Abstract
Sinusoidal dedifferentiation is a complicated process induced by several factors, and exists in early stage of diverse liver diseases. The mechanism of sinusoidal dedifferentiation is poorly unknown. In this study, we established a NaAsO2-induced sinusoidal dedifferentiation mice model. Liver sinusoidal endothelial cells were isolated and isobaric tag for relative and absolute quantitation (iTRAQ) based proteomic approach was adopted to globally examine the effects of arsenic on liver sinusoidal endothelial cells (LSECs) during the progression of sinusoidal dedifferentiation. In all, 4205 proteins were identified and quantified by iTRAQ combined with LC-MS/MS analysis, of which 310 proteins were significantly changed in NaAsO2 group, compared with the normal control. Validation by western blot showed increased level of clathrin-associated sorting protein Disabled 2 (Dab2) in NaAsO2 group, indicating that it may regulate receptor endocytosis, which served as a mechanism to augment intracellular VEGF signaling. Moreover, we found that knockdown of Dab2 reduced the uptake of VEGF in LSECs, furthermore blocking VEGF-mediated LSEC dedifferentiation and angiogenesis.
Introduction
Liver sinusoidal endothelial cell (LSEC) is a type of liver specific microvascular cell, which characterizes unique phenotype and function. In normal liver, differentiated LSECs form capillaries of microvasculature and facilitate filtration by fenestrae as a selectively permeable barrier between liver parenchyma and sinusoid1. Upon liver injury (e.g., fibrosis2–4, hepatitis5,6, alcoholic liver injury7 and arsenic exposing8), LSECs loss their highly specialized fenestration and gain an organized basement membrane, which calls LSEC dedifferentiation or capillarization. Although the mechanism of LSEC dedifferentiation has been comprehensively studied, the molecular mechanisms driving dedifferentiation have not been fully elucidated. So far, there are few ideal models to study the molecular mechanisms of LSEC dedifferentiation in vivo, as most models cause cirrhotic fibrosis simultaneously, obfuscating the real issues of LSEC dedifferentiation. However, Straub et al.9 tested the effects of sodium arsenite (NaAsO2) on dedifferentiated LSECs and proved that NaAsO2 induced LSEC dedifferentiation without fibrosis initiation. Therefore, NaAsO2-induced LSEC dedifferentiation mice model could be applied to study LSEC dedifferentiation mechanisms.
In this study, we used NaAsO2 in drinking water of mice for 5 weeks as the early injury phase to induce LSEC dedifferentiation, comparing with normal mice. Then LSECs from these two groups were isolated and lysed. Isobaric tags for relative and absolute quantification (iTRAQ) coupled with LC-MS/MS was used for relative quantification of proteins in vivo, based on a more powerful and sensitive proteomic method than traditional approaches, especially quantifying low-abundance proteins10–12. Protein identification and quantification was accurately performed using Protein Pilot Software with specifically developed algorithms.
For our experiments, iTRAQ-labeled LSECs in NaAsO2-induced LSEC dedifferentiation mice model were first used for differentially expressed proteome analysis through Protein Pilot software, and 4205 proteins were identified. Among these, there were 207 up-regulated proteins and 103 down-regulated proteins, respectively. For functional analysis in depth, we found that two significantly increased proteins, disabled homolog 2 (Dab2) and clathrin heavy chain (CLTC), were involved in VEGF receptor endocytosis, serving as a mechanism to induce intracellular receptor signaling upon the stimulation of VEGF signal, which is referred as a regulator leading to angiogenesis13.
Results
Confirmation of NaAsO2-induced LSEC dedifferentiation in mice model
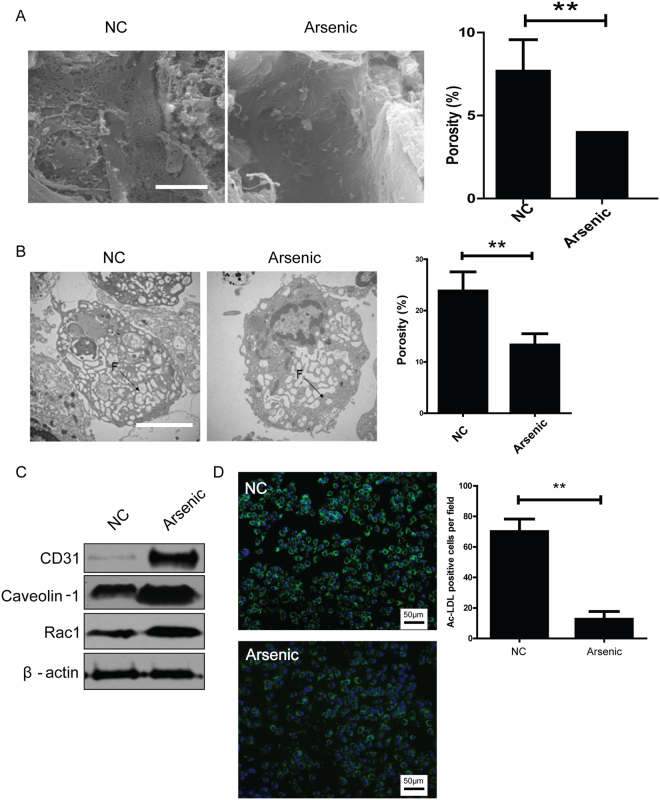
0.25 μg/mL NaAsO2 in drinking water was administered for 5 weeks in mice to induced LSEC dedifferentiation as described previously9. The open fenestrae of liver sinusoids were decreased in arsenic exposed group, compared with the normal counterparts (7.46 ± 0.41% vs 3.56 ± 0.76%) (Fig. 1A). The permeability of isolated LSECs from arsenic exposed group, identified by porosity of cell surface, was also reduced (22.64 ± 5.38% vs 13.35 ± 2.78%) (Fig. 1B). Meanwhile, the expression of LSEC dedifferentiation markers, such as CD31, Caveolin-1 and Rac1, was increased in arsenic exposed group (Fig. 1C). But LSEC differentiation marker, the uptake of acetylated low density lipoprotein (Ac-LDL), was instead reduced in arsenic exposed group (Fig. 1D). These results suggested that 5-week NaAsO2 administration successfully induced LSEC dedifferentiation in mice.
Figure 1.
NaAsO2-induced LSEC dedifferentiation. (A) Representative SEM images of liver sinusoids (n = 5) and quantitative porosity of sinusoidal fenestrae in normal and NaAsO2-induced groups. Bar = 1μm. **P < 0.01. (B) Representative TEM images of primary LSECs isolated from normal (n = 5) and NaAsO2-induced group (n = 5) and quantitative porosity of sinusoidal fenestrae. Bar = 1μm. **P < 0.01. (C) The expressions of CD31, Caveoloin-1 and Rac1 in normal and NaAsO2-induced groups were analyzed by western blot. All western blot experiments were repeated at least three times. Full-length blots are included in Supplemental Information. (D) Uptake of Ac-LDL was analyzed by fluorescence microscopy in primary LSECs isolated from normal and NaAsO2-induced groups and cultured overnight. All experiments in (C) and (D) were repeated at least three times.
Purity and characterization of primarily isolated LSECs
To generate LSEC proteome, we isolated LSECs from normal and arsenic exposed mice respectively using a modified protocol including two-step collagenase perfusion, centrifugation and magnetic beads sorting, as described previously14–16. Purity and viability of LSECs were up to 93.6 ± 1.7% and 88.4 ± 0.5% confirmed by CD146 + F4/80- and 7-AAD + flow cytometry analysis separately, and yield of LSECs was approximately (2.1 ± 0.2) × 106 per mouse, (Supplemental Fig. 1A–C). Nextly, we assessed the quality of primary LSECs to exclude the false positive analysis of flow cytometry. After 8 h culture and extensive washing, LSEC monolayer showed a typical cobblestone, sheet-like appearance (Supplemental Fig. 1D). In liver, Ac-LDL was mainly taken up by LSECs17. Therefore, fluorescently labeled Ac-LDL was used to confirm LSEC quality18. After overnight culture, above 98% LSECs were labeled by Ac-LDL, according to Ac-LDL endocytosis assay (Supplemental Fig. 1E). These results demonstrated that primary LSECs with high purity and viability were obtained.
Proteome differential analysis of normal and dedifferentiated LSECs by iTRAQ
To elucidate the molecular mechanisms of LSEC dedifferentiation, quantitative proteomic analysis based on iTRAQ labeling was executed between NaAsO2 induced LSEC dedifferentiation mice model and the counterparts. Total 7763 proteins were identified in two independent biological replicates (FDR < 1%). Among these, 54.16% (4205/7763) proteins were shared by these two experiments (Supplemental Fig. 2A and Supplemental Table 1). In addition, linear regression analysis was performed with ln [115/116 ratio] and ln [116/115 ratio] in these two independent experiments to examine the biological reproducibility, and the Pearson correlation coefficient was 0.7181 (P < 0.0001), indicating high biological reproducibility of our experiments. To identify significant up- or down-regulated proteins during LSEC dedifferentiation, the threshold values of 115/116 or 116/115 ratios were ≥1.50 or ≤0.67 (≥1.5-fold) in both two iTRAQ analyses. Accordingly, 207 and 103 proteins were significantly up- or down-regulated, respectively, in dedifferentiated LSECs (Supplemental Tables 2 and 3), suggesting dramatic alterations during LSEC dedifferentiation.
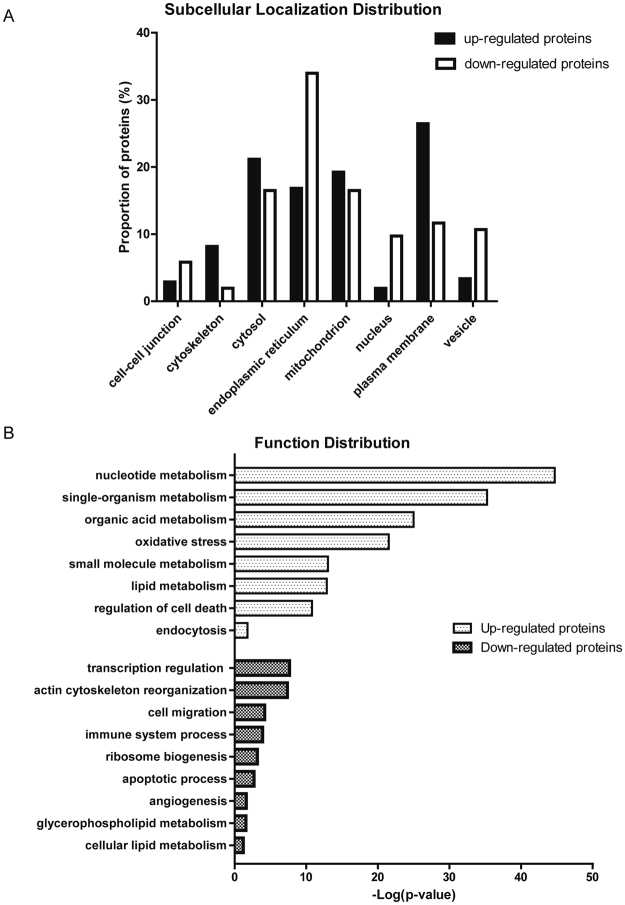
Figure 2.
Subcellular localization and biological functions of differential proteins during LSEC dedifferentiation (A) Subcellular localization of altered proteins during LSEC dedifferentiation. (B) Biological functions of differential proteins during LSEC dedifferentiation. The top biological functions of up-regulated and down-regulated proteins were determined by Gene Ontology and DAVID annotation analysis.
Bioinformatic analysis of differentially expressed proteins
The 310 differentially expressed proteins were categorized by their cellular component and biological function using Gene Ontology analysis (GO) or DAVID functional annotation. Most of up-regulated proteins were localized in plasma membrane and cytosol, while down-regulated proteins in endoplasmic reticulum and mitochondrion (Fig. 2A), indicating that differentially expressed proteins are significantly devided at the subcellular level. The enriched biological functions of up-regulated proteins were mainly associated with multiple component metabolism (such as nucleotide, single-organism, organic acid, small molecular, lipid, et al.), oxidative stress, cell survival and endocytosis (Fig. 2B), showing oxidative stress and energy metabolism are involved in LSEC dedifferentiation process, in accordance with the findings described previously8,9. Interestingly, endocytosis was the unique function with top enrichment score, suggesting endocytosis may mediate LSEC dedifferentiation. Meanwhile, proteins associated with transcription regulation, immune system process, ribosome biogenesis, apoptotic process, et al., were all down-regulated during LSEC dedifferentiation (Fig. 2B), among which the reduced immune system process might disable defense line against arsenic insult. These findings discovered that endocytosis induced LSEC dedifferentiation for the first time. In addition, LSEC were also found to lose the defense ability against injury insults.
Experimental Validation of Proteomic analysis in LSECs
Differential expressions of 5 selected proteins were further validated by western blot, focusing on those involved in receptor endocytosis and innate immune response. Compared with the normal LSECs, proteins involved in receptor endocytosis (CLTC) and clathrin coat assembly (Dab2) were significantly up-regulated in arsenic induced dedifferentiated LSECs, whereas three proteins (Galectin-3, SAMHD1, Rab10) related to innate immune response and antigen presentation showed significant down-regulation in dedifferentiated LSECs. The western blot results were in keeping with the iTRAQ data (Fig. 3A).
Figure 3.
Validation of selected proteins in LSECs from NaAsO2-induced LSEC dedifferentiation and CCl4-induced liver injury mice model. (A) Differential expressions of 2 up-regulated proteins (CLTC, Dab2) and 3 down-regulated proteins (Galectin-3, SAMHD1, Rab-10) were validated by Western blot. The relative ratios of proteins by iTRAQ analysis were shown right. All western blot experiments were repeated at least three times. Full-length blots are included in Supplemental Information. (B) Representative SEM images of liver sinusoids (n = 5) and quantitative porosity of sinusoidal fenestrae at 0 and 6th week with CCl4 administration. Bar = 1μm. (C) Masson’s trichrome staining and αSMA expression in liver at 6th week with CCl4 administration (n = 5) were analyzed by immunohistochemistry analysis, Bar = 100μm. (D) Western blot analysis of primary LSECs isolated from 0 and 6th week for the levels of CLTC, Dab2, Galectin-3, SAMHD1 and Rab-10 proteins, and GAPDH was used as loading control. All western blot experiments were repeated at least three times. Full-length blots are included in Supplemental Information. (E) CLTC expression in liver at normal control mice and mice at 6th week with CCl4 administration (n = 5) were analyzed by immunohistochemistry analysis, Bar = 100μm.
For further validation, the chronic liver injury mice model induced by carbon tetrachloride (CCl4) was established as described previously19, generating LSEC dedifferentiation at 6th week after CCl4 administration (Fig. 3B). And this model was further verified by Masson trichrome staining and αSMA immunohistochemistry, showing obvious chronic liver injury (Fig. 3C). Differential expression of selected proteins was further evaluated by Western blot in LSECs isolated from in vivo model, and confirmed CLTC and Dab 2 were increased at the 6th week after CCl4 administration, when LSECs were dedifferentiated and fibrotic septa formed. Galectin-3, SAMHD1, Rab10 showed down-regulation as displayed in Fig. 3D. The expression of CLTC, which is marker of clathrin-coat associated receptor endocytosis, was increased in liver of CCl4-treated mice at the 6th week, comparing with normal counterparts (olive given only) (Fig. 3E). These results further proved the expression levels of these proteins in chronic liver injury.
The biological significance of Dab2 in LSEC dedifferentiation
Expression of clathrin heavy chain 1 (CLTC), an important mediator regulating receptor mediated endocytosis20, was up-regulated in dedifferentiated LSECs based on our proteome data and the following protein validation. Meanwhile, disabled homolog 2 (Dab2), a clathrin-associated sorting protein21, which mediates clathrin-dependent VEGF receptor endocytosis13, was also up-regulated, supporting that VEGF receptor endocytosis may be involved in LSEC dedifferentiation.
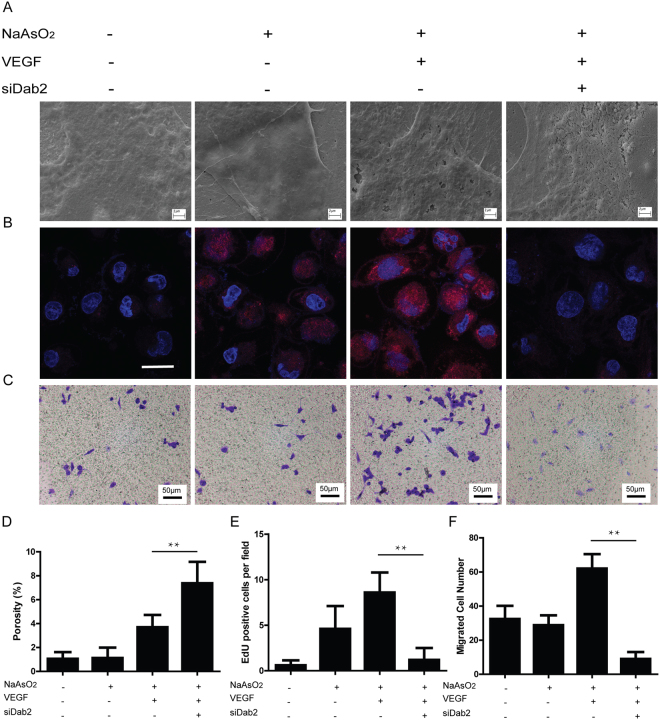
To verify this hypothesis, we explored the role of Dab2 in VEGF receptor endocytosis and LSEC dedifferentiation in SK-HEP1 human liver sinusoidal endothelial cell line. SK-HEP1 cultured 3 days or treated with 5 μM NaAsO2 for 12 h can induce SK-HEP1 dedifferentiation (Fig. 4A,D). Western blot of surface VEGFR1 and VEGFR2 and immunofluorescence images showed that NaAsO2 might induce endocytosis of VEGFR1 and VEGFR2 and 40ng/ mL VEGF can enhance this response to maintain these receptors’ steady expression on the cell surface, regardless of their total expression alteration after NaAsO2 and VEGF induction, as these responses augmented VEGF-VEGFR signaling in perinuclear localization (Supplemental Fig. 3C,D). The biological significance of Dab2 in LSECs was evaluated by knockdown experiment using siRNA (Supplemental Fig. 3A), and the expression of Dab2 was decreased after siRNA transfection (Supplemental Fig. 3B). Dab2 knockdown significantly suppressed VEGFR1 and VEGFR2 endocytosis, increased their expression on cell surface and attenuated perinuclear VEGF signaling (Supplemental Fig. 3C,D). Meanwhile, fenestration of SK-HEP1 cells was well maintained in Dab2 knockdown plus VEGF group, compared with NC, Arsenic and Arsenic plus VEGF group (Fig. 4A,D), suggesting that Dab2 plays a key role in LSEC dedifferentiation. In addition, cell proliferation and migration, two key steps in angiogenesis, was also reduced after Dab2 knockdown (Fig. 4B,E and C,F), indicating that Dab2 inhibition may suppress LSEC dedifferentiation associated angiogenesis.
Figure 4.
Involvement of Dab2 in VEGF receptor endocytosis, regulating LSEC dedifferentiation, proliferation and migration (A) Representative SEM images of fenestrae in SK-HEP1 from NaAsO2- VEGF- siDab2-, NaAsO2 + VEGF- siDab2-, NaAsO2 + VEGF + siDab2- and NaAsO2 + VEGF + siDab2 + group in vitro. Bar = 2μm. All experiments were repeated at least three times. (B) Representative EdU staining images in SK-HEP1 groups described above in vitro. Bar = 50μm. All experiments were repeated at least three times. (C) Representative crystal violet staining images of migrated cells in SK-HEP1 groups described above. Bar = 100μm. All experiments were repeated at least three times. (D) Quantitative porosity of fenestrae in SK-HEP1 groups described above in vitro. **P < 0.01. (E) Cell numbers per field of EdU stained SK-HEP1 groups described above in vitro. **P < 0.01. (F) Cell numbers per field of crystal violet stained SK-HEP1 groups described above in vitro. **P < 0.01.
Discussion
The proteomic aspect of differentially expressed proteins between normal and dedifferentiated LSECs
LSECs are morphologically unique endothelial cells, also the only mammalian sinusoidal endothelium combining non-diaphragm fenestrae without basement membrane22,23. They are also functionally unique, providing high rate and capacity to clear waste from the circulation24, and LSECs are the initial target of chemical liver injury25 and susceptible to ischemia-reperfusion injury26. In addition, LSECs maintain HSC quiescence, inhibiting intrahepatic vasoconstriction and liver fibrosis development4,27. LSEC dedifferentiation occurs following liver injury in animal models and patients5,6, then LSECs lose their protective properties and promote liver injury associated angiogenesis and vasoconstriction. LSEC dedifferentiation is an early event preceding HSC activation and the onset of liver fibrosis. Therefore, it could be a preliminary step necessary for liver fibrosis. Although some researchers have illustrated multiple mechanisms of LSEC dedifferentiation, the extensive and systematic analysis of proteome between normal and dedifferentiated LSECs has not been identified. NaAsO2 was reported by Straub et al. to cause functional changes in signaling of LSEC dedifferentiation, mainly pathogenesis of intrahepatic vascular diseases8,9. Therefore, availability of NaAsO2-induced liver chronic injury model in vivo and LSEC-resolved proteome could help to understand the global protein changes during LSEC dedifferentiation in liver disease associated angiogenesis. The SK-HEP1 cell line was proved by Heffelfinger et al. as human endothelial origin28. Cogger et al. further discovered theis cell line displayed many characteristics of LSECs such as fenestration, uptake of Ac-LDL and tube forming, making it more appropriate for specific LSEC research29. Therefore, we applied SK-HEP1 to analyze the biological effects of candidate proteins.
To our knowledge, this is the first comprehensive proteomic analysis of LSEC dedifferentiation. The present iTRAQ-based proteomic study identified a variety of novel proteins associated with LSEC dedifferentiation and dysfunction, extending our understanding of this process. Based on GO and DAVID analysis, the most enriched biological function categories of up-regulated proteins in dedifferentiated LSECs were nucleotide, organic acid metabolism, oxidative stress, small molecular and lipid metabolism, cell death regulation and endocytosis. Most function categories have been reported to associate with LSEC dedifferentiation8,9,30,31, except for receptor endocytosis. Therefore, CLTC, which is an important component of clathrin-coated vesicle32 and participate in endocytosis, was screened for further validation by western blot and immunohischemistry in our study. Our results supported that CLTC may be a potential marker of endocytosis during LSEC dedifferentiation. Instead, the top enriched biological function categories of down-regulated proteins in LSEC dedifferentiation were transcription regulation, actin cytoskeleton reorganization, cell migration, immune system process, ribosome biogenesis, apoptotic process, angiogenesis, glycerophospholipid metabolism and cellular lipid metabolism. Several proteins involved in innate immune response were simultaneously down-regulated, including Galectin-3, SAMHD1 and Rab-10, determined by proteomic analysis and western blot validation, suggesting that reduced innate immune response caused defense line damage against injury insult, leading to the initiation of liver diseases.
The role of Dab2 in LSEC dedifferentiation
The expression of Dab2, a clathrin-associated sorting protein, was induced during LSEC dedifferentiation based on our proteomic data. As described previously, Dab2 is referred as a mediator of VEGF receptor endocytosis, leading to angiogenesis13,33. We supposed that Dab2 contributed to LSEC dedifferentiation through VEGF receptor endocytosis. With our results, Dab2 expression was increased by NaAsO2 administration, resulting in VEGFR1 and VEGFR2 endocytosis and localization at the perinuclear region, and in addition, VEGF promoted these responses, furthermore augmented its intracellular signaling. Consistently, Dab2 knockdown reduced VEGFR1 and VEGFR2 endocytosis and declined VEGF signal in perinuclear, which resulted in the maintenance of LSEC fenestration and limited LSEC proliferation and migration rate to inhibit angiogenesis process, suggesting that Dab2 knockdown may contribute to anti- LSEC dysfunction and anti- angiogenesis therapy. It is reported that VEGF maintains LSEC differentiation and prevents liver fibrosis progression4,27, but it is confused that VEGF also induces angiogenesis, which may lead to liver disease progression34,35, but so far, this contradiction has not yet been clearly clarified. In this study, we supposed that the effects of VEGF on LSECs depended on the receptor endocytosis instead of VEGF concentration. Dab2, regulating clathrin-coated receptor endocytosis, induced VEGFR1 and VEGFR2 endocytosis, and enhanced VEGF signaling which has been linked to angiogenesis. Therefore, Dab2 may be a mediator initiating LSEC dedifferentiation and multiple liver injury process. Targeting drugs inhibiting Dab2 expression might provide a novel therapy that improves LSEC homeostasis and blocks angiogenesis associated with multiple liver diseases.
In brief, proteome changes between normal and dedifferentiated LSECs using iTRAQ provided the comprehensive database of differentially expressed proteins. Bioinformatic analysis of proteome data promoted our understanding of the characteristics of dedifferentiated LSECs, such as receptor endocytosis, multiple compound metabolisms, and reduced innate immune response. Moreover, this study revealed the role of Dab2 in VEGF receptor endocytosis and provided insight into LSEC dedifferentiation and angiogenesis. In conclusion, as the first comprehensive proteomic analysis of dedifferentiated LSECs, the data provided here will enhance our understanding of LSEC effects on liver injury and concomitant angiogenesis, and will accelerate the development of diagnostic and therapeutic strategies for multiple liver diseases.
Materials and Methods
Animal studies
Mice were housed at the Institutional Animal Care Facility of Beijing Proteome Research Center. All experiments were performed in accordance with relevant guidelines and regulations for laboratory animals. 6-8 week old male C57BL/6J mice (purchased from Vital River Co, Beijing, China) were used for NaAsO2-induced LSEC dedifferentiation and CCl4-induced chronic liver injury model. The animal use protocol was approved by the Animal Care Committee of Beijing Proteome Research Center. For NaAsO2-induced LSEC dedifferentiation model, standard mouse chow and drinking water solutions were fed freely for 5 weeks to normal control mice (n = 12) housed for three per box. Fresh drinking water solutions with 250 ng/ mL NaAsO2 and standard mouse chow were prepared 3 times per week using commercially bottled drinking water for arsenic-exposed mice (n = 12), as described previously9. For CCl4-induced chronic liver injury mouse model (n = 12), CCl4 in olive oil was intraperitoneal administration twice per week for 6 weeks, mice administrated with olive oil was referred as normal controls (n = 12), according to a previous study36.
Cell isolation and culture
LSECs were isolated from male C57BL/6 J mice via protocols adapted from modified method14,15. Briefly, after mice anaesthetized by pelltobarbitalum natricum, the liver was perfused in situ with two steps of Hanks buffer and collagenase solution, respectively, and then excised and digested in perfusion buffer. The resulting supernatant was centrifuged at 50 g for 3 min to eliminate hepatocytes. The resting supernatant enriched in HSCs, KCs and LSECs was separated by OptiprepTM density gradient medium. The cell fraction between the 8.2 and 17.6% OptiprepTM enriched in LSECs and KCs was further separated by mouse LSEC binding magnetic beads (Miltenyi, Germany). Purity and viability of isolated LSECs were confirmed by CD146 + F4/80- and 7-AAD + flow cytometry, respectively, and quality of LSECs was detected by fluorescently labeled Ac-LDL.
Human LSEC line SK-HEP1 was purchased from American Type Culture Collection (Manassas, VA). Cells were cultured in 24-well plates with DMEM (Hyclone, South Logan, UT) supplemented with 10% fetal bovine serum (FBS) (Gibco, Grand Island, NY) and antibiotics.
Differential proteome analysis based on iTRAQ labeling and Triple TOF MS
A total of 100 μg proteins of primary LSECs in normal and LSEC dedifferentiated samples were labeled with iTRAQ according to the Applied Bio systems iTRAQ labeling protocol (Foster city, CA). The digested peptides of each sample were labeled with 115 (normal LSECs) and 116 (dedifferentiated LSECs) iTRAQ reagents. The labeled peptides were mixed together, and then cleaned up the excess trypsin and iTRAQ reagents by Hamilton PRP™-C18 reversed phase HPLC column (Reno, NV) using Thermo DINOEX Ultimate 3000 BioRS system high-performance liquid chromatography (HPLC) (Grand Island, NY) before mass spectrometry. Replicate analyses were performed using 116 (normal LSECs) and 115 (dedifferentiated LSECs) iTRAQ reagents.
Mass spectrometric analysis was performed with an AB SCIEX Triple TOF 5600 System (Concord, Canada). Samples were chromatographed by a 90 min gradient from 2-30% (mobile phase A 0.1% (v/v) formic acid, 5% (v/v) acetonitrile; mobile phase B 0.1% (v/v) formic acid, 95% (v/v) acetonitrile) injected onto a 20μm Pico Frit emitter (New Objective) packed to 12 cm with Magic C18 AQ 3μm 120 Å stationary phase. MS1 spectra were collected in the range 350-1500 m/z for 250ms. The 20 most intense precursors with charge state 2-5 were selected for fragmentation, and MS2 spectra were collected in the range 50-2000 m/z for 100ms; precursor ions were excluded from reselection for 15 s.
In this study, the original MS/MS file data was analyzed by Protein Pilot software (version 4.0, AB SCIEX), the following parameters were used: the instrument was Triple TOF 5600, iTRAQ quantification, cysteine modified with iodoacetamide; biological modifications were selected as ID focus, the Quantitate, and trypsin digestion. Bias Correction and Background Correction was checked for protein quantification and normalization. Proteins with at least 95% confidence determined by Protein Pilot Unused scores (≥1.3) were reported, and the false discovery rate (FDR) was calculated and set up less than 1%. Fold changes ≥1.5 or ≤0.67 were considered significant.
Bioinformatic Analysis of Differential Proteins
The bioinformatic analysis of differential proteins was performed with Gene Ontology Terms (http://www.geneontology.org/) and DAVID Annotation (https://david.ncifcrf.gov/). The lists of differential proteins were input into these platforms for identification of Subcellular localization and biological functional distribution. The false discovery rate was set less than 0.05.
Statistical Analysis
The sample size (n) of each experimental group is described in each corresponding Figure legends, and all experiments were repeated at least three times. Data was expressed as the mean ± standard error with at least 3 independent experiments. To compare values between groups, the ANOVA or Student’s t test was used. P value < 0.05 was considered significant.
Data availability statement
All data generated or analysed during this study are included in this published article and its Supplementary Information files.
Ethical approval
All animal experiments were reviewed and approved by the Animal Care and Use Committee at the Academy of Military Medical Sciences to ensure the ethical and humane treatment of the animals.
Electronic supplementary material
Acknowledgements
The authors thank Shanshan Du and Yanfei Hu (Tsinghua University), Quancheng Zhao, Juanjuan Shang and Ping Wu (National Center for Protein Sciences, Beijing) for their invaluable assistance with scanning electron microscopy; Miss. Yuqing Han (National Center for Protein Sciences, Beijing) for her invaluable assistance with immunofluorescence analysis; Dr. Hongyu Zhang (Tsinghua University) for kindly providing her invaluable suggestions with our study. This work was partially supported by Chinese State Key Projects for Basic Research (“973 Program”) (Nos. 2014CBA02001 and 2013CB910502), National Key Research and Development Project (2016YFC0902400, SQ2017YFSF090210), National Natural Science Foundation of China (81123001 and 81570526), Innovation project (16CXZ027), Natural Science Foundation of Beijing (7152036) and Open Project Program of the State Key Laboratory of Proteomics (Academy of Military Medical Sciences, SKLP-O201509)
Author Contributions
Yuanxiang Lao and Yanyan Li designed and conducted the experiments, acquired and analyzed the data, and wrote the manuscript; Yufang Hou and Huahai Chen conducted the experiments of LSEC sorting; Bintao Qiu and Weiran Lin conducted the experiments of proteomic analysis; Handong Wei provided reagents and conducted experiment facilities; Aihua Sun designed and supervised the experiments; and Ying Jiang and Fuchu He designed the study, supervised experiments, analyzed data, and wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Yuanxiang Lao and Yanyan Li contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-13917-9.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ying Jiang, Email: jiangying304@hotmail.com.
Fuchu He, Email: hefc@bmi.ac.cn.
References
- 1.Hang TC, Lauffenburger DA, Griffith LG, Stolz DB. Lipids promote survival, proliferation, and maintenance of differentiation of rat liver sinusoidal endothelial cells in vitro. American journal of physiology. Gastrointestinal and liver physiology. 2012;302:G375–388. doi: 10.1152/ajpgi.00288.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schaffner F, Poper H. Capillarization of hepatic sinusoids in man. Gastroenterology. 1963;44:239–242. [PubMed] [Google Scholar]
- 3.DeLeve LD, Wang X, Kanel GC, Atkinson RD, McCuskey RS. Prevention of hepatic fibrosis in a murine model of metabolic syndrome with nonalcoholic steatohepatitis. The American journal of pathology. 2008;173:993–1001. doi: 10.2353/ajpath.2008.070720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie G, et al. Role of differentiation of liver sinusoidal endothelial cells in progression and regression of hepatic fibrosis in rats. Gastroenterology. 2012;142:918–927 e916. doi: 10.1053/j.gastro.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu B, et al. Capillarization of hepatic sinusoid by liver endothelial cell-reactive autoantibodies in patients with cirrhosis and chronic hepatitis. The American journal of pathology. 2003;163:1275–1289. doi: 10.1016/S0002-9440(10)63487-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warren A, et al. Marked changes of the hepatic sinusoid in a transgenic mouse model of acute immune-mediated hepatitis. J Hepatol. 2007;46:239–246. doi: 10.1016/j.jhep.2006.08.022. [DOI] [PubMed] [Google Scholar]
- 7.Horn T, Junge J, Christoffersen P. Early alcoholic liver injury: changes of the Disse space in acinar zone 3. Liver. 1985;5:301–310. doi: 10.1111/j.1600-0676.1985.tb00253.x. [DOI] [PubMed] [Google Scholar]
- 8.Straub AC, et al. Arsenic-stimulated liver sinusoidal capillarization in mice requires NADPH oxidase-generated superoxide. The Journal of clinical investigation. 2008;118:3980–3989. doi: 10.1172/JCI35092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Straub AC, et al. Arsenic stimulates sinusoidal endothelial cell capillarization and vessel remodeling in mouse liver. Hepatology. 2007;45:205–212. doi: 10.1002/hep.21444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ross PL, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Molecular & cellular proteomics: MCP. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Wu WW, Wang G, Baek SJ, Shen RF. Comparative study of three proteomic quantitative methods, DIGE, cICAT, and iTRAQ, using 2D gel- or LC-MALDI TOF/TOF. Journal of proteome research. 2006;5:651–658. doi: 10.1021/pr050405o. [DOI] [PubMed] [Google Scholar]
- 12.Herbrich SM, et al. Statistical inference from multiple iTRAQ experiments without using common reference standards. Journal of proteome research. 2013;12:594–604. doi: 10.1021/pr300624g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakayama M, et al. Spatial regulation of VEGF receptor endocytosis in angiogenesis. Nature cell biology. 2013;15:249–260. doi: 10.1038/ncb2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu W, et al. Sample preparation method for isolation of single-cell types from mouse liver for proteomic studies. Proteomics. 2011;11:3556–3564. doi: 10.1002/pmic.201100157. [DOI] [PubMed] [Google Scholar]
- 15.Ding C, et al. A Cell-type-resolved Liver Proteome. Molecular & cellular proteomics: MCP. 2016;15:3190–3202. doi: 10.1074/mcp.M116.060145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azimifar SB, Nagaraj N, Cox J, Mann M. Cell-type-resolved quantitative proteomics of murine liver. Cell metabolism. 2014;20:1076–1087. doi: 10.1016/j.cmet.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Katz SC, Pillarisetty VG, Bleier JI, Shah AB, DeMatteo RP. Liver sinusoidal endothelial cells are insufficient to activate T cells. Journal of immunology. 2004;173:230–235. doi: 10.4049/jimmunol.173.1.230. [DOI] [PubMed] [Google Scholar]
- 18.Seckert CK, et al. Liver sinusoidal endothelial cells are a site of murine cytomegalovirus latency and reactivation. Journal of virology. 2009;83:8869–8884. doi: 10.1128/JVI.00870-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang L, et al. Vascular endothelial growth factor promotes fibrosis resolution and repair in mice. Gastroenterology. 2014;146:1339–1350 e1331. doi: 10.1053/j.gastro.2014.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vieira AV, Lamaze C, Schmid SL. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274:2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- 21.Traub LM. Sorting it out: AP-2 and alternate clathrin adaptors in endocytic cargo selection. The Journal of cell biology. 2003;163:203–208. doi: 10.1083/jcb.200309175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wisse E, De Zanger RB, Charels K, Van Der Smissen P, McCuskey RS. The liver sieve: considerations concerning the structure and function of endothelial fenestrae, the sinusoidal wall and the space of Disse. Hepatology. 1985;5:683–692. doi: 10.1002/hep.1840050427. [DOI] [PubMed] [Google Scholar]
- 23.DeLeve LD. Liver sinusoidal endothelial cells and liver regeneration. The Journal of clinical investigation. 2013;123:1861–1866. doi: 10.1172/JCI66025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elvevold K, Smedsrod B, Martinez I. The liver sinusoidal endothelial cell: a cell type of controversial and confusing identity. American journal of physiology. Gastrointestinal and liver physiology. 2008;294:G391–400. doi: 10.1152/ajpgi.00167.2007. [DOI] [PubMed] [Google Scholar]
- 25.DeLeve LD, et al. Embolization by sinusoidal lining cells obstructs the microcirculation in rat sinusoidal obstruction syndrome. American journal of physiology. Gastrointestinal and liver physiology. 2003;284:G1045–1052. doi: 10.1152/ajpgi.00526.2002. [DOI] [PubMed] [Google Scholar]
- 26.Caldwell-Kenkel JC, Currin RT, Tanaka Y, Thurman RG, Lemasters JJ. Kupffer cell activation and endothelial cell damage after storage of rat livers: effects of reperfusion. Hepatology. 1991;13:83–95. [PubMed] [Google Scholar]
- 27.Deleve LD, Wang X, Guo Y. Sinusoidal endothelial cells prevent rat stellate cell activation and promote reversion to quiescence. Hepatology. 2008;48:920–930. doi: 10.1002/hep.22351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heffelfinger SC, Hawkins HH, Barrish J, Taylor L, Darlington GJ. SK HEP-1: a human cell line of endothelial origin. In vitro cellular & developmental biology: journal of the Tissue Culture Association. 1992;28A:136–142. doi: 10.1007/BF02631017. [DOI] [PubMed] [Google Scholar]
- 29.Cogger VC, et al. The response of fenestrations, actin, and caveolin-1 to vascular endothelial growth factor in SK Hep1 cells. American journal of physiology. Gastrointestinal and liver physiology. 2008;295:G137–G145. doi: 10.1152/ajpgi.00069.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Straub AC, Klei LR, Stolz DB, Barchowsky A. Arsenic requires sphingosine-1-phosphate type 1 receptors to induce angiogenic genes and endothelial cell remodeling. The American journal of pathology. 2009;174:1949–1958. doi: 10.2353/ajpath.2009.081016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyao M, et al. Pivotal role of liver sinusoidal endothelial cells in NAFLD/NASH progression. Laboratory investigation; a journal of technical methods and pathology. 2015;95:1130–1144. doi: 10.1038/labinvest.2015.95. [DOI] [PubMed] [Google Scholar]
- 32.Collinet C, et al. Systems survey of endocytosis by multiparametric image analysis. Nature. 2010;464:243–249. doi: 10.1038/nature08779. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, et al. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature. 2010;465:483–486. doi: 10.1038/nature09002. [DOI] [PubMed] [Google Scholar]
- 34.Taura K, et al. Hepatic stellate cells secrete angiopoietin 1 that induces angiogenesis in liver fibrosis. Gastroenterology. 2008;135:1729–1738. doi: 10.1053/j.gastro.2008.07.065. [DOI] [PubMed] [Google Scholar]
- 35.Corpechot C, et al. Hypoxia-induced VEGF and collagen I expressions are associated with angiogenesis and fibrogenesis in experimental cirrhosis. Hepatology. 2002;35:1010–1021. doi: 10.1053/jhep.2002.32524. [DOI] [PubMed] [Google Scholar]
- 36.Constandinou C, Henderson N, Iredale JP. Modeling liver fibrosis in rodents. Methods in molecular medicine. 2005;117:237–250. doi: 10.1385/1-59259-940-0:237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its Supplementary Information files.