Abstract
In order for the first genomes to evolve, independent replicators had to act cooperatively, with some reducing their own replication rate to help copy others. It has been argued that limited diffusion explains this early cooperation. However, social evolution models have shown that limited diffusion on its own often does not favour cooperation. Here we model early replicators using social evolution tools. We show that: (i) replicators can be considered to be cooperating as a result of kin selection; (ii) limited diffusion on its own does not favour cooperation; and (iii) the addition of overlapping generations, probably a general trait of molecular replicators, promotes cooperation. These results suggest key life-history features in the evolution of the genome and that the same factors can favour cooperation across the entire tree of life.
Keywords: molecular replicators, cooperation, kin selection, social evolution, limited diffusion, genome origin
1. Introduction
Genomes are made up of genes, or replicators, which work together to produce an organism. These genes specialize in different tasks—for example, some produce replication machinery to copy the other genes, while others focus on acquiring energy for this process. However, life began with independent replicators, whose sole purpose was to copy themselves [1–3]. Thus, at some point, before the last universal common ancestor, independent replicators came together to form a rudimentary genome. At least some of these replicators had to focus on the task of replicating others, reducing their own replication rate in the process. Thus, to get from the first replicators to the first genome, independent replicators must have acted cooperatively [2–4]. This poses a problem, because we expect natural selection to favour individuals that selfishly replicate themselves. For example, imagine a mutant replicator that, rather than help copy others, replicated itself instead, and therefore had a higher replication rate than its neighbours. All else being equal, this mutant should prevail. So why would early replicators cooperate?
It has been argued that limited diffusion or dispersal could explain cooperation between early replicators [5–12]. The existence of early replicators on surfaces, such as rocks, would have led to relatively limited diffusion [3,13]. A number of simulation studies have examined this possibility, assuming that replicators exist on connected nodes in a two dimensional lattice. In these lattice models, when replicators replicate, their offspring can move one, two, or many nodes away. How far offspring can travel in a given step determines whether the system has limited diffusion or is well mixed. These simulations have shown that limited diffusion can favour the evolution of cooperative replicators, who help others replicate [5–12]. Limited diffusion keeps cooperators together, and so their cooperation is directed towards other cooperators, allowing them to outcompete non-cooperators.
However, this suggested role of limited diffusion raises two further questions. First, limited dispersal has previously been argued to explain cooperation in organisms ranging from bacteria to birds because it keeps relatives together [14–20]. In these cases, cooperation is favoured because it is directed at relatives who share the same genes, termed kin selection [14]. Can we think of this early cooperation between replicators as being favoured by kin selection, analogous to that in higher organisms? If so, we could make broad generalizations about the factors that have favoured cooperation, across different biological levels, as life on earth evolved.
Second, theoretical kin selection models have shown that limited dispersal, on its own, does not favour cooperation (reviewed by [21,22]). Although limited dispersal increases the likelihood that cooperation can be directed towards relatives, it also increases competition between relatives. Taylor [23,24] showed that in the simplest case, these effects exactly cancel, and that the rate of dispersal does not influence selection for cooperation. Since then, a number of models have shown that limited dispersal can favour cooperation, but only if additional factors are added, such as overlapping generations, or dispersal in groups (buds), which allow the benefits of increased relatedness to outweigh the extra competition (e.g. [25–27]). Consequently, we must ask how limited diffusion manages to favour cooperation in these replicator models.
We develop theoretical models to address these two questions. We focus on a specific example of replicator biology, termed the trans-replicase system, because it is one of the simplest forms of molecular cooperation. First, we develop a simple kin selection model to examine whether we can think of limited diffusion as favouring cooperation in trans-replicases by kin selection [28]. This model allows us to compare the evolution of cooperation in a simple replicator with models developed to explain cooperation across a range of other taxa. Second, we develop a more spatially explicit model, to ask how limited diffusion might favour cooperation among replicators. We develop a relatively simple model to capture the key features of the previous simulations, but where we can obtain an analytical solution [5–12].
2. Heuristic overview
We start by developing the simplest possible, heuristic model. The purpose of this is to try to capture the biology of an early replicator using the tools of social evolution theory [29–31]. This kind of streamlined model aims to cut out all but the most essential biological and biochemical details, to capture a wider range of possible biologies, and identify key parameters. We sacrifice realism for generality and insight.
We model one possible system for cooperation in replicators: the trans-acting replicase or trans-replicase (figure 1) [12]. There are a variety of possible biologies for early replicators, but the trans-replicase model is one of the simplest. Trans-replicases are replicating molecules that, upon replicating, through mechanisms such as alternate folding, can express one of two phenotypes: (i) replicases, which are molecules that can copy other replicators, or (ii) templates, which can be copied by a replicase but do not act as replicases (figure 1). The replicase phenotype can be considered to be cooperative, because it reduces its own replication rate in order to increase the replication rate of others, and the template phenotype to be relatively selfish. An individual maintains its phenotype of being either a replicase or a template for life, such that any given individual is either a replicase or a template.
Figure 1.
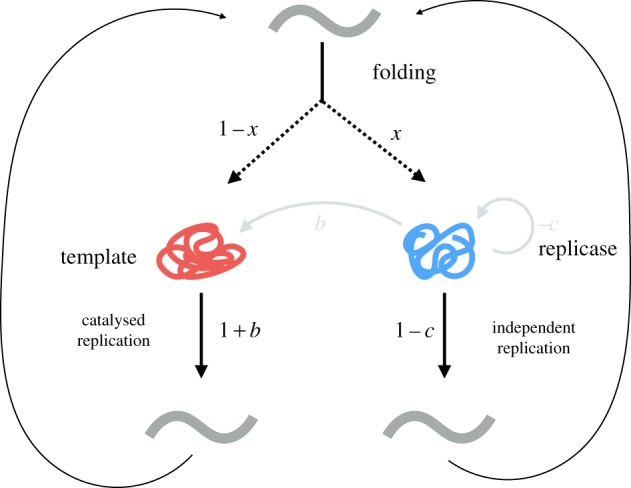
Life cycle for a trans-replicase. Individuals replicate and can express one of two phenotypes (e.g. through alternative folding patterns). With probability x they become a replicase, otherwise, with probability 1 − x, they become a template. All individuals have a baseline replication rate of 1. Replicases incur a cost, c, in terms of replication rate, and enzymatically increase the replication rate of templates by a factor of b. Replicases replicate at a rate of 1 − c, and templates replicate at a rate of 1, unless they interact with a replicase, in which case they replicate at rate 1 + b.
We assume that when a replicator is copied, the new copy (offspring) folds to become a replicase with probability x and a template with probability 1 − x. Mutations could cause individuals to express the cooperative replicase phenotype with higher or lower probability—creating more or less cooperative strategies. We are envisaging phenotypic variation generated by alternate folding, but our model captures other ways to generate variable phenotypes. Although a trans-replicase capable of self-sustaining in a pre-biotic world has yet to be identified, the development of simple RNA molecules capable of template directed synthesis suggest their plausibility [32–37].
Individual replicators have a baseline replication rate, or fitness, of 1. A replicase experiences a replication rate reduced by c, which can be considered the cost of helping or cooperating with templates. If replicases cannot replicate, then c = 1. The presence of replicases increase the replication rate of a template by a factor of by, where y is the average proportion of replicators which are replicases over the scale at which replicators can interact (the social group). This increase in the template replication rate can be considered the benefit of the cooperative or helpful act. When there are a higher fraction of replicases in the social group (y), there is a greater likelihood of any template being helped. Replicases do not catalyse the replication of other replicases, and so do not provide a benefit to replicases.
We can write the expected fitness of a focal individual (w) as the summed fitness of its replicase (1 − c) and template (1 + by) offspring, multiplied by their relative frequency, which is x and 1 − x, respectively, giving
| 2.1 |
We are considering an asexual population, and so ignoring mutation, the strategy or phenotype of a replicator and the copies (offspring) that it produces will be the same, x. Consequently, equation (2.1) can also be conceptualized as the sum of the likelihood that a replicator developed as a replicase multiplied by its fitness in that scenario, and the likelihood that a replicator developed as a template multiplied by its fitness in that scenario. More traditionally, equation (2.1) is conceptualized as the average reproductive value of the focal offspring [38].
We seek the evolutionary stable strategy (ESS), x*, which cannot be beaten by any other strategy [39]. Taylor [29] and Frank [31] developed an approach for determining the ESS in social models. Assuming selection is weak, candidate ESSs occur where the derivative, with respect to deviations in x (also known as the ‘inclusive fitness effect’) equals zero:
| 2.2 |
The dy/dx term is the slope of the regression of a random partner's phenotype on the focal individual's, and can be replaced with r [29], the standard coefficient of relatedness [40–42]. Relatedness, r, is a measure of genetic similarity between our focal individual and the other individuals on the patch. In our model, r is the likelihood that our focal individual shares the same gene at a given locus with another individual on their patch, relative to a random member of the population. Relatedness can arise for a number of reasons, and r represents a summary of all details about the population structure. This kind of approach has proved useful for linking data with theory, because a simple model can then be applied to a variety of different biological cases, where a positive relatedness arises for different reasons [43–47].
Replacing dy/dx with r, we calculate the ESS (x*) to be
| 2.3 |
From the above equation, we can see that increasing relatedness increases the ESS value of x. Our model is agnostic to how relatedness between interacting individuals arises and therefore captures a variety of ways through which relatedness could be positive. One way to achieve higher r is through limited diffusion, because this leads to identical copies of the gene being more likely to find themselves near each other [14]. Thus, our result captures previous claims that limited diffusion would favour cooperation between replicators. Equation (2.3) is analogous to the ESS identified in Frank's [30] paired suicide model, but with an arbitrary cost of cooperation.
We found that cooperation evolves (x* > 0) when rb − c > 0, which is the classic result known as Hamilton's [14] rule. Hamilton's rule is a relatively general result stating that a cooperative trait will evolve if the cost, c, is outweighed by the benefit, b, weighted by relatedness, r [48]. This analysis, therefore, confirms that we can think of kin selection as the reason limited diffusion favours cooperation between replicators, in exactly the same way as kin selection is usually applied to explain cooperation in other taxa, such as bacteria and animals.
3. Population structure
(a). Island model
Our above model showed that high relatedness favours cooperation, but left open the mechanism by which high relatedness is generated. One possibility is through limited diffusion of offspring copies [14], as has been argued for a wide range of organisms, including trans-replicases [12]. We test this idea by explicitly modelling population structure in an infinite island model. This is a standard approach to modelling population structure in evolutionary biology and is slightly different from a lattice model. In a lattice model we explicitly track distance, such that individuals might be further or closer apart. In an island model, we do not track distance, but instead allow individuals to stay in one place or move arbitrarily far away. The island model has been shown to give similar results to more explicit lattice and stepping stone structures [49].
Our infinite population is now subdivided into an infinite number of patches, or islands. For example, we can imagine that groups of replicators are isolated in crevices, on separate rocks, or even in droplets [4,50]. These patches have limited resources (e.g. nucleotides), such that they contain N individuals. Individual replicators interact within these patches, and these interactions determine their fitnesses or the number of offspring copies they produce.
Offspring are produced in a single generation, or round, of replication, and offspring copies diffuse to a distant patch with probability (1 − s). Otherwise, with probability s, they stay on the same patch. Offspring that remain compete randomly among themselves and with new arrivees from other patches for the N available spots, and those that do not secure a spot die. Thus, an individual's fitness determines the chances that the next generation is made up of its offspring. Dispersers, similarly, compete on their new patch with other dispersers and residents for the N spots on that patch. ‘Dispersal’ and ‘diffusion’ are usually used in the kin selection and replicator literature, respectively, to mean the same thing ((1 − s))—we use diffusion for replicators.
Biological models often assume that generations are discrete. This means that when offspring copies are produced, parent copies die, such that each new generation is made up exclusively of offspring. This may not be realistic for simple replicators. Thus, we allow some proportion, k, of parent individuals to survive into the next generation. Survivors maintain their spot on a patch (for example, because they are bound to one of the free binding sites). As a result, all offspring individuals are competing for the 1 − k fraction of free spots on a patch.
In some ways, the patches in our model are similar to cells, in that they allow associations to build up between individuals. However, they are distinct from cells in that offspring disperse independently. The diffusion rate is fixed in this model, which is justified if it is a function of an extrinsic factor (e.g. displacement by movement of the surrounding water), or a non-varying intrinsic factor (e.g. a chemical bond that is independent of mutation). k is fixed for similar reasons.
An individual's fitness now depends on whether it diffuses (with probability (1 − s)) or stays (s), because this will determine the individual's competitive arena. If an individual disperses, its fitness is proportional to the population average fitness, which we assume to be 1 (the population is neither growing or shrinking). If it stays, its fitness is relative to the average fitness on the patch. To determine the average fitness on the patch, it will be necessary to take into account the average phenotype of the whole patch, including the focal individual, which is equal to (y(N − 1) + x)/N, but, which, for simplicity we will denote Z. After diffusion, the number of individuals on a patch is equal to the number of individuals produced on a patch that stay (with probability s) plus the number of individuals arriving from elsewhere ((1 − s)N). So the total number of offspring on a patch after diffusion is
| 3.1 |
These offspring then compete for the available (1 − k) fraction of places on the patch. This allows us to write the fitness of an individual in terms of whether it stays or disperses, as a function of x and y (remember that Z is a function of x and y):
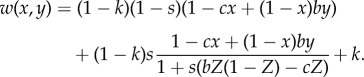 |
3.2 |
From this equation, we can use the Taylor–Frank approach to calculate the equilibrium strategy. However, the resulting equation is not analytically tractable. Instead, if we assume b and c are small, we can write a first-order approximation of this function that can be solved analytically. We also solved the Taylor–Frank equilibrium equation determined from equation (3.2) numerically, and found that relaxing the assumption of small b and c does not change the results (figures 2 and 3). The first-order approximation is
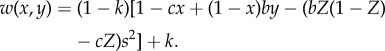 |
3.3 |
Figure 2.
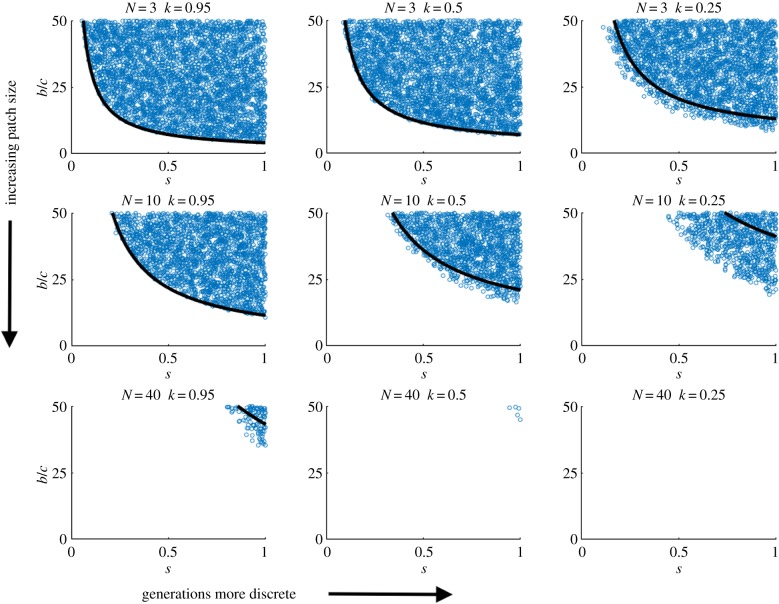
The condition for the evolution of cooperation when generations overlap (k, s > 0.) The y-axis shows the benefit to cost ratio of cooperation (b/c), and the x-axis shows the staying rate (s). The region above the solid black line is where cooperation can evolve. Open blue circles show ESS values identified from numerically solving the equilibrium determined from equation (3.2) in the main text. The benefit to cost ratio captures the degree to which a replicase can increase the replication rate of a template, relative to the associated decrease in autocatalytic replication that results from acting as an enzyme. Cooperation is more likely to be favoured by lower diffusion (increasing s); greater overlap in generations (increasing k) and smaller patch sizes (decreasing N).
Figure 3.
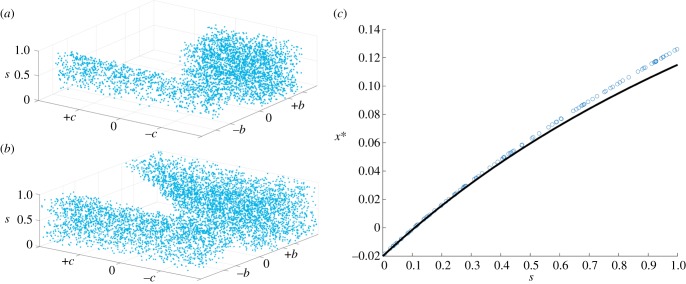
Possible ESS values determined numerically. The figure shows the results from numerically solving the equilibrium determined from equation (3.2) in the main text. (a) Discrete generations (k = 0, N = 20). Blue dots indicate candidate ESSs for different parameter values. If (b,c) are both positive, there are no possible ESS values and so cooperation cannot evolve. (b) The case of overlapping generations (k = 0.9, N = 20). Now, in the region where (b) and (c) are both positive, if s is high (limited diffusion), there are positive candidate ESS values, and so cooperation can evolve. (c) Quantitative fit between numerical (open circles) and analytical (solid line) solutions for small b,c. k = 0.9, N = 5, b = 0.5, c = 0.01.
The fitness components in this equation have a simple biological interpretation. The terms on the left (inside the square brackets) capture the primary consequences of exhibiting the cooperative behaviour, as in the simpler model (equation (2.1)). Specifically, given an individual is cooperative, it incurs a cost, c (−cx) and given it is not cooperative, it receives a benefit, b from cooperative partners ((1 − x)by). The terms on the right capture how cooperation leads to an increase in the local competition. Specifically, extra offspring produced by the average of the trait (Z) on the patch displace the focal individual, given both the extra offspring and the focal individual remain on the patch (with probability s2) ((bZ(1 − Z) − cZ)s2). This model is analogous to a haploid asexual model of others-only cooperation like that found in Taylor & Irwin [25]. As replicases cannot receive benefits, we are modelling what has been called a negatively synergistic game [51].
Using the Taylor–Frank approach, we can write the inclusive fitness effect as
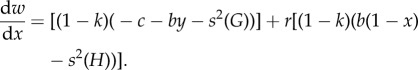 |
3.4 |
The terms in the first set of square brackets are the direct effects of cooperation and the terms in the second set capture the indirect effects mediated through relatives. G = − c/N + b(1 − Z)/N − bZ/N and H = (N − 1)(c + b(2Z − 1))/N capture the secondary effects of extra offspring that stay on the natal patch, and are decreasing functions of Z. From this, we can calculate the ESS to be
| 3.5 |
This gives a solution in terms of relatedness (r) and diffusion rate (1 − s), but we expect r to depend on s. Limited diffusion (increasing s) should increase relatedness (r). Specifically, r is determined by the diffusion rate, the survival rate, k and the patch size, N. We can calculate r in terms of these parameter values, and plug this value for r back into equation (3.5) (see appendix A for details). This closes the model [23,26] to give the equilibrium value
| 3.6 |
(b). Discrete generations
First, we consider the specific case of discrete generations (k = 0), which is the simplest possible case. When generations are discrete, we find that
| 3.7 |
This equation shows that, in the case of discrete generations, diffusion has no effect on the ESS value of cooperation—the parameter s is not in equation (3.3). Furthermore, that under limited diffusion, cooperation cannot evolve (c and b are positive, and the direction of selection at x* = 0 is negative, making pure templates the stable boundary condition). This result echoes the classic result by Taylor [23,24], which showed that, while limited dispersal increases relatedness, this effect is exactly offset by the corresponding increase in local competition. This can be seen in our equation (3.4), by the two ways in which s determines the inclusive fitness effect of cooperation. Increasing s raises r, and therefore increases the indirect benefits gained by cooperating. However, increasing s also leads to the losses owing to H and G (extra offspring on the patch) being more heavily weighted. In the case of k = 0, these two effects exactly cancel.
(c). Overlapping generations
We now consider when there is some degree of overlapping generations (k > 0). In this case, the condition for cooperation to evolve becomes
| 3.8 |
If k and s are both greater than zero—that is, if there is some degree of overlapping generations and limited diffusion—cooperation can evolve. This is because increasing k raises r, relatedness, and therefore increases the indirect benefits of cooperation without increasing the competitive effects of extra offspring (equation (3.4)). Consequently, decreasing diffusion rate (1 − s) and increasing survivorship (k) tend to favour cooperation (figure 2). Increasing the benefit of cooperation, b, and decreasing the cost of cooperation, c, make it easier for cooperation to evolve.
Decreasing patch size (N) makes it easier for cooperation to evolve. This is because the larger the patch size, the lower the average relatedness on a patch (equation (A 1)). One caveat is that we assume, deterministically, that each patch contains both templates and replicases. As N gets smaller, stochastic variation in the patch composition make this less likely to hold. In the extreme, if N = 1, then the patch could only contain a template or a replicase, but not both. Consequently, replicases would be paying the cost of cooperation, when there are no templates to gain the benefit. This stochastic effect would be reduced or removed if cooperation is conditional upon being in a patch where there are templates. An analogous problem of stochasticity in small patch sizes has been considered with sex allocation in structured populations (local mate competition) [45].
In our above model, we assumed that survivors maintain their spot on a patch. This is reasonable if, for example, once a replicator finds a binding site it remains there until death. Alternatively, we might allow survivors to remain on the patch but to compete equally with offspring for a place. This would be reasonable if, for example, offspring can ‘bump’ adults from a patch. A third possibility is that survivors can disperse along with offspring, and compete globally—this might occur if during each replication event replicators are dislodged from their binding site. We show in appendix A that neither allowing for survivors to compete for sites nor allowing survivors to disperse qualitatively alters the results, although both changes make cooperation more difficult to evolve.
4. Discussion
We have used the analytical tools of social evolution theory to model a simple replicating molecule scenario: transacting replicases. We have shown that cooperation between replicators can be understood as evolving via the process of kin selection through limited diffusion. However, we have also shown that limited diffusion on its own does not favour cooperation (equation (3.7)). Instead, an additional life-history detail of simple replicators is needed—that of overlapping generations (figure 2).
Our social evolution model illustrates two points about replicators. First, we can view limited diffusion as favouring cooperation in simple replicators via kin selection. Consequently, the factor favouring cooperation in trans-replicases: (i) links to a large existing theoretical literature [14,21–23], and (ii) is the same factor that has been previously shown to favour cooperation in a range of organisms including birds, mammals, insects and microbes [15–20,52,53]. By clarifying these links across taxa, we can simplify our understanding of life, rather than having to provide different explanations for different cases. We are not saying cooperation in replicators has to be conceptualized via kin selection, just that it can be useful to do so.
Second, both limited diffusion and overlapping generations are required to favour cooperation. Limited diffusion leads to a build-up of relatedness, which favours cooperation [14]. But at the same time, limited diffusion leads to increased competition between relatives, which disfavours cooperation [23,31,54]. Overall, in the simplest possible scenario, these two effects exactly cancel (equation (3.7)). However, we found that the addition of overlapping generations allows limited diffusion to favour cooperation (figure 2). When generations overlap, this increases relatedness within patches, but without increasing competition between relatives, because offspring still diffuse to the same extent [25]. Specifically, increasing overlap (k) raises relatedness ((r), and therefore increases the indirect benefits of cooperation without increasing the competitive effects of extra offspring (equation (3.4)). Consequently, when there is both limited diffusion and overlapping generations, the build-up of relatedness outweighs the increased competition between relatives, such that cooperation is favoured (figure 2).
There are many ways to model social behaviours. One decision is whether to assume discrete strategies, such as ‘cooperators’ and ‘cheats’, or to allow for continuous strategies, ranging, for example, from completely selfish to completely cooperative [29,39,55]. The assumption of continuous strategies is clearly valid for animals, where traits are determined by multiple genes, but simple replicators might only have a limited number of strategies open to them by mutation. Another decision is whether to develop explicit simulations or analytical models. Simulations allow greater detail to be incorporated, which can be especially useful when considering specific systems or species. By contrast, the analytical approach usually used in kin selection models tends to be more streamlined, sacrificing details and precision for clarity and generality [29,31]. Further, the kin selection approach offers a heuristic which non-mathematicians can apply across a range of organisms [45,47]. Rather pleasingly, in the replicator scenario examined here, the different approaches make the same qualitative prediction [12].
The route from independent replicators to the first genomes probably involved two kinds of cooperation. Early cooperation could have been between genetic relatives, or replicators of the same type. However, early genomes were probably too simple to copy themselves accurately, and yet inaccurate replication prevented genomes from getting large enough to improve their accuracy [1]. In order for the genome to overcome this ‘error threshold in replication’, it is probable that different types of replicators needed to cooperatively copy each other. Individual replicators could remain small enough to be copied accurately, but the collection of replicators could become large enough to produce the kind of enzyme machinery needed for accuracy [2]. We have modelled the first kind of cooperation—between replicators of the same ‘type’—and have shown that this can be understood as evolving via kin selection [28]. Cooperation between different types, however, would have required some factor to align the interests of unrelated replicators.
To conclude, although we have phrased our model in terms of a specific replicator system, the trans-replicase, we expect our predictions to hold more generally for other types of replicators. We do not yet know the actual biology of the earliest life forms. But, while many higher organisms may adopt a system of discrete generations, we would expect overlapping generations to be a feature of all simple replicators. Our results, then, would apply to various possible routes through which simple replicators could come together to cooperate.
Acknowledgements
We thank Miguel dos Santos, Guy Cooper, Matishalin Patel, Tom Scott, Asher Leeks, Paul Higgs, Peter Taylor, Geoff Wild and one anonymous reviewer for very helpful discussion and/or comments; and Magdalen College for emergency housing. This paper was inspired by Shay et al. [12].
Appendix A
(a) Writing relatedness in terms of model parameters
We start by determining the relatedness, at equilibrium, of a focal individual to a random member of its patch, drawn with replacement. This is known as whole-group relatedness (denoted by R), because it includes the focal individual, in contrast with others-only relatedness, which does not include the focal individual [42]. Note that in our model, we are dealing with r, others-only relatedness, because y is the average of the individuals on the patch, excluding the focal individual. We can write R (whole-group relatedness), the relatedness between two individuals drawn randomly from a patch with replacement, as the probability that those two individuals are the same individual (1/N), and thus have relatedness 1, plus the probability that those two individuals are not the same ((N − 1)/N), and thus have the relatedness of two random individuals drawn without replacement, or others-only relatedness, r:
| A 1 |
Now we take two individuals (without replacement) on the same patch with relatedness r, and determine the relatedness of their representatives in the previous generation. With chance k2 they are both survivors from the previous generation, in which case their relatedness is the same (r). With chance 2k(1 − k) one is a survivor and the other is a new offspring, which is native with probability s, in which case their relatedness is R. Else, with chance (1 − k)2 they are both new offspring, are both native with probability s2, and thus have relatedness R. We can write others-only relatedness between two individuals in the current generation as equal to
| A 2 |
Here rt is relatedness in the current generation, or time step, and rt−1 and Rt−1 are others-only and whole-group relatednesses, respectively, in the previous one. Setting rt = rt−1 we find the equilibrium others-only relatedness. Plugging into equation (A 1), we find the equilibrium value of whole-group relatedness, R*, to be
| A 3 |
This equation for relatedness was identified by Taylor & Irwin [25]. However, here we are modelling an others only-trait, and thus require others-only relatedness, r. RN gives us the number of relatives on our patch. Subtracting the focal individual, and dividing by the total number of remaining individuals (N − 1), gives us r*:
| A 4 |
Plugging into equation (3.5) gives us equation (3.6).
(b) Allowing survivors to remain and compete for patch sites or disperse globally
Our original model assumed that surviving parents maintained their places on a patch, meaning offspring competed for the remaining 1 − k fraction of available sites. Here we relax this assumption. First, we allow survivors to remain on their patch, but compete equally with offspring for available sites. Using the relatedness recursion in equation (A 2), we calculate the ESS (assuming small b and c) to be
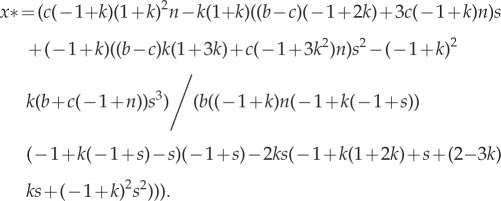 |
A 5 |
Next, we allow survivors to disperse along with offspring. This requires a new relatedness recursion, which we write as
| A 6 |
We now determine the ESS to be
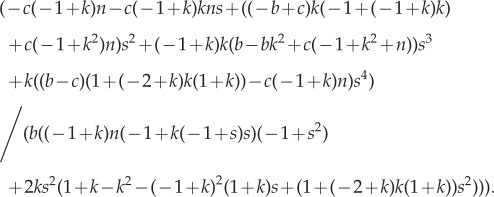 |
A 7 |
If k = 0 (discrete generations), both equations (A 5) and (A 7) revert to equation (3.7) in the text, and cooperation cannot evolve. However, given some degree of overlapping generations and limited diffusion, cooperation can evolve, although the condition is now more stringent.
Data accessibility
This article has no additional data.
Author's contributions
S.R.L. and S.A.W. contributed to conception, modelling and write up of the manuscript. All authors gave final approval for publication.
Competing interests
We have no competing interests.
Funding
S.R.L. is funded by The Clarendon Fund, Hertford College and the Natural Environment Research Council.
References
- 1.Eigen M. 1971. Self-organization of matter and the evolution of biological macromolecules. Naturwissenschaften 58, 465–523. ( 10.1007/BF00623322) [DOI] [PubMed] [Google Scholar]
- 2.Eigen M, Schuster P. 1977. A principle of natural self-organization. Naturwissenschaften 64, 541–565. ( 10.1007/BF00450633) [DOI] [PubMed] [Google Scholar]
- 3.Smith JM, Szathmary E. 1995. The major transitions in evolution. Oxford, UK: Oxford University Press. [Google Scholar]
- 4.Higgs PG, Lehman N. 2015. The RNA world: molecular cooperation at the origins of life. Nat. Rev. Genet. 16, 7–17. ( 10.1038/nrg3841) [DOI] [PubMed] [Google Scholar]
- 5.Boerlijst MC, Hogeweg P. 1991. Spiral wave structure in pre-biotic evolution: hypercycles stable against parasites. Phys. D Nonlinear Phenom. 48, 17–28. ( 10.1016/0167-2789(91)90049-F) [DOI] [Google Scholar]
- 6.Boerlijst MC, Hogeweg P. 1995. Spatial gradients enhance persistence of hypercycles. Phys. D Nonlinear Phenom. 88, 29–39. ( 10.1016/0167-2789(95)00178-7) [DOI] [Google Scholar]
- 7.Cronhjort MB, Blomberg C. 1997. Cluster compartmentalization may provide resistance to parasites for catalytic networks. Phys. D Nonlinear Phenom. 101, 289–298. ( 10.1016/S0167-2789(97)87469-6) [DOI] [Google Scholar]
- 8.McCaskill JS, Füchslin RM, Altmeyer S. 2001. The stochastic evolution of catalysts in spatially resolved molecular systems. Biol. Chem. 382, 1343–1363. ( 10.1515/BC.2001.167) [DOI] [PubMed] [Google Scholar]
- 9.Szabó P, Scheuring I, Czárán T, Szathmáry E. 2002. In silico simulations reveal that replicators with limited dispersal evolve towards higher efficiency and fidelity. Nature 420, 340–343. ( 10.1038/nature01187) [DOI] [PubMed] [Google Scholar]
- 10.Sardanyés J, Solé RV. 2007. Spatio-temporal dynamics in simple asymmetric hypercycles under weak parasitic coupling. Phys. D Nonlinear Phenom. 231, 116–129. ( 10.1016/j.physd.2007.04.009) [DOI] [Google Scholar]
- 11.Bianconi G, Zhao K, Chen IA, Nowak MA. 2013. Selection for replicases in protocells. PLoS Comput. Biol. 9, e1003051 ( 10.1371/journal.pcbi.1003051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shay JA, Huynh C, Higgs PG. 2015. The origin and spread of a cooperative replicase in a prebiotic chemical system. J. Theor. Biol. 364, 249–259. ( 10.1016/j.jtbi.2014.09.019) [DOI] [PubMed] [Google Scholar]
- 13.Wächtershäuser G. 1988. Before enzymes and templates: theory of surface metabolism. Microbiol. Rev. 52, 452–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamilton WD. 1964. The genetical theory of social behavior. I and II. J. Theor. Biol. 7, 1–52. ( 10.1016/0022-5193(64)90038-4) [DOI] [PubMed] [Google Scholar]
- 15.Griffin AS, West SA, Buckling A. 2004. Cooperation and competition in pathogenic bacteria. Nature 430, 1024–1027. ( 10.1038/nature02744) [DOI] [PubMed] [Google Scholar]
- 16.Hughes WOH, Oldroyd BP, Beekman M, Ratnieks FLW. 2008. Ancestral monogamy shows kin selection is key to the evolution of eusociality. Science 320, 1213–1216. ( 10.1126/science.1156108) [DOI] [PubMed] [Google Scholar]
- 17.Diggle SP, Gardner A, West SA, Griffin AS. 2007. Evolutionary theory of bacterial quorum sensing: when is a signal not a signal? Phil. Trans. R. Soc. B 362, 1241–1249. ( 10.1098/rstb.2007.2049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuzdzal-Fick JJ, Fox SA, Strassmann JE, Queller DC. 2011. High relatedness is necessary and sufficient to maintain multicellularity in dictyostelium. Science 334, 1548–1551. ( 10.1126/science.1213272) [DOI] [PubMed] [Google Scholar]
- 19.Lukas D, Clutton-Brock T. 2012. Cooperative breeding and monogamy in mammalian societies. Proc. R. Soc. B 279, 2151–2156. ( 10.1098/rspb.2011.2468) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher RM, Cornwallis CK, West SA. 2013. Group formation, relatedness, and the evolution of multicellularity. Curr. Biol. 23, 1120–1125. ( 10.1016/j.cub.2013.05.004) [DOI] [PubMed] [Google Scholar]
- 21.West SA, Pen I, Griffin AS. 2002. Cooperation and competition between relatives. Science 296, 72–75. ( 10.1126/science.1065507) [DOI] [PubMed] [Google Scholar]
- 22.Lehmann L, Rousset F. 2010. How life history and demography promote or inhibit the evolution of helping behaviours. Phil. Trans. R. Soc. B 365, 2599–2617. ( 10.1098/rstb.2010.0138) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor PD. 1992. Altruism in viscous populations—an inclusive fitness model. Evol. Ecol. 6, 352–356. ( 10.1007/BF02270971) [DOI] [Google Scholar]
- 24.Taylor PD. 1992. Inclusive fitness in a homogeneous environment. Proc. R. Soc. Lond. B 249, 299–302. ( 10.1098/rspb.1992.0118) [DOI] [Google Scholar]
- 25.Taylor PD, Irwin AJ. 2000. Overlapping generations can promote altruistic behavior. Evolution 54, 1135–1141. ( 10.1111/j.0014-3820.2000.tb00549.x) [DOI] [PubMed] [Google Scholar]
- 26.Gardner A, West SA. 2006. Demography, altruism, and the benefits of budding. J. Evol. Biol. 19, 1707–1716. ( 10.1111/j.1420-9101.2006.01104.x) [DOI] [PubMed] [Google Scholar]
- 27.Lehmann L, Perrin N, Rousset F, Day T. 2006. Population demography and the evolution of helping behaviors. Evolution 60, 1137–1151. ( 10.1111/j.0014-3820.2006.tb01193.x) [DOI] [PubMed] [Google Scholar]
- 28.Frank SA. 1994. Kin selection and virulence in the evolution of protocells and parasites. Proc. R. Soc. Lond. B 258, 153–161. ( 10.1098/rspb.1994.0156) [DOI] [PubMed] [Google Scholar]
- 29.Taylor PD, Frank SA. 1996. How to make a kin selection model. J. Theor. Biol. 180, 27–37. ( 10.1006/jtbi.1996.0075) [DOI] [PubMed] [Google Scholar]
- 30.Frank SA. 1997. Multivariate analysis of correlated selection and kin selection, with an ESS maximization method. J. Theor. Biol. 189, 307–316. ( 10.1006/jtbi.1997.0516) [DOI] [PubMed] [Google Scholar]
- 31.Frank SA. 1998. Foundations of social evolution. Princeton, NJ: Princeton University Press. [Google Scholar]
- 32.Inoue T, Orgel LE. 1983. A nonenzymatic RNA polymerase model. Science 219, 859–862. ( 10.1126/science.6186026) [DOI] [PubMed] [Google Scholar]
- 33.Johnston WK, Unrau PJ, Lawrence MS, Glasner ME, Bartel DP. 2001. RNA-catalyzed RNA polymerization: accurate and general RNA-templated primer extension. Science 292, 1319–1325. ( 10.1126/science.1060786) [DOI] [PubMed] [Google Scholar]
- 34.Zaher HS, Unrau PJ. 2007. Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA 13, 1017–1026. ( 10.1261/rna.548807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajamani S, Ichida JK, Antal T, Treco DA, Leu K, Nowak MA, Szostak JW, Chen IA. 2010. Effect of stalling after mismatches on the error catastrophe in nonenzymatic nucleic acid replication. J. Am. Chem. Soc. 132, 5880–5885. ( 10.1021/ja100780p) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wochner A, Attwater J, Coulson A, Holliger P. 2011. Ribozyme-catalyzed transcription of an active ribozyme. Science 332, 209–212. ( 10.1126/science.1200752) [DOI] [PubMed] [Google Scholar]
- 37.Attwater J, Wochner A, Holliger P. 2013. In-ice evolution of RNA polymerase ribozyme activity. Nat. Chem. 5, 1011–1018. ( 10.1038/nchem.1781) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fisher Ronald A. 1930. The genetical theory of natural selection. Oxford, UK: University Press Google Scholar. [Google Scholar]
- 39.MaynardSmith J, Price GR. 1973. The logic of animal conflict. Nature 246, 15–18. ( 10.1038/246015a0) [DOI] [Google Scholar]
- 40.Hamilton WD. 1970. Selfish and spiteful behaviour in an evolutionary model. Nature 228, 1218–1220. ( 10.1038/2281218a0) [DOI] [PubMed] [Google Scholar]
- 41.Grafen A. 1985. A geometric view of relatedness. Oxf. Surv. Evol. Biol. 2, 28–89. [Google Scholar]
- 42.Pepper JW. 2000. Relatedness in trait group models of social evolution. J. Theor. Biol. 206, 355–368. ( 10.1006/jtbi.2000.2132) [DOI] [PubMed] [Google Scholar]
- 43.Sachs JL, Mueller UG, Wilcox TP, Bull JJ. 2004. The evolution of cooperation. Q. Rev. Biol. 79, 135–160. ( 10.1086/383541) [DOI] [PubMed] [Google Scholar]
- 44.West SA, Griffin AS, Gardner A. 2007. Evolutionary explanations for cooperation. Curr. Biol. 17, R661–R672. ( 10.1016/j.cub.2007.06.004) [DOI] [PubMed] [Google Scholar]
- 45.West S. 2009. Sex allocation. Princeton, NJ: Princeton University Press. [Google Scholar]
- 46.Abbot P, et al. 2011. Inclusive fitness theory and eusociality. Nature 471, E1–E4. ( 10.1038/nature09831) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bourke AFG. 2014. Hamilton's rule and the causes of social evolution. Phil. Trans. R. Soc. B 369, 20130362 ( 10.1098/rstb.2013.0362) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gardner A, West SA, Wild G. 2011. The genetical theory of kin selection. J. Evol. Biol. 24, 1020–1043. ( 10.1111/j.1420-9101.2011.02236.x) [DOI] [PubMed] [Google Scholar]
- 49.Comins HN, Hamilton WD, May RM. 1980. Evolutionarily stable dispersal strategies. J. Theor. Biol. 82, 205–230. ( 10.1016/0022-5193(80)90099-5) [DOI] [PubMed] [Google Scholar]
- 50.Zwicker D, Seyboldt R, Weber CA, Hyman AA, Jülicher F. 2016. Growth and division of active droplets provides a model for protocells. Nat. Phys. 13, 408–413. ( 10.1038/nphys3984) [DOI] [Google Scholar]
- 51.Queller DC. 1985. Kinship, reciprocity and synergism in the evolution of social behaviour. Nature 318, 366–367. ( 10.1038/318366a0) [DOI] [Google Scholar]
- 52.Cornwallis CK, West SA, Griffin AS. 2009. Routes to indirect fitness in cooperatively breeding vertebrates: kin discrimination and limited dispersal. J. Evol. Biol. 22, 2445–2457. ( 10.1111/j.1420-9101.2009.01853.x) [DOI] [PubMed] [Google Scholar]
- 53.Cornwallis CK, West SA, Davis KE, Griffin AS. 2010. Promiscuity and the evolutionary transition to complex societies. Nature 466, 969–972. ( 10.1038/nature09335) [DOI] [PubMed] [Google Scholar]
- 54.Queller DC. 1994. Genetic relatedness in viscous populations. Evol. Ecol. 8, 70–73. ( 10.1007/BF01237667) [DOI] [Google Scholar]
- 55.Grafen A. 1979. The hawk-dove game played between relatives. Anim. Behav. 27, 905–907. ( 10.1016/0003-3472(79)90028-9) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.