Abstract
Synergists can counteract metabolic insecticide resistance by inhibiting detoxification enzymes or transporters. They are used in commercial formulations of insecticides, but are also frequently used in the elucidation of resistance mechanisms. However, the effect of synergists on genome-wide transcription in arthropods is poorly understood. In this study we used Illumina RNA-sequencing to investigate genome-wide transcriptional responses in an acaricide resistant strain of the spider mite Tetranychus urticae upon exposure to synergists such as S,S,S-tributyl phosphorotrithioate (DEF), diethyl maleate (DEM), piperonyl butoxide (PBO) and cyclosporin A (CsA). Exposure to PBO and DEF resulted in a broad transcriptional response and about one third of the differentially expressed genes (DEGs), including cytochrome P450 monooxygenases and UDP-glycosyltransferases, was shared between both treatments, suggesting common transcriptional regulation. Moreover, both DEF and PBO induced genes that are strongly implicated in acaricide resistance in the respective strain. In contrast, CsA treatment mainly resulted in downregulation of Major Facilitator Superfamily (MFS) genes, while DEGs of the DEM treatment were not significantly enriched for any GO-terms.
Introduction
Insecticide resistance is a major threat for the agricultural productivity of commercial crops1, and understanding the mechanisms underlying insecticide resistance is a high priority for the design and implementation of effective resistance management programs2. Resistance mechanisms can generally be classified into either (1) changes in sensitivity of the target-site due to point mutations, or to (2) increased metabolic detoxification through qualitative or quantitative changes in enzymes involved in the detoxification process. The latter process typically occurs in 3 phases. In phase I the insecticide is functionalized with nucleophilic groups (a hydroxyl, carboxyl or amine group) to make it more reactive and water soluble. In phase II, conjugation occurs with endogenous molecules (such as glutathione (GSH) or sugars), further increasing the compound’s polarity. Ultimately, in phase III, the phase II conjugated product is excreted by cellular transporters. Cytochrome P450 monooxygenases (P450s) and carboxyl/choline esterases (CCEs) are well-known examples of enzymes that are responsible for phase I reactions while glutathione-S-transferases (GSTs) and UDP-glycosyltransferases (UGTs) are enzymes that typically operate during phase II. Finally, in phase III, metabolites are often transported out of the cell by ATP-binding cassette (ABC) transporters and solute carrier proteins, of which a major class are proteins of the Major Facilitator Superfamily (MFS)3,4.
Insecticide synergists are defined as “compounds that greatly enhance the toxicity of an insecticide, although they are usually practically nontoxic on their own”5. They can either act as a surrogate substrate or an inhibitor of detoxification enzymes and transporters and, as such, are a powerful tool to investigate insecticide resistance mechanisms. Synergists are also of commercial interest as combining them with insecticides increases efficacy and aids in keeping pesticide use to a minimum6–8. As a result of the fast and widespread development of resistance, coupled with the slowdown in the number of registrations of new pesticides and a new trend towards “greener” and more “sustainable” pest management9, a renewed interest has arisen in the identification and development of plant-based synergists10–12. However, as of yet relatively few of these new synergists have made the transition from the laboratory to the field or greenhouse.
The most well-known and commonly used commercial insecticide synergist is the methylene dioxyphenyl compound piperonyl butoxide (PBO), an inhibitor of P450s. PBO has been commercially used since 1940, mainly in combination with pyrethroid insecticides. Its lack of specificity in P450 inhibition has contributed to its success as a synergist13. The inhibition mechanism of PBO consists of two phases, starting with the binding of PBO to the active site of the P450, followed by the formation of a quasi-irreversible inhibitor complex between the electrophilic carbene moiety of PBO and the ferrous iron of the P450. This results in a decreased metabolic activity of the P450 enzyme7,14,15. Synergists such as the defoliant S,S,S-tributyl phosphorotrithioate (also known as tribufos, DEF or TBPT), the fungicide iprobenfos (IBP) and triphenyl phosphate (TPP) are well-known carboxyl esterase inhibitors16,17. These organophosphorus compounds (OPs) behave like the natural substrate of esterases and enter the active site where they covalently bind to the serine –OH group. Subsequently, the OP is split, with the enzyme being non–reversibly phosphorylated, and regeneration of the free enzyme by hydrolyzation not possible18,19. An additional major synergist is the carbonyl compound diethyl maleate (DEM) that is known to conjugate reduced glutathione (GSH), thereby depleting cells of this tripeptide. As a consequence, it reduces the ability of GSTs to utilize GSH for conjugation with insecticides or with the oxidative stress products they induce20–22. Finally, verapamil and cyclosporin A are well-known first generation modulators (competitive inhibitors) of vertebrate P-glycoproteins (ABC transporters of the B subfamily)23–25. Human P-glycoproteins are well-known for their role in protecting tissues from toxic xenobiotics and endogenous metabolites26 and in the last decade their counterparts in arthropods have also been linked to insecticide transport and/or resistance27,28. For example, pretreatment with verapamil has been shown to markedly enhance the toxicity of DDT or abamectin in Drosophila 29,30.
Synergists, however, do not always act as intended or expected. A frequently reported unanticipated effect is an altered cuticular penetration of the insecticide after pretreatment with a synergist31–34. In some cases, synergists might also inhibit non-target enzyme systems. PBO has been reported to act as an inhibitor of esterases in Helicoverpa armigera, Frankliniella occidentalis and Bemisia tabaci 35–37 and as an inhibitor of mammalian UGTs38, while DEF was shown to act, albeit to a much lower extent compared to PBO, as an inhibitor of P450s in Blatella germanica 34. These studies suggest that in some cases caution should be applied in interpreting results of synergist application as they are not entirely specific to a single detoxification enzyme class37,39,40. However, inhibition of these non-target enzyme systems typically occur in a non-specific way at high synergist concentrations41,42, and results can be cross-checked. For example, confirmation of P450 rather than esterase inhibition by PBO is straightforward and can be done with another class of P450 inhibitors13.
Insects and mites are known to display a massive and rapid reprogramming of gene expression in response to xenobiotic exposure43–45. While synergist compounds are mostly used at concentrations that cause no or very low mortality, they are nonetheless used at high enough concentrations to cause maximal inhibition of the targeted detoxification enzymes46. Hence, one might expect that synergists also induce gene expression changes on their own6. Little is known, however, regarding the genome-wide transcriptional changes in arthropods exposed to synergists. Using a full genome and a custom “detox” microarray it was found that a subset of P450s and GSTs, and to a lesser extent UGTs, were induced in Drosophila melanogaster upon exposure to PBO47, while, using Illumina RNA sequencing, a P450 was shown to be upregulated in the whitefly B. tabaci upon exposure to PBO + cypermethrin as compared to cypermethrin alone48. However, genome-wide transcriptional changes upon exposure to synergist compounds other than PBO have not been investigated in any herbivorous arthropod pest.
The two-spotted spider mite, Tetranychus urticae (Arthropoda: Chelicerata: Acari: Tetranychidae), is a highly polyphagous agricultural pest that is able to colonize more than 1100 plant species49. Further, among arthropods T. urticae is also considered to be the ‘‘pesticide resistance champion” based on the total number of different pesticides to which populations have become resistant50,51. Synergists such as PBO, DEF and DEM have been frequently used for the identification of metabolic resistance pathways in T. urticae 52–61, and a high quality Sanger genome assembly is available for this mite species62. T. urticae is therefore an ideal arthropod herbivore to study the impact of synergists at the transcriptional level. In this study, we used Illumina HiSeq 2500 technology to generate deep paired-end, strand-specific RNA-seq reads from adult T. urticae females that were exposed for 24 hours to either DEF, DEM, PBO, CsA or formulation only. Subsequent differential expression (DE) analyses allowed us to identify genes for which expression was significantly altered upon exposure to synergists. A selection of differentially expressed genes was validated by qPCR and for each comparison (synergist compared to formulation) a Gene Ontology (GO) analysis was performed to shed light on the types of genes and pathways that respond to synergist exposure (DEGs).
Results
Synergist bioassays and RNA-seq
Adult female mites of the JP-R strain were either not sprayed (CON) or sprayed with PBO (1000 ppm), DEF (500 ppm), DEM (2000 ppm), CsA (50ppm) or formulation (FORM; N,N-dimethylformamide and emulsifier W (3:1 w/w) diluted in deionized water 100-fold) and collected after 24 hours, a commonly used time point in synergist studies56,63–65. Mortality for each treatment was scored, and found to be in line with those of previous reports for that strain56. Mites alive at 24 hours were collected for each treatment (PBO, DEF, DEM, CsA) and the controls (CON, FORM) and used for RNA extraction and RNA-seq library generation. Illumina sequencing generated ~82–92 million strand-specific paired end reads per sample. Alignment of RNA-seq reads against the T. urticae annotation resulted in an overall mapping rate of uniquely mapped reads of 83.01 ± 0.19 SE% across samples (Supplementary Table S1).
Principal Component Analysis (PCA)
A PCA revealed that about forty percent (36.8%) of the total variation could be explained by PC1 while PC2 accounted for 25.7% of the variation (Fig. 1). Except for replicate 4 of the CsA treatment, biological replicates clustered by treatment on PC1 relatively well. On the other hand, all replicates of the CsA treatment clustered relatively well on PC2. Interestingly, in the PCA the expression data of PBO treatments clustered most closely with those of the DEF treatments, those of the FORM treatments grouped with those of unsprayed mites (CON), and those of the CsA treatments grouped with those of the DEM treatments.
Figure 1.
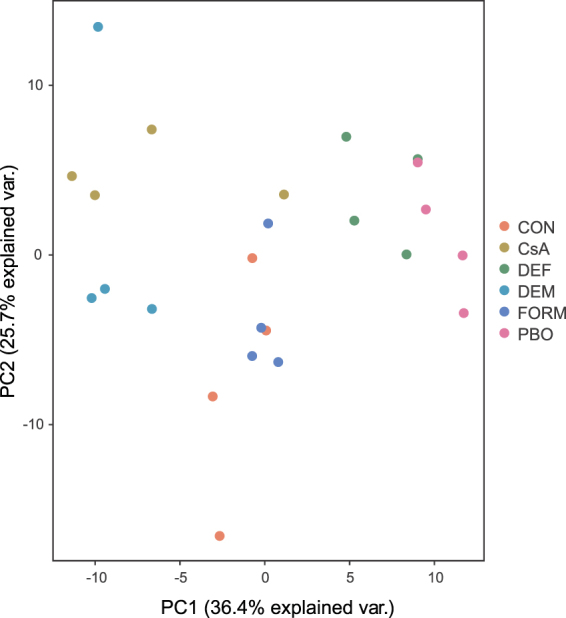
Gene expression relationships among synergist treatments and controls. PCA plot of gene expression levels in untreated adult T. urticae females (CON), adult T. urticae females sprayed with formulation only (FORM) or adult T. urticae females exposed to synergist compounds CsA, DEF, DEM or PBO.
Differential expression analysis and qPCR validation
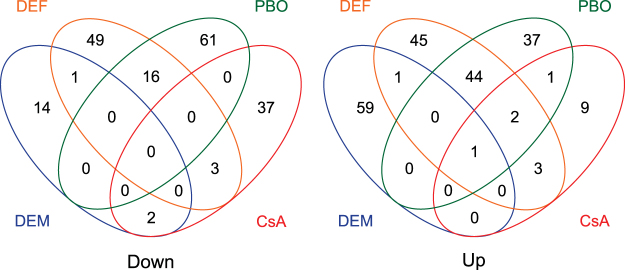
We used DESeq2 to perform differential gene expression (DEG) analyses between treatments and controls (fold change (FC) ≥ 1.5 and a Benjamini-Hochberg adjusted p-value < 0.05)66. Between mites sprayed with formulation and unsprayed mites, only one gene (tetur13g00990, coding for an “orphan secreted protein”) was differentially expressed (FC of 1.53), indicating that the formulation on its own had no significant effect on gene expression. Next, we compared gene expression levels between mites treated with one of the synergists and mites that were treated with formulation only. One hundred and sixty-two genes were differentially expressed between mites treated with PBO and mites treated with formulation, of which 77 were downregulated and 85 upregulated (Supplementary Table S2). The top twenty down- and upregulated genes had a FC of −2.11 to −1.74 and 1.86 to 3.33 respectively. For the DEF treatment, 174 genes were differentially expressed, of which 69 were downregulated and 105 were upregulated (Supplementary Table S3). The expression level of the 20 most down- and upregulated genes varied from a FC of −2.33 to −1.73 and 1.88 to 3.10, respectively. For the DEM treatment, 78 genes were differentially expressed, with 17 genes being downregulated and 61 upregulated and the expression level of the 20 most down- and upregulated genes varied from a FC of −2.3 to −1.5 and 1.5. to 2.1, respectively (Supplementary Table S4). Finally, for the CsA treatment, 58 genes were differentially expressed. Forty-two of them were downregulated, while 16 were upregulated and the expression level of the 20 most downregulated genes varied from −3.87 to −1.72 (Supplementary Table S5). For a selection of genes, the differential expression analyses based on RNA-seq were consistent with differential expression as validated independently by qPCR (Fig. 2). As shown in Fig. 3, the majority of DEGs were not shared between the different treatments. Only one upregulated gene was in common for all treatments (tetur01g06580, a sodium dependent glucose transporter), while two upregulated genes (tetur16g03490 and tetur11g05520, coding for an antigen B membrane protein and CYP385C4, respectively) were shared between the PBO, DEF and CsA treatments. In contrast, 60 DEGs (37% and 35% of the total number of DEGs for the PBO and DEF treatment, respectively) were shared between PBO and the DEF treatment and had the same direction of fold change, with 44 genes being upregulated and 16 being downregulated.
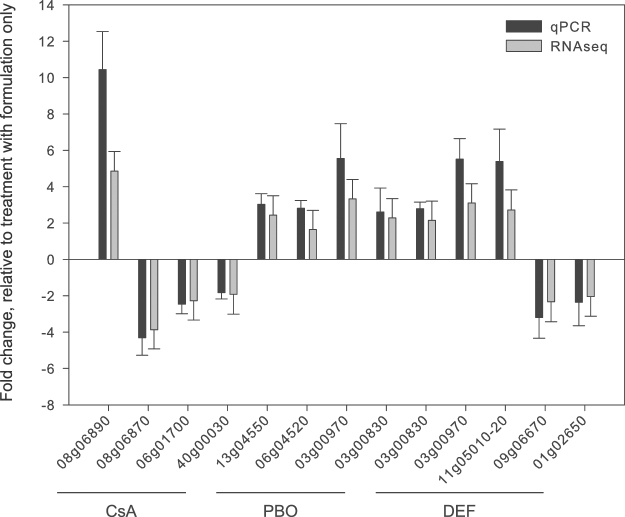
Figure 2.
qPCR validation of differentially expressed genes in adult T. urticae females after PBO, DEF or CsA treatment. Eight up- and five downregulated genes as assessed by differential gene expression (DESeq2) analysis of RNA-seq data were selected for qPCR analysis. Error bars represent the standard error of the calculated mean. Except for tetur40g00030 (CsA treatment), each gene was significantly differentially expressed (based on an unpaired t-test) compared to the reference condition (FORM). The ‘‘tetur’’ prefix was removed from T. urticae gene IDs for figure clarity.
Figure 3.
Venn diagrams depicting overlap among differentially expressed genes of adult T. urticae females exposed to either PBO, DEF, DEM or CsA compared to adult T. urticae females treated with formulation only. Left panel: downregulated genes, right panel: upregulated genes.
GO enrichment and cluster analysis
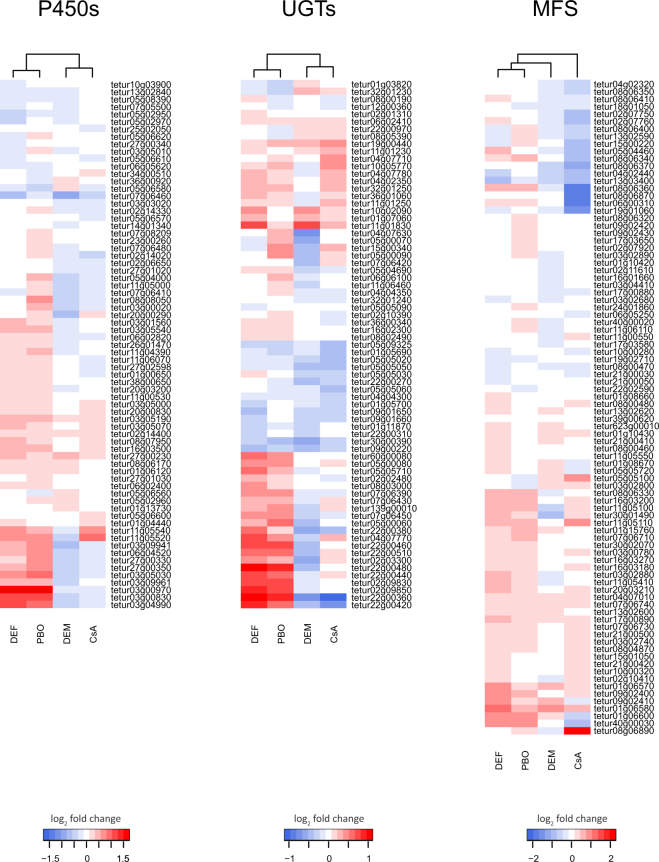
We next performed GO enrichment analyses for the various differentially expressed gene sets. For the DEGs of the DEM treatment, no significantly (Benjamini-Hochberg adjusted p-value < 0.05) enriched GO terms were identified, while for the CsA treatment DEGs were significantly enriched in the GO terms “transmembrane transport” (GO:0055085) and “integral component of membrane” (GO:0016021) (Table 1). Inspecting the DEGs in more detail revealed that these GO terms were mainly present in genes coding for transporters of the major facilitator superfamily (MFS, InterPro domain IPR020846) (20 out of 58 DEGS, 34.5%), and the majority of these MFS genes (15 out of 20) were downregulated (Supplementary Table S5, Supplementary Table S6, Fig. 4). Based on their best BLASTp hit in the Transporter Classification database67, these MFS genes belong to either the Oxalate:Formate Antiporter Family (1/20), Proton Coupled Folate Transporter/Heme Carrier Protein Family (4/20), Fucose H + Symporter (5/20) or the Anion/Cation Symporter (10/20) MFS subfamily. The GO:0016021 term was also associated with genes coding for P450s (3 DEGs), UGTs (2 DEGs), cation-proton exchanger proteins (InterPro domain IPR018422, 2 DEGs) and a protein with an Apple-like domain (tetur25g02030, InterPro domain IPR003609). Interestingly, genes coding for ABC transporters, which are known to be inhibited by CsA68, were not found among the DEGs of the CsA treatment.
Table 1.
GO enrichment analysis of differentially expressed genes (absolute fold change ≥1.5 and Benjamini-Hochberg adjusted p-value < 0.05) in adult T. urticae females of the JP-R strain exposed to either PBO, DEF or CsA compared to adult T. urticae females treated with formulation only.
| Treatment | GO Category | Description | adjusted p-value | Subontology* |
|---|---|---|---|---|
| CsA | GO:0055085 | transmembrane transport | 0 | BP |
| CsA | GO:0016021 | integral component of membrane | 6.72E-05 | CC |
| PBO | GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | 2.68E-09 | MF |
| PBO | GO:0020037 | heme binding | 3.05E-08 | MF |
| PBO | GO:0004497 | monooxygenase activity | 3.05E-08 | MF |
| PBO | GO:0005506 | iron ion binding | 3.08E-08 | MF |
| PBO | GO:0055114 | oxidation-reduction process | 1.28E-05 | BP |
| PBO | GO:0016999 | antibiotic metabolic process | 8.83E-04 | BP |
| PBO | GO:0016758 | transferase activity, transferring hexosyl groups | 2.21E-02 | MF |
| DEF | GO:0016758 | transferase activity, transferring hexosyl groups | 2.86E-05 | MF |
| DEF | GO:0016999 | antibiotic metabolic process | 4.00E-05 | BP |
| DEF | GO:0005506 | iron ion binding | 1.45E-04 | MF |
| DEF | GO:0055114 | oxidation-reduction process | 3.43E-04 | BP |
| DEF | GO:0020037 | heme binding | 4.93E-04 | MF |
| DEF | GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | 5.10E-04 | MF |
| DEF | GO:0004497 | monooxygenase activity | 7.96E-04 | MF |
| DEF | GO:0005179 | hormone activity | 1.18E-03 | MF |
| DEF | GO:0016021 | integral component of membrane | 1.66E-03 | CC |
*BP, Biological Process; MF, Molecular Function; CC, Cellular Component.
Figure 4.
Expression heatmaps of genes coding for P450s, UGTs or MFS members in adult T. urticae females exposed to either DEF, PBO, DEM or CsA. The log2 transformed fold changes are relative to adult T. urticae females treated with formulation only and were clustered using a Euclidean distance metric and Ward’s method. T. urticae gene IDs are shown on the right.
DEGs of both the PBO and DEF treatment were significantly enriched in the GO terms “oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen” (GO:0016705), “heme binding” (GO:0020037), “monooxygenase activity” (GO:0004497), “iron ion binding” (GO:0005506), “oxidation-reduction process” (GO:0055114), “antibiotic metabolic process” (GO:0016999) and “transferase activity, transferring hexosyl groups” (GO:0016758). The first five GO terms were mainly present in genes coding for P450s (14 and 11 DEGs for the PBO and DEF treatments, respectively) while the latter two terms were found in UGTs (7 and 12 DEGs for the PBO and DEF treatments, respectively). Interestingly, almost all P450 and UGT genes (13/14 P450s and 7/7 UGTs (PBO treatment) and 10/11 P450s and 12/12 UGTs (DEF treatment)) were upregulated for both the PBO and DEF treatment. The majority of the upregulated P450 genes (12/13 and 8/10 for the PBO and DEF treatments, respectively) belonged to the CYP392 family within the CYP2 clan for which several members were shown previously to respond strongly to acaricide selection and feeding on different hosts44. GO:0055114 was also found in genes coding for short chain dehydrogenases (4 and 3 DEGs for the PBO and DEF treatments, respectively) and intradiol ring-cleavage dioxygenases (2 and 4 DEGs for the PBO and DEF treatments, respectively) while for the DEF treatment this term was also present in genes coding for a superoxide dismutase (tetur02g11180), a fatty acid synthase (tetur04g02370) and a glucose dehydrogenase (tetur03g09330). In addition, DEGs of the DEF treatment were also significantly enriched in the GO terms “hormone activity” (GO:0005179) and “integral component of membrane” (GO:0016021). The “hormone activity” term was present in four DEGs coding for neuropeptides (InterPro domain IPR001955 or IPR016179), while “integral component of membrane” was present in more than 45 DEGs, including, amongst others, genes coding for P450s (10 DEGs), MFS proteins (10 DEGs), UGTs (7 DEGs), ABC transporters (3 DEGs), neuropeptides (2 DEGs), innexins (InterPro domain IPR000990, 2 DEGs) and proteins with a DUF3421 domain (InterPro domain IPR024518, 2 DEGs) (Table 1 and Supplementary Tables S2 and S3). A cluster analysis of expression data of P450, UGT and MFS genes complemented the findings of our GO analyses. For both the P450 and UGT gene expression data the PBO and DEF treatment clustered together, while for MFS gene expression data, the CsA treatment fell outside the clade with all other treatments (Fig. 4).
Discussion
Gene expression changes are a major component of stress responses, and contribute to or complement alterations in metabolism, cell cycle progression, protein homeostasis, cytoskeletal organization, vesicular trafficking and modification of enzymatic activities69. Expression changes can be grouped into generic responses shared by many stresses, and stress-specific adaptive responses. In contrast to post-translational effects, which provide immediate responses, regulation of gene expression is essential for the slower, long term acclimation to and recovery from a stress factor69. At present, our understanding of gene expression changes in arthropods to synergist exposure is very limited in spite of their importance in insecticide- and acaricide-mediated control. In fact, only two studies have investigated genome-wide transcriptional changes in an insect upon synergist exposure. Willoughby et al. and Zimmer et al.47,48 studied PBO-induced gene expression changes in D. melanogaster and B. tabaci, respectively. Gene expression changes upon exposure to other synergist compounds have not yet been determined.
In this study, we examined transcriptional responses in a polyphagous arthropod pest, T. urticae, upon exposure to the synergist compounds PBO, DEF, DEM and CsA. The strain that was used for our experiments (JP-R, see Material & Methods) was shown to be resistant to the mitochondrial complex II inhibitors cyenopyrafen and cyflumetofen (both compounds are beta-keto nitrile derivatives belonging to IRAC Mode of Action Classification 25 A 2,70), with cyenopyrafen and cyflumetofen toxicity strongly synergized by PBO and DEM, respectively, while DEF moderately synergized cyenopyrafen toxicity64. Because acaricides are typically applied 24 h after synergist treatment53,56,64,65, transcriptional changes were captured 24 h after exposure. In future, it would be interesting to also capture the transcription levels at those synergist exposure periods that have been used in previous synergism studies with T. urticae (1 h and 4 h)53,55,57–61. Finally, the observed fold changes of up- and downregulated T. urticae genes (ranging from −3.8 to 3.3 across all synergist treatments) were in line with those observed for B. tabaci by Zimmer et al.48. In contrast, fold changes of PBO induced Drosophila genes were considerably higher (ranging from 2 to 32)47. However, in our study a 24 h synergist pretreatment was used, while in Drosophila expression measurements were taken after 4 hours of synergist exposure, and with the Drosophila 16 K full genome array. Briefly, the Drosophila 16 K full genome array contained only one 70mer probe per gene47, confounding reliable estimation of fold changes71, while in our study and that of Zimmer et al.48 gene-expression levels were captured by Illumina RNA-sequencing.
Stress responses can be induced by a variety of stressors such as extreme temperatures, elevated ion concentrations or toxic substances, all of which usually result in excessive amounts of denatured proteins72. The general stress response typically consists of an increased production or activation of antioxidant proteins (e.g. Cu/Zn superoxide dismutase and GSTs) and chaperone proteins (e.g. heat shock proteins)73,74, together with an increased mobilization of energy from storage tissues75. The latter is associated with the overexpression of numerous genes involved in energy production or in cellular catabolism such as NADH dehydrogenase, ATP synthase, trypsin and lipases76. Only one GST gene was significantly upregulated in the PBO and DEF treatment while a gene coding for a superoxide dismutase was downregulated in the DEF treatment. A GO enrichment analysis did not reveal a GO category that was in common between the different synergist treatments and that was related to the above-mentioned proteins. Even more, only one upregulated gene, coding for a putative sodium-dependent glucose transporter (tetur01g06580, member of the MFS-gene family), was in common for all treatments. Hence, we assume that the observed gene expression changes are not general stress responses but rather specific for the synergist compounds to which the mites had been exposed. However, based on PC1 of our principal component analysis gene expression levels of PBO and DEF treatment replicates did cluster together (Fig. 1). Even more, about 35% of the T. urticae DEGs that were identified for both these treatments were shared and had fold changes in the same direction (Fig. 3). Interestingly, about one third (15/44) of the upregulated shared DEGs coded for members of the P450 and UGT gene families (Fig. 4, Supplementary Table S2). For D. melanogaster, it has been shown that the transcription factor CncC plays an important role in xenobiotic-inducible gene expression of P450s, GSTs, UGTs and membrane transporters77. However, none of the T. urticae orthologs of D. melanogaster CncC (tetur07g06850 and tetur07g04600 44) were differentially expressed after 24 h (Supplementary Tables S2,S3,S4 and S5). In addition, T. urticae genes related to the gene coding for Drosophila Keap1 (Kelch-associated protein 1), a negative regulator of CncC, and that are downregulated in acaricide resistant and host plant adapted mite lines (OrthoMCL group 10254)44,45 were also not found among the DEGs of both the PBO and DEF treatments. However, expression changes of T. urticae transcription factors genes78 did correlate for these treatments (Supplementary Figure S1), suggesting a common transcriptional regulation. Ten of these transcription factor genes had a fold change of more than 1.25 in both the PBO and DEF treatments and had an opposite direction of fold change compared to the CsA and DEM treatments (Supplementary Table S7) and, hence, might be good candidates to be investigated for their role in xenobiotic resistance regulation in T. urticae.
The induction of P450s upon exposure by PBO has been reported by other groups47,48,79–82. As hypothesized by Chan et al.81, it might be a compensation strategy for an enzyme to be upregulated upon contact with an inhibitor in order to minimize the effect of reduced enzyme activity. In general, several studies have shown that xenobiotics are able to induce expression of arthropod P450s11,44,45,48,77,83–86. In Drosophila for example, about a third of the P450s can be induced following xenobiotic substance feeding or topical application87. For T. urticae it has been shown that the application of barbital, phenobarbital and geraniol does increase P450 activity in a dose-dependent way and that pretreatment with these inducers resulted in decreased acaricide toxicity88. Willoughby et al. suggested that the increase of detoxification enzymes by PBO might speed up insecticide tolerance if these detoxification enzymes could metabolize insecticides. Hence, one might question whether induction of T. urticae detoxification genes by synergist compounds could lead to a decreased toxicity of acaricides. In this study, CYP392A11 and CYP392A12 were both upregulated upon exposure of the JP-R strain by PBO. In 2015 it was shown that the upregulation of these genes was associated with cyenopyrafen resistance of the JP-R strain64 and that CYP392A11 could metabolize cyenopyrafen89. Similarly, only three carboxyl/choline esterases, TuCCE06 (tetur01g10800), TuCCE07 (tetur01g010810) and TuCCE25 (tetur04g06670), were upregulated upon exposure of the JP-R strain by DEF and recently it was shown that TuCCE25 was highly upregulated in the JP-R strain and responded to cyenopyrafen selection64. In summary, both PBO and DEF seem to induce genes coding for detoxification enzymes that are strongly associated/implicated in cyenopyrafen resistance of the JP-R strain. On the other hand, both PBO and DEF, at the same concentration and exposure period that was used in this study, have previously been shown to synergize cyenopyrafen toxicity64. Hence, the direct inhibition of these P450s and CCEs by PBO and DEF, respectively, seems to outweigh the impact of the synergists on their induction. In contrast, in instances where P450s or CCEs may not be involved in acaricide resistance, PBO or DEF induction of genes encoding enzymes that are not inhibited by the synergist compound and that are capable of metabolizing acaricides might interfere and lead to misinterpretation of synergism experiments (for example antagonism instead of synergism).
Both the PBO and DEF treatments resulted in an upregulation of TuGSTd14 (tetur29g00220). Of particular note, TuGSTd14 was also upregulated in multi-resistant T. urticae strains44 and showed affinity toward abamectin and a competitive type of inhibition90. In contrast, the DEM treatment did not result in the overexpression of GSTs. Even more, significantly enriched GO terms could not be identified for the DEM treatment. This result was somewhat unanticipated as it is known that depletion of GSH by DEM results in oxidative stress and destabilized proteins in vitro 91 and that the response to oxidative stress typically consists of highly coordinated changes in gene expression (for example, in human carcinoma cells92). In the study by Casey et al.92, exposure to DEM resulted in GSH depletion below control levels from 1 h to 24 h after DEM treatment. However, although the expression of about 800 genes had a significant change (over two-fold at more than one time point) during this 24 h time course experiment, log2 fold changes of almost all genes returned to near zero at the 24 h time point92. Previously it was shown that a 24 h treatment with DEM strongly synergized cyflumetofen toxicity in the JP-R strain64 and that a GST (TuGSTd05) that was upregulated in this strain could metabolize cyflumetofen. Hence, the 24 h DEM exposure most likely resulted in depleted GSH levels in mites of the JP-R strain but the expression changes that are associated with the onset of depletion may not have been captured in our study.
The Major Facilitator Superfamily (MFS) is, along with ATP-binding cassette (ABC) transporters, one of the largest transporter families present in all living organisms. MFS transporters, as opposed to ABC transporters, use an existing electrochemical gradient instead of ATP to transport substrates (either by symport, uniport or antiport) across membranes27,93. Numerous studies have pointed to the importance of ABC transporters in xenobiotic resistance in arthropods27,28. In the case of T. urticae, several of its 103 ABC transporter genes were differentially expressed in multi-pesticide resistant strains and/or in mites transferred to a more challenging host plant94. MFS transporters, however, have been less studied in arthropods and it is only recently that evidence for their role in xenobiotic resistance in arthropods has been reported44,95. Consistent with this, 37 T. urticae MFS genes (belonging to either OrthoMCL group arthro10032, arthro10082 or arthro10236) were differentially expressed upon long term transfer of mites from bean to tomato44.
None of the 103 ABC T. urticae transporter genes were differentially expressed upon exposure to the ABC transporter modulator CsA. In contrast, about one third of the DEGS of the CsA treatment coded for MFS transporters, including five sugar transporters. The differential expression of these sugar transporters might be caused by interference of cyclosporine in glucose metabolism96,97. On the other hand, the majority of the remaining differentially expressed MFS genes showed similarity with members of the anion/cation superfamily that can transport a variety of substrates98. Finally, it should be mentioned that CsA inhibition of ABC transporters is not well characterized in arthropods compared to vertebrates27, and the transcriptional changes that were observed might not reflect those that are caused by the inhibition of ABC transporters but rather by inhibition of another target like calcineurin (the T. urticae subunits of this target are tetur04g07540, tetur04g07560 and tetur03g06410 as assessed by a blastp search, E-values between E-86 and 0 with human calcineurin subunits as queries), as occurs in the vertebrate immune system99.
To conclude, transcriptional changes in arthropod pests exposed to synergist compounds have only been marginally studied. In this work, we exposed a polyphagous arthropod pest, T. urticae, for 24 hours to four different synergist compounds - PBO, DEF, DEM and CsA - and measured genome-wide gene expression changes. The CsA treatment resulted primarily in the downregulation of T. urticae MFS genes, while T. urticae DEGs of the DEM treatment were not significantly enriched for a GO term. Exposure to PBO and DEF resulted in a broad transcriptional response and about one third of the DEGs, including cytochrome P450 monooxygenases and UDP-glycosyltransferases, were shared between both treatments, suggesting modulation of a common transcriptional program. Moreover, both DEF and PBO induced genes that are strongly implicated in acaricide resistance of the strain used in this study. Based on previous synergism toxicity studies, however, the induction of these detoxification genes seems not to interfere with the outcome of synergism assays, although the effects of induction might be relatively more important when resistance is not synergized.
Methods
Mite strains and chemicals
The JP-R strain has previously been described by Khalighi et al.64. Briefly, this strain is resistant to both cyenopyrafen (LC50 of 291 mg L−1) and cyflumetofen (LC50 of 146 mg L−1). In addition, cyenopyrafen toxicity in the JP-R strain is synergized by PBO and DEF, while DEM synergizes cyflumetofen toxicity64. The JP-R strain was maintained on potted bean plants, Phaseolus vulgaris L. var. Prelude, sprayed with 200 mg a.i. cyenopyrafen L−1 (STARMITE, 30% SC) until run-off. For the week prior to collection of RNA, the strain was maintained on bean plants without cyenopyrafen selection pressure. The synergists DEF (97% purity), DEM (97% purity) and PBO (90% purity) were of analytical grade and purchased from Sigma-Aldrich (Belgium). CsA had a purity of more than 98% and was purchased from Enzo Life Sciences (Belgium).
Synergist bioassays
The synergist bioassays were performed as described earlier by Van Pottelberge et al. 2009. Briefly, synergists were dissolved in a mixture of N,N-dimethylformamide and emulsifier W (alkylarylpolyglycolether), 3:1 w/w, respectively, and subsequently diluted with deionized water 100-fold. The concentrations used for PBO (1000 ppm), DEF (500 ppm) and DEM (2000 ppm) were identical to those in Khalighi et al. and Van Pottelberge et al.53,56 and are known to cause between 5 and 10% mortality. Based on preliminary experiments a concentration of 50 ppm CsA was used, resulting in maximum 5 to 10% mortality. About 30 3–5 day old adult females were transferred to the upper (adaxial) side of a 9 cm2 square-cut kidney bean leaf discs placed on wet cotton wool. Leaf discs were sprayed at 1 bar pressure in a Cornelis spray tower with 650 µl spray fluid (1.56 ± 0.04 mg fluid deposit cm−2) containing one of the synergists (DEF, DEM, PBO, CsA) or formulation (FORM; N,N-dimethylformamide and emulsifier W (3:1 w/w) diluted in deionized water 100-fold) only. Unsprayed mites (CON) served as an additional control. About 600 mites (20 leaf disks with mites) were used for each treatment (DEF, DEM, PBO or CsA) and for the controls (FORM, CON). Next, leaf disks were placed in a climatically controlled room at 26 °C, 60% RH with a 16:8 h light:dark photoperiod. After 24 hours, living mites were scored and collected for RNA extraction. Mites were scored as being alive if they could walk normally after being prodded with a camel’s hair brush.
RNA-seq
Total RNA was extracted from about 100 adult female mites (collected from at least four different leaf disks) using the RNeasy mini kit (Qiagen, Belgium) with four-fold biological replication for each treatment (PBO, DEF, DEM, CsA) and the controls (FORM, CON). The quality and quantity of the total RNA was analyzed by a DeNovix DS-11 spectrophotometer (DeNovix, USA) and by running an aliquot on a 1% agarose gel. From the RNA samples, Illumina libraries were constructed with the TruSeq Stranded mRNA Library Preparation Kit with polyA selection (Illumina, USA), and the resulting libraries were sequenced on an Illumina HiSeq 2500 to generate strand-specific paired reads of 2 × 125 bp (library construction and sequencing was performed at the High-Throughput Genomics Core of the Huntsman Cancer Institute, University of Utah, Utah, USA). Prior to read-mapping, the quality of the reads was verified using FASTQC version 0.11.4100 (no reads flagged as poor quality or as containing adapter sequences were used in downstream analyses).
Expression quantification and principal component analysis (PCA)
All reads were aligned to the T. urticae genome62 using the two-pass alignment mode of STAR 2.5.0a101 with a maximum intron size set to 20 kb. Resulting BAM files were subsequently sorted on read name by using SAMtools 0.1.19102. Read counts per gene using the most recent T. urticae annotation (version of June 23, 2016) were then obtained using the default settings of HTSeq. 0.6.0103 with the “STRANDED” flag set to “yes” and the “feature” flag set to “exon”. A PCA was created as described by Love et al.104. Briefly, read counts were first normalized using the regularized-logarithm (rlog) transformation implemented in the DESeq2 (version 1.12.2) R-package105. A PCA was then performed using the stats (version 3.3.0) and ggplot2 (version 2.2.0) R-packages with the 5000 most variable genes across all RNA-seq samples.
Differential expression (DE) analysis and gene ontology (GO) enrichment analysis
A differential expression (DE) analysis was performed using DESeq2 (version 1.12.2)105 and the read count data obtained by HTSeq (see above). Differentially expressed genes (DEGs, fold change (FC) ≥ 1.5 and Benjamini-Hochberg106 adjusted p-value < 0.05) were determined between unsprayed mites (CON), mites treated with DEF, DEM, PBO or CsA and mites treated with formulation (FORM) (five DE comparisons in total: FORM vs. CON, DEF vs. FORM, DEM vs. FORM, PBO vs. FORM and CsA vs. FORM). For the GO enrichment analysis, the complete T. urticae proteome (19087 sequences, version of June 23, 2016) was first used as query in a blastp search against the non-redundant protein database in NCBI (version of October 31, 2016) using the following settings “ -outfmt 5 -evalue 1e-5 -word_size 3 -show_gis -num_alignments 20 -max_hsps_per_subject 20”. The resulting blastp output was then loaded into the Blast2GO (version 4.0.7) program107 and T. urticae proteins were annotated using the default parameters107. InterProScan 5 and ANNEX were used to augment the annotation of GO terms. GO terms were condensed using the generic GO Slim subset. For the DESeq2 output of four DE comparisons (DEF vs. FORM, DEM vs. FORM, PBO vs. FORM and CsA vs. FORM), a GO enrichment analysis was performed using the Bioconductor package GOSeq (version 1.24.0)108, which explicitly takes into account gene selection bias due to differences in transcript length109. The resulting p-values from GOSeq were corrected using the Benjamini-Hochberg method106 and only those GO categories with an adjusted p-value of less than 0.05 were considered significantly enriched. Gene expression heatmaps were generated using the relative transcript levels (fold changes) of four DE comparisons (DEF vs. FORM, DEM vs. FORM, PBO vs. FORM and CsA vs. FORM) and the limma (version 3.28.21) and gplots (version 3.0.1) packages in the R environment. Transcription factor, P450 and UGT gene lists were obtained from previous studies62,78,110 while the MFS gene list consisted of those from Dermauw et al.44 and those that were differentially expressed in the CsA treatment (Supplementary Table S6). Genes with no read counts in all four DE comparison were not included in the heatplot. Finally, genes were clustered using a Euclidean distance metric and Ward’s method.
qPCR
To validate the RNA-seq results, gene specific primers were designed for differentially expressed T. urticae genes (8 up- and 5 downregulated genes) using Primer 3 v.4.0.0111. The qPCR primers used, including the primers for the genes of interest, as well as those for the two reference genes, ribosomal protein gene RP49 and ubiquitin, can be found in Supplementary Table S8. Total RNA was extracted as described above and cDNA was synthesized with the Maxima First Strand cDNA kit (Fermentas Life Sciences, Aalst, Belgium) and 1.5 µg of total RNA. Three biological and two technical replicates were used and non-template controls were added to exclude sample contamination. The qPCR analysis was performed on a Mx3005 P qPCR thermal cycler (Stratagene, Agilent Technologies, Diegem, Belgium) with Maxima SYBR Green qPCR Master Mix and ROX solution (Fermentas Life Sciences) according to the manufacturer’s instructions. The run conditions were as follows: 95 °C for 10 m followed by 35 cycles of 95 °C for 15 s, 55 °C for 30 s, 72 °C for 30 s. At the end of these cycles, a melting curve was generated (from 65 °C to 95 °C, 1 °C per 2 s) to check the presence of a single amplicon. Fourfold dilution series of pooled cDNA were used to determine the standard curves and amplification efficiencies for every gene-specific primer pair. The efficiencies were incorporated in the calculations of the expression values. Relative expression levels and significant gene expression differences (one-sided unpaired t-test) were calculated with qbase+ version 3.0112.
Image processing
CorelDRAW Home & Student ×7 and SigmaPlot 12.0 software was used for processing of images.
Data availability
The RNA-seq expression data generated during the current study are available in the Gene-Expression Omnibus (GEO) repository with accession number GSE98293.
Electronic supplementary material
Acknowledgements
W.D. is a postdoctoral fellow of the Research Foundation Flanders (FWO). R.G. was supported by the National Institutes of Health Genetics Training Grant TM32GM007464. This project was supported by the Research Foundation Flanders (FWO) (grant G009312N to L.T. and T.V.L. and grant G053815N to L.T., T.V.L. and W.D.).
Author Contributions
T.V.L., W.D. and S.S. designed the experiment. S.S. conducted the experiments. Analysis and interpretation of the results was done by S.S., R.G., W.D., R.M.C. and T.V.L. The manuscript was written by S.S. and W.D., all figures were prepared by S.S. and W.D. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-13397-x.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Thomas Van Leeuwen, Email: thomas.vanleeuwen@ugent.be.
Wannes Dermauw, Email: wannes.dermauw@ugent.be.
References
- 1.Pimentel D. ‘Environmental and economic costs of the application of pesticides primarily in the United States’. Environ. Dev. Sustain. 2005;7:229–252. doi: 10.1007/s10668-005-7314-2. [DOI] [Google Scholar]
- 2.Sparks TC, Nauen R. IRAC: Mode of action classification and insecticide resistance management. Pestic. Biochem. Physiol. 2015;121:122–128. doi: 10.1016/j.pestbp.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 3.Feyereisen R, Dermauw W, Van Leeuwen T. Genotype to phenotype, the molecular and physiological dimensions of resistance in arthropods. Pestic. Biochem. Physiol. 2015;121:61–77. doi: 10.1016/j.pestbp.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Van Leeuwen T, Dermauw W. The molecular evolution of xenobiotic metabolism and resistance in chelicerate mites. Annu. Rev. Entomol. 2016;61:475–498. doi: 10.1146/annurev-ento-010715-023907. [DOI] [PubMed] [Google Scholar]
- 5.Matsumura, F. In Toxicology of Insecticides 203–298 (Springer US, 1985).
- 6.Bernard CB, Philogène BJ. Insecticide synergists: role, importance, and perspectives. J. Toxicol. Environ. Health. 1993;38:199–223. doi: 10.1080/15287399309531712. [DOI] [PubMed] [Google Scholar]
- 7.Casida JE. Mixed-function oxidase involvement in the biochemistry of insecticide synergists. J. Agric. Food Chem. 1970;18:753–772. doi: 10.1021/jf60171a013. [DOI] [PubMed] [Google Scholar]
- 8.Raffa K, Priester T. Synergists as research tools and control agents in agriculture. J. Agric. Entomol. 1985;2:27–45. [Google Scholar]
- 9.Beck B, Steurbaut W, Spanoghe P. How to define green adjuvants. Pest Manag. Sci. 2012;68:1107–1110. doi: 10.1002/ps.3308. [DOI] [PubMed] [Google Scholar]
- 10.Hill, N. A novel plant-based synergist for pyrethrum and pyrethroids against urban public health pests. In Sixth International Conference on Urban Pests (2008).
- 11.Liu SQ, et al. Dillapiol: a pyrethrum synergist for control of the Colorado potato beetle. J. Econ. Entomol. 2014;107:797–805. doi: 10.1603/EC13440. [DOI] [PubMed] [Google Scholar]
- 12.Joffe T, et al. Investigating the potential of selected natural compounds to increase the potency of pyrethrum against houseflies Musca domestica (Diptera: Muscidae) Pest Manag. Sci. 2012;68:178–184. doi: 10.1002/ps.2241. [DOI] [PubMed] [Google Scholar]
- 13.Feyereisen R. Insect P450 inhibitors and insecticides: Challenges and opportunities. Pest Manag. Sci. 2015;71:793–800. doi: 10.1002/ps.3895. [DOI] [PubMed] [Google Scholar]
- 14.Feyereisen R, Langry KC, Ortiz de Montellano PR. Self-catalyzed destruction of insect cytochrome P-450. Insect Biochem. 1984;14:19–26. doi: 10.1016/0020-1790(84)90079-9. [DOI] [Google Scholar]
- 15.Correia, M. A. & O de Montellano, P. R. In Cytochrome P450: Structure, mechanism, and biochemistry 247–322 (2005).
- 16.Plapp FW, Bigley WS, Chapmen GA, Eddy GW. Synergism of malathion against resistant house flies and mosquitoes. J. Econ. Entomol. 1963;56:643–649. doi: 10.1093/jee/56.5.643. [DOI] [Google Scholar]
- 17.Yeoh CL, Kuwano E, Eto M. Effects of the fungicide IBP as a synergist on the metabolism of malathion in insects. J. Pestic. Sci. 1982;7:31–40. doi: 10.1584/jpestics.7.31. [DOI] [Google Scholar]
- 18.Sogorb MA, Vilanova E. Enzymes involved in the detoxification of organophosphorus, carbamate and pyrethroid insecticides through hydrolysis. Toxicol. Lett. 2002;128:215–228. doi: 10.1016/S0378-4274(01)00543-4. [DOI] [PubMed] [Google Scholar]
- 19.World Health Organization, United Nations Environment Programme, Internalional Labour Organization & International Programme on Chemical safety. Organophosphorus insecticides: a general introduction/published under the joint sponsorship of the United Nations Environment Programme, the International Labour Organisation, and the World Health Organization. (World Health Organization; Geneva, 1986).
- 20.Fujioka K, Casida JE. Glutathione S -transferase conjugation of organophosphorus pesticides yields S -phospho-, S -aryl-, and S -alkylglutathione derivatives. Chem. Res. Toxicol. 2007;20:1211–1217. doi: 10.1021/tx700133c. [DOI] [PubMed] [Google Scholar]
- 21.Che-Mendoza A, Penilla R, Rodríguez D. Insecticide resistance and glutathione S-transferases in mosquitoes: A review. African J. Biotechnol. 2009;8:1386–1397. [Google Scholar]
- 22.Boyland E, Chasseaud LF. The effect of some carbonyl compounds on rat liver glutathione levels. Biochem. Pharmacol. 1970;19:1526–1528. doi: 10.1016/0006-2952(70)90075-4. [DOI] [PubMed] [Google Scholar]
- 23.Yusa K, Tsuruo T. Reversal mechanism of multidrug resistance by verapamil: direct binding of verapamil to p-glycoprotein on specific sites and transport of verapamil outward across the plasma membrane of K562/ADM cells. Cancer Res. 1989;49:5002–5006. [PubMed] [Google Scholar]
- 24.Litman T, Zeuthen T, Skovsgaard T, Stein WD. Competitive, non-competitive and cooperative interactions between substrates of P-glycoprotein as measured by its ATPase activity. Biochim. Biophys. Acta - Mol. Basis Dis. 1997;1361:169–176. doi: 10.1016/S0925-4439(97)00027-6. [DOI] [PubMed] [Google Scholar]
- 25.Foxwell BM, Mackie A, Ling V, Ryffel B. Identification of the multidrug resistance-related P-glycoprotein as a cyclosporine binding protein. Mol. Pharmacol. 1989;36:543–6. [PubMed] [Google Scholar]
- 26.Sharom, F. J. The P-glycoprotein multidrug transporter. Essays Biochem. 50 (2011). [DOI] [PubMed]
- 27.Dermauw W, Van Leeuwen T. The ABC gene family in arthropods: Comparative genomics and role ininsecticide transport and resistance. Insect Biochem. Mol. Biol. 2014;45:89–110. doi: 10.1016/j.ibmb.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Merzendorfer H. ABC transporters and their role in protecting insects from pesticides and their metabolites. Adv. In Insect Phys. 2014;46:1–72. doi: 10.1016/B978-0-12-417010-0.00001-X. [DOI] [Google Scholar]
- 29.Luo L, Sun Y-J, Yang L, Huang S, Wu Y-J. Avermectin induces P-glycoprotein expression in S2 cells via the calcium/calmodulin/NF-κB pathway. Chem. Biol. Interact. 2013;203:430–9. doi: 10.1016/j.cbi.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 30.Strycharz JP, et al. Resistance in the highly DDT-resistant 91-R strain of Drosophila melanogaster involves decreased penetration, increased metabolism, and direct excretion. Pestic. Biochem. Physiol. 2013;107:207–217. doi: 10.1016/j.pestbp.2013.06.010. [DOI] [Google Scholar]
- 31.Sun YP, Johnson ER. Quasi-synergism and penetration of insecticides. J. Econ. Entomol. 1972;65:349–53. doi: 10.1093/jee/65.2.349. [DOI] [PubMed] [Google Scholar]
- 32.Kennaugh L, Pearce D, Daly JC, Hobbs AA. A piperonyl butoxide synergizable resistance to permethrin in Helicoverpa armigera which is not due to increased detoxification by cytochrome P450. Pesticide Biochemistry and Physiology. 1993;45:234–241. doi: 10.1006/pest.1993.1026. [DOI] [Google Scholar]
- 33.Gunning RV, Devonshire AL, Moores GD. Metabolism of Esfenvalerate by Pyrethroid-Susceptible and -Resistant Australian Helicoverpa armigera (Lepidoptera: Noctuidae) Pestic. Biochem. Physiol. 1995;51:205–213. doi: 10.1006/pest.1995.1020. [DOI] [Google Scholar]
- 34.Sanchez-Arroyo H, Koehler PG, Valles SM. Effects of the synergists piperonyl butoxide and S,S,S-tributyl phosphorotrithioate on propoxur pharmacokinetics in Blattella germanica (Blattodea: Blattellidae) J. Econ. Entomol. 2001;94:1209–16. doi: 10.1603/0022-0493-94.5.1209. [DOI] [PubMed] [Google Scholar]
- 35.Young SJ, Gunning RV, Moores GD. The effect of piperonyl butoxide on pyrethroid-resistance-associated esterases in Helicoverpa armigera (Hübner) (Lepidoptera: Noctuidae) Pest Manag. Sci. 2005;61:397–401. doi: 10.1002/ps.996. [DOI] [PubMed] [Google Scholar]
- 36.Young SJ, Gunning RV, Moores GD. Effect of pretreatment with piperonyl butoxide on pyrethroid efficacy against insecticide-resistant Helicoverpa armigera (Lepidoptera: Noctuidae) and Bemisia tabaci (Sternorrhyncha: Aleyrodidae) Pest Manag. Sci. 2006;62:114–119. doi: 10.1002/ps.1127. [DOI] [PubMed] [Google Scholar]
- 37.López-Soler N, et al. Esterase inhibition by synergists in the western flower thrips Frankliniella occidentalis. Pest Manag. Sci. 2011;67:1549–56. doi: 10.1002/ps.2211. [DOI] [PubMed] [Google Scholar]
- 38.Lucier GW, McDaniel OS, Matthews HB. Microsomal rat liver UDP glucuronyltransferase: Effects of piperonyl butoxide and other factors on enzyme activity. Arch. Biochem. Biophys. 1971;145:520–530. doi: 10.1016/S0003-9861(71)80012-7. [DOI] [PubMed] [Google Scholar]
- 39.Espinosa PJ, et al. Metabolic mechanisms of insecticide resistance in the western flower thrips, Frankliniella occidentalis (Pergande) Pest Manag. Sci. 2005;61:1009–1015. doi: 10.1002/ps.1069. [DOI] [PubMed] [Google Scholar]
- 40.Gunning, R. V., Moores, G. D. & Devonshire, A. L. In Piperonyl Butoxide (Academic Press, 1998).
- 41.Valles S, Koehler P, Brenner R. Antagonism of fipronil toxicity by piperonyl butoxide and S,S,S-tributyl phosphorotrithioate in the German cockroach (Dictyoptera: Blattellidae) J. Econ. Entomol. 1997;90:1254–1258. doi: 10.1093/jee/90.5.1254. [DOI] [Google Scholar]
- 42.Philippou D, et al. The interactions between piperonyl butoxide and E4, a resistance-associated esterase from the peach-potato aphid, Myzus persicae Sulzer (Hemiptera: Aphididae) Pest Manag. Sci. 2013;69:499–506. doi: 10.1002/ps.3400. [DOI] [PubMed] [Google Scholar]
- 43.Perry T, Batterham P, Daborn PJ. The biology of insecticidal activity and resistance. Insect Biochemistry and Molecular Biology. 2011;41:411–422. doi: 10.1016/j.ibmb.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 44.Dermauw W, et al. A link between host plant adaptation and pesticide resistance in the polyphagous spider mite Tetranychus urticae. Proc. Natl. Acad. Sci. USA. 2013;110:E113–22. doi: 10.1073/pnas.1213214110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wybouw N, et al. Adaptation of a polyphagous herbivore to a novel host plant extensively shapes the transcriptome of herbivore and host. Mol. Ecol. 2015;24:4647–4663. doi: 10.1111/mec.13330. [DOI] [PubMed] [Google Scholar]
- 46.Scott, J. G. In Pesticide Resistance in Arthropods 39–57 (Springer US, 1990).
- 47.Willoughby L, Batterham P, Daborn PJ. Piperonyl butoxide induces the expression of cytochrome P450 and glutathione S-transferase genes in Drosophila melanogaster. Pest Manag. Sci. 2007;63:803–808. doi: 10.1002/ps.1391. [DOI] [PubMed] [Google Scholar]
- 48.Zimmer CT, et al. Use of the synergist piperonyl butoxide can slow the development of alpha‐cypermethrin resistance in the whitefly Bemisia tabaci. Insect Mol. Biol. 2017;26:152–163. doi: 10.1111/imb.12276. [DOI] [PubMed] [Google Scholar]
- 49.Migeon, A., Nouguier, E. & Dorkeld, F. In Trends in Acarology SE - 96 557–560 (2010).
- 50.Van Leeuwen T, Vontas J, Tsagkarakou A, Dermauw W, Tirry L. Acaricide resistance mechanisms in the two-spotted spider mite Tetranychus urticae and other important Acari: A review. Insect Biochemistry and Molecular Biology. 2010;40:563–572. doi: 10.1016/j.ibmb.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 51.Van Leeuwen T, Tirry L, Yamamoto A, Nauen R, Dermauw W. The economic importance of acaricides in the control of phytophagous mites and an update on recent acaricide mode of action research. Pestic. Biochem. Physiol. 2014;121:12–21. doi: 10.1016/j.pestbp.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 52.Van Leeuwen T, Tirry L. Esterase-mediated bifenthrin resistance in a multiresistant strain of the two-spotted spider mite, Tetranychus urticae. Pest Manag. Sci. 2007;63:150–156. doi: 10.1002/ps.1314. [DOI] [PubMed] [Google Scholar]
- 53.Van Pottelberge S, Van Leeuwen T, Nauen R, Tirry L. Resistance mechanisms to mitochondrial electron transport inhibitors in a field-collected strain of Tetranychus urticae Koch (Acari: Tetranychidae) Bull. Entomol. Res. 2009;99:23–31. doi: 10.1017/S0007485308006081. [DOI] [PubMed] [Google Scholar]
- 54.Van Leeuwen T, Stillatus V, Tirry L. Genetic analysis and cross-resistance spectrum of a laboratory-selected chlorfenapyr resistant strain of two-spotted spider mite (Acari: Tetranychidae) Exp. Appl. Acarol. 2004;32:249–261. doi: 10.1023/B:APPA.0000023240.01937.6d. [DOI] [PubMed] [Google Scholar]
- 55.Rauch N, Nauen R. Spirodiclofen resistance risk assessment in Tetranychus urticae (Acari: Tetranychidae): A biochemical approach. Pestic. Biochem. Physiol. 2002;74:91–101. doi: 10.1016/S0048-3575(02)00150-5. [DOI] [Google Scholar]
- 56.Khalighi M, Tirry L, Van Leeuwen T. Cross-resistance risk of the novel complex II inhibitors cyenopyrafen and cyflumetofen in resistant strains of the two-spotted spider mite Tetranychus urticae. Pest Manag. Sci. 2014;70:365–368. doi: 10.1002/ps.3641. [DOI] [PubMed] [Google Scholar]
- 57.Kim Y-J, Park H-M, Cho J-R, Ahn Y-J. Multiple resistance and biochemical mechanisms of pyridaben resistance in Tetranychus urticae (Acari: Tetranychidae) J. Econ. Entomol. 2006;99:954–8. doi: 10.1093/jee/99.3.954. [DOI] [PubMed] [Google Scholar]
- 58.Kim Y-J, Lee S-H, Lee S-W, Ahn Y-J. Fenpyroximate resistance in Tetranychus urticae (Acari: Tetranychidae): cross-resistance and biochemical resistance mechanisms. Pest Manag. Sci. 2004;60:1001–1006. doi: 10.1002/ps.909. [DOI] [PubMed] [Google Scholar]
- 59.Tsagkarakou A, Pasteur N, Cuany A, Chevillon C, Navajas M. Mechanisms of resistance to organophosphates in Tetranychus urticae (Acari: Tetranychidae) from Greece. Insect Biochem. Mol. Biol. 2002;32:417–24. doi: 10.1016/S0965-1748(01)00118-7. [DOI] [PubMed] [Google Scholar]
- 60.Yang X, Yan Zhu K, L. Buschman L, Margolies C. D. Comparative susceptibility and possible detoxification mechanisms for selected miticides in Banks grass mite and two-spotted spider mite (Acari: Tetranychidae) Exp. Appl. Acarol. 2001;25:293–299. doi: 10.1023/A:1017926920389. [DOI] [PubMed] [Google Scholar]
- 61.Stumpf N, Nauen R. Biochemical markers linked to abamectin resistance in Tetranychus urticae (Acari: Tetranychidae) Pestic. Biochem. Physiol. 2002;72:111–121. doi: 10.1006/pest.2001.2583. [DOI] [Google Scholar]
- 62.Grbić M, et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature. 2011;479:487–92. doi: 10.1038/nature10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Pottelberge S, Khajehali J, Van Leeuwen T, Tirry L. Effects of spirodiclofen on reproduction in a susceptible and resistant strain of Tetranychus urticae (Acari: Tetranychidae) Exp. Appl. Acarol. 2009;47:301–309. doi: 10.1007/s10493-008-9226-y. [DOI] [PubMed] [Google Scholar]
- 64.Khalighi M, et al. Molecular analysis of cyenopyrafen resistance in the two-spotted spider mite Tetranychus urticae. Pest Manag. Sci. 2015;72:103–112. doi: 10.1002/ps.4071. [DOI] [PubMed] [Google Scholar]
- 65.Van Leeuwen T, Tirry L, Nauen R. Complete maternal inheritance of bifenazate resistance in Tetranychus urticae Koch (Acari: Tetranychidae) and its implications in mode of action considerations. Insect Biochem. Mol. Biol. 2006;36:869–877. doi: 10.1016/j.ibmb.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 66.Wang C, et al. The concordance between RNA-seq and microarray data depends on chemical treatment and transcript abundance. Nat. Biotechnol. 2014;32:926–932. doi: 10.1038/nbt.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saier, M. H., Reddy, V. S., Tamang, D. G. & Västermark, Å. The transporter classification database. Nucleic Acids Res. 42 (2014). [DOI] [PMC free article] [PubMed]
- 68.Qadir M, et al. Cyclosporin A is a broad-spectrum multidrug resistance modulator. Clin. Cancer Res. 2005;11:2320–2326. doi: 10.1158/1078-0432.CCR-04-1725. [DOI] [PubMed] [Google Scholar]
- 69.de Nadal E, Ammerer G, Posas F. Controlling gene expression in response to stress. Nat. Rev. Genet. 2011;12:833–45. doi: 10.1038/nrg3055. [DOI] [PubMed] [Google Scholar]
- 70.IRAC. IRAC Mode of Action Classification Scheme. at <www.irac-online.org> (2017).
- 71.Chou C-C, Chen C-H, Lee T-T, Peck K. Optimization of probe length and the number of probes per gene for optimal microarray analysis of gene expression. Nucleic Acids Res. 2004;32:e99. doi: 10.1093/nar/gnh099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stetler RA, et al. Heat shock proteins: Cellular and molecular mechanisms in the central nervous system. Progress in Neurobiology. 2010;92:184–211. doi: 10.1016/j.pneurobio.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Santoro MG. Heat shock factors and the control of the stress response. Biochem. Pharmacol. 2000;59:55–63. doi: 10.1016/S0006-2952(99)00299-3. [DOI] [PubMed] [Google Scholar]
- 74.Takeda K, Noguchi T, Naguro I, Ichijo H. Apoptosis signal-regulating kinase 1 in stress and immune response. Annu. Rev. Pharmacol. Toxicol. 2008;48:199–225. doi: 10.1146/annurev.pharmtox.48.113006.094606. [DOI] [PubMed] [Google Scholar]
- 75.Vermetten E, Bremner JD. Circuits and systems in stress. I. Preclinical studies. Depress. Anxiety. 2002;15:126–147. doi: 10.1002/da.10016. [DOI] [PubMed] [Google Scholar]
- 76.David J-P, et al. Transcriptome response to pollutants and insecticides in the dengue vector Aedes aegypti using next-generation sequencing technology. BMC Genomics. 2010;11:216. doi: 10.1186/1471-2164-11-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Misra JR, Horner MA, Lam G, Thummel CS. Transcriptional regulation of xenobiotic detoxification in. Drosophila. Genes Dev. 2011;25:1796–1806. doi: 10.1101/gad.17280911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Phuong, C. T. N. Genome annotation and evolution of chemosensory receptors in spider mites Cao Thi Ngoc Phuong. (Ghent university, 2014).
- 79.Watanabe T, et al. Comparison of the induction profile of hepatic drug-metabolizing enzymes between piperonyl butoxide and phenobarbital in rats. J. Toxicol. Pathol. 1998;11:1–10. doi: 10.1293/tox.11.1. [DOI] [Google Scholar]
- 80.Skrinjaric-Spoljar M, Matthews HB, Engel JL, Casida JE. Response of hepatic microsomal mixed-function oxidases to various types of insecticide chemical synergists administered to mice. Biochem. Pharmacol. 1971;20:1607–1618. doi: 10.1016/0006-2952(71)90289-9. [DOI] [PubMed] [Google Scholar]
- 81.Chan HHA, Wajidi MFadzilF, Zairi J. Molecular cloning and xenobiotic induction of seven novel cytochrome P450 monooxygenases in Aedes albopictus. J. Insect Sci. 2014;14:163. doi: 10.1093/jisesa/ieu025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ellinger-Ziegelbauer H, Stuart B, Wahle B, Bomann W, Ahr HJ. Comparison of the expression profiles induced by genotoxic and nongenotoxic carcinogens in rat liver. Mutat. Res. Mol. Mech. Mutagen. 2005;575:61–84. doi: 10.1016/j.mrfmmm.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 83.Poupardin R, et al. Cross-induction of detoxification genes by environmental xenobiotics and insecticides in the mosquito Aedes aegypti: Impact on larval tolerance to chemical insecticides. Insect Biochem. Mol. Biol. 2008;38:540–551. doi: 10.1016/j.ibmb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 84.Poupardin R, Riaz MA, Vontas J, David JP, Reynaud S. Transcription profiling of eleven cytochrome p450s potentially involved in xenobiotic metabolism in the mosquito aedes aegypti. Insect Mol. Biol. 2010;19:185–193. doi: 10.1111/j.1365-2583.2009.00967.x. [DOI] [PubMed] [Google Scholar]
- 85.Bautista MAM, Tanaka T, Miyata T. Identification of permethrin-inducible cytochrome P450s from the diamondback moth, Plutella xylostella (L.) and the possibility of involvement in permethrin resistance. Pestic. Biochem. Physiol. 2007;87:85–93. doi: 10.1016/j.pestbp.2006.06.004. [DOI] [Google Scholar]
- 86.Willoughby L, et al. A comparison of Drosophila melanogaster detoxification gene induction responses for six insecticides, caffeine and phenobarbital. Insect Biochem. Mol. Biol. 2006;36:934–942. doi: 10.1016/j.ibmb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 87.Giraudo M, Unnithan GC, Le Goff G, Feyereisen R. Regulation of cytochrome P450 expression in Drosophila: Genomic insights. Pestic. Biochem. Physiol. 2010;97:115–122. doi: 10.1016/j.pestbp.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Van Pottelberge S, Van Leeuwen T, Van Amermaet K, Tirry L. Induction of cytochrome P450 monooxygenase activity in the two-spotted spider mite Tetranychus urticae and its influence on acaricide toxicity. Pestic. Biochem. Physiol. 2008;91:128–133. doi: 10.1016/j.pestbp.2008.03.005. [DOI] [Google Scholar]
- 89.Riga M, et al. Functional characterization of the Tetranychus urticae CYP392A11, a cytochrome P450 that hydroxylates the METI acaricides cyenopyrafen and fenpyroximate. Insect Biochem. Mol. Biol. 2015;65:91–99. doi: 10.1016/j.ibmb.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 90.Pavlidi N, et al. Functional characterization of glutathione S-transferases associated with insecticide resistance in Tetranychus urticae. Pestic. Biochem. Physiol. 2015;121:53–60. doi: 10.1016/j.pestbp.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 91.Freeman ML, Huntley SA, Meredith MJ, Senisterra GA, Lepock J. Destabilization and denaturation of cellular protein by glutathione depletion. Cell Stress Chaperones. 1997;2:191–8. doi: 10.1379/1466-1268(1997)002<0191:DADOCP>2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Casey W, et al. Transcriptional and physiological responses of HepG2 cells exposed to diethyl maleate: time course analysis. Physiol. Genomics. 2002;8:115–122. doi: 10.1152/physiolgenomics.00064.2001. [DOI] [PubMed] [Google Scholar]
- 93.Pao SS, Paulsen IT, Saier MH. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dermauw W, et al. A burst of ABC genes in the genome of the polyphagous spider mite Tetranychus urticae. BMC Genomics. 2013;14:317. doi: 10.1186/1471-2164-14-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.de la Paz Celorio-Mancera M, et al. Mechanisms of macroevolution: polyphagous plasticity in butterfly larvae revealed by RNA-Seq. Mol. Ecol. 2013;22:4884–4895. doi: 10.1111/mec.12440. [DOI] [PubMed] [Google Scholar]
- 96.Dresner, L. S., Andersen, D. K., Kahng, K. U., Munshi, I. A. & Wait, R. B. Effects of cyclosporine on glucose metabolism. Surgery106, 163–9, discussion 170 (1989). [PubMed]
- 97.Christians U, et al. Alterations in glucose metabolism by cyclosporine in rat brain slices link to oxidative stress: interactions with mTOR inhibitors. Br. J. Pharmacol. 2004;143:388–396. doi: 10.1038/sj.bjp.0705939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Reimer RJ. SLC17: A functionally diverse family of organic anion transporters. Mol. Aspects Med. 2013;34:350–359. doi: 10.1016/j.mam.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu J, et al. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-H. [DOI] [PubMed] [Google Scholar]
- 100.Andrews, S. FastQC: A quality control tool for high throughput sequence data. (2010).
- 101.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Anders S, Pyl PT, Huber W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Love MI, Anders S, Kim V, Huber W. RNA-Seq workflow: gene-level exploratory analysis and differential expression. F1000Research. 2015;4:1070. doi: 10.12688/f1000research.7035.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Anders S, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 2013;8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 106.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 107.Conesa A, et al. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 108.Young, M. D., Wakefield, M. J. & Smyth, G. K. goseq: Gene ontology testing for RNA-seq datasets reading data. Gene 1–21 (2010).
- 109.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ahn SJ, Dermauw W, Wybouw N, Heckel DG, Van Leeuwen T. Bacterial origin of a diverse family of UDP-glycosyltransferase genes in the Tetranychus urticae genome. Insect Biochem. Mol. Biol. 2014;50:43–57. doi: 10.1016/j.ibmb.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 111.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 112.Hellemans J, Mortier G, De PA, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq expression data generated during the current study are available in the Gene-Expression Omnibus (GEO) repository with accession number GSE98293.