Abstract
Rapid and reliable detection of disease-associated DNA methylation patterns has major potential to advance molecular diagnostics and underpin research investigations. We describe the development and validation of minimal methylation classifier (MIMIC), combining CpG signature design from genome-wide datasets, multiplex-PCR and detection by single-base extension and MALDI-TOF mass spectrometry, in a novel method to assess multi-locus DNA methylation profiles within routine clinically-applicable assays. We illustrate the application of MIMIC to successfully identify the methylation-dependent diagnostic molecular subgroups of medulloblastoma (the most common malignant childhood brain tumour), using scant/low-quality samples remaining from the most recently completed pan-European medulloblastoma clinical trial, refractory to analysis by conventional genome-wide DNA methylation analysis. Using this approach, we identify critical DNA methylation patterns from previously inaccessible cohorts, and reveal novel survival differences between the medulloblastoma disease subgroups with significant potential for clinical exploitation.
Introduction
Altered DNA methylation patterns have emerged as a common feature of disease pathogenesis, showing clear potential in diagnostics, sub-classification and prediction of therapeutic response/ disease course1–7. In contrast to current high-throughput, genome-wide research methodologies (e.g. whole-genome bisulfite sequencing8, DNA methylation arrays9), particular challenges exist in the clinical application of disease-associated methylation patterns. These include derivation and validation of representative DNA methylation signatures from genome-scale datasets, and their assessment using platform-independent assays that can be applied rapidly to single samples, including low quality and/or quantity biopsies, in routine diagnostics. To address this, we have developed and validated MIMIC (minimal methylation classifier), a novel polymerase chain reaction (PCR)-based assay for the multiplexed assessment of bisulfite-induced methylation-dependent DNA sequence changes10 at multiple signature CpG loci. Sequence-specific single-base variants are exposed by primer extension and, here, coupled to detection by MALDI-TOF mass spectrometry commonly used in clinical DNA diagnostics11, enabling the investigation of samples that were not suitable for analysis using existing methods.
We focussed assay development on medulloblastoma, the most common malignant brain tumour of childhood12, where DNA methylation signatures have clear potential for use in routine clinical sub-classification13,14. Medulloblastoma comprises four primary molecular subgroups – WNT, SHH, Grp3 and Grp4 - defined by distinct methylomic, transcriptomic and genomic features13–15. These subgroups display characteristic clinical features, drug targets and outcomes, and have significantly contributed to the 2016 World Health Organisation (WHO) classification of brain tumours16. Following design and validation of a MIMIC assay for molecular subgrouping, we assessed its efficacy in limited archival tumour biopsies previously refractory to subgrouping using current research methods, taken from the pan-European HIT-SIOP-PNET4 medulloblastoma clinical trial (2000–2006)17,18. This trial enrolled patients negative for all established clinico-molecular risk-factors (termed ‘standard-risk (SR)’ disease12), a group for which there is an urgent unmet need to develop biomarker-driven treatment strategies.
Results
Derivation of minimal DNA methylation signatures to identify the four medulloblastoma molecular subgroups
We first identified a DNA methylation signature of 17 CpG loci, established detection methods and developed a Support Vector Machine (SVM) classification model for distinction of the four medulloblastoma molecular subgroups (WNT, SHH, Grp3 and Grp4; Fig. 1a). Non-negative matrix factorisation (NMF) consensus clustering13,19,20 was used to identify subgroup membership of a training cohort comprising genome-scale Illumina 450k DNA methylation microarray data for 220 medulloblastomas (Fig. 1b; Fig. 2). The 50 most discriminatory CpG loci for each subgroup (i.e. 200 in total) were considered as signature candidates. These were triaged using (i) a 10-fold cross validated classification fusion algorithm, (ii) a reiterative primer design process where amenability to primer design and multiplex bisulfite PCR was assessed in silico (Supplemental experimental methods), and (iii) in vitro PCR validation (Fig. 1a; Fig. 3). Candidate signature CpG loci were assayed by the development of a novel application of the Agena iPlex assay21, whereby methylation-dependent SNPs representative of CpG methylation status were induced by initial treatment of DNA with sodium bisulfite10, followed by multiplexed PCR and single base extension of probe oligonucleotides. The resultant products were quantified by MALDI-TOF MS (Matrix-assisted laser desorption/ionization-time of flight mass spectrometry; Supplementary Fig. 1). The accuracy and precision of methylation estimates from multiplexed extension reactions were tested using incremental proportions of bisulfite-treated methylated:unmethylated DNA (Supplementary Fig. 2). Using these techniques, our optimal, multiply-redundant 17-CpG locus signature was generated. Finally, the training cohort was used to generate an optimised SVM classifier for the signature using 450k DNA methylation array data.
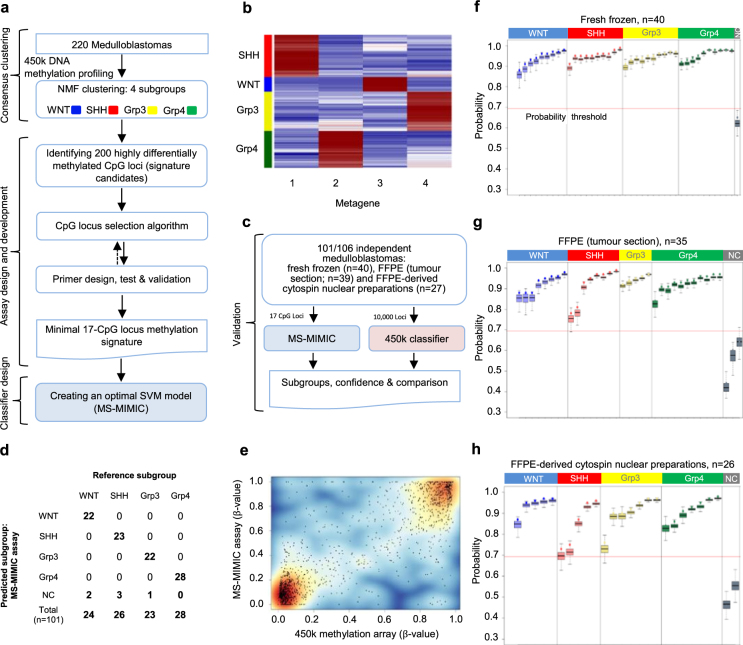
Figure 1.
MS-MIMIC, a novel assay to assess minimal DNA methylation signatures, and its application in a proof of concept study for the identification of molecular disease subgroups in medulloblastoma. (a) Summary of assay design and development including derivation and validation of minimal DNA methylation signatures. (b) Non-negative matrix factorisation (NMF) consensus clustering identified four molecular subgroups (WNT, SHH, Grp3 and Grp4), defined by four metagenes. (c) Assessing MS-MIMIC performance against 450k DNA methylation microarray using 101/106 independent medulloblastoma samples. 5/106 samples failed QC (>6/17 CpG locus fails). (d) Subgroup classification from MS-MIMIC showed total concordance with the reference subgroup (from 450k and GoldenGate DNA methylation arrays and CTNNB1 mutation status) after applying a classification confidence probability threshold (NC –non classifiable). (e) Estimates of methylation (β-values) using MS-MIMIC assay correlated closely with gold-standard 450k DNA methylation microarray (n = 91). (f–h) Subgroup assignment and probability estimates (dots) along with 95% confidence intervals (boxplots) for 101 medulloblastoma samples for which DNAs were derived from three types of materials: Fresh frozen biopsies (n = 40), formalin-fixed paraffin-embedded biopsies (FFPE, tumour section; n = 35) and FFPE-derived cytospin nuclear preparations (n = 26). Samples which did not exceed a classification confidence probability threshold of 0.69 (red-line; empirically derived, Supplementary Fig. 5) were deemed nonclassifiable.
Figure 2.
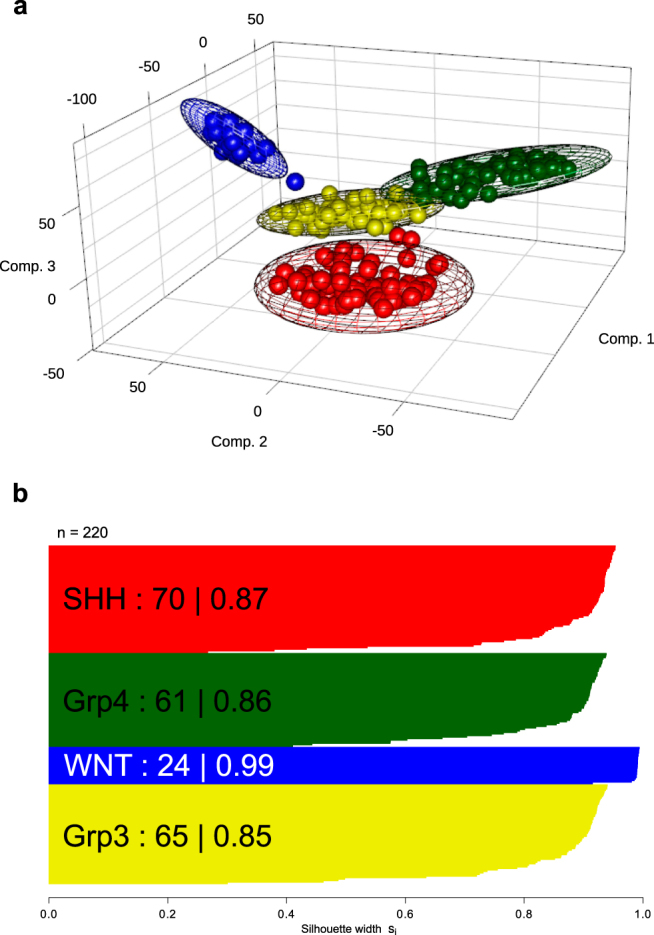
Derivation of the four consensus medulloblastoma molecular subgroups in a training cohort (n = 220) using genome-wide Illumina 450k DNA methylation microarray data. (a) Principal Component Analysis (PCA) visualization of groups identified using consensus NMF clustering. Subgroup members are shown in their consensus colours (blue (WNT); red (SHH); yellow (Grp3) and green (Grp4)). Covariance spheroids were plotted at 95% confidence intervals. (b) Silhouette plot demonstrates robustness of each group (number and average silhouette width are shown).
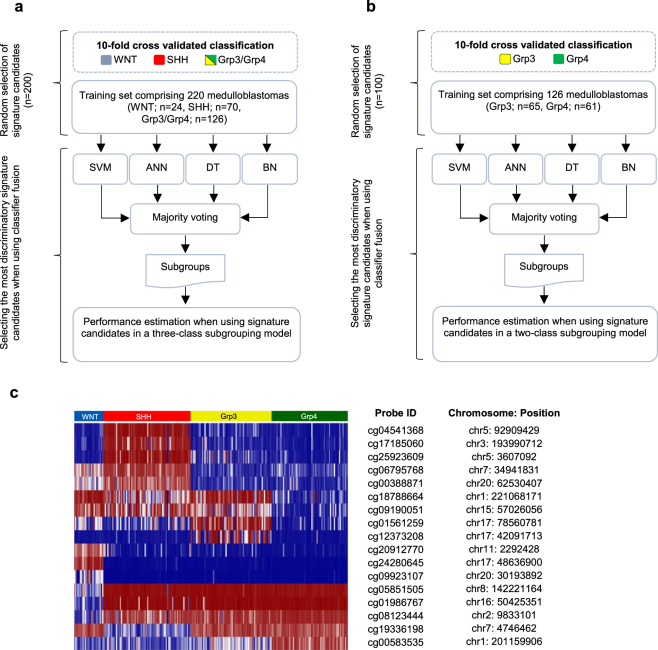
Figure 3.
Derivation of a minimal, multiply-redundant, methylation signature for medulloblastoma subgroup identification. Using a three-class (a) and two-class (b) subgrouping model, a minimal multiply-redundant methylation signature was identified by selecting a subset of signature candidates with the highest ranking in a classifier fusion model (support vector machine (SVM), artificial neural network (ANN), decision tree (DT) and Bayesian network (BN). (c) Heatmap of the most discriminatory signature candidates (n = 17) recapitulates the four known medulloblastoma subgroups. Chromosomal locations (hg19 assembly) and identifiers of the 17 CpG loci are reported in the adjacent table.
Development and validation of MS-MIMIC
We next assessed mass spectrometry-minimal methylation classifier (MS-MIMIC) performance against Illumina 450k methylation microarrays in an independent validation cohort of 106 medulloblastoma DNA samples (Fig. 1c) containing all four medulloblastoma subgroups (Supplementary Table 1). DNAs were derived from tissue samples reflecting different clinical fixation methods; fresh-frozen biopsies (n = 40), formalin-fixed paraffin-embedded biopsies (FFPE, tumour section; n = 39), or FFPE-derived cytospin nuclear preparations18 (n = 27). QC measures for CpG locus-specific assay failure were established; up to 6 failed CpGs per sample were tolerated within the multiply-redundant signature/classifier without impacting performance and this was the case for 101/106 validation cohort samples (Supplementary Figs 3 and 4). A probability threshold for confident subgroup classification was derived empirically, below which samples were deemed non-classifiable (6/101 samples; 5.7%) (Supplementary Fig. 5). Respective subgroup classifications were compared; MS-MIMIC classifications showed complete concordance with the reference subgroup (as determined by DNA methylation array and/or CTNNB1 mutation status for WNT tumours15; Fig. 1d). Furthermore, CpG-level methylation estimates (β-values) were equivalent between methods (R2 = 0.79, p < 0.00001; Fig. 1e). As expected, fresh-frozen derivatives performed best (n = 39/40; 98% successfully subgrouped), with 92% success (n = 56/61) using FFPE-derived DNA (Fig. 1f–h).
Application of MS-MIMIC to the HIT-SIOP-PNET4 clinical trials cohort
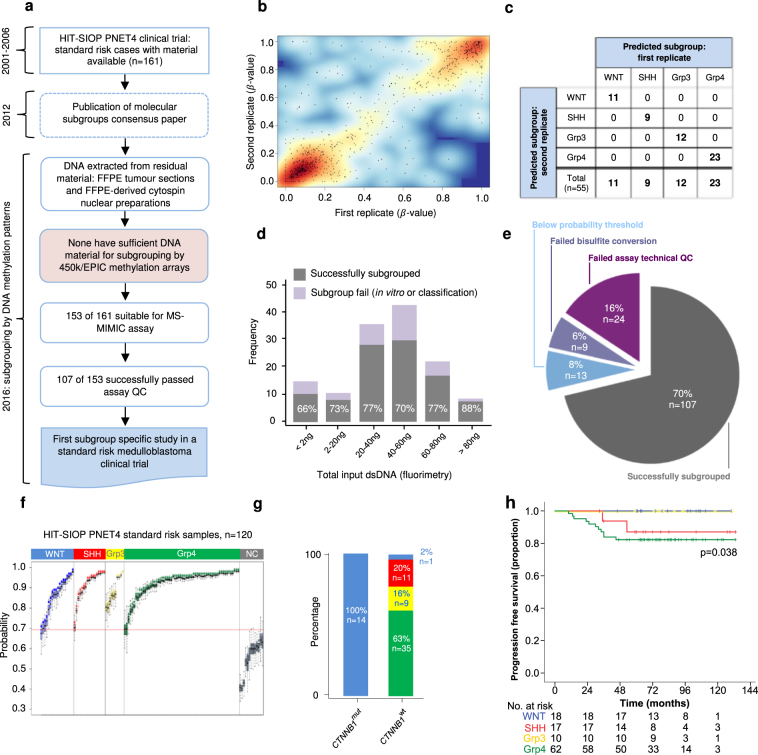
Following successful assay development and validation, we next wished to test the application of MS-MIMIC methylation signature detection in limited, poor quality, archival, clinical biopsies. Analysis of remnant material from the HIT-SIOP-PNET4 cohort offers the first opportunity to determine the potential utility of molecular subgroup status to predict disease outcome in a clinical trial of risk-factor negative (SR) medulloblastoma (Fig. 4a, Supplementary Table 2). Only FFPE sections (n = 42/153 available tumour samples) and cytospin nuclear preparations (approximately 30,000 nuclei isolated and centrifuged onto microscope slides18; n = 111/153) remained, whose DNA derivatives all fell below quality and quantity thresholds (>200 ng double-stranded DNA (dsDNA)) required for methylation profiling using conventional research methods (Illumina 450k and MethylationEpic arrays14). Using MS-MIMIC, 70% (107/153) of samples were successfully subgrouped, and subgroup assignments and β-value estimations were consistent across duplicate determinations (Fig. 4b,c). Assay performance was equivalent across the input DNA range (<2 ng (limit of detection) to 100ng dsDNA, 41.4ng median DNA input, p = 0.852, chi-squared test) (Fig. 4d,e,f). Reasons for assay failure included unsuccessful bisulfite conversion/PCR (6%; 9/153), and inability to classify due to assay QC failure (24%; 37/153) (Fig. 4e). These findings from HIT-SIOP-PNET4 reveal important subgroup-dependent molecular pathology in SR medulloblastoma. Grp4 was most common (n = 62; 58%), with approximately equivalent numbers of WNT (18/170; 16%), SHH (17/107; 16%) and Grp3 (10/107; 9%) tumours observed. As expected, all tumours with CTNNB1 mutations (n = 14) were classified as WNT (Fig. 4g). The majority of events (defined as disease recurrence or progression following treatment) observed (11/13) affected Grp4 patients (82% 5-year progression-free survival (PFS)), with > 95% PFS in non-Grp4 patients (p = 0.038, log-rank test; Fig. 4h). Subgroup assignment will thus be essential to inform future clinical and research studies in SR medulloblastoma.
Figure 4.
Application of MS-MIMIC to remnant, archival materials from the HIT-SIOP-PNET4 clinical trial cohort enables the first subgroup-specific characterisation of standard-risk medulloblastoma. (a) The HIT-SIOP-PNET4 clinical trial ran prior to the discovery of medulloblastoma molecular subgroups (2000–2007) and remnant tumour sample materials available were old, scant and unsuitable for array-based DNA methylomic subgrouping. (b,c) First and second replicates of individual samples show concordance both at the level of estimate of methylation (β-value; R2 = 0.59; n = 55 replicates, 17 CpG loci) and subgroup assigment calls (55/55 replicates; 100%). (d) Bar plot of MS-MIMIC assay input dsDNA amounts. The proportions of samples that were successfully subgrouped are shown grey and were consistent across different dsDNA input quantities. Lilac represents samples for which subgrouping was not possible due to QC failure in vitro or in classification. (e) Proportion of attempted samples (153/161; 95%) successfully subgrouped (107/153; 70%) or failing the assay due to failure to 1) bisulfite convert (9/153; 6%) 2) meet QC citerion for CpG-locus specific failure (>6/17 CpG-locus fails, 24/153 samples; 16%), 3) meet probablility threshold in classification confidence (13/153; 8%). (f) Subgroup assignment and probability estimates (dot) along with 95% confidence intervals (boxplots) for 120 standard risk HIT-SIOP-PNET4 tumours after applying the confidence probability threshold (red-line). (g) All samples which were CTNNB1 mutated (CTNNB1 mut; a well-established marker of WNT medulloblastoma) were assigned as WNT by MS-MIMIC. No non-WNT tumours were CTNNB1 mut. (h) Progression free survival (PFS) Kaplan-Meier curves for MS-MIMIC derived subgroups reveal that standard-risk Grp4 medulloblastomas show a significantly worse disease outcome compared to other subgroups (p = 0.038, log-rank test). Numbers below x-axis represent patients at risk of event.
Discussion
We have provided a blueprint for defining minimal, multiply-redundant disease-associated DNA methylation signatures from genome-wide datasets, and have developed MS-MIMIC as a validated assay for their assessment, including open-source classification tools for data interpretation (http://medulloblastomadiagnostics.ncl.ac.uk; Supplementary Fig. 6). Unlike research methodologies (e.g. Illumina 450k and MethylationEPIC arrays) which require batched assessments (≥8 samples per run), MS-MIMIC exploits detection technologies in common clinical use (MALDI-TOF) to enable rapid (<3 days from DNA extraction to result), low-cost (<$200 per sample in 2017), routine assessment in single or multiple samples. The assay format allows a flexible, modular approach, in which multiplex PCRs can be straightforwardly added or removed, offering the ability to adapt or extend panels to evolving clinical needs. Moreover, its low DNA input requirements and applicability to archival sample collections has the potential to unlock previously inaccessible molecular information from informative cohorts, as demonstrated for HIT-SIOP-PNET4. This assessment of DNA methylation signatures in the clinical setting holds rich promise for molecular sub-classification and prognostication across diverse diseases.
Methods
Cohorts and sample collection
Three cohorts (training; n = 220, validation; n = 106 and test; n = 153) were used in this study and are described in Supplementary Table 1. The training and validation cohorts comprised archival non-trial medulloblastoma DNA samples and included tumour samples provided by the UK Children’s Cancer and Leukaemia Group (CCLG) biobank as part of CCLG-approved biological study BS-2007–04. The validation cohort consisted of samples with varying DNA quality, to assess assay performance. The test cohort included samples from the HIT-SIOP-PNET4 clinical trial (2001–2006)17. All tumours assayed had a confirmed histopathological diagnosis of medulloblastoma, with a high tumour cell content. Informed consent was obtained from all participants and/or their legal guardians. All experiments were performed in accordance with relevant guidelines and regulations.
Identification of a minimal methylation signature for discrimination of medulloblastoma molecular subgroups
Non-negative matrix factorisation (NMF) consensus clustering19 was used to identify the four recognised medulloblastoma consensus molecular subgroups13 using 220 training cohort samples run on the Illumina 450k methylation microarray platform (Fig. 2). The 50 most differentially-methylated CpG loci for each subgroup were selected as potential signature candidates using limma22. An iterative CpG locus selection algorithm was used to select signature gene candidates. To optimise signature loci redundancy in each level, up to 6 loci were repeatedly removed at random and classification performance evaluated. The 17 signature loci with the highest ranking in classification were identified (Fig. 3).
Assay methodology
The MS-MIMIC assay is based on an adaptation of Agena Biosciences’ iPLEX assay and the MassARRAY platform21. In order to determine methylation status at each signature CpG locus, bisulfite treatment of DNA was used to induce methylation-dependent SNPs. These regions were amplified by multiplex PCR, followed by single base extension using mass-modified dideoxynucleotide terminators. MALDI-TOF mass spectrometry then identifies the proportions of the induced-SNP alleles, from which methylation status can be inferred.
Primer design and validation
PCR and extension primers were designed for multiplex assessment of methylation in 17 signature loci (Supplemental experimental methods) across three multiplexes. Plex 1 contained an additional bisulfite conversion control, targeting an invariably unmethylated locus which undergoes complete conversion to uracil. The multiplexes were validated in vitro using a triplicate mixture series of control DNAs ranging from 0–100% methylation (Supplementary Fig. 2). The correlation between the input and estimated DNA methylation was calculated, and amplicons with a poor correlation were discarded and replaced with a new CpG locus as part of the iterative redesign process (Figs 1a and 3). All signature loci had good linear correlation (average correlation coefficient R2 = 0.86.)
Assay implementation
Where possible, 100ng of DNA was bisulfite converted and purified using the Qiagen EpiTect Bisulfite kit, according to manufacturer’s protocol. To ensure that template was not too fragmented for analysis, a test bisulfite PCR, targeting a 200 bp amplicon, was performed (Supplementary Table 3). Reaction mixtures and thermal cycling parameters used to amplify the 17 signature loci in multiplex are shown in the Supplementary Table 4. Successful amplification was confirmed by gel electrophoresis. The multiplex primer iPLEX extension assay was performed as previously described23. Primer sequences for multiplex signature loci PCR and iPLEX extension PCR are shown in the Supplementary Table 5 and 6. Mass spectra for the multiplexes were acquired on a MALDI-TOF mass spectrophotometer (Voyager DE; PerSeptive Biosystems).
Classifier design and validation
Two support vector machine (SVM) classifier models were created to assign subgroup and corresponding probability24, one trained on 450k array data using the 10,000 most variably methylated CpG loci, the second with the 17-CpG locus signature from the training cohort (Fig. 1c). Subsequently, 101/106 validation cohort samples were used to assess MS-MIMIC concordance with 450k-derived data, at the level of molecular subgroup call (Fig. 1d) and estimates of methylation β-value (Fig. 1e). It was anticipated that when applying MS-MIMIC to poor quality samples, certain loci would not be assessable. Using bootstrapped datasets (n = 10,000), a threshold of 6 was established for a maximum acceptable number of missing loci. Missing loci were imputed using expectation maximisation. Subgroup assignments using MS-MIMIC classifier were compared against corresponding subgroup calls from the 450k classifier. A threshold for probability of assignment by the MS-MIMIC classifier was empirically set to 0.69, below which samples were non-classifiable (Supplementary Fig. 5).
Application to HIT-SIOP-PNET4 clinical trial cohort
Following successful assay development and validation, we applied MS-MIMIC to remnant, poor quality, archival, biopsies from the HIT-SIOP-PNET4 clinical trial of risk-factor negative medulloblastoma (Fig. 4a, Supplementary Table 2). Only FFPE sections (n = 42/153 available tumour samples) and cytospin nuclear preparations (approximately 30,000 nuclei isolated and centrifuged onto microscope slides18; n = 111/153) remained, whose DNA derivatives all fell below quality and quantity thresholds (>200 ng double-stranded DNA (dsDNA)) required for methylation profiling using conventional research methods (Illumina 450k and MethylationEpic arrays14). We assessed differential survival of the MS-MIMIC subgroup assignments using log-rank tests.
Further technical details are provided in the Supplemental experimental methods.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
This work was supported by Cancer Research UK (Programme grants C8464/A13457 and C8464/A23391; S.C.C., D.W. and S.B.) and the UK Children’s Cancer and Leukaemia Group (CCLG)/Tom Grahame Trust (Project grant to S.C.C, D.W. and S.B.). The HIT-SIOP-PNET4 clinical trial was supported by Cancer Research UK, Swedish Childhood Cancer Foundation, French Ministry of Health/French National Cancer Institute and German Children’s Cancer Foundation; informed consent was obtained from all subjects. Tumour investigations were conducted with approval from Newcastle and North Tyneside Research Ethics Committee (Study reference 07/Q0905/71).
Author Contributions
E.C.S., D.H., G.R., and S.C.C. designed and supervised the study and wrote the manuscript. E.C.S., G.R., A.E., M.B. and D.W. developed and implemented the in silico methods and computational algorithms. D.H., E.C.S., H.G., S.P., M.M., J.M., P.T., J.C.L, S.C. and A.J.S. performed the laboratory experimentation and analysis. R.C., K.S., A.S. and A.C. developed the clinical diagnostic test. T.G., F.D., S.R., B.L., T.P. and S.B. provided clinical trial samples, data and interpretation. All authors contributed to and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
E. C. Schwalbe, D. Hicks, and G. Rafiee contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-13644-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Besingi W, Johansson A. Smoke-related DNA methylation changes in the etiology of human disease. Human molecular genetics. 2014;23:2290–2297. doi: 10.1093/hmg/ddt621. [DOI] [PubMed] [Google Scholar]
- 2.Wahl S, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541:81–86. doi: 10.1038/nature20784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehta D, et al. Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8302–8307. doi: 10.1073/pnas.1217750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ong ML, Holbrook JD. Novel region discovery method for Infinium 450 K DNA methylation data reveals changes associated with aging in muscle and neuronal pathways. Aging Cell. 2014;13:142–155. doi: 10.1111/acel.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bacalini MG, et al. Identification of a DNA methylation signature in blood cells from persons with Down Syndrome. Aging. 2015;7:82–96. doi: 10.18632/aging.100715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Portales-Casamar E, et al. DNA methylation signature of human fetal alcohol spectrum disorder. Epigenetics Chromatin. 2016;9:1–20. doi: 10.1186/s13072-016-0074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sturm D, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 8.Harris RA, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotech. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bibikova M, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Wang RY, Gehrke CW, Ehrlich M. Comparison of bisulfite modification of 5-methyldeoxycytidine and deoxycytidine residues. Nucleic Acids Research. 1980;8:4777–4790. doi: 10.1093/nar/8.20.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffin TJ, Smith LM. Single-nucleotide polymorphism analysis by MALDI-TOF mass spectrometry. Trends In Biotechnology. 2000;18:77–84. doi: 10.1016/S0167-7799(99)01401-8. [DOI] [PubMed] [Google Scholar]
- 12.Pizer BL. & Clifford, S.C. The potential impact of tumour biology on improved clinical practice for medulloblastoma: progress towards biologically driven clinical trials. British Journal Of Neurosurgery. 2009;23:364–375. doi: 10.1080/02688690903121807. [DOI] [PubMed] [Google Scholar]
- 13.Schwalbe EC, et al. DNA methylation profiling of medulloblastoma allows robust subclassification and improved outcome prediction using formalin-fixed biopsies. Acta Neuropathologica. 2013;125:359–371. doi: 10.1007/s00401-012-1077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hovestadt V, et al. Robust molecular subgrouping and copy-number profiling of medulloblastoma from small amounts of archival tumour material using high-density DNA methylation arrays. Acta Neuropathologica. 2013;125:913–916. doi: 10.1007/s00401-013-1126-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor MD, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathologica. 2012;123:465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Louis, D. N., Cavenee, W. K., Ohgaki, H. & Wiestler, O. D. WHO classification of tumours of the central nervous system, Edn. 4th revised, 184–200 (IARC Press, Lyon; 2016).
- 17.Lannering B, et al. Hyperfractionated Versus Conventional Radiotherapy Followed by Chemotherapy in Standard-Risk Medulloblastoma: Results From the Randomized Multicenter HIT-SIOP PNET 4 Trial. Journal of Clinical Oncology. 2012;30:3187–3193. doi: 10.1200/JCO.2011.39.8719. [DOI] [PubMed] [Google Scholar]
- 18.Clifford SC, et al. Biomarker-driven stratification of disease-risk in non-metastatic medulloblastoma: Results from the multi-center HIT-SIOP-PNET4 clinical trial. Oncotarget. 2015;6:38827–38839. doi: 10.18632/oncotarget.5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill RM, et al. Combined MYC and P53 defects emerge at medulloblastoma relapse and define rapidly progressive, therapeutically targetable disease. Cancer Cell. 2015;27:72–84. doi: 10.1016/j.ccell.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gabriel, S., Ziaugra, L. & Tabbaa, D. SNP genotyping using the sequenom massARRAY iPLEX platform. Current Protocols in Human Genetics, 2.12.1–2.12.18 (John Wiley & Sons, Inc., 2009). [DOI] [PubMed]
- 22.Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research43, e47 (2015). [DOI] [PMC free article] [PubMed]
- 23.Ross, P., Hall, L., Smirnov, I. & Haff, L. High level multiplex genotyping by MALDI-TOF mass spectrometry. Nat Biotech16, 1347–1351 (1998). [DOI] [PubMed]
- 24.Chih-Chung, C. & Chih-Jen, L. LIBSVM: A library for support vector machines. ACM Trans Intell Syst Technol. 2(3), 1–27 (2011).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.