in our analysis. Furthermore, we were not able to consider evolution of the phage separately for this as our resistance measurements with only contemporary phage did not allow detection of phage coevolution.
We tested for differences in the contribution of ecology and evolution (Eco, Evo) between treatments (Ciliate, Phage, All) and time (Sampling day) using geeGLM models. To further disentangle differences in the ecological and evolutionary contribution of the ciliate and phage in the ‘All’ treatment, we used geeGLM models with consumer (Ciliate, Phage), contributor (Eco, Evo) as an explanatory variable.
3. Results
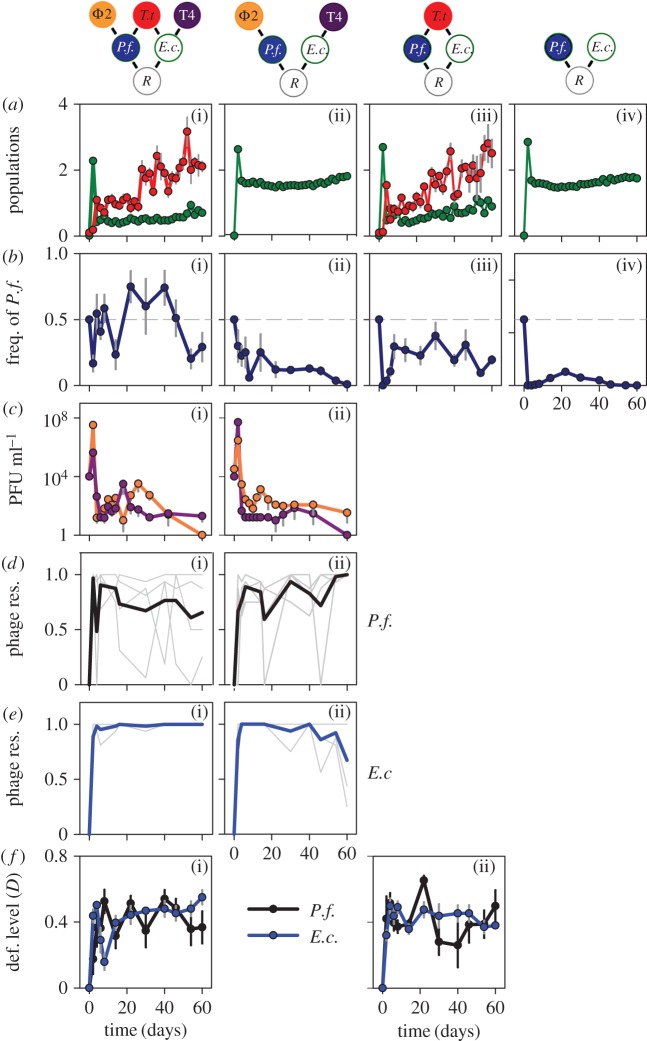
Population and evolutionary dynamics differed significantly depending on community structure (figure 1). Most importantly, increasing community complexity, in the presence of ciliates, facilitated coexistence of the bacteria P. fluorescens and E. coli over time (geeGLM, treatment: χ2 = 521, d.f. = 3, p < 2 × 10−16, treatment × days: χ2 = 22, d.f. = 3, p < 5.2 × 10−5; figure 1b(i–iv); for additional statistical results, see electronic supplementary material, table S2), with the lowest average frequency of P. fluorescens over time when there was no consumer and the highest frequencies when all consumer types were present. Overall bacterial densities were also lowest in the ‘All’ and highest in the ‘Bacteria-only’ treatments (geeGLM, treatment: χ2 = 507, d.f. = 3, p < 2 × 10−16; figure 1a(i–iv); electronic supplementary material) and changed over time depending on the treatment (geeGLM, treatment × days: χ2 = 11, d.f. = 3, p = 0.013). Ciliate densities were similar between treatments (geeGLM: χ2 = 0, d.f. = 1, p = 0, figure 1a(i)) and changed over time (geeGLM: days: χ2 = 173.6, d.f. = 1, p < 2 × 10−16) depending on the treatment (treatment × days: χ2 = 3.9, d.f. = 3, p = 0.049). Phage ϕ2 numbers differed significantly between treatments and over time between treatments (geeGLM, treatment: χ2 = 49.2, d.f. = 1, p < 2.4*−12, treatment × days: χ2 = 4.3, d.f. = 1, p = 0.039) with lower ϕ2 densities in the presence of the ciliate. Phage T4 numbers were not different between the ‘All’ and ‘Phage’ treatments (geeGLM: treatment χ2 = 2.87, d.f. = 1, p = 0.09). Even though phage densities became very low in some populations, extinctions were not observed.
Figure 1.
Ecological and evolutionary dynamics of two bacteria when grown without consumers (a(iv), b(iv)), with the ciliate T. thermophila (a(iii), b(iii), f(ii)), two phages, ϕ2 and T4 (a(ii)–e(ii)), or with all consumers (a(i)–f(i)). Average prey population size and stability of the P. fluorescens proportion are presented in electronic supplementary material, table S1. Community structure (top row): R = shared resource for both bacteria, E.c. = E. coli, P.f. = P. fluorescens, T.t. = T. thermophila. (a) Combined bacterial biomass (OD × 10) and T.t. cells ml−1 × 105 (mean ± s.e.). (b) Frequency of P. fluorescens over time (mean ± s.e.). Dashed horizontal line indicates equal proportions. (c) Phage densities (plaque forming unit, PFU ml−1): ϕ2 (orange line), T4 (purple line) (mean ± s.e.). (d) Pseudomonas fluorescens resistance (res.) against contemporary ϕ2 (black line: average; grey lines: individual replicates). (e) Escherichia coli resistance against contemporary T4 (blue line: average; light grey lines: individual replicates). (f) Bacterial defence (def.) against ciliates (mean ± s.e.): E. coli (blue line) and P. fluorescens (black line).
To follow the evolutionary response to ciliate predation, we measured the fitness of the ancestral predator when grown on ancestral and evolved bacteria isolated from different time points. From this, we calculated the defence level D, with 0 meaning that the evolved bacteria had the same level of defence as the ancestor and values close to 1 signifying a very high level of defence compared with the ancestor. We performed this assay separately for each bacterium. Both bacteria evolved defence against consumption by the ciliate (associated with notable increase in cell aggregation) within the first two to four transfers (approx. 10–20 generations, figure 1f(i,ii)). Overall, we found no difference in the levels of defence between E. coli and P. fluorescens (geeGLM for bacterial species: χ2 = 0.42, d.f. = 1, p = 0.52). The defence levels of E. coli and P. fluorescens did not differ between treatments (geeGLM: E. coli χ2 = 1, d.f. = 0.15, p = 0.68: P. fluorescens: χ2 = 1, d.f. = 0.28, p = 0.6). However, we observed significantly lower temporal stability of the defence level D compared with E. coli (measured as coefficient of variation: CV mean ± s.d.; ciliate: E. coli 0.19 ± 0.03, P. fluorescens 0.62 ± 0.31; all: E. coli 0.31 ± 0.10, P. fluorescens 0.44 ± 0. 13; Kruskal–Wallis test for bacteria: χ2 = 8.04, d.f. = 1, p = 0.005) and independent of treatment (Kruskal–Wallis: χ2 = 0.17, d.f. = 1, p = 0.67).
In the treatments with phages, E. coli rapidly evolved resistance against the phage and this depended on the presence of ciliates (geeGLM: treatment χ2 = 1.27 × 1016, d.f. = 1, p < 2 × 10−16; days: χ2 = 1 × 1029, d.f. = 1, p < 2 × 10−16, treatment × days: χ2 = 1. 37 × 1012, d.f. = 1, p < 2 × 10−16; figure 1e(i,ii)). We also found significant differences between treatments for the evolution of resistance of P. fluorescens against its phage ϕ2, with lower levels of resistance in the presence of the predator (geeGLM: treatment χ2 = 6.52, d.f. = 1, p = 0.011; days: χ2 = 16.33, d.f. = 1, p = 5.3 × 10−5; figure 1d(i,ii)).
There were no evolutionary changes in bacterial resource use based on the growth of P. fluorescens in an E. coli filtrate from the start and endpoint of the experiment; the same applied to E. coli growth in a P. fluorescens filtrate (see electronic supplementary material, methods; Kruskal–Wallis rank test E. coli: treatment χ2 = 1.23, d.f. = 3, p = 0.75; time: χ2 = 0.75, d.f. = 1, p = 0.39; P. fluorescens: treatment: χ2 = 4.72, d.f. = 3, p = 0.19; time: χ2 = 0.01, d.f. = 1, p = 0.92).
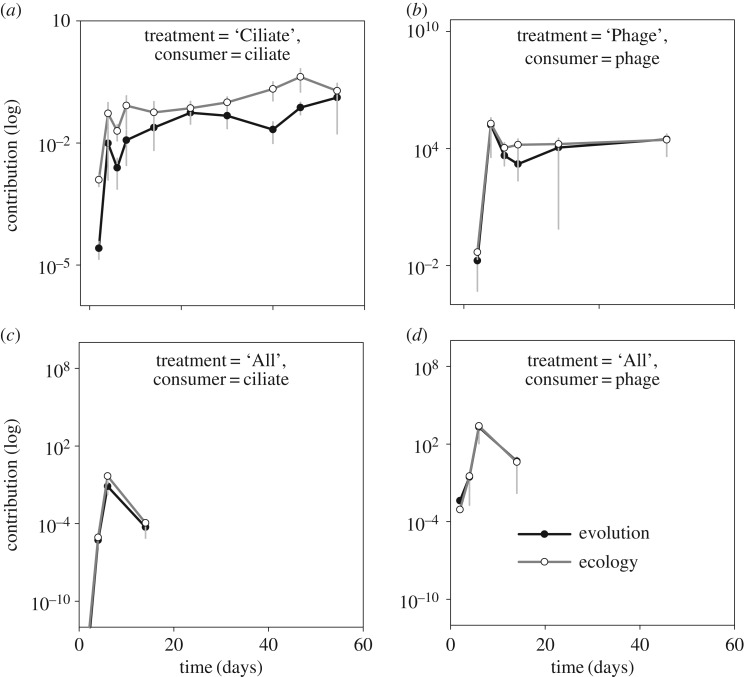
We tested how the consumer affected the roles of evolution and ecology in the frequency of P. fluorescens and compared the contributions of evolutionary and ecological change between the ‘Ciliate’, ‘Phage’ and ‘All’ treatments (figure 2). We found that the contributions of ecology and evolution to the change in the frequency of P. fluorescens differed between treatments (geeGLM treatment: χ2 = 10.69, d.f. = 2, p = 0.005). Specifically, the contribution of ecology was greater than the evolutionary contribution in the ‘Ciliate’ and ‘Phage’ treatments, whereas both had equal contributions in the ‘All’ treatment. We further found differences in contributions between treatments with ciliates (‘Ciliate’ and ‘All’ treatments: geeGLM treatment: χ2 = 4.83, d.f. = 1, p = 0.028). Within the ‘All’ treatment, the ciliate and phage had significantly different contributions to the changes in P. fluorescens frequency (geeGLM, interaction between consumer and contribution: χ2 = 5.98, d.f. = 1, p = 0.015) with very low overall contributions by the phage.
Figure 2.
Evolutionary and ecological contribution (absolute values) to the frequency of P. fluorescens over time when growing (a) with phages, (b) with ciliates, (c) with phages and ciliates (phage effect) and (d) with phages and ciliates (ciliate effect). Contributions were estimated, using the Geber method, from consumer densities (ecology) and the resistance/defence trait of P. fluorescens (evolution). Black lines with filled symbols indicate the contribution of evolution and grey lines with open symbols the effect of ecology.
4. Discussion
Consumer-mediated coexistence is a highly important and classic notion in ecology. However, little is known about whether rapid evolutionary changes in traits affecting species interactions contribute to the diversity and whether and how the presence of multiple consumers plays a role. Here, we tested how rapid trait evolution affects the frequencies of two bacterial species, E. coli and P. fluorescens, as a proxy for coexistence, using bacterial communities with none, one generalist, two specialists, or all three consumers together. Frequency of P. fluorescens was lowest in the treatment without consumers, and the generalist consumers allowed a higher frequency of P. fluorescens than the specialist consumers. Furthermore, we observed the strongest consumer effect on coexistence when both specialist and the generalist consumers were present in the communities. However, temporal changes in the frequency of the two bacterial species were explained not solely by changes in consumer densities over time (ecology) but also by changes in consumer-avoidance traits (evolution). Interestingly, the role of trait evolution differed across the treatments: the strongest alteration of competition by adaptive evolution was observed in the treatments with a generalist consumer (‘Ciliates’, ‘All’).
In the community without consumers, E. coli was the dominant species (figure 1b(iv)). The frequency of P. fluorescens decreased very rapidly, suggesting that neither stabilizing nor equalizing processes were present for ancestral bacteria. Pseudomonas fluorescens frequencies continued to decline until the end of the experiment. As we found no evidence for the evolution of resource use over time, rapid evolution did not lead to equalizing or stabilizing effects in this treatment. It is possible that the transfers between microcosms led to low competition for resources after the transfer to a new vial and before growing to the stationary phase. This might have allowed P. fluorescens to be maintained in the systems at low frequencies.
Phages as specialist consumers had only a small effect on the outcome of competition between the bacteria. Both E. coli and P. fluorescens populations evolved almost complete resistance to their contemporary phages after approximately 10–20 generations and we did not observe any cost of resistance in our resource use measurements. On the basis of these results, we conclude that because both species reacted similarly to specialist consumers they had no or a minor effect on the coexistence pattern over time. We, however, also acknowledge that the resource use is potentially not the only form of cost of resistance. The role of resistance evolution was thus important only during the initial bacterial generations (figure 2). Changes in the frequency of P. fluorescens were later mostly driven by changes in phage densities (ecology). Bacteria and phage most likely continued coevolving through arms race dynamics (for P. fluorescens, see [25]) and fluctuating selection (for E. coli, see [44]), but we did not investigate this further. Because there was no difference in resource use over time and both bacterial populations evolved resistance very rapidly and seemingly at a similar pace, there were no stabilizing evolutionary effects for coexistence. Differences in the frequency of P. fluorescens compared with the communities without consumers are most probably the result of reduced overall bacterial population sizes and thus reduced competition for resources.
Compared with specialist parasites, the generalist predator had a more substantial effect on the coexistence of the two bacteria. However, as opposed to the ‘Phage’ treatment, evolution played a significant role in coexistence as the frequency of P. fluorescens changed continuously through temporal changes in the defence trait (figure 2). Pseudomonas fluorescens frequencies dropped initially to comparably low frequencies with the ‘Bacteria’ and ‘Phage’ treatments, suggesting that the presence of the predator alone did not change the conditions for coexistence. Frequencies only started to increase again when the bacteria evolved a defence (figures 1 and 2), suggesting that the evolutionary change in both bacteria augmented the initial differences in growth and defence (electronic supplementary material, figures S1 and S2) and had a stabilizing effect.
The highest frequency of P. fluorescens was observed when both the specialist and the generalist consumers were present in the community and temporal changes in frequencies were mainly driven by ecology, i.e. changes in the densities of the ciliate and the phages. The role of evolution (defence and resistance) was only important during the initial transfers when bacteria evolved resistance against phage and defence against the ciliate. Interestingly, the frequency of phage-resistant P. fluorescens was significantly lower in the ‘All’ treatment compared with the ‘Phage’ treatment, and resistance levels decreased over time. A likely explanation for the lower levels of resistance in P. fluorescens is the reduced bacterial population size compared with the ‘Phage’ treatment. Lower population sizes reduce encounter rates and can limit the supply of mutations within a population in a given number of generations, restraining coevolutionary dynamics of the bacteria and phage [32] and generally slowing down adaptive evolution in consumer–resource systems [45]. Moreover, the evolution of costly anti-predator defences might hinder the evolution of costly resistance against viral parasites. Furthermore, the generalist consumer is able to consume resistant cells as long as there is no positive correlation between phage resistance and anti-predator defence [46] directly affecting the frequency of resistant types in the population.
An examination of the relative contribution of evolution to changes in the frequencies of the two different bacteria demonstrates that the relative contributions of evolution and ecology vary, as indicated by earlier findings [17,35]. Interestingly, this means that the relative roles of ecology and evolution depend not only on the complexity of the food web but also other factors, a finding that requires further consideration in future studies.
The Geber method allowed us to disentangle the effects of ecology and evolution over time, but the relative speed of ecological and evolutionary changes cannot be predicted from this. The speed of evolution compared with ecological change has been shown in theory to be decisive for the outcome of eco-evolutionary dynamics [47,48]. In our experiment, we found that the evolution of phage resistance was much faster than defence against ciliate grazing. The rate of evolution of resistance and defence generally depends on mutation rate, population size, generation time and the number of mutations that are required. Concerning the last of these, phage resistance might require only a single mutation, for example one that alters cell-surface receptors (for P. fluorescens SBW25, see [49]), whereas defence against grazing by the ciliate might require several mutations (e.g. for the production of mucus in P. fluorescens, see [50]). Another difference between resistance and defence evolution, we observed here and elsewhere [21], is that the bacteria population can evolve lower levels of defence than the ancestor at certain time points, most probably as a result of eco-evolutionary dynamics, which favour fast growing but highly undefended prey types at low predator densities. Further studies are needed to explore whether and how differences in the speed of evolution and the presence of coevolution alter the contributions of ecology and evolution.
We observed no changes in the resource use of bacteria when comparing the growth of the other species in the medium used (filtrate). This finding contrasts with a study where adaptive evolution in bacterial communities with five species led to differential resource use over time [51]. Potential explanations for this different observation include differences in properties of species used, a higher number of species (i.e. more potential for metabolic products), differences in growth media and the presence of consumers.
In summary, we find that coexistence is promoted in the presence of multiple consumers, which is in line with other studies (e.g. [28,52–54]). Previous theoretical work has already shown that eco-evolutionary dynamics can alter coexistence, for example, by reversing apparent competition, through neighbour-dependent selection [54] or trait adjustments in response to selection that enables a temporally variable convergence and divergence of species traits [59]. We further show that rapid evolution can alter the co-occurrence over time, but that the role of evolution might depend on the consumer types and their combination. Most importantly, our findings show that eco-evolutionary feedback is also crucial with increasing community complexity. This is in agreement with other studies on coexistence at one trophic level [51]. The results of our study indicate also that making predictions based on findings from single-interaction studies by extrapolation might not be possible when exploring eco-evolutionary dynamics in communities with higher complexity and multiple simultaneous interactions. Finally, these findings can be considered important for understanding eco-evolutionary dynamics in natural communities.
Supplementary Material
Acknowledgements
We thank Tuulia Niska, Saara Suomalainen, Tuulia Virolainen, Heikki Kiheri and Johannes Cairns for their help with data collection, Johannes Cairns for editing the language, and Lasse Ruokolainen and an anonymous reviewer for advice on data analyses.
Data accessibility
The data used for this study are available in Dryad: http://dx.doi.org/10.5061/dryad.f306q [56].
Authors' Contributions
T.H. designed and performed research; L.B. analysed data; L.B. and T.H. wrote the paper; V.K. and J.L. provided laboratory materials; all authors contributed to the final version of the manuscript.
Competing interests
We have no competing interests.
Funding
The study was funded by the Academy of Finland to T.H. (project no. 106993), to J.L. (project no. 1255572) and to V.K. (project no. 1267541), DFG to L.B. (BE 4135/3-1) and the Academy of Finland & DAAD bilateral mobility program.
References
- 1.Holt RD. 1977. Predation, apparent competition, and the structure of prey communities. Theor. Popul. Biol. 12, 197–229. ( 10.1016/0040-5809(77)90042-9) [DOI] [PubMed] [Google Scholar]
- 2.Gause GF. 1934. Experimental analysis of Vito Volterra's mathematical theory of the struggle for existence. Science 79, 16–17. ( 10.1126/science.79.2036.16-a) [DOI] [PubMed] [Google Scholar]
- 3.Hutchinson GE. 1961. The paradox of the plankton. Am. Nat. 95, 137–145. ( 10.1086/282171) [DOI] [Google Scholar]
- 4.Chesson P. 2000. Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Evol. Syst. 31, 343–366. ( 10.1146/annurev.ecolsys.31.1.343) [DOI] [Google Scholar]
- 5.Chase JM, Abrams PA, Grover JP, Diehl S, Chesson P, Holt RD, Richards SA, Nisbet RM, Case TJ. 2002. The interaction between predation and competition: a review and synthesis. Ecol. Lett. 5, 302–315. ( 10.1046/j.1461-0248.2002.00315.x) [DOI] [Google Scholar]
- 6.Paine RT. 1966. Food web complexity and species diversity. Am. Nat. 100, 65–75. ( 10.1086/282400) [DOI] [Google Scholar]
- 7.Neill WE. 1975. Experimental studies of microcrustacean competition, community composition and efficiency of resource utilization. Ecology 56, 809–826. ( 10.2307/1936293) [DOI] [Google Scholar]
- 8.Porter JW. 1972. Predation by Acanthaster and its effect on coral species diversity. Am. Nat. 106, 487–492. ( 10.1086/282789) [DOI] [Google Scholar]
- 9.Abrams PA, Matsuda H. 1997. Prey adaptation as a cause of predator-prey cycles. Evolution 51, 1742–1750. ( 10.1111/j.1558-5646.1997.tb05098.x) [DOI] [PubMed] [Google Scholar]
- 10.Jürgens K, Güde H. 1994. The potential importance of grazing-resistant bacteria in planktonic systems. Mar. Ecol. Prog. Ser. 112, 169–188. ( 10.3354/meps112169) [DOI] [Google Scholar]
- 11.Bohannan BJM, Lenski RE. 2000. The relative importance of competition and predation varies with productivity in a model community. Am. Nat. 156, 329–340. ( 10.1086/303393) [DOI] [PubMed] [Google Scholar]
- 12.Friman V-P, Hiltunen T, Laakso J, Kaitala V. 2008. Prey resource availability drives evolution of predator–prey interaction. Proc. R. Soc. B 275, 1625–1633. ( 10.1098/rspb.2008.0174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiltunen T, Laakso J, Kaitala V, Suomalainen LR, Pekkonen M. 2008. Temporal variability in detritus resource maintains diversity of bacterial communities. Acta. Oecol. 33, 291–299. ( 10.1016/j.actao.2007.12.002) [DOI] [Google Scholar]
- 14.Ellner SP. 2013. Rapid evolution: from genes to communities, and back again? Funct. Ecol. 27, 1087–1099. ( 10.1111/1365-2435.12174) [DOI] [Google Scholar]
- 15.Yoshida T, Jones LE, Ellner SP, Fussmann GF, Hairston NG Jr. 2003. Rapid evolution drives ecological dynamics in a predator–prey system. Nature 424, 303–306. ( 10.1038/nature01767) [DOI] [PubMed] [Google Scholar]
- 16.Becks L, Ellner SP, Jones LE, Hairston NG Jr. 2010. Reduction of adaptive genetic diversity radically alters eco-evolutionary community dynamics. Ecol. Lett. 13, 989–997. ( 10.1111/j.1461-0248.2010.01490.x) [DOI] [PubMed] [Google Scholar]
- 17.Becks L, Ellner SP, Jones LE, Hairston NG Jr. 2012. The functional genomics of an eco-evolutionary feedback loop: linking gene expression, trait evolution, and community dynamics. Ecol. Lett. 15, 492–501. ( 10.1111/j.1461-0248.2012.01763.x) [DOI] [PubMed] [Google Scholar]
- 18.Kasada M, Yamamichi M, Yoshida T. 2014. Form of an evolutionary tradeoff affects eco-evolutionary dynamics in a predator–prey system. Proc. Natl Acad. Sci. USA 111, 16 035–16 040. ( 10.1073/pnas.1406357111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaret TM. 1972. Predators, invisible prey, and the nature of polymorphism in the Cladocera (class Crustacea). Limnol. Oceanogr. 17, 171–184. ( 10.4319/lo.1972.17.2.0171) [DOI] [Google Scholar]
- 20.Meyer JR, Kassen R. 2007. The effects of competition and predation on diversification in a model adaptive radiation. Nature 446, 432–435. ( 10.1038/nature05599) [DOI] [PubMed] [Google Scholar]
- 21.Hiltunen T, Becks L. 2014. Consumer co-evolution as an important component of the eco-evolutionary feedback. Nat. Comm. 5, 5226 ( 10.1038/ncomms6226) [DOI] [PubMed] [Google Scholar]
- 22.Hiltunen T, Ayan GB, Becks L. 2015. Environmental fluctuations restrict eco-evolutionary dynamics in predator–prey system. Proc. R. Soc. B 282, 20150013 ( 10.1098/rspb.2015.0013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friman V-P, Buckling A. 2012. Effects of predation on real-time host–parasite coevolutionary dynamics. Ecol. Lett. 16, 39–46. ( 10.1111/ele.12010) [DOI] [PubMed] [Google Scholar]
- 24.Buckling A, Rainey PB. 2002. Antagonistic coevolution between a bacterium and a bacteriophage. Proc. R. Soc. Lond. B 269, 931–936. ( 10.1098/rspb.2001.1945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brockhurst MA, Morgan AD, Fenton A, Buckling A. 2007. Experimental coevolution with bacteria and phage: the Pseudomonas fluorescens—Φ2 model system. Infect. Genet. Evol. 7, 547–552. ( 10.1016/j.meegid.2007.01.005) [DOI] [PubMed] [Google Scholar]
- 26.Frickel J, Sieber M, Becks L. 2016. Eco-evolutionary dynamics in a coevolving host–virus system. Ecol. Lett. 19, 450–459. ( 10.1111/ele.12580) [DOI] [PubMed] [Google Scholar]
- 27.Coloma S, Dienstbier A, Bamford D, Sivonen K, Roine E, Hiltunen T. 2017. Newly isolated Nodularia phage influences cyanobacterial community dynamics. Env. Microbiol. 19, 273–286. ( 10.1111/1462-2920.13601) [DOI] [PubMed] [Google Scholar]
- 28.Brockhurst MA, Fenton A, Roulston B, Rainey PB. 2006. The impact of phages on interspecific competition in experimental populations of bacteria. BMC Ecol. 6, 19 ( 10.1186/1472-6785-6-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janzen DH. 1980. When is it coevolution. Evolution 34, 611–612. ( 10.1111/j.1558-5646.1980.tb04849.x) [DOI] [PubMed] [Google Scholar]
- 30.Tenaillon O, Toupance B, Le Nagard H, Taddei F, Godelle B. 1999. Mutators, population size, adaptive landscape and the adaptation of asexual populations of bacteria. Genetics 152, 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friman VP, Dupont A, Bass D, Murrell DJ, Bell T. 2016. Relative importance of evolutionary dynamics depends on the composition of microbial predator–prey community. ISME J. 10, 1352–1362. ( 10.1038/ismej.2015.217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gómez P, Buckling A. 2011. Bacteria-phage antagonistic coevolution in soil. Science 332, 106–109. ( 10.1126/science.1198767) [DOI] [PubMed] [Google Scholar]
- 33.Rainey PB, Travisano M. 1998. Adaptive radiation in a heterogeneous environment. Nature 394, 69–72. ( 10.1038/27900) [DOI] [PubMed] [Google Scholar]
- 34.Hairston NG, Ellner SP, Geber MA, Yoshida T, Fox JA. 2005. Rapid evolution and the convergence of ecological and evolutionary time. Ecol. Lett. 8, 1114–1127. ( 10.1111/j.1461-0248.2005.00812.x) [DOI] [Google Scholar]
- 35.Ellner SP, Geber MA, Hairston NG. 2011. Does rapid evolution matter? measuring the rate of contemporary evolution and its impacts on ecological dynamics. Ecol. Lett. 14, 603–614. ( 10.1111/j.1461-0248.2011.01616.x) [DOI] [PubMed] [Google Scholar]
- 36.Rainey PB, Bailey MJ. 1996. Physical and genetic map of the Pseudomonas fluorescens SBW25 chromosome. Mol. Microbiol. 19, 521–533. ( 10.1046/j.1365-2958.1996.391926.x) [DOI] [PubMed] [Google Scholar]
- 37.Brockhurst MA, Morgan AD, Rainey PB, Buckling A. 2003. Population mixing accelerates coevolution. Ecol. Lett. 6, 975–979. ( 10.1046/j.1461-0248.2003.00531.x) [DOI] [Google Scholar]
- 38.Brockhurst MA, Rainey PB, Buckling A. 2004. The effect of spatial heterogeneity and parasites on the evolution of host diversity. Proc. R. Soc. Lond. B 271, 107–111. ( 10.1098/rspb.2003.2556) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kassen R, Buckling A, Bell G, Rainey PB. 2000. Diversity peaks at intermediate productivity in a laboratory microcosm. Nature 406, 508–512. ( 10.1038/35020060) [DOI] [PubMed] [Google Scholar]
- 40.Halekoh U, Højsgaard S, Yan J. 2006. The R package geepack for generalized estimating equations. J. Stat. Softw. 15, 1–11. ( 10.18637/jss.v015.i02) [DOI] [Google Scholar]
- 41.Yan J, Fine J. 2004. Estimating equations for association structures. Stat. Med. 23, 859–874. ( 10.1002/sim.1650) [DOI] [PubMed] [Google Scholar]
- 42.Yan J. 2002. Geepack: yet another package for generalised estimating equations. R-News 2/3, 12–14. [Google Scholar]
- 43.Bohannan BJ, Lenski RE. 2000. Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol. Lett. 3, 362–377. ( 10.1046/j.1461-0248.2000.00161.x) [DOI] [Google Scholar]
- 44.Mellard JP, de Mazancourt C, Loreau M. 2015. Evolutionary responses to environmental change: trophic interactions affect adaptation and persistence. Proc. R. Soc. B 282, 20141351 ( 10.1098/rspb.2014.1351) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Örmälä-Odegrip A-M, et al. 2015. Protist predation can select for bacteria with lowered susceptibility to infection by lytic phages. BMC Evol. Biol. 15, 81 ( 10.1186/s12862-015-0341-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cortez MH. 2015. Coevolution-driven predator-prey cycles: predicting the characteristics of eco-coevolutionary cycles using fast-slow dynamical systems theory. Theor. Ecol. 8, 369–382. ( 10.1007/s12080-015-0256-x) [DOI] [Google Scholar]
- 47.Klauschies T, Vasseur DA, Gaedke U. 2016. Trait adaptation promotes species coexistence in diverse predator and prey communities. Ecol. Evol. 6, 4141–4159. ( 10.1002/ece3.2172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scanlan PD, Hall AR, Blackshields G, Friman VP, Davis MR, Goldberg JB, Buckling A. 2015. Coevolution with bacteriophages drives genome-wide host evolution and constrains the acquisition of abiotic-beneficial mutations. Mol. Biol. Evol. 32, 1425–1435. ( 10.1093/molbev/msv032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hinsa SM, O'Toole GA. 2006. Biofilm formation by Pseudomonas fluorescens WCS365: a role for LapD. Microbiology 152, 1375–1383. ( 10.1099/mic.0.28696-0) [DOI] [PubMed] [Google Scholar]
- 50.Lawrence D, Fiegna F, Behrends V, Bundy JG, Phillimore AB, Bell T, Barraclough TG. 2012. Species interactions alter evolutionary responses to a novel environment. PLoS Biol. 10, e1001330 ( 10.1371/journal.pbio.1001330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schreiber SJ, Bürger R, Bolnick DI. 2011. The community effects of phenotypic and genetic variation within a predator population. Ecology 92, 1582–1593. ( 10.1890/10-2071.1) [DOI] [PubMed] [Google Scholar]
- 52.Schreiber SJ, Patel S. 2015. Evolutionarily induced alternative states and coexistence in systems with apparent competition. arXiv (http://arXiv.org/abs/1504.00906)
- 53.Vasseur DA, Amarasekare P, Rudolf VH, Levine JM. 2011. Eco-evolutionary dynamics enable coexistence via neighbor-dependent selection. Am. Nat. 178, E96–E109. ( 10.1086/662161) [DOI] [PubMed] [Google Scholar]
- 54.De Roos AM, Schellekens T, Van Kooten T, Persson L. 2008. Stage-specific predator species help each other to persist while competing for a single prey. Proc. Natl Acad. Sci. USA 105, 13 930–13 935. ( 10.1073/pnas.0803834105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao L, Zhang QG, Zhang DY. 2015. Evolution alters ecological mechanisms of coexistence in experimental microcosms. Funct. Ecol. 30, 1440–1446 ( 10.1111/1365-2435.12611) [DOI] [Google Scholar]
- 56.Hiltunen T, Kaitala V, Laakso J, Becks L.. 2017. Data from: Evolutionary contribution to coexistence of competitors in microbial food webs Dryad Digital Repository. ( 10.5061/dryad.f306q) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Hiltunen T, Kaitala V, Laakso J, Becks L.. 2017. Data from: Evolutionary contribution to coexistence of competitors in microbial food webs Dryad Digital Repository. ( 10.5061/dryad.f306q) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The data used for this study are available in Dryad: http://dx.doi.org/10.5061/dryad.f306q [56].