Abstract
Segmented worms (Annelida) are among the most successful animal inhabitants of extreme environments worldwide. An unusual group of enchytraeid oligochaetes of genus Mesenchytraeus are abundant in the Pacific northwestern region of North America and occupy geographically proximal ecozones ranging from low elevation rainforests and waterways to high altitude glaciers. Along this altitudinal transect, Mesenchytraeus representatives from disparate habitat types were collected and subjected to deep mitochondrial and nuclear phylogenetic analyses. Our data identify significant topological discordance among gene trees, and near equivalent interspecific divergence levels indicative of a rapid radiation event. Collectively, our results identify a Mesenchytraeus ‘explosion’ coincident with mountain building in the Pacific northwestern region that gave rise to closely related aquatic, ice, snow and terrestrial worms.
Keywords: adaptive, glacier, evolution, discordance, annelid
1. Introduction
Among Animalia, representatives of the phylum Annelida occupy a continuum of diverse habitats, ranging from geothermal black smokers on the ocean floor to glacier ice [1]. At the latter end of this spectrum, oligochaetes of the genus Mesenchytraeus are exceptionally well represented along coastal regions of the Pacific Northwest, from northern California to central Alaska [2–7]. This geographically complex region is characterized by vast temperate rainforests accommodating many of the world's largest trees, snow-fed waterways, and rugged mountains harbouring lower elevation transient snowfields and higher altitude glacier ice [8]. These maritime peaks are altitudinally stratified into zones, each characterized by distinct thermal maxima and minima, vegetation, and faunal communities [9]. Within this geographical ecozone, species of the Holarctic genus Mesenchytraeus have evolved to occupy remarkably diverse environmental niches.
Comprising more than 700 species and 33 genera, Enchytraeidae are predominantly small, inconspicuous soil-dwelling worms with a worldwide distribution [10,11]. Yet Mesenchytraeus, one of its most speciose genera, is distinguished by extraordinary intra-generic variation, not only in habitat (aquatic, terrestrial, snow, glaciers), but also in size (4 to >60 mm long), and colouration (transparent, white, yellow, red, tawny, brown, grey, black, uniform, variegated) [2–4,6,12]. Mesenchytraeus solifugus [13], the glacier-obligate ice worm, was the first enchytraeid described from the Pacific Northwest, and over the next two decades numerous Mesenchytraeus species/subspecies, occupying the full spectrum of ecozones, were formally described [2–4]. Mesenchytraeus dominance, in terms of species richness and ecological specialization, within the Beringian coastal region suggests an unusual adaptive capacity for this lineage [6,14]. Although other Enchytraeidae genera were likewise discovered in the Pacific Northwest, none are as diverse or extreme in habitat choice.
That M. solifugus was the focus of virtually all recent investigations can be ascribed to their rare and compelling status as glacier-obligate macrofauna. For most animals, extended exposure to temperatures approximately 0°C results in cellular dysfunction and metabolic failure [15–17]. Thus, the ability of ice worms to overcome this physiological barrier garnered intense interest. Unfortunately, its congeners have been virtually ignored and little is known about lifestyles and genealogical relationships among these worms. Modern biology recognizes that comparative trait analyses, informed by a credible phylogenetic framework, provide the best methodology for identification of ecologically adaptive traits [18–21]. To that end, our focus in the current study was to establish the evolutionary relationships between the glacier ice worm, M. solifugus, and Pacific Northwest congeners from diverse habitats, with the longer-term goal of understanding the genetic basis for transitioning between thermal regimes (e.g. terrestrial ↔ ice habitats).
We employed Illumina high-throughput sequencing to generate full, normal-expression transcriptomes and the polymerase chain reaction (PCR) to amplify overlapping segments of the mitochondrial genome from genomic DNA. Phylogenetic analyses employed more than 24 000 base pairs (bp) of sequences from 30 genes/species using both maximum likelihood and Bayesian frameworks. Herein, we present the first examination of genetic relationships among select Mesenchytraeus worms of the Pacific Northwest and the first molecular sequence data for two ingroup members (M. antaeus, M. hydrius).
2. Material and methods
(a). Specimens
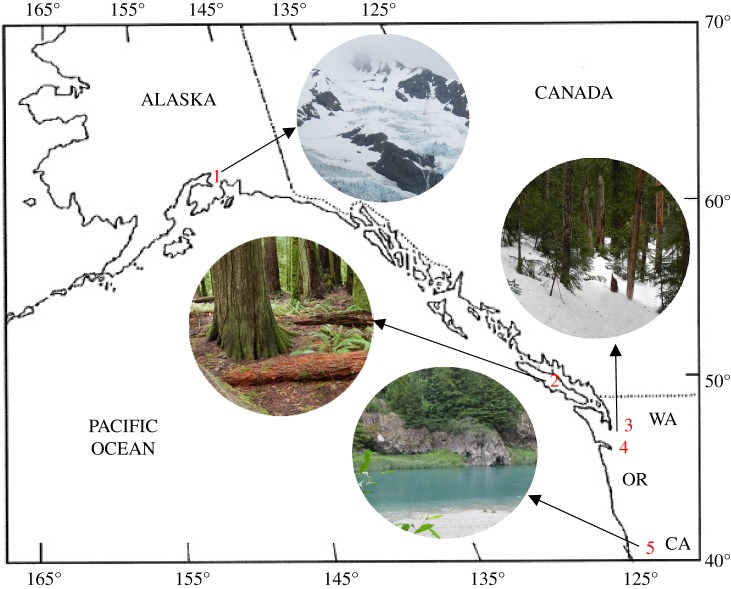
Mesenchytraeus specimens were collected from four distinct habitat types along the Pacific northwestern coast of North America between 2011 and 2014. These comprised (i) cold, outflow rivers from coastal mountains (M. cf pedatus [2]; aquatic), (ii) low elevation temperate rainforests (M. antaeus [6]; terrestrial), (iii) transient snowfields shaded by coniferous stands (M. cf gelidus [3], M. hydrius [4]; snow), and (iv) coastal, maritime glaciers (M. solifugus; ice) (figure 1, table 1). Additionally, two confamilials, Enchytraeus albidus [23] and E. cf crypticus [22], both well-studied European species, were included as outgroups. Specimens were express-shipped live to Rutgers University, and maintained according to their endogenous habitat/temperature (e.g. soil, refrigerated, etc.) as described [24]. Enchytraeus crypticus were from laboratory-maintained stock, and E. albidus was an ethanol-preserved sample used for DNA analysis. Species were identified by comparison to molecular data, when available, and/or morphological examination. Further details on species identification can be found in electronic supplementary material, materials and methods. Ethanol-preserved samples of all species are stored at Rutgers University and are available for examination.
Figure 1.
Mesenchytraeus collection sites and habitats. Site 1: Chugach Mountains, Alaska; Mesenchytraeus solifugus, glacier-obligate. Site 2: Vancouver Island, BC; M. antaeus, temperate forests. Sites 3, 4: Cascade Mountains, WA; M. gelidus (3) and M. hydrius (4), transient snow beneath conifers. Site 5: Sacramento River, CA; M. pedatus, aquatic. (Online version in colour.)
Table 1.
Description and collection sites of Enchytraeidae species.
| species | overt phenotype (colour, length x diameter) | collection location and date | habitat description (at time of collection) |
|---|---|---|---|
| Mesenchytraeus solifugus [13] | dark brown to black 10–20 mm × 0.5–0.7 mm |
60.762003° N, 148.846545° W July 2012 |
0°C (constant), snow overlying glacier ice; alt: 122 m |
| Mesenchytraeus pedatus [2] | white 10–20 mm × 0.3–0.5 mm |
40.5937222° N, 122.39919° W June 2011 |
8–14°C, cold mountain streams/rivers; alt: 148 m |
| Mesenchytraeus gelidus [3] | dark red-brown to black 20–38 mm × 1–2 mm |
47.288126° N, 121.263348° W June 2014 |
0–2°C, transient snow shaded by coniferous trees, alt: 1353 m |
| Mesenchytraeus hydrius [4] | white 15–35 mm × 0.5–0.9 mm |
46.746679° N, 121.790266° W June 2014 |
0–2°C, transient snow shaded by coniferous trees, alt: 1332 m |
| Mesenchytraeus antaeus [6] | white to yellowish 30–60 mm × 1.8–2.5 mm |
48.874057° N, 124.701034° W May 2014 |
10–15°C, temperate rainforest, alt: 149 m |
| Enchytraeus crypticus [22] | white 3–12 mm × 0.2–0.4 mm |
commercially purchased lab stock |
22°C, moist, acidic, peat-enriched soil; alt: 5 m |
(b). DNA processing and analysis
Genomic DNA was extracted from a single worm/species using an E.Z.N.A. Tissue DNA Isolation Kit (Omega Bio-tek, Inc.) following the tissue protocol. Single worms were used in an effort to reduce sequencing uncertainty, a concern only for M. pedatus with 0–15% variability at the cytochrome oxidase 1 (cox1) locus (n = 20; data not shown). Cryptic speciation is likely within this population but since all haplotypes proved equidistant to the considered congeners, their utility as an aquatic Mesenchytraeus was not compromised. For E. albidus and M. hydrius, only a single worm was available for DNA extraction, and population analyses of the remaining species showed very low intra-population genetic diversity (unpublished data). With the exception of E. albidus, extractions were from freshly sacrificed worms. Nuclear 28S ribosomal RNA was PCR-amplified using annelid-specific primers designed for this study: 28sC1-F (5′-ACCCGYTGAAYTTAAGCATAT-3′)/28sC4-R (5′-TTCGATTRGTCTTTCGCCCCT-3′) amplified a 5′-end fragment of approximately 1180 bp. For some species, additional internal primers were necessary: 28sC2-F (5′-CAAGTACCGTGAGGGAAAGTTG-3′)/28sC3-R (5′-CCGTGTTTCAAGACGGGTCG-3′). For mitochondrial genes, primer pairs were designed to amplify six overlapping mitochondrial genome (mt-genome) fragments, 2–4 Kb in length, based on gene size and order as found in Lumbricus terrestris Linnaeus, 1758 [25]. Additional species-specific primers were designed as needed based on sequenced PCR products to fill genome gaps (see electronic supplementary material, table S1 for mitochondrial primers). Purified PCR products were cloned into pMiniT vectors (New England Biolabs (NEB)), transformed into NEB 10-beta chemically competent E. coli, and 3–5 clones/fragment/species were selected for purification using Zyppy Plasmid Miniprep (Zymo Research). Sanger DNA sequencing was performed by GENEWIZ, Inc. (South Plainfield, NJ, USA). Transcriptomes were generated and processed as detailed in electronic supplementary material, materials and methods.
(c). Phylogenetic analyses
Individual gene sequences were aligned using Muscle [26] under default parameters, as implemented in MEGA v. 6 [27], with protein coding genes (PCGs) aligned by codons. Statistical analyses to assess phylogenetic informativeness and divergence were also performed in MEGA (electronic supplementary material, table S2). Gene trees were inferred using Maximum-likelihood (ML) and Bayesian inference (BI) for each gene. Prior to ML analysis, jModelTest 2.1.7 [28,29] was used to select the best-fitting evolutionary model for each locus according to the Akaike information criterion (AIC; [30]). ML analyses were conducted with PhyML v. 3.1 [31] using the recommended best-fit substitution model. The subtree pruning and regrafting (SPR) option was used to more thoroughly explore the space of tree topologies and five random starting trees were generated to avoid local maxima. Branch support was estimated using the non-parametric Shimodaira–Hasegawa-like approximate likelihood ratio test [31], as this method provides superior performance, relative to bootstrapping, for datasets with short branches [31] and topological conflict [32]. Analyses were run with, and without, outgroup designation to test their suitability.
Bayesian analyses were performed in MrBayes v. 3.2 [33] using Bayes-block nexus files created in Mesquite v. 3.04 [34], with default parameters modified as described below. Each PCG dataset was analysed partitioned (by codon position) and unpartitioned; non-coding genes were unpartitioned. Model settings for each gene were: nst = mixed, rates = gamma. The ‘mixed’ model allows the analysis to integrate across the full general time-reversible (GTR, [35]) model space and avoids specifying a particular, possibly inappropriate, evolutionary model of nucleotide substitution [36]. The ‘gamma’ parameter models per site mutation rate heterogeneity with the assumption that it is drawn from a random statistical distribution [37]. For partitioned analyses, partition parameters were unlinked and the ‘ratepr’ parameter of the ‘prset’ command was set to variable to allow rates across partitions to differ. All analyses were run for ≥1 000 000 generations (x2) with a relative burn-in fraction of 25%, sample frequency = 1000, and diagnostic frequency = 1000. All other parameters were default values. The ‘sum parameters’ (sump) command was used to assess run convergence by examining the generation versus log likelihood plot, average deviation of split frequencies (less than 0.01), the Potential Scale Reduction Factor (PSRF, approx. 1), and the Effective Sample Size (ESS, more than 200). Consensus trees were visualized and annotated using FigTree v. 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree), and DensiTree2 v. 2.2.4 [38] was used to analyse and portray the degree of uncertainty present.
Phylogenetic reconstruction was further explored using various combinations of genes concatenated into supermatrices using Mesquite v. 3.04 [34]. Five combined-gene datasets were examined: (i) HK6 = nuclear housekeeping genes (actin, α-tubulin, EF-1α, histone H3, GAPDH, 28S rRNA), (ii) nuASU = nuclear-encoded ATP synthase subunits (alpha, beta, gamma, delta, b, c, d, OSCP), (iii) MtPCGs = mitochondrial protein-coding genes (cox 1–3, cytb, nad 1–6, nad 4 L, atp6, atp8), (iv) Mt13 + RNAs = 13 MtPCGs + 12S-(tRNA-Val)-16S, and (v) Mt4 = mitochondrial subset (cox1, cytb, 12S, 16S) (electronic supplementary material, table S3). These were subjected to ML and BI analyses as described above with the following changes. For ML analyses, the designated model was GTR + I + G as this was the most commonly recommended for the individual genes, and PhyML does not currently support partitioning. Prior to Bayesian analysis, PartitionFinder v. 1.1.0 [39] was used to find the best-fit partitioning scheme for each dataset under the following criteria: branch lengths = linked, models = MrBayes, model selection = BIC, search = greedy. This scheme was then incorporated into the Bayes blocks and analyses proceeded as described above (electronic supplementary material, table S4).
Next, species trees were inferred using the BEST multi-species coalescent (MSC) algorithms [40] as implemented in MrBayes v. 3.2. [33]. The MSC models the inherent stochasticity in genetic drift to infer a probabilistic species tree by reconciliation of topological discordance among gene trees [41]. Gene tree discordance is particularly problematic in phylogenetic analyses of closely related species due to incomplete lineage sorting (ILS) [42], thus likely a more realistic fit given our taxa. We used two datasets, one comprising 18 genes (Coal set 1) and the second with 15 genes (Coal set 2), with overlap between them (electronic supplementary material, table S3). For set 1, 18 gene trees were estimated (1/gene), whereas for set 2, the five mitochondrial genes were grouped as a single locus, thus 11 gene trees were estimated. Prior settings were: brlenspr = clock:speciestree, topologypr = speciestree, popvarpr = variable, and popsizepr = gamma (1100). Model and rate were as above (mixed/gamma) with topologies unlinked; mitochondrial gene ploidy was set to haploid. Four independent analyses of 30 million generations/set, with burn-in fraction = 0.33, were run via the CIPRES-Portal v. 3.3 [43] and convergence was assessed as previously described. Coalescence analyses excluded E. albidus. See electronic supplementary material, materials and methods for additional analyses employing divergence time algorithms.
3. Results
Illumina sequencing produced more than 49 million paired-end reads per transcriptome, resulting in more than 102 000 contigs with an average size of 934 bp. The average G + C content (approx. 41%) was uniform across species, including the outgroup. All gene queries were recovered, and raw-read mapping indicated that depth of coverage was more than sufficient. This description does not apply to the E. albidus transcriptome which was not generated for this investigation. Of 30 genes extracted for analysis (representing more than 24 000 alignment positions), 29 were full length, with 28S rRNA the only exception (electronic supplementary material, table S2). Note that exhaustive GenBank and literature searches for relevant Enchytraeid species/sequences (e.g. [11]) yielded overlapping fragments totalling only approximately 1400 positions, and thus too shallow to generate any meaningful comparisons based on consistently unresolved tree topologies (not shown). Sequence alignment was unambiguous for PCGs, but 12S and 16S rRNA required trimming. PCR-amplified mitochondrial genes matched those extracted from the respective species transcriptome. Neither ATP synthase subunit (ASU) gamma, nor epsilon, were recovered from the E. albidus transcriptome and were treated as missing data in analyses. GenBank accession numbers for sequences are in electronic supplementary material, table S5.
Among Mesenchytraeus species, the average nucleotide per cent identity within concatenated datasets was (i) housekeeping genes: 87.17 (±1.93 s.d.), (ii) ATP synthase subunits: 85.14 (±2.05 s.d.), (iii) mitochondrial PCGs: 76.87 (±1.52 s.d.), and (iv) mitochondrial subset cox1-cytb-rRNAs: 83.8 (±1.1 s.d.). Pairwise divergence values were highly similar within each dataset, displaying no obvious interspecific relationship trends. Per cent identity at the protein level was approximately 10% greater (electronic supplementary material, table S6).
Expectedly, the housekeeping (HK) genes were the least parsimony informative (PI), with a range of 2.2 (28S) to 11.5% (histone H3) (electronic supplementary material, table S2). Interestingly, histone H3 is the only gene in the dataset that is 100% conserved at the protein level. Furthermore, the low phylogenetic signal for 28S within the genus jumps to more than 21% when comparing inter-generic data, indicating its utility for delimiting higher level taxa. Likewise, the ATP synthase subunits (ASU) had a PI range of 6.5–11.9% among Mesenchytraeus, but a few were three- to fourfold higher when Enchytraeus was included, and may prove useful in resolving annelid phylogeny questions. The mitochondrial PCGs had an average PI of approximately 16%, with nad6 highest at 21.1%, and cox1 and nad3 lowest at 13%. Ribosomal RNA genes, 12S and 16S, were the most highly conserved and least phylogenetically informative in the mitochondrial genome with PIs of 7.6 and 6.8%, respectively.
(a). Phylogenetic relationships
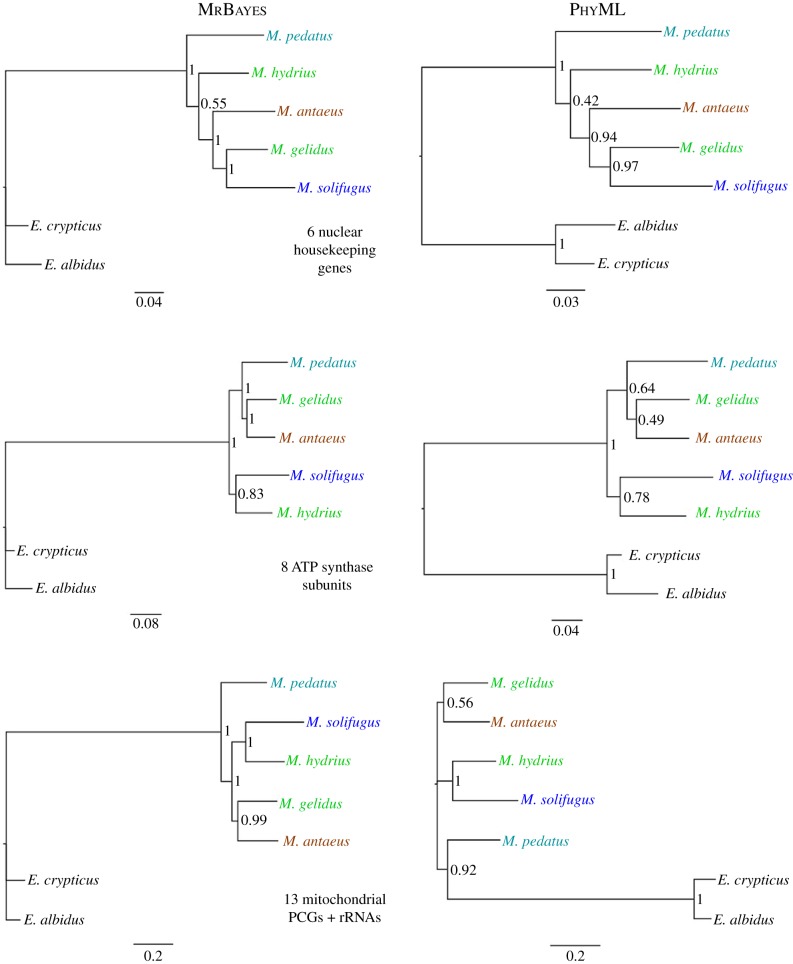
Individual gene tree results were comparable with regard to partitioning scheme (partitioned versus un-partitioned) and framework (Bayesian versus likelihood), but topological discord was significant (electronic supplementary material, table S7). Of the 30 gene trees generated, 23 distinct topologies were present: HK genes (6/6), ASU genes [8/9; (alpha, delta)], mitochondrial genes [11/15; (cytb, atp8) (nad2, 16S) (nad3, nad6, 12S)]. The Enchytraeus worms consistently appeared outside the Mesenchytraeus clade, even when outgroup status was not specified, supporting their utility in these analyses. Mesenchytraeus monophyly was violated only for ASU gamma and the ML analysis of the 15 mitochondrial genes (Mt13 + RNAs), placing M. pedatus as an outgroup. The Bayesian analysis of the latter dataset placed M. pedatus as sister taxon to its four congeners with 100% support (figure 2). Indeed, this sister taxon positioning was the second most frequently observed grouping, occurring in five gene trees and the HK dataset. By far, the most common sister taxa pair was the ice worm, M. solifugus, and the white snow worm, M. hydrius, present in 16 individual gene trees, the ASU tree, and the coalescent tree (figure 3). The two largest species, M. antaeus and M. gelidus, appeared in four gene trees and the ASU tree as sister taxa, making them the third most common pair. The concatenated datasets, HK6, nuASU, and Mt13 + RNA, did little to resolve interspecific relationships, each producing topologically different consensus trees (the mitochondrial subsets, MtPCGs and Mt4, reiterated the Mt13 + RNA tree). Unlike the individual gene trees, partitioning significantly affected topological outcomes. While no difference was seen for the HK dataset, un-partitioned analyses of the ASU and Mt13 + RNA resulted in lower probability values and polytomies (data not shown).
Figure 2.
Bayesian and maximum-likelihood consensus trees for concatenated datasets. Upper: housekeeping genes, actin, EF-1α, histone H3, α-tubulin, GAPDH, and 28s rRNA (5916 bp). Middle: nuclear ATP synthase subunit genes, alpha, beta, gamma, delta, b, c, d, OSCP (6894 bp). Lower: mitochondrial protein-coding genes, cox1–3, cytb, nad1–6 + 4 L, atp6, atp8, and RNA contig, 12S-tRNA-val-16S (13 381 bp). Branch support for Bayesian trees (left) are posterior probabilities and for ML trees (right) Shimodaira–Hasagawa approximate likelihood ratio test. In-group taxa (Mesenchytraeus) names are colour-coded by habitat: aquatic = aqua, blue = glacier, green = snow, and brown = terrestrial; outgroup enchytraeids are in black. (Online version in colour.)
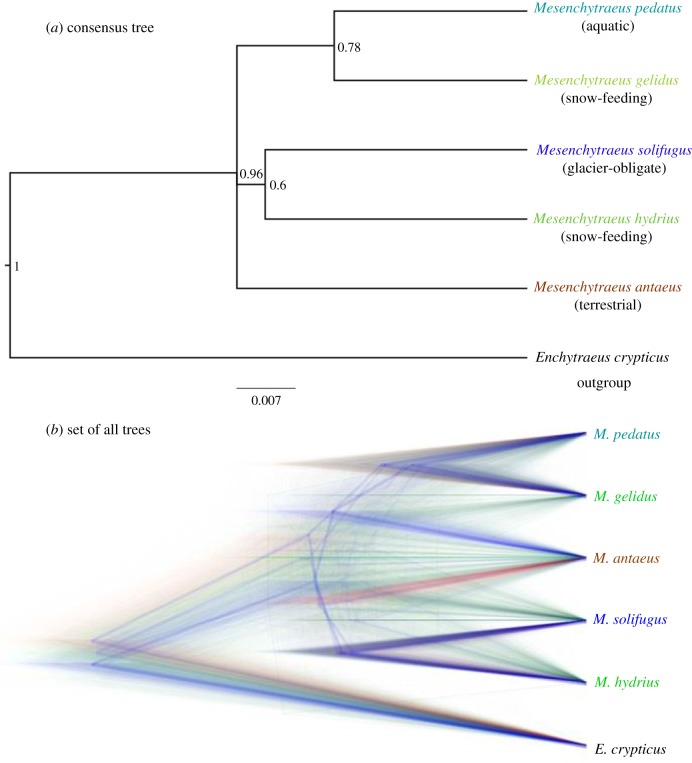
Figure 3.
Species delimitation analysis for genus Mesenchytraeus using a multi-species coalescent paradigm in MrBayes. Shown are results for Coal_set 2 (Suppl. Table 3) comprising 15 genes: five mitochondrial (cytb, nad2/4/5, 12S), five ATP synthase subunits (alpha, gamma, b, c, delta), and five housekeeping (actin, EF-1α, α-tubulin, histone H3, 28S). For this analysis, the mitochondrial genes were considered as a single locus. (a) Depicts the consensus tree with posterior probabilities. (b) Shows a DensiTree2 [38] visualization of the ambiguity present in the consensus tree. This programme uses the full set of trees generated in a Bayesian analysis and overlays them using transparent coloured lies. Branches with greater support among the trees appear darker and heavier while areas of greater topological conflict are more diffuse. Colours are hierarchical with blue lines having highest support, followed by red, and then green.
The multi-species coalescent (MSC) analyses resulted in similar consensus trees for datasets 1 and 2, both with polytomies and low posterior probabilities (Set 2, figure 3a). Figure 3b displays a DensiTree [38] visualization of the uncertainty present in the consensus tree. By overlaying the set of consensus trees (i.e. the average of all trees with the same topology) with the sum total of all trees generated in the analysis, areas of topological conflict became apparent. Blue lines represent the highest probability topology, thus explaining the consensus tree pairing of M. pedatus/M. gelidus and M. solifugus/M. hydrius. However, red lines, used for the second highest probability trees, and green, third highest, show that a significant number of trees conflict with those groupings, hence the low posterior probabilities. The nearly equal-intensity red and blue lines for M. antaeus indicate considerable relationship ambiguity.
Attempts to establish divergence times using a fossil-calibrated root node were unsuccessful for both BEAST and MrBayes analyses. Output file assessments indicated that convergence was not achieved and ESS values were frequently below 200, even for combined runs, thus the resulting consensus trees were deemed unreliable.
4. Discussion
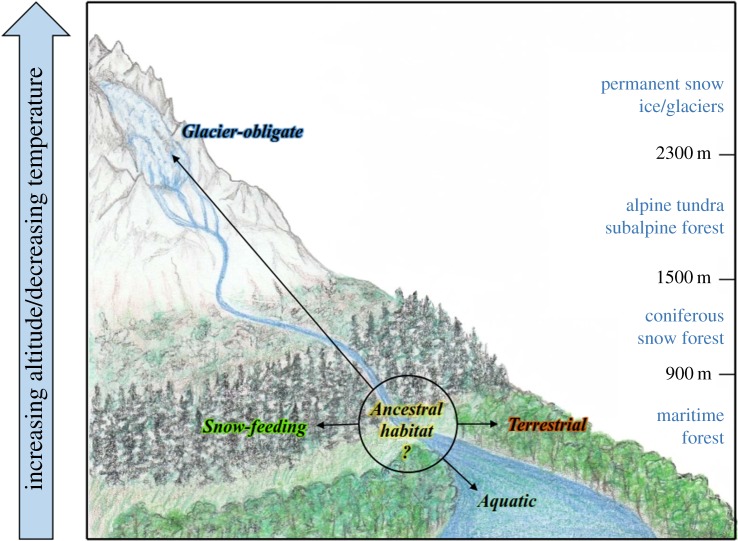
This is the first molecular phylogenetic reconstruction of an unusually diverse clade (i.e. spanning aquatic, terrestrial, snow, and ice ecozones) of Pacific Northwest-endemic Mesenchytraeus worms. Note that the respective ecozones occur throughout the Pacific Northwest and that, for example, a transect within the Mt. Rainier region would yield ecologically representative species comprising the current analysis (i.e. aquatic, ice, snow, and terrestrial species; figure 4). Our approach employed tree estimates of 30 individual genes, five concatenated supermatrices, and two multi-species coalescent datasets. We did not know a priori divergence rates among the considered species, thus the genes chosen for analyses span a wide conservation range. Clearly, our results indicate that the housekeeping genes are too highly conserved to provide resolution for the examined species. Individually, these six genes (actin, α-tubulin, EF-1α, GAPDH, histone H3, 28S) produced six topologically distinct trees, often with very high branch support, and the concatenated dataset yielded yet a seventh topology (figure 2). Gene tree discord is a commonly observed phenomenon attributed to incomplete lineage sorting (ILS) [42,44,45], mitochondrial DNA introgression [46,47], horizontal gene transfer [42,48], gene duplication/loss [49,50], and adaptive selection [51,52]. Incomplete lineage sorting occurs when DNA sequence coalescence is temporally out of step with speciation, a situation more often found in species separated by relatively few genetic changes [42]. Thus, given the short branch lengths observed in our data, and the extraordinary level of discordance, the failure of the housekeeping gene dataset to resolve these interspecific relationships is likely attributable to ILS.
Figure 4.
Proposed radiation of Mesenchytraeus species along an altitudinal gradient. Ecozones with elevations are indicated to the right. Unknown ancestral habitat depicted by a circle from which extant Mesenchytraeus lineages (arrows) radiated into diverse ecological niches. (Online version in colour.)
Likewise, the ASU genes produced discordant gene trees—eight topologies for nine genes—and ILS may underlie this lack of agreement as well, but other factors merit consideration. Generally, gene products functioning in critical metabolic pathways are under strong negative (purifying) selection because almost all mutations that arise are deleterious. This is certainly the case for ASU alpha and beta with more than 78% amino acid identity across diverse metazoan phyla. However, nuclear-encoded oxidative phosphorylation gene products that functionally interact with faster-evolving mitochondrial-encoded proteins experience additional selective pressure. This ‘mito-nuclear’ association creates an ‘arms race’ whereby the nuclear genes must adapt to, and compensate for, mutations in mitochondrial genes to maintain optimum energy production [53–55]. Therefore, ASU gene comparisons from species evolving in ecologically diverse niches may support relationships incongruent with true phylogenetic relationships [56].
Mitochondrial genomes have been considered the best dataset for discernment of species-level relationships because of their approximately 10-fold greater mutation rate, lack of recombination, conserved gene content, and the perception that they evolve under a neutral equilibrium model. While the first three attributes are true in most cases, it has become clear that selection plays a non-trivial role in mt-genome evolution and these effects can confound phylogenetic interpretations [51,52,57,58]. Although our BI analysis of the concatenated mitochondrial genes (Mt13 + RNAs, figure 2) produced a highly supported (posterior probabilities ≥0.99) bifurcating tree, 10 of the 15 individual gene trees had polytomies and much lower support values (electronic supplementary material, table S7).
This dichotomy between a well-supported concatenated multi-gene tree and discordant individual gene trees is not surprising. Numerous studies have demonstrated that phylogenetic inference based on concatenation of genes evolving under different coalescent properties can produce incorrect, but well supported trees [59–61]. This effect is particularly prevalent in species-level analyses where few genetic changes separate the taxa [61]. Recognition of this problem has led to coalescent-based methodologies that model emergent relationship properties within a group of individual gene trees to produce a more realistic species tree [60,62,63]. Unfortunately, even this framework failed to resolve relationships among Mesenchytraeus, as demonstrated by the multifurcating consensus tree (figure 3a), and the considerable ambiguity present in the full set of trees (figure 3b). This failure may be due to limitations of the MSC, in that gene tree incongruities due to factors other than ILS (e.g. hybridization, adaptive selection) are not accounted for.
Among the proposed causative factors of gene tree/species tree discord, ILS is a likely agent, given the short branch lengths within the Mesenchytraeus clade, and their extraordinary habitat diversity suggests adaptive selection may also play a role. Horizontal gene transfer, thought to be primarily confined to prokaryotes, is increasingly found to be the source of novel traits in a variety of eukaryotes [64] and cannot be dismissed. Finally, mitochondrial introgression resulting from gene flow (i.e. hybridization) between species may explain the considerable incongruities seen in the mitochondrial datasets and the inefficacy of the MSC to resolve species relationships. Interestingly, mitochondrial introgression has a putative role in rapid adaptive radiations [65] and energy metabolism [66]. Indeed, the overwhelmingly discordant trees, short branch lengths, and equidistant divergence rates among the Mesenchytraeus worms examined here argue persuasively for a recent and rapid adaptive radiation event.
Mountain building in the Pacific northwestern region occurred 5–10 Ma (late Miocene to early Pliocene; [67–69]), thus the worms considered in this study are unlikely to predate this time period. Periodic glacial cycles and volcanic activity [70] characterize the region, and the continuity of glacier ice/snow is supported by the persistence of ice worms [71], which are a stenothermic, glacially obligate species [72–75]. Further, documented refugia have been described throughout this region and time period, providing a genetic pool from which the diverse species could have arisen [76–79]. Previous analyses estimate ice worm origins between approximately 4.9 and 9 Ma [71], which may approximate the time frame of the Mesenchytraeus radiation proposed here. We hypothesize that geological uplifting in the region created the diverse ecozones into which these worms invaded, and thus these two events (i.e. mountain building/Mesenchytraeus radiation) are likely coincident. Regardless of timing, however, our data support a short time window of rapid speciation in which environmental conditions were favourable for a cold adapted species to invade a wide variety of newly created habitats. Clearly, our use of a large dataset has served to enhance the phylogenetic signal, but increasing the number of taxa as data becomes available may provide finer resolution in future analyses.
Their origins notwithstanding, extant Mesenchytraeus species in the Pacific Northwest offer a unique animal group to dissect the relationship between genes and the environment, particularly in the context of habitat diversity and thermal regimes. We anticipate that life histories of other regional Mesenchytraeus worms, and possibly other psychrophilic fauna in the Pacific Northwest, may share a similar evolutionary paradigm.
Supplementary Material
Acknowledgements
We thank K. Vincents Fisker for providing E. albidus specimens, C. Erséus for access to the E. albidus transcriptome data, and P. Wimberger and M. Tetraeu for fieldwork contributions. For assistance with RNA-seq data generation and assembly, we thank K. Halanych and members of his lab (P. Brannock, K. Kocot, and D. Waits) at Auburn University, and R. Fernández Garcia at the Giribet lab, Harvard University.
Ethics
No vertebrate animals were used in this research.
Data accessibility
DNA sequences: GenBank accession numbers KU728801-KU728933.
Authors' contributions
S.A.L. conducted molecular, phylogenetic and computational analyses and contributed to writing; N.S. examined and dissected specimens for species identification; J.K. conducted computational analyses to process transcriptomes; D.H.S. conducted fieldwork and contributed to writing the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This study was supported by NSF IOS0820505 and Busch grants to D.H.S.
References
- 1.Shain DH. 2009. Annelids in modern biology. Hoboken, NJ: Wiley-Blackwell. [Google Scholar]
- 2.Eisen G. 1904. Enchytraeidae of the West Coast of North America. In Harriman Alaska expedition series. Vol. XII. Annelida, pp. 1–166. New York, NY: Smithsonian Institution. [Google Scholar]
- 3.Welch PS. 1916. Snow-field and glacier Oligochaeta from Mt. Rainier, Washington. Trans. Am. Microscop. Soc. 35, 85–124. ( 10.2307/3221561) [DOI] [Google Scholar]
- 4.Welch PS. 1919. Further studies on North American Mesenchytraeids (Oligochaeta). Trans. Am. Microscop. Soc. 38, 175–188. ( 10.2307/3221532) [DOI] [Google Scholar]
- 5.Altman LC. 1936. Oligochaeta of Washington. University of Washington Publications in Biology 4, 1–137. [Google Scholar]
- 6.Rota E, Brinkhurst RO. 2000. Mesenchytraeus antaeus, a new giant enchytraeid (Annelida, Clitellata) from the temperate rainforest of British Columbia, Canada, with a revised diagnosis of the genus Mesenchytraeus. J. Zool. 252, 27–40. ( 10.1111/j.1469-7998.2000.tb00817.x) [DOI] [Google Scholar]
- 7.Healy B, Fend S. 2002. The occurrence of Mesenchytraeus (Enchytraeidae: Oligochaeta) in riffle habitats of north-west American rivers, with description of a new species. J. Nat. Hist. 36, 15–23. ( 10.1080/713833842) [DOI] [Google Scholar]
- 8.Littell JS, McGuire Elsner M, Whitely Binder LC, Snover AK (eds). 2009. The Washington climate change impacts assessment: evaluating Washington's future in a changing climate. Seattle, Washington, DC: Climate Impacts Group, University of Washington. [Google Scholar]
- 9.Taylor WP. 1922. A distributional and ecological study of Mount Rainier, Washington. Ecology 3, 214–236. ( 10.2307/1929036) [DOI] [Google Scholar]
- 10.Schmelz RM, Collado R. 2015. Checklist of taxa of Enchytraeidae (Oligochaeta): an update. Soil. Organisms 87, 149–152. [Google Scholar]
- 11.Erséus C, Rota E, Matamoros L, De Wit P. 2010. Molecular phylogeny of Enchytraeidae (Annelida, Clitellata). Mol. Phylogen. Evol. 57, 849–858. ( 10.1016/j.ympev.2010.07.005) [DOI] [PubMed] [Google Scholar]
- 12.Schmelz RM, Collado R. 2010. A guide to European terrestrial and freshwater species of Enchytraeidae (Oligochaeta). Soil. Organisms 82, 1–176. [Google Scholar]
- 13.Emery C. 1898. Diagnosi di un nuovi genore e nuovi specie di Annellidi della famiglia deli Enchytraeidae. R. Accademia nazionale dei Lincei. Classe di scienze fisishe matematiche e naturali.
- 14.Piper SR, MacLean SF, Christensen B. 1982. Enchytraeidae (Oligochaeta) from the taiga and tundra habitats of northeastern U.S.S.R. Can. J Zool. 60, 2594–2609. ( 10.1139/z82-334) [DOI] [Google Scholar]
- 15.Thauer R, Brendel W. 1962. Hypothermie. Prog. Surg. 2, 73–271. ( 10.1159/000386265) [DOI] [PubMed] [Google Scholar]
- 16.Kruuv J, Glofcheski D, Cheng KH, Campbell SD, Al-Qysi HM, Nolan WT, Lepock JR. 1983. Factors influencing survival and growth of mammalian cells exposed to hypothermia. I. Effects of temperature and membrane lipid perturbers. J. Cell. Physiol. 115, 179–185. ( 10.1002/jcp.1041150212) [DOI] [PubMed] [Google Scholar]
- 17.Zachariassen KE. 1991. Hypothermia and cellular physiology. Arctic. Med. Res. 50, 13–17. [PubMed] [Google Scholar]
- 18.Felsenstein J. 1985. Phylogenies and the comparative method. Am. Nat. 125, 1–15. ( 10.1086/284325) [DOI] [Google Scholar]
- 19.Martins EP, Hansen TF. 1997. Phylogenies and the comparative method: a general approach to incorporating phylogenetic information into the analysis of interspecific data. Am. Nat. 149, 646–667. ( 10.1086/286013) [DOI] [Google Scholar]
- 20.Maddison WP, Midford PE, Otto SP. 2007. Estimating a binary character's effect on speciation and extinction. Syst. Biol. 56, 701–710. ( 10.1080/10635150701607033) [DOI] [PubMed] [Google Scholar]
- 21.Rezende EL, Diniz-Filho JFA. 2012. Phylogenetic analyses: comparing species to infer adaptations and physiological mechanisms. Compr. Physiol. 2, 639–674. ( 10.1002/cphy.c100079) [DOI] [PubMed] [Google Scholar]
- 22.Westheide W, Graefe U. 1992. Two new terrestrial Enchytraeus species (Oligochaeta, Annelida). J. Nat. Hist. 26, 479–488. ( 10.1080/00222939200770311) [DOI] [Google Scholar]
- 23.Henle FGJ. 1837. Ueber Enchytraeus, eine neue Anneliden-Gattung. Müllers Archiv für Anatomie, Physiologie, und Wissenschaftliche Medizin, Berlin 1837: 74–90.
- 24.Napolitano MJ, Shain DH. 2004. Four kingdoms on ice: convergent energetic processes boost energy levels at low physiological temperatures. Proc. R. Soc. Lond. B 271(Suppl.), S273–S276. ( 10.1098/rsbl.2004.0180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boore JL, Brown WM. 1995. Complete sequence of the mitochondrial DNA of the Annelid worm Lumbricus terrestris. Genetics 141, 305–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. ( 10.1093/nar/gkh340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6, molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. ( 10.1093/molbev/mst197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772 ( 10.1038/nmeth.2109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guindon S, Gascuel O. 2003. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst. Biol. 52, 696–704. ( 10.1080/10635150390235520) [DOI] [PubMed] [Google Scholar]
- 30.Akaike H. 1974. A new look at the statistical model identification . IEEE Trans. Autom. Control 19, 716–723. ( 10.1109/TAC.1974.1100705) [DOI] [Google Scholar]
- 31.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. ( 10.1093/sysbio/syq010) [DOI] [PubMed] [Google Scholar]
- 32.Simmons MP, Norton AP. 2014. Divergent maximum-likelihood-branch-support values for polytomies. Mol. Phylogenet. Evol. 73, 87–96. ( 10.1016/j.ympev.2014.01.018) [DOI] [PubMed] [Google Scholar]
- 33.Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. ( 10.1093/sysbio/sys029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maddison WP, Maddison DR.2015. Mesquite: a modular system for evolutionary analysis, version 3.04. See http://mesquiteproject.org .
- 35.Tavare S. 1986. Some probabilistic and statistical problems in the analysis of DNA sequences. Lectures on Mathematics in the Life Sciences (American Mathematical Society) 17, 57–86. [Google Scholar]
- 36.Huelsenbeck JP, Larget B, Alfaro ME. 2004. Bayesian phylogenetic model selection using reversible jump Markov chain Monte Carlo. Mol. Biol. Evol. 21, 1123–1133. ( 10.1093/molbev/msh123) [DOI] [PubMed] [Google Scholar]
- 37.Yang Z. 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J. Mol. Evol. 39, 306–314. ( 10.1007/BF00160154) [DOI] [PubMed] [Google Scholar]
- 38.Bouckaert RR, Heled J.2014. DensiTree 2: Seeing Trees Through the Forest. See . . [DOI]
- 39.Lanfear R, Calcott B, Ho S, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. ( 10.1093/molbev/mss020) [DOI] [PubMed] [Google Scholar]
- 40.Liu L, Pearl DK. 2007. Species trees from gene trees: reconstructing Bayesian posterior distributions of a species phylogeny using estimated gene tree distributions. Syst. Biol. 56, 504–514. [DOI] [PubMed] [Google Scholar]
- 41.Rannala BH, Yang Z. 2003. Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics 164, 1645–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maddison WP. 1997. Gene trees in species trees. Syst. Biol. 46, 523–536. ( 10.1093/sysbio/46.3.523) [DOI] [Google Scholar]
- 43.Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, 14 November 2010, pp. 1–8. [Google Scholar]
- 44.Pamilo P, Nei M. 1988. Relationships between gene trees and species trees. Mol. Biol. Evol. 5, 568–583. [DOI] [PubMed] [Google Scholar]
- 45.Degnan JH, Rosenberg NA. 2006. Discordance of species trees with their most likely gene trees. PLOS Genet. 2, 762–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaw KL. 2002. Conflict between nuclear and mitochondrial DNA phylogenies of a recent species radiation: what mtDNA reveals and conceals about modes of speciation in Hawaiian crickets. Proc. Natl Acad. Sci. USA 99, 16 122–16 127. ( 10.1073/pnas.242585899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan KM, Levin SA. 2005. Leaky prezygotic isolation and porous genomes: rapid introgression of maternally inherited DNA. Evolution 59, 720–729. ( 10.1111/j.0014-3820.2005.tb01748.x) [DOI] [PubMed] [Google Scholar]
- 48.Kidwell MG. 1993. Lateral transfer in natural populations of eukaryotes. Annu. Rev. Genet. 27, 235–256. ( 10.1146/annurev.ge.27.120193.001315) [DOI] [PubMed] [Google Scholar]
- 49.Fitch WM. 1970. Distinguishing homologous from analogous proteins. Syst. Zool. 19, 99–113. ( 10.2307/2412448) [DOI] [PubMed] [Google Scholar]
- 50.Goodman M, Czelusniak J, Moore GW, Romero-Herrera AE, Matsuda G. 1979. Fitting the gene lineage into its species lineage, a parsimony strategy illustrated by cladograms constructed from globin sequences. Syst. Zool. 28, 132–163. ( 10.2307/2412519) [DOI] [Google Scholar]
- 51.Bazin E, Glemin S, Galtier N. 2006. Population size does not influence mitochondrial genetic diversity in animals. Science 312, 570–572. ( 10.1126/science.1122033) [DOI] [PubMed] [Google Scholar]
- 52.Toews DPL, Brelsford A. 2012. The biogeography of mitochondrial and nuclear discordance in animals. Mol. Ecol. 21, 3907–3930. ( 10.1111/j.1365-294X.2012.05664.x) [DOI] [PubMed] [Google Scholar]
- 53.Bayona-Bafaluy MP, Muller S, Moraes CT. 2005. Fast adaptive evolution of nuclear and mitochondrial subunits of ATP synthase in Orangutan. Mol. Biol. Evol. 22, 716–724. ( 10.1093/molbev/msi059) [DOI] [PubMed] [Google Scholar]
- 54.Mishmar D, Ruiz-Pesini E, Mondragon-Palomino M, Procaccio V, Gaut B, Wallace DC. 2006. Adaptive selection of mitochondrial complex I subunits during primate radiation. Gene 378, 11–18. ( 10.1016/j.gene.2006.03.015) [DOI] [PubMed] [Google Scholar]
- 55.Bar-Yaacov D, Blumberg A, Mishmar D. 2012. Mitochondrial-nuclear co-evolution and its effects on OXPHOS activity and regulation. Biochim. Biophys. Acta 1819, 1107–1111. [DOI] [PubMed] [Google Scholar]
- 56.Hill GE. 2016. Mitonuclear coevolution as the genesis of speciation and the mitochondrial DNA barcode gap. Ecol. Evol. 6, 5831–5842. ( 10.1002/ece3.2338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dowling DK, Friberg U, Lindell J. 2008. Evolutionary implications of non-neutral mitochondrial genetic variation. Trends Ecol. Evol. 23, 546–554. ( 10.1016/j.tree.2008.05.011) [DOI] [PubMed] [Google Scholar]
- 58.Galtier N, Nabholz B, Glemin S, Hurst GDD. 2009. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol. Ecol. 18, 4541–4550. ( 10.1111/j.1365-294X.2009.04380.x) [DOI] [PubMed] [Google Scholar]
- 59.Degnan JH, Rosenberg NA. 2006. Discordance of species trees with their most likely gene trees. PLoS Genet. 2, 762–768. ( 10.1371/journal.pgen.0020068) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edwards SV, Liu L, Pearl DK. 2007. High-resolution species trees without concatenation. Proc. Natl. Acad. Sci. USA 104, 5936–5941. ( 10.1073/pnas.0607004104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kubatko LS, Degnan JH. 2007. Inconsistency of phylogenetic estimates from concatenated data under coalescence. Syst. Biol. 56, 17–24. ( 10.1080/10635150601146041) [DOI] [PubMed] [Google Scholar]
- 62.Heled J, Drummond AJ. 2010. Bayesian inference of species trees from multilocus data. Mol. Biol. Evol. 27, 570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang Z, Rannala B. 2010. Bayesian species delimitation using multilocus sequence data. Proc. Natl Acad. Sci. USA, 107, 9264–9269. ( 10.1073/pnas.0913022107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boto L. 2014. Horizontal gene transfer in the acquisition of novel traits by metazoans. Proc. R. Soc. B 281, 20132450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grant PR, Grant BR, Petren K. 2005. Hybridization in the recent past. Amer. Nat. 166, 56–67. ( 10.1086/430331) [DOI] [PubMed] [Google Scholar]
- 66.Boratyński Z, Alves PC, Berto S, Koskela E, Mappes T, Melo-Ferreira J. 2011. Introgression of mitochondrial DNA among Myodes voles: consequences for energetics? BMC Evol. Biol. 11, 355 ( 10.1186/1471-2148-11-355) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hammond PE. 1979. A tectonic model for evolution of the Cascade Range. In The Cenozoic paleogeography of the Western United States (eds Armentrout JM, et al.), pp. 219–237. Pacific coast paleogeography symposium 3, Anaheim, CA, Society of Economic Paleontologists and Mineralogists. [Google Scholar]
- 68.Parrish RR. 1983. Cenozoic thermal evolution and tectonics of the Coast Mountains of British Columbia: 1. Fission track dating, apparent uplift rates, and patterns of uplift. Tectonics 2, 601–631. ( 10.1029/TC002i006p00601) [DOI] [Google Scholar]
- 69.Montgomery DR. 2000. Coevolution of the Pacific salmon and Pacific Rim topography. Geology 28, 1107–1110. ( 10.1130/0091-7613(2000)28%3C1107:COTPSA%3E2.0.CO;2) [DOI] [Google Scholar]
- 70.Bjornstad BN. 2006. On the trail of the Ice Age floods: a geological field guide to the mid-Columbia basin, p. 4 Sandpoint, Idaho: Keokee Books. [Google Scholar]
- 71.Dial CR, et al. 2012. Historical biogeography of the North American glacier ice worm, Mesenchytraeus solifugus (Annelida: Oligochaeta: Enchytraeidae). Mol. Phylogenet. Evol. 63, 577–584. ( 10.1016/j.ympev.2012.01.008) [DOI] [PubMed] [Google Scholar]
- 72.Tynen MJ. 1970. Worms on ice. Nature 225, 587 ( 10.1038/225587a0) [DOI] [PubMed] [Google Scholar]
- 73.Tynen MJ. 1970. The geographical distribution of ice worms (Oligochaeta: Enchytraeidae). Can. J. Zool. 48, 1363–1367. ( 10.1139/z70-233) [DOI] [Google Scholar]
- 74.Goodman D. 1971. Ecological investigations of ice worms on Casement Glacier, Southeast Alaska. Ohio State Univ. Res. Foundation, Institute of Polar Studies, Report 39, 1–59.
- 75.Shain DH, Mason TA, Farrell AH, Michalewicz LA. 2001. Distribution and behavior of ice worms (Mesenchytraeus solifugus) in south-central Alaska. Can. J. Zool. 79, 1813–1821. ( 10.1139/z01-143) [DOI] [Google Scholar]
- 76.Bernatchez L, Wilson CC. 1998. Comparative phylogeography of nearctic and palearctic fishes. Mol. Ecol. 7, 431–452. ( 10.1046/j.1365-294x.1998.00319.x) [DOI] [Google Scholar]
- 77.Wilke T, Duncan N. 2004. Phylogeographical patterns in the American Pacific Northwest: lessons from the arionid slug Prophysaon coeruleum . Mol. Ecol. 13, 2303–2315. ( 10.1111/j.1365-294X.2004.02234.x) [DOI] [PubMed] [Google Scholar]
- 78.Steele CA, Storfer A. 2006. Coalescent-based hypothesis testing supports multiple Pleistocene refugia in the Pacific Northwest for the Pacific giant salamander (Dicamptodon tenebrosus). Mol. Ecol. 15, 2477–2487. ( 10.1111/j.1365-294X.2006.02950.x) [DOI] [PubMed] [Google Scholar]
- 79.Shafer ABA, Cullingham CI, Cote SD, Coltman DW. 2010. Of glaciers and refugia: a decade of study sheds new light on the phylogeography of northwestern North America. Mol. Ecol. 19, 4589–4621. ( 10.1111/j.1365-294X.2010.04828.x) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences: GenBank accession numbers KU728801-KU728933.