Abstract
Chondrocyte hypertrophy makes important contributions to bone development and growth. We have investigated a number of novel cartilage genes identified in a recent transcriptomic study to determine whether they are differentially expressed between different zones of equine foetal growth cartilage. Twelve genes (ATP6V0D2, BAK1, DDX5, GNB1, PIP4K2A, RAP1B, RPS7, SRSF3, SUB1, TMSB4, TPI1 and WSB2) were found to be more highly expressed in the zone of hypertrophic chondrocytes than in the reserve or proliferative zones, whereas FOXA3 and SERPINA1 were expressed at lower levels in the hypertrophic zone than in the reserve zone. ATP6V0D2, which encodes vacuolar H+ ATPase (V-ATPase) V0 subunit d2 (ATP6V0D2), was selected for further study. Immunohistochemical analysis of ATP6V0D2 in growth cartilage showed stronger staining in hypertrophic than in reserve zone or proliferative chondrocytes. Expression of ATP6V0D2 mRNA and protein was up-regulated in the mouse chondrocytic ATDC5 cell line by conditions inducing expression of hypertrophy-associated genes including Col10a1 and Mmp13 (differentiation medium). In ATDC5 cells cultured in control medium, knockdown of Atp6v0d2 or inhibition of V-ATPase activity using bafilomycin A1 caused a decrease in Col2a1 expression, and in cells cultured in differentiation medium the two treatments caused a decrease in nuclear area. Inhibition of V-ATPase, but not Atp6v0d2 knockdown, prevented the upregulation of Col10a1, Mmp13 and Vegf by differentiation medium, while Atp6v0d2 knockdown, but not inhibition of V-ATPase, caused an increase in the number of ATDC5 cells cultured in differentiation medium. These observations identify ATP6V0D2 as a novel chondrocyte hypertrophy-associated gene. The results are consistent with roles for V-ATPase, both ATP6V0D2-dependent and -independent, in supporting chondrocyte differentiation and hypertrophy.
Keywords: Chondrocyte, Hypertrophy, Endochondral ossification, ATP6V0D2, Vacuolar H+-ATPase
Abbreviations: ABH, alcian blue/haematoxylin/eosin/acid fuchsin stain; ATP6V0D2, vacuolar H+ ATPase V0 subunit d2; DAPI, 4′,6-diamidino-2-phenylindole; DMEM, Dulbecco's modified Eagle's medium; FCS, foetal calf serum; MMP-13, matrix metalloproteinase-13; MNE, mean normalised expression; PBS, phosphate-buffered saline; qPCR, quantitative polymerase chain reaction; V-ATPase, vacuolar H+ ATPase
Highlights
-
•
Twelve novel chondrocyte hypertrophy-associated genes have been identified.
-
•
One of these, Atp6v0d2, supports differentiation of chondrocytic ATDC5 cells.
-
•
V-ATPase activity contributes to expression of hypertrophy-associated genes.
-
•
The V-ATPase V0 subunit d2 is not required for this up-regulation.
1. Introduction
Most bones develop through the process of endochondral ossification, which involves the initial formation of a cartilage anlage and its replacement by bone tissue (Mackie et al., 2011). As part of this process, chondrocytes proliferate, secrete cartilage matrix and undergo hypertrophy. Chondrocyte hypertrophy plays critical roles in bone elongation and in preparation of the cartilage matrix for invasion by the complex mixture of cells that comprise the ossification front.
The orderly progression of chondrocyte proliferation, matrix secretion and hypertrophy in growth cartilage is regulated by the coordinated actions of a variety of systemic and locally secreted factors, acting through stage-specific transcription factors. Thyroid hormones are important circulating inducers of hypertrophy, and their actions appear to be mediated by pathways including the WNT/β-catenin, insulin-like growth factor 1 and fibroblast growth factor receptor 3-mediated pathways (Mackie et al., 2011, Shao et al., 2006). The transcription factor RUNX2 induces expression of hypertrophy-associated genes including that encoding collagen type X (COL10A1), and stimulates hypertrophy (Lefebvre and Smits, 2005).
While much has been learnt in recent years about regulation of chondrocyte hypertrophy, there are still many aspects of this process that remain unclear. For example, while remodelling of the extracellular matrix is clearly required to accommodate the swelling of individual cells, no matrix-degrading enzyme has yet been identified as being critical for this process (see discussion in Mackie et al. (Mackie et al., 2011)).
Chondrocytes undergoing hypertrophy not only express molecules that contribute to their own morphological and functional changes, but also secrete factors that regulate behaviour of cells in the invading ossification front. For example, hypertrophic chondrocytes promote vascular invasion of the growth plate through expression of vascular endothelial growth factor (Zelzer et al., 2001, Zelzer et al., 2004), and osteoclast differentiation through expression of receptor activator of NFκB ligand (Usui et al., 2008).
In a recent unbiased transcriptomic study of cartilage from lesions of equine osteochondrosis (a developmental orthopaedic disease), we identified a number of genes that had not previously been described in cartilage (Mirams et al., 2016). The current study was undertaken to determine whether expression of any of these genes is regulated in association with chondrocyte hypertrophy, in order to shed light on molecular mechanisms of this process. These studies made use of equine foetal growth cartilage and ATDC5 cells, a mouse chondrocyte line that can be induced to express hypertrophy-associated genes including those encoding collagen type X and matrix metalloproteinase-13 (MMP13) (Shukunami et al., 1997, Wang et al., 2004). Following the identification of several novel hypertrophy-associated genes, one of these, ATP6V0D2, was selected for further study.
2. Materials and methods
2.1. Tissue samples
Samples were collected from the distal end of the right third metatarsal bones of 22 foetal horses obtained from pregnant mares killed at a Melbourne knackery. Gestational age of foetuses was estimated using the crown-rump length method and ranged from 120 to 300 days. Foetuses with gross skeletal deformities were excluded. The specimens included some collected before the secondary ossification centre was formed in the distal epiphysis (designated as early samples; n = 8), and some collected after formation of the secondary ossification centre (late samples; n = 14). The distal end of the bone (4 cm in length) was excised and bisected longitudinally; one piece was used for histology and the other for gene expression studies. All samples included part of the primary ossification centre. The collection of these samples met the requirements of the University of Melbourne Animal Ethics Committee.
2.2. Histology
At the time of collection, samples were fixed in 4% w/v paraformaldehyde in phosphate-buffered saline (PBS), pH 7.4 at 4 °C overnight, then rinsed and demineralised using 0.33 M ethylenediaminetetraacetic acid, pH 7.4. Demineralised specimens were incubated in 25% sucrose in PBS overnight then trimmed to remove all except the growth plate and a small amount of primary ossification centre and secondary ossification centre, if present; for specimens lacking a secondary ossification centre, the region remaining after trimming extended from the primary ossification centre to the articular surface. Specimens were then embedded in Tissue-Tek® OCT compound (ThermoFisher Scientific, Waltham USA). Frozen sections (10 μm) were cut using a cryostat (Leica Microsystems, Germany). Some sections were stained with alcian blue and counterstained with haematoxylin, eosin and acid fuchsin (ABH) using standard procedures; other sections were stained by immunohistochemistry.
2.3. Immunohistochemistry
Sections to be stained for the presence of collagen type X were first treated with bovine testicular hyaluronidase (1000 U/ml in 0.15 M sodium chloride and 20 mM sodium acetate, pH 5.0). All sections were permeabilised using 0.1% v/v Triton-X100 in PBS before being blocked for one hour using either 10% v/v foetal calf serum (FCS; for anti-collagen type X) or 2.5% FCS (for anti-ATP6V0D2) in PBS. Sections were incubated overnight at 4 °C with rabbit anti-human collagen type X (1:100 in 5% FCS in PBS; Abcam, Cambridge, UK), rabbit anti-human ATP6V0D2 (0.02 μg/μl in PBS containing 0.5% Triton X100 and 1% w/v bovine serum albumin; Abcam) or normal rabbit serum (control for anti-collagen type X) or purified rabbit Ig (control for anti ATP6V0D2) diluted appropriately. Sections were rinsed with PBS prior to incubation with AlexaFluor 488-conjugated goat anti-rabbit immunoglobulin (1:300 in PBS; ThermoFisher Scientific). After being washed, sections were mounted in gelvatol containing 4′,6-diamidino-2-phenylindole (DAPI; 1 μg/ml), then examined by standard fluorescence microscopy.
2.4. Microdissection of growth cartilage
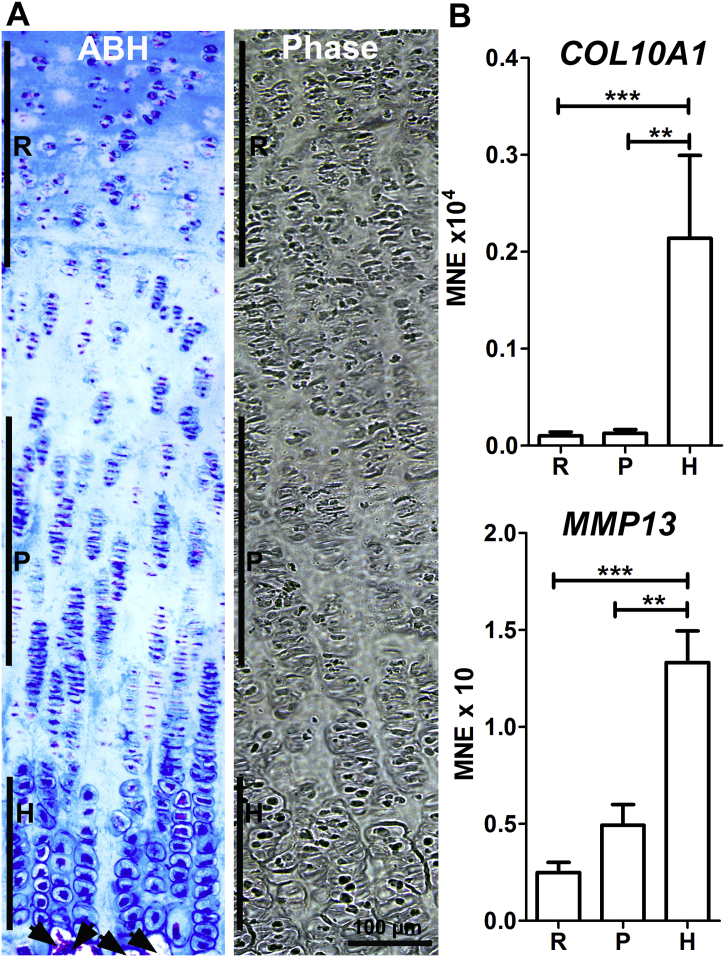
Specimens harvested for analysis of gene expression were wrapped in aluminium foil, frozen in liquid nitrogen and stored at − 80 °C. Specimens were trimmed to remove any bone and embedded frozen in OCT compound; sections (15 μm) were collected on glass slides then held on dry ice until they were microdissected. The region dissected was the growth cartilage giving rise to the primary centre of ossification, whether or not a secondary ossification centre was present. Under an inverted phase contrast microscope, the reserve, proliferative and hypertrophic zones were identified by reference to morphological features observed in ABH-stained sections (Fig. 1A), and the hypertrophic zone in collagen type X-stained sections. For each section, tissue from each of the reserve, proliferative and hypertrophic zones was collected using a scalpel and placed in separate microcentrifuge tubes. For each sample, the tissue for each zone was pooled from 20 to 30 sections.
Fig. 1.
Microdissection of equine foetal growth cartilage.
A: Representative sections of growth cartilage collected from horse foetuses. ABH staining (left) and phase contrast image (right) show growth cartilage zones: reserve zone (R), zone of proliferative chondrocytes (P), zone of hypertrophic chondrocytes (H) and ossification front (arrowheads in alcian blue staining). The magnification is the same for both figure parts. B: Mean normalised expression of known hypertrophy-associated genes (COL10A1 and MMP13) was used to verify the microdissection of the hypertrophic zone; **P < 0.01, ***P < 0.001 for comparisons between zones indicated by lines; Kruskal-Wallis test and Dunn's post test (n = 22). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.5. Cell culture and immunocytochemistry
Cells of the ATDC5 mouse chondrocyte cell line (kindly provided by Prof. A. Fosang, Murdoch Childrens Research Institute, Melbourne) were maintained in Dulbecco's modified Eagle's medium (DMEM) and Ham's F12 nutrient mix (1:1; DMEM/F12; ‘control medium’). For induction of hypertrophy-associated gene expression, the medium of confluent cells was changed to differentiation medium consisting of α minimum essential medium containing insulin-transferrin‑selenium (1% w/v), ascorbate-2-phosphate (100 μg/ml) and triiodothyronine (100 nM), in which cells were cultured for a further 4 days. This medium was selected as a combination of factors used in previous studies (Siebler et al., 2002, Temu et al., 2010), following optimisation experiments (data not shown). Some cultures (in control medium or in differentiation medium) were treated with bafilomycin A1 (1 nM; Sigma Aldrich, St Louis, USA) over this 4-day period.
For immunocytochemistry, cells on glass coverslips were fixed with 4% w/v paraformaldehyde in PBS for 5 min, then rinsed, blocked, stained with anti-collagen type X or anti-ATP6V0D2, and mounted in gelvatol containing DAPI as described above for tissue sections. In some cases, cells on glass coverslips were incubated with Lysotracker® Red DND-99 (1:20,000; ThermoFisher Scientific) or wheat germ agglutinin, AlexaFluor® 488 conjugate (1:2000; ThermoFisher Scientific) for 30 min before fixation (as described above) or incubated with AlexaFluor® 488/568 phalloidin (1:300; ThermoFisher Scientific; 30 min) after fixation. For cell counting and morphometry, coverslips were examined by standard fluorescence microscopy. Images were taken of 8 fields per coverslip, and cell number, nuclear area and cell area were determined using Image Pro software (Media Cybernetics, Rockville, USA); nuclear area was assessed as the area of DAPI staining, and to measure cell area phalloidin-stained cells were outlined manually. Mitotic figures (visualised with DAPI staining) were counted and expressed as a percentage of total nuclei. Some coverslips were examined by confocal microscopy using a Zeiss LSM 510 Meta microscope (Carl Zeiss, Germany).
2.6. Analysis of gene expression
Microdissected tissues were homogenised in Tri-Reagent (Sigma Aldrich) and total RNA prepared, then further purified using the SV total RNA isolation kit (Promega, Madison, USA) and eluted in nuclease-free water, according to the manufacturers' instructions. Complementary DNA was synthesised using 0.5 μg purified RNA with the VILO SuperScript II reverse transcriptase system (ThermoFisher Scientifc) then diluted 1:5 in nuclease-free water. ATDC5 cell monolayers were washed in cold PBS, then RNA was extracted using the SV total RNA isolation kit and used to synthesise cDNA using the GoScript reverse transcription system (Promega) according to the manufacturer's instructions.
Quantitative polymerase chain reaction (qPCR) was performed using an MX3000P qPCR machine (Stratagene, La Jolla, USA) and Sybr Green chemistry (ThermoFisher Scientific). Oligonucleotide primers as described (Mirams et al., 2016, Ahmed et al., 2007a, Ahmed et al., 2007b) or designed using Primer 3 (Table 1) were synthesised by Geneworks (Thebarton, Australia). Data are presented as mean normalised expression (MNE) (Simon, 2003) whereby RPS23 is the gene used for normalisation.
Table 1.
Oligonucleotide primers.
| Gene | Forward | Reverse |
|---|---|---|
| Horse1 | ||
| FREM1 | GTCCTTTTGGATAGGCGATG | CCAAAACACAGTTCTTTCCAGA |
| H3F3B | CTGACTTGAGGTTTCAGAGTGC | GGGCATGATGGTGACCTCT |
| SUB1 | AGCAGCAGCAGAGATGACAA | ATTTCACCTTCCGGATCCAT |
| THAP5 | GACGTCAGATGGGGTATTCG | TGGCTTTTAGGCACACTTCTT |
| Mouse | ||
| Atp6v0d2 | AAGCCTTTGTTTGACGCTGT | ATGCCAGCACATTCATCTGT |
| Col10a1 | TTCCCTGGATCTAAGGGTGA | CTCTGTCCGCTCTTTGTGAA |
| Col2a1 | AGAACTGGTGGAGCAGCAAG | CGGAGGAAAGTCATCTGGAC |
| Ddx5 | TCATCGAATTGGAAGAACTGC | CCCGAAGCACAGAGATAAGG |
| Foxa3 | TCAGTGAAGATGGAGGCTCA | GGCACAGGATTCACTGGAGA |
| Mmp13 | ACATCCATCCCGTGACCTTA | GCGCTCAGTCTCTTCACCTC |
| RpS23 | ACCAGAAGTGGCATGACAAAC | GATGGCAGAATTTGGCTGTT |
| Tmsb4 | ACAAACCCGATATGGCTGAG | GCCAGCTTGCTTCTCTTGTT |
| Tpi1 | TCTGTGACTGGAGCAACCTG | TTGGCATTGATGATGTCCAC |
| Vegf | ATCTTCAAGCCGTCCTGT | GCATTCACAATCTGCTGTG |
1: These four genes were identified in the subtractive hybridisation study described by Mirams et al. (2016), but unlike the other genes investigated in the current study, were not further analysed by qPCR in the earlier study.
2.7. Knockdown of Atp6v0d2 in ATDC5 cells
Cells were cultured in maintenance medium until 70% confluent, then transfected with either 20 nM Silencer Select siAtp6v0d2 and 20 nM Silencer siAtp6v0d2 siRNA duplexes or 40 nM scrambled (negative control) siRNA duplexes (ThermoFisher Scientific) using Lipofectamine 2000 (1 μl/well; ThermoFisher Scientific) in maintenance medium for 48 h. Medium was then replaced with fresh maintenance or differentiation medium and cells were cultured for a further 4 days before being processed for RNA extraction, immunocytochemistry, cell counting or morphometry.
2.8. Statistical analysis
Data are expressed as mean ± standard error of the mean. Quantitative PCR data involving ≤ 20 samples/group were analysed for significant differences by a pairwise fixed reallocation randomisation test, using REST-384 software (Pfaffl et al., 2002). For qPCR data sets containing > 20 samples/group, data were analysed by Kruskal-Wallis test and Dunn's post test using GraphPad Prism software. For cell morphometry, data were analysed by Student's t-test using GraphPad Prism.
3. Results
3.1. Verification of separation of zones by microdissection
Equine growth cartilages were dissected microscopically for the collection of different chondrocyte zones (Fig. 1A). In order to confirm that the zone of hypertrophic chondrocytes was effectively separated from the proliferative and reserve zones, the expression of two hypertrophy-associated genes, COL10A1 and MMP13, was determined by qPCR. It was observed that both of these genes were significantly more highly expressed in the region dissected as hypertrophic zone than in the reserve and proliferative zones (Fig. 1B).
3.2. In vivo expression of genes of interest
We recently published the results of a transcriptomic study of equine osteochondrosis, which identified a number of genes that had not previously been described or were poorly characterised in cartilage (Mirams et al., 2016). For the current study, a subset of 27 of these genes was selected (Table 2) as representing a variety of functions of potential relevance to chondrocyte hypertrophy, including gene transcription and extracellular matrix turnover.
Table 2.
Genes investigated for differential expression in equine growth cartilage.
| Gene1 | Protein | Regulation2 |
|---|---|---|
| ANKRA2 | Ankyrin repeat family A2 | No |
| ATP6V0D2 | Vacuolar H+ ATPase subunit V0 D2 | + |
| BAK 1 | BCL2-antagonist/killer 1 | + |
| CDK11A | Cyclin-dependent kinase 11A | No |
| DDX5 | DEAD (Asp-Glu-Ala-Asp) | + |
| EEF1G | Eukaryotic Translation elongation factor I Gamma | No |
| FOXA3 | Forkhead Box A3 | − |
| FREM1 | Fras1 related ECM protein 1 | No |
| GJA1 | Gap Junction Protein, Alpha1 | No |
| GNB1 | Guanine nucleotide binding protein beta polypeptide 1 | + |
| H3F3B | Histone 3.3B | No |
| MFAP2 | Microfibrillar Associated Protein 2 | No |
| MRRF | Mitochondrial Ribosome Recycling Factor | No |
| NACA | Nascent Polypeptide Associated Complex Alpha Subunit | No |
| PIP4K2A | Phosphatidylinositol-5-phosphate 4- kinase type II alpha | + |
| PURB | Purine Rich Element Binding Protein | No |
| RAP1B | Ras oncogene family member | + |
| RERE | Arginine-glutamic acid dipeptide (RE) repeats | No |
| RPS7 | Ribosomal Protein S7 | + |
| SERPINA1 | Serpin peptidase inhibitor, clade A member 1 | − |
| SRSF3 | Splicing factor, arginine/serine-rich3 | + |
| SSR4 | Signal Sequence Receptor-Delta | No |
| SUB1 | SUB1 homolog | + |
| THAP5 | Thap domain containing 5 | No |
| TMSB4 | Thymosin β4 | + |
| TPI1 | Triose phosphate isomerase 1 | + |
| WSB2 | WD Repeat and socs box-containing 2 | + |
1: Genes selected as novel or poorly characterised cartilage genes identified in a previous subtractive hybridisation study (Mirams et al., 2016).
2: ‘no’ - not differentially expressed between zones of growth cartilage; ‘+’ - more highly expressed in zone of hypertrophic chondrocytes than in other zones; ‘-‘ - less highly expressed in zone of hypertrophic chondrocytes than in other zones.
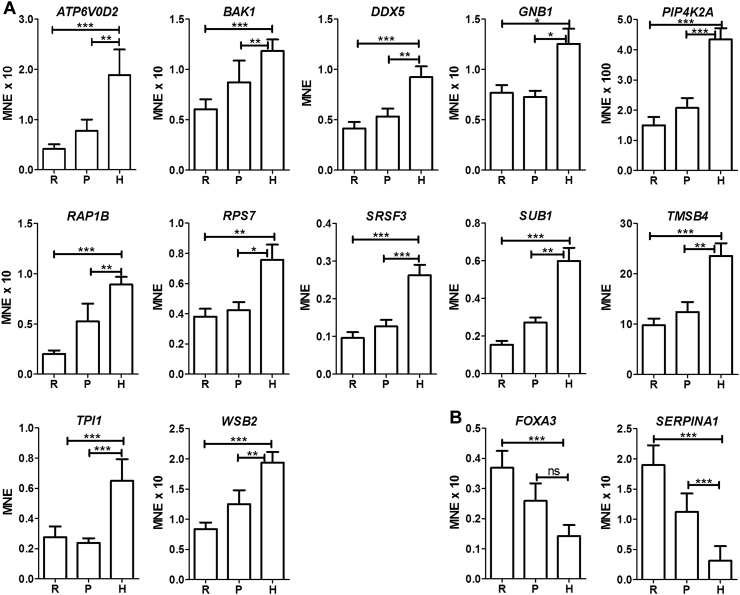
Expression of 12 of the selected genes was found to be significantly higher in the hypertrophic zone than in the reserve or proliferative zones of equine foetal growth cartilage when all 22 samples were analysed together (Table 2, Fig. 2A). These genes were ATP6V0D2, BAK1, DDX5, GNB1, PIP4K2A, RAP1B, RPS7, SRSF3, SUB1, TMSB4, TPI1 and WSB2. Expression of two genes, FOXA3 and SERPINA1, was significantly lower in the hypertrophic zone than in the reserve zone of all samples (Fig. 2B). When early and late specimens (based on the absence or presence of a secondary centre of ossification) were analysed separately, for four of the genes, some but not all of the comparisons were found to be significant (Table 3).
Fig. 2.
Expression of genes of interest in equine foetal growth cartilage.
Mean normalised expression of genes differentially expressed in the hypertrophic zone (H) compared to the reserve (R) and proliferative (P) zones of growth cartilage. *P < 0.05, **P < 0.01, ***P < 0.001, ns - no significant difference, for comparisons between zones indicated by lines; Kruskal-Wallis test and Dunn's post test (n = 22).
Table 3.
Comparative gene expression between zones of growth cartilage: separate P values for early and late specimens.
| Early (n = 8) |
Late (n = 14) |
|||
|---|---|---|---|---|
| Gene | H vs R1 | H vs P1 | H vs R | H vs P |
| Genes up-regulated in the hypertrophic zone: | ||||
| ATP6V0D2 | P = 0.004 | P = 0.045 | P = 0.002 | P = 0.008 |
| BAK1 | P = 0.004 | ns2 | P = 0.005 | P = 0.004 |
| DDX5 | P = 0.006 | P = 0.045 | P = 0.001 | P = 0.001 |
| GNB1 | P = 0.045 | P = 0.017 | P = 0.03 | P = 0.018 |
| PIP4K2A | P = 0.001 | P = 0.009 | P = 0.001 | P = 0.001 |
| RAP1B | P = 0.001 | ns | P = 0.001 | P = 0.001 |
| RPS7 | P = 0.005 | ns | P = 0.014 | P = 0.035 |
| SRSF3 | P = 0.001 | P = 0.013 | P = 0.001 | P = 0.001 |
| SUB1 | P = 0.001 | P = 0.038 | P = 0.001 | P = 0.001 |
| TMSB4 | P = 0.001 | P = 0.012 | P = 0.004 | P = 0.007 |
| TPI1 | P = 0.001 | P = 0.006 | P = 0.003 | P = 0.001 |
| WSB2 | P = 0.001 | P = 0.021 | P = 0.001 | P = 0.024 |
| Genes down-regulated in the hypertrophic zone: | ||||
| FOXA3 | P = 0.001 | ns | P = 0.049 | ns |
| SERPINA1 | P = 0.001 | P = 0.002 | P = 0.001 | P = 0.001 |
1: R - reserve zone; P - zone of proliferative chondrocytes; H - zone of hypertrophic chondrocytes.
2: ns - not significantly different.
The remaining thirteen genes presented in Table 2 were not differentially expressed between zones of growth cartilage, whether specimens were analysed as a whole or as separate early and late groups.
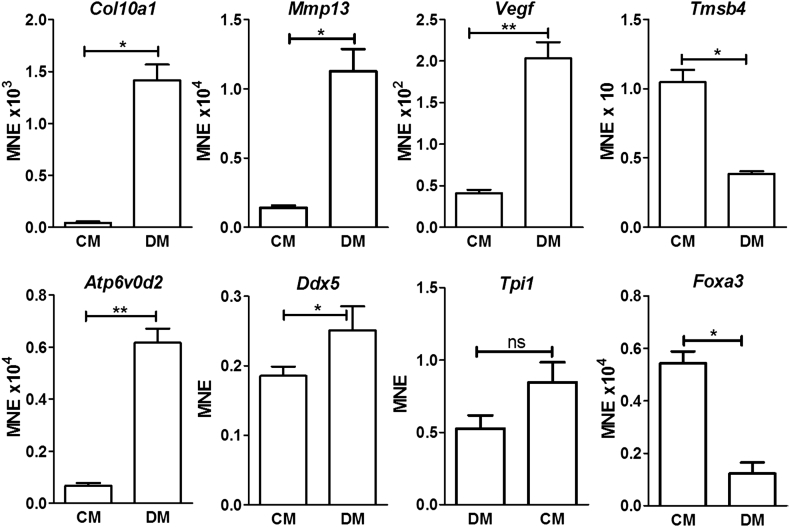
3.3. In vitro expression of genes of interest
A subset of five of the 14 regulated genes (Atp6v0d2, Ddx5, Tmsb4, Tpi1 and Foxa3) were selected for further study using ATDC5 cells. Expression of these genes was investigated in cells cultured in control medium or differentiation medium for 4 days (Fig. 3). Differentiation medium caused the expected up-regulation of known hypertrophy-associated genes (Col10a1, Mmp13 and Vegf). It also stimulated expression of Atp6v0d2 and Ddx5, and down-regulated Foxa3, as predicted on the basis of the in vivo expression studies. Contrary to expectations, however, Tmsb4 expression was down-regulated by treatment with differentiation medium; moreover, Tpi1 was not significantly up-regulated.
Fig. 3.
Expression of genes of interest in ATDC5 cells.
Mean normalised expression of established and novel chondrocyte hypertrophy-regulated genes in ATDC5 cells cultured in control medium (CM) or differentiation medium (DM). *P < 0.05, **P < 0.01, ns - no significant difference, for comparisons between treatments; pairwise fixed reallocation randomisation test (n = 4).
3.4. Expression of vacuolar H+ ATPase V0 subunit d2
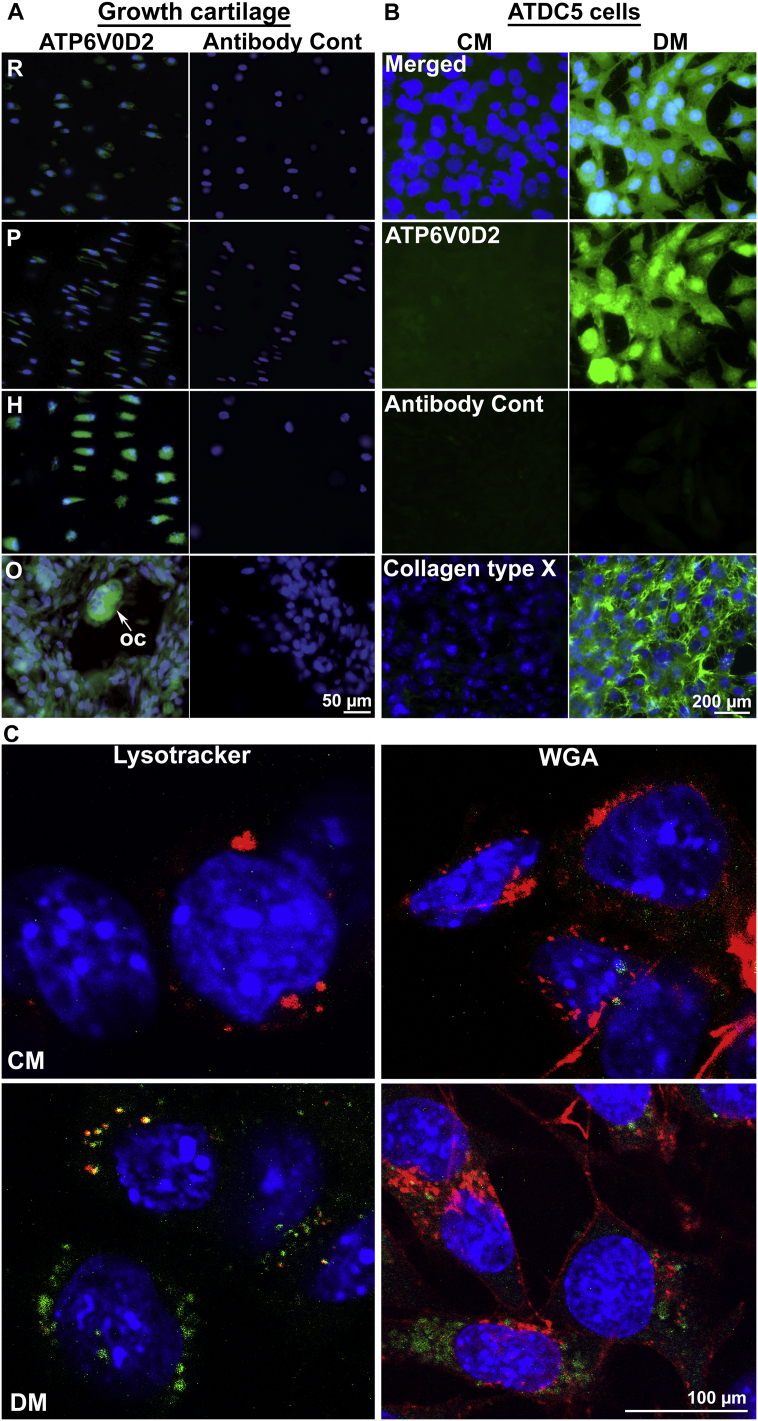
Atp6v0d2 was selected for further study. Expression of the protein encoded by this gene was investigated in equine growth cartilage using immunohistochemistry (Fig. 4A). ATP6V0D2 staining was stronger in hypertrophic chondrocytes than in reserve zone or proliferative chondrocytes. As expected, staining was also intense in osteoclasts (observed at the ossification front), which are known to express ATP6V0D2.
Fig. 4.
Expression of ATP6V0D2 in growth cartilage and ATDC5 cells.
A: Sections of an equine foetal bone stained with anti-ATP6V0D2 or rabbit Ig (Antibody Cont) and counterstained with DAPI; growth cartilage including the reserve zone (R), zone of proliferative chondrocytes (P), zone of hypertrophic chondrocytes (H) and ossification front (O); oc - osteoclast. B: ATDC5 cells cultured in control medium (CM) or differentiation medium (DM), stained with anti-ATP6V0D2, rabbit Ig or anti-collagen type X and counterstained with DAPI. C: Confocal microscopy images of ATDC5 cells cultured in control or differentiation medium, stained with anti-ATP6V0D2 (green fluorescence) and Lysotracker® (red fluorescence) or wheat germ agglutinin (red fluorescence) and counterstained with DAPI. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Expression of ATP6V0D2 was investigated in ATDC5 cells using immunocytochemistry (Fig. 4B). Staining for collagen type X was absent from cells cultured in control medium, but present in cells cultured in differentiation medium. Staining for ATP6V0D2 was weak in control cells but intense in cells cultured in differentiation medium. Confocal microscopy was used to examine the subcellular localisation of ATP6V0D2 staining in ATDC5 cells treated under control or differentiation conditions (Fig. 4C). ATP6V0D2 staining was found to be co-localised with some but not all lysosomes (stained with Lysotracker); in some cells, the staining was closely adjacent to the nucleus. There was no co-localisation between ATP6V0D2 staining and wheat germ agglutinin binding, indicating that it was not present in the plasma membrane or in the Golgi apparatus. There was some ATP6V0D2 staining of discrete cytoplasmic structures that were not labelled by either of these markers.
3.5. Knockdown of Atp6v0d2 in ATDC5 cells
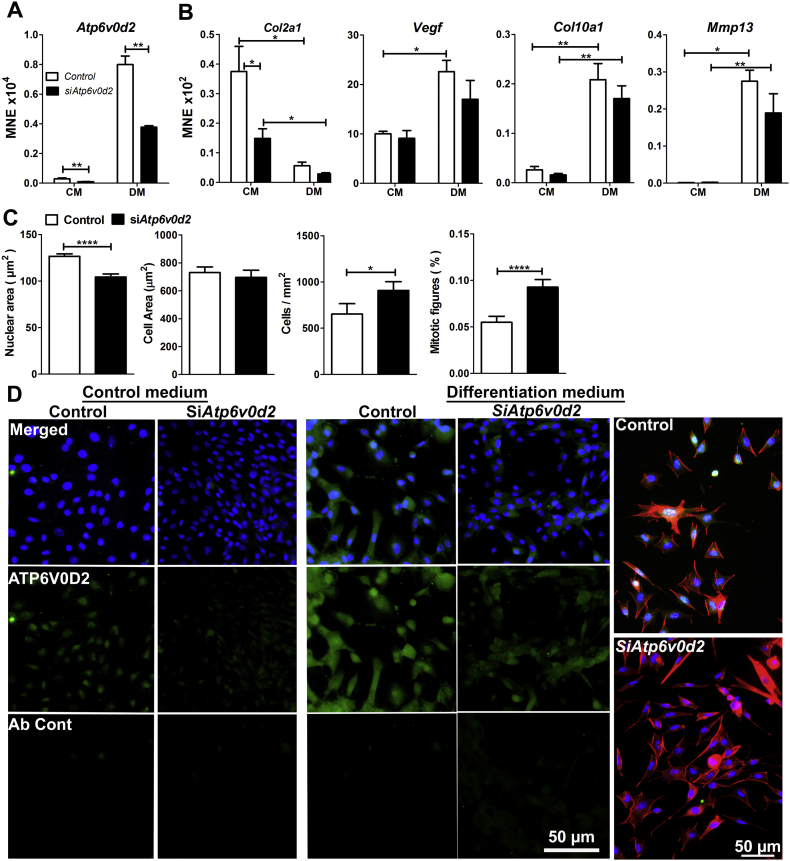
In order to identify possible roles of ATP6V0D2 in chondrocytes, ATDC5 cells were transfected with Atp6v0d2-targetted or control siRNA. The siAtp6v0d2 significantly down-regulated Atp6v0d2 mRNA expression in cells cultured in either control or differentiation medium (Fig. 5A), and immunostaining for ATP6V0D2 was weaker in siAtp6v0d2-transfected than in control siRNA-transfected cells cultured under both conditions (Fig. 5D).
Fig. 5.
Knockdown of Atp6v0d2 in ATDC5 cells.
A, B: Mean normalised expression of Atp6v0d2 (A) and Col2a1, Vegf, Col10a1, and Mmp13 (B) in ATDC5 cells cultured in control medium (CM) or differentiation medium (DM) and transfected with siAtp6v0d2 or control siRNA. C: Morphometry of ATDC5 cells cultured in differentiation medium and transfected with siAtp6v0d2 or control siRNA. D: ATDC5 cells cultured in control medium or differentiation medium, stained with anti-ATP6V0D2 (green fluorescence) or rabbit Ig (Ab Cont), or with phalloidin (red fluorescence) and counterstained with DAPI. *P < 0.05, **P < 0.01, ****P < 0.0001, for comparisons indicated by lines; pairwise fixed reallocation randomisation test (A, B) and Student's t-test (C); n = 4. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The cartilage-specific gene Col2a1 was down-regulated by siAtp6v0d2 in cells cultured in control medium, but not in differentiation medium (Fig. 5B). For the hypertrophy-associated genes Col10a1, Mmp13 and Vegf, there was no significant difference between values for siAtp6v0d2 and control siRNA.
The effect of Atp6v0d2 knockdown on morphology and number of ATDC5 cells cultured in differentiation medium was also assessed (Fig. 5C). Cell area was not affected by transfection with siAtp6v0d2, but nuclear area was significantly reduced. There were significantly more cells and mitotic figures in siAtp6v0d2-transfected than in control siRNA-transfected cultures.
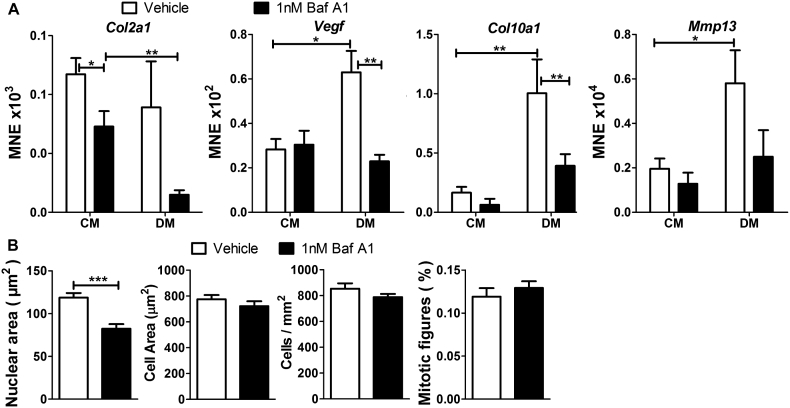
3.6. Inhibition of V-ATPase activity in ATDC5 cells
ATDC5 cells treated with the V-ATPase inhibitor bafilomycin A1 were assessed in the same assays as for siAtp6v0d2-transfected cells. Bafilomycin A1 caused a decrease in expression of Col2a1 in cells cultured in control medium, and prevented the up-regulation of Vegf, Col10a1 and Mmp13 by differentiation medium (Fig. 6A). Bafilomycin caused a significant decrease in nuclear area in ATDC5 cells cultured in differentiation medium, but did not affect cell number or the number of mitotic figures (Fig. 6B).
Fig. 6.
Inhibition of V-ATPase activity in ATDC5 cells.
A: Mean normalised expression of Col2a1, Vegf, Col10a1, and Mmp13 in ATDC5 cells cultured in control medium (CM) or differentiation medium (DM) and treated with bafilomycin A1 (Baf A1) or vehicle. B: Morphometry of ATDC5 cells cultured in differentiation medium and treated with bafilomycin A1 or vehicle. *P < 0.05, **P < 0.01, ***P < 0.001, for comparisons indicated by lines; pairwise fixed reallocation randomisation test (A) and Student's t-test (B); n = 6.
3.7. Other V-ATPase subunits
Since there was some difference between the effects of V-ATPase inhibition and Atp6v0d2 knockdown on ATDC5 cells, expression of two additional V-ATPase subunits was investigated in ATDC5 cells cultured in control and differentiation medium. Expression of Atp6v0d1, the gene encoding the alternative subunit d isoform, and Atp6v1a, the gene encoding the catalytic ATP binding site, was examined. Expression of these genes was detected in control medium and up-regulated by differentiation medium, although not to the same extent as Atp6v0d2 (Fig. 7). Atp6v0d2 knockdown had no effect on expression of these genes.
Fig. 7.
Expression of other V-ATPase subunits.
Mean normalised expression of Atp6v0d2, Atp6v0d1 and Atp6v1a in ATDC5 cells cultured in control medium (CM) or differentiation medium (DM) and transfected with siAtp6v0d2 or control siRNA. *P < 0.05, **P < 0.01, for comparisons indicated by lines; pairwise fixed reallocation randomisation test; n = 4.
4. Discussion
Here we present evidence for 12 novel chondrocyte hypertrophy-regulated genes, based on expression patterns in equine growth cartilage. Of these, 10 were found to be up-regulated during hypertrophy, and two down-regulated. The regulated genes are known to have a wide variety of functions in other cell types, but any roles they may play in regulating the behaviour of chondrocytes undergoing hypertrophy is currently unknown. ATP6V0D2 was selected for further in vitro studies, to be discussed below. In the absence of any functional studies on the other regulated genes, we speculate briefly here about the possible roles of a few of them in chondrocytes undergoing hypertrophy.
BAK1 encodes BCL2 antagonist/killer protein, which is localised in mitochondria where it induces apoptosis by accelerating voltage-dependent opening of mitochondrial anion channels, which leads to loss of mitochondrial membrane potential and release of cytochrome C (Chipuk et al., 2008). We were interested in examining the regulation of genes known to have roles in physiological cell death, since while some authors have described apoptosis of hypertrophic chondrocytes, others have suggested that these cells die by mechanisms other than apoptosis (discussed in (Mackie et al., 2008)). Moreover, recent lineage-tracking studies suggest that at least some hypertrophic chondrocytes transdifferentiate into osteoblasts and contribute to metaphyseal bone formation, rather than dying (Yang et al., 2014). The up-regulation of BAK1 demonstrates that hypertrophic chondrocytes invoke at least part of the biochemical machinery associated with apoptosis, but whether this leads to physiological death in some or all cells will require further investigation.
Another of the genes found to be up-regulated with hypertrophy, DDX5, encodes a transcriptional co-activator of steroid hormone receptors and other transcription factors, including RUNX2 (Fuller-Pace and Ali, 2008). The interaction between DDX5 and RUNX2, as well as the enhancement of RUNX2 transcriptional activity, were first identified in osteoblast-like cells (Jensen et al., 2008). Such an interaction has not been described in the context of cartilage, however RUNX2 is also up-regulated with chondrocyte hypertrophy and is essential for this process (Lefebvre and Smits, 2005). It seems likely, therefore, that DDX5 enhances RUNX2 transcriptional activity in chondrocytes and thus enhances RUNX2’s stimulation of hypertrophy.
Mutations of RPS7, which encodes ribosomal subunit protein 7, are responsible for a small proportion (1–2%) of cases of Diamond Blackfan anaemia; about 60% of cases of this condition can be attributed to mutations in genes encoding large (RPL) and small (RPS) ribosomal subunit proteins, all of which result in defects in rRNA processing (Farrar and Dahl, 2011). In addition to anaemia, craniofacial, musculoskeletal, cardiovascular, ophthalmologic and genitourinary malformations are observed in some patients, but there is no consistent association between specific mutations and these malformations. An Rps7 mouse mutant has recently been described, in which ribosomal biogenesis is defective and, among other phenotypic abnormalities, decreased body size and skeletal patterning defects are observed (Watkins-Chow et al., 2013). No histological data are presented in the paper describing this study, however alizarin red and alcian blue-stained skeletal preparations demonstrate a delay in ossification of the vertebral bodies; such an observation could potentially result from failure of chondrocytes to undergo hypertrophy or failure of hypertrophic chondrocytes to secrete factors that favour ossification (such as vascular endothelial growth factor). It will be interesting to examine the skeletons of these mice more closely to determine whether Rps7 is required for normal hypertrophic chondrocyte behaviour.
ATP6V0D2 was selected for further study because it was one of the most highly up-regulated genes of those examined in equine growth cartilage, as well as in ATDC5 cells. This gene encodes the d2 subunit of the V-ATPase V0 domain. V-ATPase is a proton pump; it is a macromolecular complex, which is comprised of two domains (V0 and V1) each of which contains multiple subunits (reviewed by Qin et al., 2012). The multimeric enzyme is ubiquitously expressed, however some of its subunits have multiple isoforms, some of which are selectively expressed in a subset of tissues. ATP6V0D2 expression has been described in kidney, epididymis, dendritic cells and osteoclasts, but not in chondrocytes, although evidence for expression of V-ATPase in chondrocytes has been described (Yocum et al., 1995). V-ATPase can be located in a variety of intracellular membranes, including lysosomes, endosomes and Golgi-derived vesicles, or in the plasma membrane, depending on cell type (Toei et al., 2010). The compartment-specific acidification caused by V-ATPase allows it to contribute to a wide variety of functions, including activation of proteases within lysosomes, and dissolution of bone mineral as well as activation of proteases within the extracellular compartment between osteoclasts and the bone surface. In primary cultures of articular chondrocytes, V-ATPase causes lysosomal acidification and contributes to interleukin-1-stimulated acid extrusion, indicating that it is present in both the lysosomal and plasma membranes (Yocum et al., 1995, Tattersall et al., 2005). Here we present the results of experiments designed to investigate the role of V-ATPase in chondrocytes undergoing hypertrophy.
The up-regulation of Atp6v0d2 by differentiation medium in ATDC5 cells confirmed that this culture system was appropriate for studies investigating functions of this gene in chondrocytes undergoing hypertrophy. Two approaches were taken to investigating the hypothesis that ATP6V0D2 is required for chondrocyte hypertrophy: expression of Atp6v0d2 was knocked down using siRNA and V-ATPase activity was inhibited using bafilomycin A1. The two approaches yielded overlapping, but partially distinct results. Both Atp6v0d2 knockdown and V-ATPase inhibition led to suppression of Col2a1 expression in control but not in differentiation medium. This suggests that V-ATPase activity is required for maintaining differentiation of non-hypertrophic chondrocytes, and that ATP6V0D2 is required for this activity. Both treatments also caused a decrease in nuclear area in cells cultured in differentiation medium. The volume of chondrocyte nuclei increases as the cells undergo hypertrophy (Buckwalter et al., 1986), thus this observation could be in keeping with a role for V-ATPase (and specifically ATP6V0D2) in chondrocyte hypertrophy. We measured cell area with the idea that this would reflect hypertrophy (or lack thereof), and neither treatment had any effect; it is possible that the monolayer culture system used here is inappropriate for the morphological detection of chondrocyte hypertrophy and that a three-dimensional system such as a pellet culture system would be more appropriate. Either or both of these roles for ATP6V0D2 (maintenance of Col2a1 expression in non-hypertrophic chondrocytes and nuclear area in hypertrophic chondrocytes) may be mediated by lysosomal acidification, given the presence of ATP6V0D2 in these organelles.
In contrast, while inhibition of V-ATPase prevented up-regulation of hypertrophy-associated genes in cells cultured in differentiation medium, no such effect was observed in cells in which Atp6v0d2 expression was knocked down, suggesting that V-ATPase is required for the up-regulation of these genes, but that the ATP6V0D2 subunit is not essential for this effect. We therefore investigated the expression of Atp6v1a, the gene encoding the catalytic subunit, and Atp6v0d1, the gene encoding the alternative subunit d isoform; both of these genes were expressed in control medium, up-regulated in differentiation medium, and not influenced by Atp6v0d2 knockdown, explaining how V-ATPase activity may occur independently of Atp6v0d2 expression.
A recent study of the role of V-ATPase in long bone growth yielded results that appear to contradict our observations of a role for this enzyme in expression of hypertrophy-associated genes (Newton et al., 2015). In this study, organ cultures of metatarsal bones from 3.5-day-old mice provided the source of chondrocytes; bafilomycin A1 was found to cause an increase in the length of the bones, the size of hypertrophic chondrocytes and the length of the zone positive for collagen type X. Unfortunately, since these metatarsals contain a primary centre of ossification, it is not possible to determine which of these effects can be attributed to V-ATPase expressed by hypertrophic chondrocytes as opposed to osteoclasts. Since osteoclast V-ATPase is required for the resorption of cartilage matrix surrounding hypertrophic chondrocytes, inhibition of osteoclast V-ATPase would be expected to cause an increase in the length of the collagen type X-expressing zone, and by allowing the cells to survive for longer within this matrix before its replacement by bone, may allow further expansion in their size. It is difficult to predict what effect inhibition of osteoclast V-ATPase would have on total length of the cultured bones. In order to attribute any effects of V-ATPase inhibition to V-ATPase expressed by chondrocytes, it would be necessary to culture epiphyseal cartilage without the ossification centre.
The results presented here suggest that ATP6V0D2 is required for some but not all V-ATPase actions in chondrocytes. One possible explanation for this observation could be that ATP6V0D2 is only present in a subset of the locations in which V-ATPase is found within chondrocytes. Indeed, while evidence for the presence of V-ATPase in lysosomes and the plasma membrane of cultured articular chondrocytes has previously been described (Yocum et al., 1995, Tattersall et al., 2005), we observed staining for ATP6V0D2 in lysosomes, but not in the plasma membrane. Given the diversity of functions of V-ATPase in different subcellular membrane locations, this explanation seems feasible.
Another difference between the two treatments was that Atp6v0d2 knockdown, but not bafilomycin A1 treatment, in cells cultured in differentiation medium caused an increase in cell number that could be attributed to proliferation. A mouse in which the Atp6v0d2 gene has been deleted has been described (Lee et al., 2006). This mouse shows a defect in fusion of osteoclast precursors, resulting in a reduced number of multinucleate osteoclasts and a consequent elevated bone mass. The mouse is described as showing normal growth rates, but no description of the morphology of growth plate chondrocytes is provided, thus it is not possible to determine whether chondrocyte hypertrophy is abnormal. It is interesting to note that inactivation of Atp6v0d2 did not affect the v-ATPase activity of osteoclasts, suggesting that ATP6V0D2 may have a function independent of a contribution to V-ATPase activity. Such a function may help to explain the fact that, in chondrocytes, knockdown of Atp6v0d2 exerted effects not mimicked by bafilomycin A1 treatment.
In conclusion, we have identified a number of novel chondrocyte hypertrophy-associated genes, and conducted further studies on one of them, ATP6V0D2, which encodes the V-ATPase subunit ATP6V0D2. The results demonstrate that V-ATPase activity is required for maintenance of expression of the chondrocyte-specific Col2a1 gene and contributes to various features of chondrocyte hypertrophy, but that isoform 2 of the V0 domain subunit d (ATP6V0D2) is not required for all of these actions.
Acknowledgments
Acknowledgments
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. The authors thank Dr. Helen Davies and Mr. Brendan Kehoe for assistance with collection of specimens.
Declaration of interest
Conflicts of interest: none.
References
- Ahmed Y.A., Tatarczuch L., Pagel C.N., Davies H.M., Mirams M., Mackie E.J. Hypertrophy and physiological death of equine chondrocytes in vitro. Equine Vet. J. 2007;39(6):546–552. doi: 10.2746/042516407X223699. [DOI] [PubMed] [Google Scholar]
- Ahmed Y.A., Tatarczuch L., Pagel C.N., Davies H.M., Mirams M., Mackie E.J. Physiological death of hypertrophic chondrocytes. Osteoarthr. Cartil. 2007;15(5):575–586. doi: 10.1016/j.joca.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Buckwalter J.A., Mower D., Ungar R., Schaeffer J., Ginsberg B. Morphometric analysis of chondrocyte hypertrophy. J. Bone Joint Surg. Am. 1986;68(2):243–255. [PubMed] [Google Scholar]
- Chipuk J.E., Fisher J.C., Dillon C.P., Kriwacki R.W., Kuwana T., Green D.R. Mechanism of apoptosis induction by inhibition of the anti-apoptotic BCL-2 proteins. Proc. Natl. Acad. Sci. U. S. A. 2008;105(51):20327–20332. doi: 10.1073/pnas.0808036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar J.E., Dahl N. Untangling the phenotypic heterogeneity of Diamond Blackfan anemia. Semin. Hematol. 2011;48(2):124–135. doi: 10.1053/j.seminhematol.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller-Pace F.V., Ali S. The DEAD box RNA helicases p68 (Ddx5) and p72 (Ddx17): novel transcriptional co-regulators. Biochem. Soc. Trans. 2008;36(Pt 4):609–612. doi: 10.1042/BST0360609. [DOI] [PubMed] [Google Scholar]
- Jensen E.D., Niu L., Caretti G., Nicol S.M., Teplyuk N., Stein G.S., Sartorelli V., van Wijnen A.J., Fuller-Pace F.V., Westendorf J.J. p68 (Ddx5) interacts with Runx2 and regulates osteoblast differentiation. J. Cell. Biochem. 2008;103(5):1438–1451. doi: 10.1002/jcb.21526. [DOI] [PubMed] [Google Scholar]
- Lee S.H., Rho J., Jeong D., Sul J.Y., Kim T., Kim N., Kang J.S., Miyamoto T., Suda T., Lee S.K., Pignolo R.J., Koczon-Jaremko B., Lorenzo J., Choi Y. v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat. Med. 2006;12(12):1403–1409. doi: 10.1038/nm1514. [DOI] [PubMed] [Google Scholar]
- Lefebvre V., Smits P. Transcriptional control of chondrocyte fate and differentiation. Birth Defects Res. C Embryo Today. 2005;75(3):200–212. doi: 10.1002/bdrc.20048. [DOI] [PubMed] [Google Scholar]
- Mackie E.J., Ahmed Y.A., Tatarczuch L., Chen K.S., Mirams M. Endochondral ossification: how cartilage is converted into bone in the developing skeleton. Int. J. Biochem. Cell Biol. 2008;40(1):46–62. doi: 10.1016/j.biocel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Mackie E.J., Tatarczuch L., Mirams M. The skeleton: a multi-functional complex organ: the growth plate chondrocyte and endochondral ossification. J. Endocrinol. 2011;211(2):109–121. doi: 10.1530/JOE-11-0048. [DOI] [PubMed] [Google Scholar]
- Mirams M., Ayodele B.A., Tatarczuch L., Henson F.M., Pagel C.N., Mackie E.J. Identification of novel osteochondrosis—associated genes. J. Orthop. Res. 2016;34(3):404–411. doi: 10.1002/jor.23033. [DOI] [PubMed] [Google Scholar]
- Newton P.T., Vuppalapati K.K., Bouderlique T., Chagin A.S. Pharmacological inhibition of lysosomes activates the MTORC1 signaling pathway in chondrocytes in an autophagy-independent manner. Autophagy. 2015;11(9):1594–1607. doi: 10.1080/15548627.2015.1068489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W., Horgan G.W., Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30(9) doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin A., Cheng T.S., Pavlos N.J., Lin Z., Dai K.R., Zheng M.H. V-ATPases in osteoclasts: structure, function and potential inhibitors of bone resorption. Int. J. Biochem. Cell Biol. 2012;44(9):1422–1435. doi: 10.1016/j.biocel.2012.05.014. [DOI] [PubMed] [Google Scholar]
- Shao Y.Y., Wang L., Ballock R.T. Thyroid hormone and the growth plate. Rev. Endocr. Metab. Disord. 2006;7(4):265–271. doi: 10.1007/s11154-006-9012-2. [DOI] [PubMed] [Google Scholar]
- Shukunami C., Ishizeki K., Atsumi T., Ohta Y., Suzuki F., Hiraki Y. Cellular hypertrophy and calcification of embryonal carcinoma-derived chondrogenic cell line ATDC5 in vitro. J. Bone Miner. Res. 1997;12(8):1174–1188. doi: 10.1359/jbmr.1997.12.8.1174. [DOI] [PubMed] [Google Scholar]
- Siebler T., Robson H., Shalet S.M., Williams G.R. Dexamethasone inhibits and thyroid hormone promotes differentiation of mouse chondrogenic ATDC5 cells. Bone. 2002;31(4):457–464. doi: 10.1016/s8756-3282(02)00855-4. [DOI] [PubMed] [Google Scholar]
- Simon P. Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics. 2003;19(11):1439–1440. doi: 10.1093/bioinformatics/btg157. [DOI] [PubMed] [Google Scholar]
- Tattersall A.L., Browning J.A., Wilkins R.J. Modulation of H + transport mechanisms by interleukin-1 in isolated bovine articular chondrocytes. Cell. Physiol. Biochem. 2005;16(1–3):43–50. doi: 10.1159/000087730. [DOI] [PubMed] [Google Scholar]
- Temu T.M., Wu K.Y., Gruppuso P.A., Phornphutkul C. The mechanism of ascorbic acid-induced differentiation of ATDC5 chondrogenic cells. Am. J. Physiol. Endocrinol. Metab. 2010;299(2):E325–34. doi: 10.1152/ajpendo.00145.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toei M., Saum R., Forgac M. Regulation and isoform function of the V-ATPases. Biochemistry. 2010;49(23):4715–4723. doi: 10.1021/bi100397s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui M., Xing L., Drissi H., Zuscik M., O'Keefe R., Chen D., Boyce B.F. Murine and chicken chondrocytes regulate osteoclastogenesis by producing RANKL in response to BMP2. J. Bone Miner. Res. 2008;23(3):314–325. doi: 10.1359/JBMR.071025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Woods A., Sabari S., Pagnotta L., Stanton L.A., Beier F. RhoA/ROCK signaling suppresses hypertrophic chondrocyte differentiation. J. Biol. Chem. 2004;279(13):13205–13214. doi: 10.1074/jbc.M311427200. [DOI] [PubMed] [Google Scholar]
- Watkins-Chow D.E., Cooke J., Pidsley R., Edwards A., Slotkin R., Leeds K.E., Mullen R., Baxter L.L., Campbell T.G., Salzer M.C., Biondini L., Gibney G., Phan Dinh Tuy F., Chelly J., Morris H.D., Riegler J., Lythgoe M.F., Arkell R.M., Loreni F., Flint J., Pavan W.J., Keays D.A. Mutation of the diamond-blackfan anemia gene Rps7 in mouse results in morphological and neuroanatomical phenotypes. PLoS Genet. 2013;9(1) doi: 10.1371/journal.pgen.1003094. (e1003094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Tsang K.Y., Tang H.C., Chan D., Cheah K.S. Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc. Natl. Acad. Sci. U. S. A. 2014;111(33):12097–12102. doi: 10.1073/pnas.1302703111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yocum S.A., Lopresti-Morrow L.L., Gabel C.A., Milici A.J., Mitchell P.G. Bafilomycin A1 inhibits IL-1-stimulated proteoglycan degradation by chondrocytes without affecting stromelysin synthesis. Arch. Biochem. Biophys. 1995;316(2):827–835. doi: 10.1006/abbi.1995.1111. [DOI] [PubMed] [Google Scholar]
- Zelzer E., Glotzer D.J., Hartmann C., Thomas D., Fukai N., Soker S., Olsen B.R. Tissue specific regulation of VEGF expression during bone development requires Cbfa1/Runx2. Mech. Dev. 2001;106(1–2):97–106. doi: 10.1016/s0925-4773(01)00428-2. [DOI] [PubMed] [Google Scholar]
- Zelzer E., Mamluk R., Ferrara N., Johnson R.S., Schipani E., Olsen B.R. VEGFA is necessary for chondrocyte survival during bone development. Development. 2004;131(9):2161–2171. doi: 10.1242/dev.01053. [DOI] [PubMed] [Google Scholar]