Abstract
For endocytosis and exocytosis, membranes transition among planar, budding, and vesicular topographies through nanoscale reorganization of lipids, proteins, and carbohydrates. However, prior attempts to understand the initial stages of nanoscale bending have been limited by experimental resolution. Through the implementation of polarized localization microscopy, this article reports the inherent membrane bending capability of cholera toxin subunit B (CTxB) in quasi-one-component-supported lipid bilayers. Membrane buds were first detected with <50 nm radius, grew to >200 nm radius, and extended into longer tubules with dependence on the membrane tension and CTxB concentration. Compared to the concentration of the planar-supported lipid bilayers, CTxB was (12 ± 4)× more concentrated on the positive curvature top and (26 ± 11)× more concentrated on the negative Gaussian curvature neck of the nanoscale membrane buds. CTxB is frequently used as a marker for liquid-ordered lipid phases; however, the coupling between CTxB and membrane bending provides an alternate understanding of CTxB-induced membrane reorganization. These findings allow for the reinterpretation of prior observations by correlating CTxB clustering and diffusion to CTxB-induced membrane bending. Single-particle tracking was performed on single lipids and CTxB to reveal the correlations among single-molecule diffusion, CTxB accumulation, and membrane topography. Slowed lipid and CTxB diffusion was observed at the nanoscale bud locations, suggesting a local increase in the effective membrane viscosity or molecular crowding upon membrane bending. These results suggest inherent CTxB-induced membrane bending as a mechanism for initiating CTxB internalization in cells that could be independent of clathrin, caveolin, actin, and lipid phase separation.
Introduction
Membrane function is governed by the molecular organization, clustering, and interaction of its constituents. In particular, curvature-dependent reorganization has captured a growing interest as a mechanism for creating locally distinct membrane environments (1, 2, 3). In this study, we focus on the membrane bending effects of cholera toxin subunit B (CTxB) in a quasi-one-component model membrane. Cholera toxin is a member of the AB5 toxin family that multivalently binds to GM1 and is most frequently used as the lipid raft marker in biophysical studies (4). CTxB-GM1 partitions with order-preferring lipids (5, 6), induces lipid phase segregation (6, 7, 8), and sorts to high curvature regions (2, 3). GM1 plays a vital role in numerous biological functions including endocytosis (9), viral egress (10), Alzheimer disease (11, 12), vesicular trafficking (13), and immunological signaling (14).
CTxB and GM1 adopts a sequence of macromolecular complexes from its initial membrane binding, local clustering, and subsequent cellular internalization. Accordingly, numerous observations of multimodal diffusion and nanoscale confinement of CTxB on living cells (15) and on synthetic bilayers (16, 17) have been reported. Even in the absence of coexisting lipid phases, CTxB exhibits multiple populations of diffusion rates and transient confinement in regions as small as 20 nm in radii (16, 17). On living cells, CTxB diffusion is independent of the diffusion of caveolin, clathrin, or glycosylphosphatidylinositol-linked proteins, which suggests the internalization of CTxB is initialized distinctly from conventional endocytotic processes (18, 19, 20).
Inward membrane vesiculation and tubulation have been observed in cells and synthetic vesicles upon exposure to Cholera toxin (10, 21, 22). CTxB has been observed to sort to membranes of negative curvature for supported lipid bilayers (SLBs) on wavy glass (3), micronscale nanoparticles (23), and membrane tethers (2). The capability of CTxB to bind to membranes in which both of the local principle curvatures are negative (i.e., with a positive Gaussian curvature) is well established with CTxB-induced inward pits in giant unilamellar vesicles (GUVs) (24). This is supported by molecular dynamics simulations of the structurally similar Shiga toxin (24). However, the nanoscale detail of CTxB intrinsically inducing membrane curvature as necessary for endocytosis, and the capability of CTxB to bind to membranes with differing signs of principle curvatures, remains uncertain.
We hypothesize that CTxB aggregates and internalizes as a result of its inherent physical effects on the membrane topography. Testing this hypothesis requires the use of an examination method that is able to resolve the colocalization of nanoscale membrane bending with CTxB. Polarized localization microscopy (PLM) combines single-molecule localization microscopy (SMLM) with polarized total internal reflection fluorescence microscopy to detect nanoscale membrane orientation with super-resolution (25). This technique distinguishes between membranes of varying orientation due to the differential excitation of membrane-confined fluorophores depending on the linear polarization of the incident excitation light. In particular, indocarbocyanine dyes (e.g., DiI) are photoswitchable probes (26) that maintain their fluorescence dipole moment in the plane of the membrane (27, 28, 29), such that membranes parallel to the coverslip are preferentially excited by incident s-polarized light, and membranes vertical to the coverslip are preferentially excited by incident p-polarized light. The robust identification of nanoscale membrane bending provided by PLM enables the correlation of membrane topography and molecular sorting on physiologically relevant length scales (<50 nm) with numerous technical advantages over other super-resolution techniques (25).
The microscopy setup for PLM permits simultaneous multicolor SMLM and single-particle tracking (SPT) of lipids and proteins. For example, AlexaFluor dyes are commonly conjugated to proteins with flexible linkers, such that the fluorescence excitation of dyes has no observed dependence on the illumination polarization. Such dyes are common probes for imaging via direct stochastic optical reconstruction microscopy (dSTORM) (30). As demonstrated here, the multicolor, simultaneous combination of PLM and dSTORM enables the determination of membrane organization, molecular sorting, and single-molecule diffusion relative to membrane bending.
In this article, we report the nanoscale organization and dynamics of CTxB relative to membrane bending events on an SLB with 99.4% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 0.3% DiI, and 0.3% GM1. Using PLM, we found that the SLBs initially exhibit a flat uniform topology before the addition of CTxB. Nanoscale membrane bending and bud formation occurred within 30 s upon the addition of CTxB. The subset of CTxB coincident with membrane budding became clustered and slowed to (19 ± 9)% of the initial CTxB diffusion rate. Similarly, only the DiI coincident with the membrane budding demonstrated diffusion that was slowed to (18 ± 4)% of the diffusion rate of DiI in a planar SLB. At later times after CTxB addition (>20 min), freely diffusing CTxB on planar bilayers, small accumulations of CTxB on nanoscale membrane buds, and rings of CTxB at larger membrane protrusions were simultaneously observed (Fig. 1). Both single event analysis and spatially averaged correlation analysis demonstrated the strong interdependence of membrane structure, single-MD, and CTxB accumulation. In sum, these studies represent, to the best of our knowledge, the previously undetected phenomena of nanoscale membrane budding and tubulation by CTxB on SLBs without the apparent need of lipid phase separation. PLM has enabled observing the effects of CTxB on spontaneous molecular sorting, immobilization, curvature, and tubule formation processes.
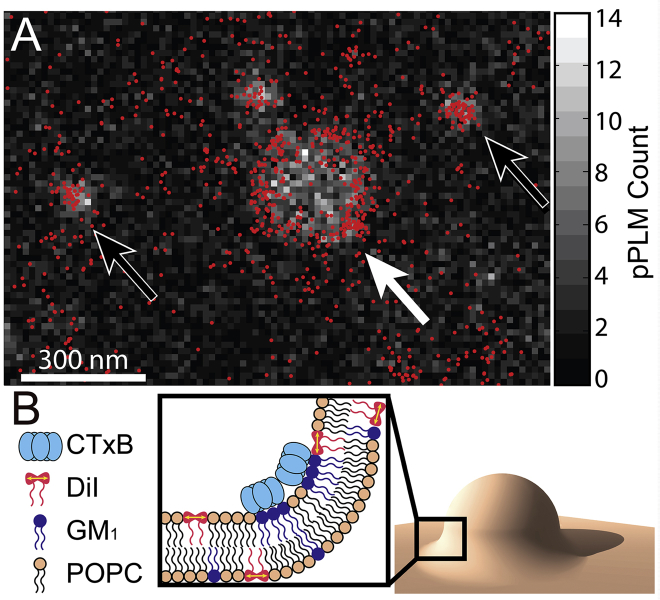
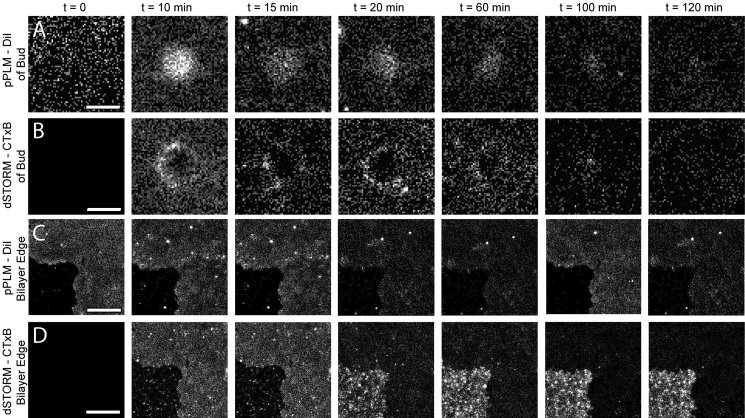
Figure 1.
Colocalization of CTxB accumulations and membrane bending on supported lipid bilayers demonstrates the inherent capability of CTxB to induce membrane curvature. (A) The 2D histogram of DiI localizations with p-polarized excitation highlights where the membrane is more perpendicular to the coverslip. This is overlaid with the single-molecule localizations of CTxB that are displayed with red points. The membrane was bent where CTxB was concentrated. Larger membrane buds, as indicated by a white arrow, showed a high concentration of CTxB preferentially at the neck of the bud, where the membrane had a negative Gaussian curvature; it is shown schematically in (B). Smaller membrane buds, as indicated by black arrows, also had a locally increased concentration of CTxB, although sorting of CTxB on the smallest buds was not resolved. To see this figure in color, go online.
Materials and Methods
SLB formation
GUVs of primarily POPC (Avanti Polar Lipids, Alabaster, AL) with 0.3 mol % 1,1′-didodecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI; Life Technologies, Carlsbad, CA) and 0.3 mol % GM1 Ganglioside (Avanti Polar Lipids) were prepared by electroformation, as described previously (31). A hydration solution of 200 mM sucrose was added to the dried lipid films and the ITO slides were connected to either side of a sine wave function generator. Further description of the GUV preparation method is included in the Supporting Material. Experiments were also repeated by using diphytanoyl phosphatidylcholine (DPhyPC; Avanti Polar Lipids) instead of POPC and DiO or DiD instead of DiI with indistinguishable results. Exposing the GUVs on plasma-cleaned coverslips resulted in their bursting open into a continuous bilayer.
CTxB addition
CTxB was labeled with AlexaFluor 647 or AlexaFluor 488 before purchase (Thermo Fisher Scientific, Waltham, MA). CTxB was added to the SLB for a final concentration of 0.25 μg/mL above the SLB to saturate all available GM1. After 0.5 min of incubation, the unbound CTxB was rinsed away. The time (t) is said to equal zero before CTxB was added, and otherwise, t reports the time because the unbound CTxB was rinsed away. CTxB-AlexaFluor 647 was used for all data as shown below and indistinguishable results were obtained with CTxB-AlexaFluor 488.
Engineered membrane curvature
For select experiments, membrane curvature was engineered before the addition of CTxB, as done previously (25). GUVs were draped over the 70 nm-radius nanoparticles and coverslip to create the engineered membrane curvature for greater consistency in membrane bud size when needed. Further description of NP sedimentation is described in the Supporting Material.
Imaging optics
PLM was performed with a conventional total internal reflection fluorescence microscope with an additional liquid crystal wave plate placed in the excitation path to control the excitation polarization. The ratio of the p-polarized to s-polarized laser power in each polarization incident upon the sample was 207:1 and 54:1, respectively, at the optimal liquid crystal waveplate voltages for each polarization (25). Image acquisition was performed with an iXon-897 Ultra EMCCD camera (Andor Technology, Belfast, UK) proceeded by an OptoSplit IILS (Cairn Research, Faversham, UK) with emission filters (BrightLine; Semrock, Rochester, NY), a 4-band notch filter (ZET405/488/561/640m; Chroma, McHenry, IL), and a 2× magnification lens.
Imaging procedure
The sample was exposed to >80 mW of excitation light with λex = 561 (DiI) and λex = 647 nm (CTxB-AF647) simultaneously to provide a steady-state blinking of the fluorophores. Between 10,000 and 30,000 frames were acquired for each time point at a frame rate of 50 Hz on a region of interest with 18 ms acquisition per frame (texp). Further description is provided in the Supporting Material.
Imaging buffer
PLM was performed on samples present in an oxygen-scavenging buffer (150 mM NaCl, 50 mM TRIS, 0.5 mg/mL glucose oxidase, 20 mg/mL glucose, 40 μg/mL catalase, and 1% β-mercaptoethanol at pH 8). Buffer proteins were purchased from Sigma-Aldrich (St. Louis, MO) and salts were purchased from Thermo Fisher Scientific. These conditions maintain a low free oxygen concentration in the buffer to minimize nonreversible fluorophore bleaching and encourage transient fluorophore blinking, as is necessary for SMLM.
Single-molecule localization
The analysis of the raw, diffraction-limited images included low-pass Gaussian filtering, multiemitter fitting routines, median background subtraction, lateral stage drift correction, and the fitting of each isolated fluorophore image via the ImageJ (National Institutes of Health, Bethesda, MD) plug-in, ThunderSTORM (32). ThunderSTORM provided the single fluorophore positions, localization uncertainty, and photon per fluorophores for further analysis. A threshold value of 100 photons per fluorophore was used to keep only the bright localizations for further analysis. The separate channel images were overlaid via a custom-made MATLAB routine (The MathWorks, Natick, MA) via the alignment of TetraSpeck nanoparticles (100 nm diameter; Life Technologies) that were visible in all channels.
Bud identification and size evaluation
The detection of buds in each color channel was performed via a custom-made MATLAB program that applies a mask and detects regions with >3× the density of the average flat surrounding background bilayer. Each bud was fitted with a 2D Gaussian function for center estimation. The size of each bud (rbud) was set equal to the mean distance of all bud-associated localizations from the bud center. This was calculated by taking into consideration the background from flat SLB localizations of uniform density (b), the distance of each localization from the bud center (ri), and a threshold distance that was significantly greater than rbud (R). Typically, R = 400 nm but the following calculation is independent of the particular R chosen. The number of extra localizations due to the presence of the bud (Nbud) is equal to the total number of localizations (Nall) within ri < R subtracted from the number of localizations expected within R if no bud was present (NSLB); NSLB = πR2b = Nall − Nbud. The mean ri expected for the flat SLB within R is 2R/3. By analyzing all collected localizations within R and subtracting the expected localizations from the flat SLB, rbud is calculated according to
| (1) |
Single-particle tracking
The sequential localizations of single fluorophores were analyzed to reveal the diffusion rate of individual molecules versus membrane topography. A camera blur was caused by the single-frame exposure time (texp) being comparable to the time between frames (Δt) (33, 34). The diffusion coefficient (D) was calculated from the Dfit according to
| (2) |
where σr = 15 nm, Δt = 20 ms, and texp = 18 ms; if Dfit = 0.5 μm2/s, then D = 0.7 μm2/s or if Dfit = 0.1 μm2/s, then D = 0.13 μm2/s. Because the microscopy methods used here reveal only the z-projection of the diffusion, D calculated from Eq. 2 is reported as Dxy to emphasize that only the diffusion through the xy plane has been measured. Diffusion coefficients from SPT are typically extracted by fitting the mean squared displacement versus Δt. However, fitting a whole trajectory to a single diffusion coefficient blurs the effects of nanoscale curvature with the lipid trajectory sampling both curved and flat membranes (34). Further description of the SPT procedure and single-step fitting is provided in the Supporting Material.
Results
CTxB induces membrane budding in SLBs
The reconstructed time-lapse dSTORM and pPLM images of CTxB and DiI revealed the initial protein accumulation and membrane budding processes, respectively. Within the first minute of CTxB addition to the membrane, some CTxB exhibited confinement on the flat bilayer, as demonstrated by a detectable accumulation of CTxB localizations without a significant increase in the local density of DiI localizations from pPLM (Fig. 2). After 1 min, the clusters of CTxB became colocalized with higher densities of DiI localizations as detected with pPLM. A local increase in DiI localizations obtained by pPLM represents areas in which the membrane would be more perpendicular to the microscopy coverslip, as would be expected for a membrane bud. Membrane buds formed at the locations in which CTxB accumulated, demonstrating the capability of CTxB to initiate and induce nanoscale membrane bending. PLM of DiI revealed both a continued growth in the size of the buds and the formation of new buds with continued CTxB exposure. PLM allowed earlier visualization of membrane bending initiated by CTxB than was detectable by epifluorescence microscopy, as described in the Supporting Material.
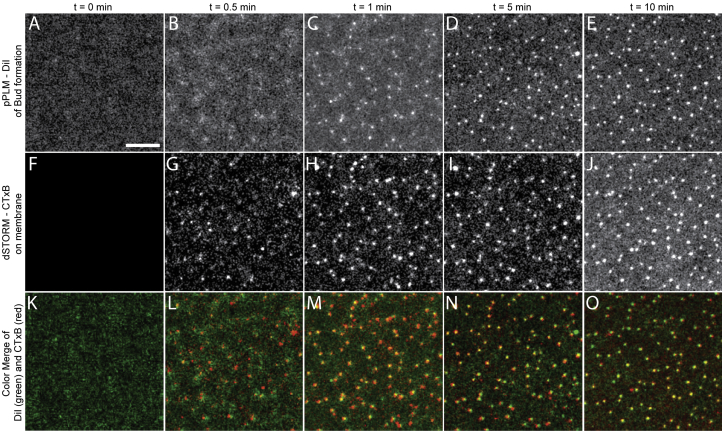
Figure 2.
Simultaneous observation of (A–E) membrane bending detected via pPLM and (F–J) CTxB clustering on the supported lipid bilayer detected via dSTORM reveals the CTxB-induced membrane bending. Before CTxB is added, (A) the DiI localizations by pPLM are uniform and (F) no CTxB localizations were found. Within the first 30 s of CTxB on the SLB, (B) slight variations in the DiI localizations were present and (G) CTxB clusters formed. At later times, (C–E) the membrane buds became increasingly apparent and (H–J) CTxB became increasingly concentrated at the buds. (K–O) Color merge of DiI in green and CTxB in red. The scale bar represents 200 nm. To see this figure in color, go online.
Regions of clustered localizations, observed in both DiI and CTxB channels, with localization density >3× that of the flat membrane were identified as membrane buds. Histograms of bud sizes versus time from the pPLM of DiI and dSTORM of CTxB show buds increasing in number and size over time (Fig. 3, A and B). Additionally, this analysis further shows that CTxB accumulations precede membrane bending because the CTxB accumulations are larger and more numerous than the DiI accumulations, as is qualitatively shown in Fig. 2. There was no clear separation of the buds into distinct stages of growth into larger structures (i.e., buds versus tubules), so there existed no apparent characteristic distances between the buds or tubules.
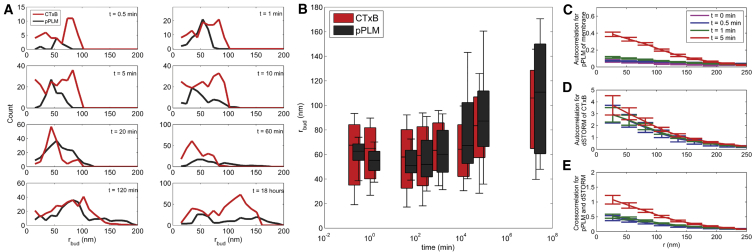
Figure 3.
The super-resolution capabilities of PLM and dSTORM revealed membrane bud size (rbud) versus CTxB incubation time. As some buds grew to a larger diameter or into tubules (Figs. 4 and S3), new small buds (rbud < 50 nm) continued to form, and the distribution of rbud widened over time. The number and size of the buds increased as the CTxB incubation time increased. This is shown by (A) histograms and (B) whisker plots of the bud sizes observed in both the pPLM and CTxB channels. (C and D) Autocorrelation analysis of pPLM and CTxB localizations as a function of incubation time also shows the bud formation dynamics without the threshold-based identification of individual buds. (E) Cross-correlation analysis of pPLM and CTxB localizations shows the correlated localization of CTxB and membrane bending. To see this figure in color, go online.
Autocorrelation analysis of bud clusters in the membrane channel and CTxB channel was plotted separately. The increased autocorrelation between membrane buds with time is clearly depicted in Fig. 3 C, where membrane buds are increasing in number with time. In the CTxB channel, a high autocorrelation between CTxB localizations exists at earlier times (t = 0.5 min), indicating the presence of CTxB clusters before the formation of membrane buds (Fig. 3 D). The mapping between clustered localizations observed in CTxB and the membrane channel was examined with cross-correlation analysis. The increasing cross-correlation analysis as a function of incubation time indicates the tight mapping between clusters observed in both channels. This indicated the strong relation between buds observed in pPLM and CTxB accumulations at bud locations observed via dSTORM (Fig. 3 E).
Some membrane buds grew into tubules
A wide distribution of bud sizes was observed with increasing bud sizes present after longer CTxB incubation times (Fig. 3, A and B). The smallest buds displayed an apparently uniform distribution of CTxB across the bud where the specific distribution of CTxB on the bud was limited by the resolution of dSTORM (i.e., Fig. 1 A, black arrows). Intermediate size buds were observed in which the DiI localizations suggest a hemispherical membrane shape greater than 100 nm radius and CTxB preferentially localized at the bud neck (Fig. 1 A, white arrow). These large hemispherical buds showed a radially decreasing density of DiI localizations rather than a ring of DiI localizations. This effect is due to the anisotropic emission of the orientationally confined DiI within the membrane (25, 35). The largest membrane bending events observed were membrane tubules with the membrane protruding away from the glass coverslip by >3 μm (Fig. 4). In these membrane tubules, the ring of DiI localizations and the ring of CTxB localizations were both apparent.
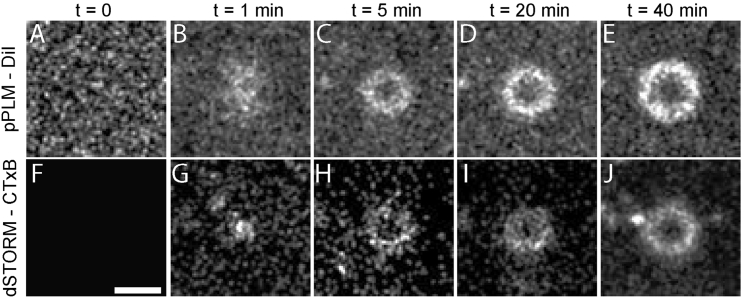
Figure 4.
Some membrane buds grew into tubules extending away from the glass coverslip (Fig. S3). In the absence of CTxB, the supported bilayer was flat as shown by the random distribution of localizations in the membrane channel (A), and the absence of localizations in the CTxB channel (F). The budding process started with (B) a membrane bud and (G) small clusters of CTxB (rbud < 100 nm). Over time, (C) a ring of DiI localizations formed as the bud top extended away from the coverslip and (D and E) the ring widened with an increasing tubule diameter. (H–J) Membrane bending was driven by CTxB accumulation at the base of the tubule where the negative Gaussian membrane curvature was present. The scale bar represents 100 nm.
Buds vanish upon CTxB depletion
CTxB concentration was a key factor in this membrane budding and tubule formation. Typically, the concentration of CTxB on the SLB was determined by saturating the 0.3% GM1 within the membrane, and the CTxB concentration was apparently constant within 2 min of CTxB addition. However, in select experiments, the glass coverslip surrounding the patch of SLB was prepared to encourage CTxB binding directly to the glass. In the first minutes after CTxB addition to the flat SLB, nanoscale membrane clusters were detected, as seen in all other experiments. However, the CTxB concentration was not constant over long times in this experiment. As CTxB laterally diffused on the membrane, it eventually came into close proximity with the perimeter of the SLB and the surrounding glass. Only in this experiment was CTxB observed to stick and accumulate on the glass surface (Fig. 5). This binding of CTxB to the glass caused a 93% decrease in CTxB concentration from 0.029 to 0.0018 localizations/nm2 on the SLB. In contrast, the rate of DiI localizations showed no significant change over time. Meanwhile, the localization density of CTxB on the glass increased to 0.014 localizations/nm2 at t = 120 min, an 8.5× increase from the density of 0.0017 localizations/nm2 at early times (t = 10 min). As a result of the decreasing CTxB concentration on the SLB, the number and size of the nanoscale buds decreased, as shown in both CTxB and DiI localizations (Fig. 5, A and B). The buds and tubules disappeared from the membrane and the SLB returned to its original flat topography upon decreasing CTxB concentration. This phenomenon demonstrated the reversibility and the CTxB dependence of the induced membrane curvature.
Figure 5.
CTxB depletion from the membrane showed that the budding process was reversibly dependent on the CTxB concentration on the membrane. In this experiment, the surrounding glass coverslip was prepared as to encourage CTxB absorption and removal from the SLB. (A) The pPLM images show the membrane buds decreasing in height and diameter with decreasing CTxB concentration. (B) The dSTORM images of CTxB show a decrease of CTxB on the SLB and a uniform concentration across where the bud had been. (C) The bilayer showed no change in the average localization rate compared to the surrounding membrane-free cover glass. (D) The concentration of CTxB on the membrane decreased whereas the concentration of CTxB on the membrane-free coverslip increased, as revealed by the relative localization rates over time. The scale bars (A and B) represent 100 nm and (C and D) 1 μm.
As a further control to confirm that the budding and tubulation processes were induced by CTxB binding to the membrane, these experiments were repeated without addition of CTxB. POPC/GM1/DiI membranes were imaged under identical conditions for 24 h and no observable membrane deformations were detected.
Single-molecule mobility varies with budding
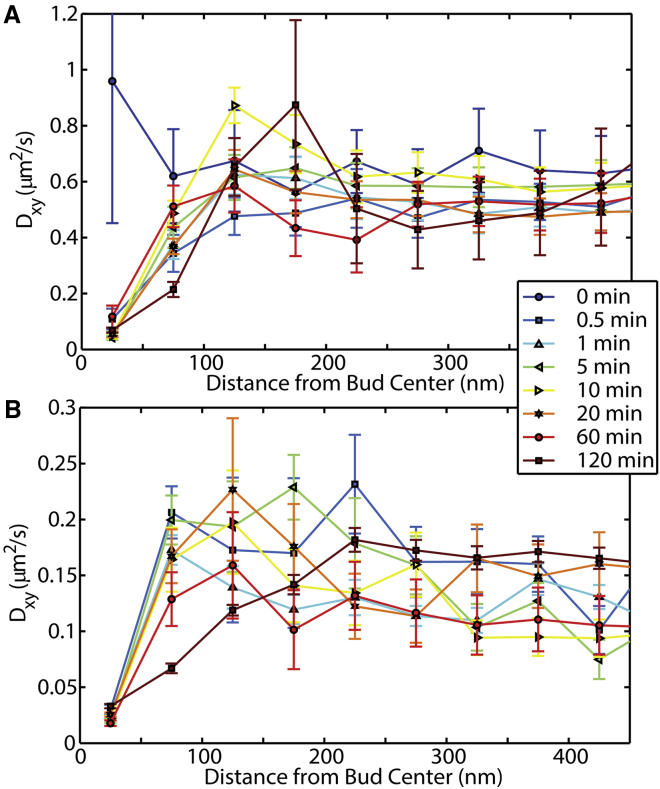
SPT was performed on both the DiI and CTxB as a function of location within the membrane buds. Single-fluorophores that stayed “on” for sequential frames in the raw data collection were individually localized and linked to reveal the single-molecule mobility. The diffusive rate of DiI and CTxB on the planar SLB were measured to be (0.55 ± 0.05) and (0.14 ± 0.03) μm2/s, respectively, with correcting for localization uncertainty in this single-step length analysis and camera blur, as described above. These diffusion rates did not vary with the duration of CTxB incubation. However, the diffusion rate did vary with distance from the center of the membrane bud (Fig. 6). Both the diffusion of DiI and CTxB demonstrated slowed diffusion rates through the xy plane by (82 ± 4) and (81 ± 9)% at the center of the membrane bud relative to the surrounding planar SLB. Although a significant component of this perceived slowing of the single-molecule diffusion could be attributed to the membrane tilt, localization uncertainty (σ = 20 nm), and frame rate (50 Hz) limit, these conditions affect at most a 60% slowing on nanoscale buds, as discussed below and in our companion article (25). The diffusion coefficients of DiI and CTxB were independent of the presence of buds for distances greater than 200 nm from bud centers.
Figure 6.
Single-particle tracking was performed on both (A) DiI and (B) CTxB as a function of position within a membrane bud. Before the CTxB is added (t = 0 min), no CTxB was located on the SLB, and no membrane buds were present. Random locations in PLM were chosen to confirm that our analysis routines demonstrated no significant variation in Dxy versus distance away from those locations. At all later times, a significant slowing of both the DiI and CTxB diffusion was observed within 50 nm of the bud center. The error bars represent at a 95% confidence interval of the fitting of Eq. 1 to the histograms of single step lengths. The step lengths were binned based on the distance from the bud center to the mean of the two linked localizations. To see this figure in color, go online.
Budding occurs with varying lipid types
All budding experiments were repeated with DPhyPC replacing POPC as the primary membrane component to confirm that the particular lipids used here were not dominating this budding observation. DPhyPC and POPC are both liquid crystalline at room temperature, yet with a highly different molecular structure of their acyl chains. The disordered acyl tails of POPC and DPhyPC decreases the possibility of obtaining observable lipid phase separation for the membrane composition conditions utilized here. Indistinguishable curvature induction by CTxB was observed on SLBs formed with 99.4% DPhyPC, 0.3% DiI, and 0.3% GM1 as with 99.4% POPC, 0.3% DiI, and 0.3% GM1. Further, experiments were also performed with varying membrane labels to ensure that probable lipid degradation or traces of imperfections were not the cause for such an observation. Membranes of 99.4% POPC, 0.3% GM1, and 0.3% DiO, DiI, or DiD were created and CTxB-AF647 or CTxB-AF488 was added to the bilayers. CTxB induced membrane budding in all cases of varying lipids, lipid dyes, and CTxB labels.
Quantifying CTxB sorting
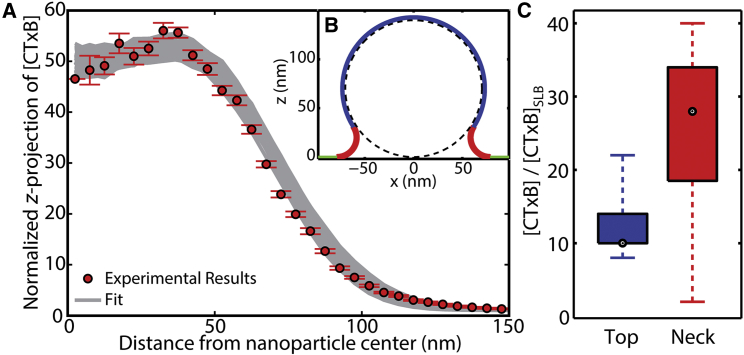
The buds formed by CTxB addition to planar SLBs were of sizes that varied from each other and varied with time (Fig. 3, A and B). To measure the partition coefficient of CTxB versus membrane curvature, a relatively static membrane curvature was engineered and the local CTxB concentration was observed. Only in this experiment, membrane buds were engineered by draping SLBs over nanoparticles of known sizes before the addition of CTxB. The ability to measure the CTxB distribution on multiple engineered buds of roughly similar known sizes enabled averaging between buds. This data averaging provided lower noise in the experimental data and the fitting of the CTxB distribution to a predicted model of the membrane topography (Figs. 7 and S2) (25). The radial density of CTxB observed on 25 separate nanoparticle-created membrane buds provides the experimental data (Fig. 7 A). A modeled CTxB distribution on a simulated membrane topography was z projected onto the xy plane, and the projection was used to fit the partition coefficients for CTxB versus membrane curvature (Fig. 7 B). Fitting the model to the experimental results required incorporating the single-fluorophore localization imprecision, nanoparticle-induced inaccuracy, imprecision in identifying the center of the nanoparticle, and the curvature-dependent CTxB concentration. The curvature-dependent CTxB sorting was simplified to consider just three concentrations: on the SLB ([CTxB]SLB), over the top of the nanoparticle of rNP = 70 nm ([CTxB]Top), and on the membrane neck with one principal radius of curvature of 20 nm ([CTxB]Neck). Many combinations of these fitting parameters yielded similar quality fits to the experimental data. The mean and SD of adequate fits to the experimental data yielded [CTxB]Top/[CTxB]SLB = (12 ± 4) and [CTxB]Neck/[CTxB]SLB = (26 ± 11) (Fig. 7 C).
Figure 7.
When membrane buds were formed by draping SLBs over nanoparticles of known size (70 nm radius), the sorting of CTxB versus membrane curvature could be determined. (A) Many combinations of the fitting parameters yielded quality fitting to the experimental data. (B) The membrane topography over the nanoparticle could be approximated to connect the concentration of CTxB per membrane area to the acquired z-projected data. The distribution of adequate model fits to the experimental data yielded a mean and SD of CTxB concentration on the membrane top and neck relative to the planar SLB; these were (12 ± 4) and (26 ± 11), respectively, with the median, quartiles, and range of CTxB concentrations shown in (C). To see this figure in color, go online.
Membrane curvature is generated in unsupported bilayers
CTxB-induced budding was reproduced on unfused GUVs. CTxB was introduced to POPC/GM1/DiI GUVs placed on an agarose-coated coverslip to prevent their rupture. Within 2 min of CTxB addition, the suspended membranes bent and formed inward tubules and vesicles. Most commonly, small vesicular invaginations coated with CTxB were observed (Fig. S1), similar to features reported previously (10, 21). Bending away from the leaflet exposed to CTxB was observed when possible; however, bending in this analogous direction was not possible for SLBs due to the close proximity (∼2 nm) of the bilayer to the glass. When unable to bend away from the CTxB, the membrane buds and tubules grew outward, toward the CTxB, demonstrating a preference for CTxB to bind to curved membranes more than planar membranes.
Discussion
PLM is, to our knowledge, a novel microscopy technique that enables imaging membrane dynamics, organization, and topography simultaneously (25). Because PLM requires no modification to the fluorescence emission path, it is trivially coupled to other super-resolution techniques, such as multicolor STORM and PALM. Membrane buds were detected by PLM with higher signal-to-noise than any other comparable diffraction-limited or super-resolution optical technique (25).
In this article, PLM was used to reveal nanoscale membrane curvature induced by membrane-bound CTxB. PLM provided direct, super-resolution time-lapse imaging of bud initiation and growth. Each step in the progression was detected: 1) binding of CTxB to the GM1 in a planar, quasi-one-component SLB; 2) clustering of CTxB in the planar membrane; 3) inducing of nanoscale membrane buds; and 4) formation of the nanoscale membrane tubules protruding away from the coverslip. PLM has enabled detection of nanoscale bud formation and the inherent membrane bending capability of CTxB that has previously gone unnoticed.
Super-resolution images of the smallest CTxB-induced membrane buds (<100 nm diameter) reveal Gaussian-like distributions of CTxB in the imaging xy plane. These small membrane buds of radius (rbud) equal 50 ± 9 nm displayed CTxB apparently bound upon the whole curved membrane of the bud, although the distribution of CTxB on the small bud was limited by the resolution of dSTORM. As the size of the buds increased, CTxB became most concentrated at the neck of the bud and yielded a ringlike structure of CTxB localizations when rbud > 100 nm (Figs. 1, 7, and S2).
PLM distinguishes between buds and tubules
Reconstructed pPLM images were able to distinguish between membrane buds and membrane tubules by the distribution of the DiI localizations; a heterogeneous population of bud sizes was calculated at each time point (Fig. 3, A and B). The confinement of the DiI within the lipid bilayers prevents free tumbling of the fluorophore, which is critical for PLM polarization sensitivity. However, it also yields an anisotropic emission from the DiI and a systematic shift of the single-fluorophore image that is dependent on the membrane orientation and height (35). In particular, when the membrane is tilted 45° relative to the coverslip and 100 nm out of focus, the single-fluorophore image can be shifted by up to 50 nm. On membrane buds, the anisotropic emission effects on single DiI images systematically shift the localizations toward the center of the bud and reduces any ringlike distribution of DiI localizations (Figs. 1 and 2) (25). However, in the case of membrane tubules, the dominant orientations are of 0° or 90° relative to the coverslip. These orientations result in no anisotropic emission and allow the observation of clear ringlike distributions (Fig. 4). Further description of CTxB-induced membrane tubulation and the expected CTxB localization densities are given in Figs. S3 and S4.
Membrane tension affects bud formation
CTxB-induced membrane buds initially formed at the central part of the SLB patches (Fig. S5). With increasing CTxB incubation time, more buds formed at increasing distances from the SLB patch center. The bud-forming region of the SLB typically extended >10 μm away from the center of the SLB batch, and the perimeter of the patch without bud formation was commonly 5 ± 4 μm wide. Membrane buds were most likely to form in the center of the SLB patch rather than close to the edge of the SLB and the exposed glass coverslip (Fig. S5). We hypothesize this observation stems from varying membrane tension across the SLB patch, imparted by the GUV fusion process.
Variations in SLB tension could occur during two distinct stages of SLB creation by GUV fusion. The first stage would consist of the initial GUV-glass contact, before or immediately after the GUV has ruptured. The rupture of the GUV may have exposed a loose or floppy bilayer to the glass coverslip and initiated a membrane-glass contact that trapped nanoscale undulations and decreased lateral tension in the SLB. Over time, the bilayer would spread across the glass with Marangoni flow, as encouraged by the membrane-glass adhesion, and yield higher lateral membrane tension, similar to as seen previously (36).
The rate of GUV fusion was controlled by the duration of the plasma cleaning and the occasional presence of a coverslip cushion. When no GUV fusion was wanted, a thin film of agarose was used to minimize the membrane-substrate adhesion (Fig. S1). Further discussion is provided in the Supporting Material.
Membrane budding slows CTxB and DiI diffusion
The diffusion of CTxB on membranes has been reported with widely varying rates, including rates that range from 0.04 to 2.44 μm2/s within a single cell, and spatially confined in regions that are 100–1800 nm in diameter (37, 38). Even in the absence of coexisting lipid phases such as >99% DOPC model membranes, CTxB exhibited multiple diffusion populations, having one population of D = (0.18 ± 0.04) μm2/s and a second population of D = (0.06 ± 0.02) μm2/s with transient confinement in regions as small as 20 nm radii (16, 17). These prior measurements had no means of detecting changes to membrane topography or correlating topography with mobility, which is a focus of this article. The diffusion measurements reported here are consistent with both the previously reported diffusion rates and confinement sizes. In this study, the diffusion rates are presented while demonstrating local membrane curvature as a mechanism for varying CTxB behavior in a single membrane. This curvature-dependent analysis of CTxB diffusion and accumulation has the potential to explain prior measurements of both distinct populations of CTxB diffusion rates (Fig. 6) and the intermembrane molecular sorting that is independent of lipid phase.
The mechanisms by which membrane bending slows CTxB and DiI diffusion are most likely to be the result of molecular crowding, local phase separation, and/or a curvature-dependent membrane viscosity. If CTxB becomes dense enough in a local region of the bilayer, it would be expected that this crowding would slow the diffusion of CTxB and/or lipids (i.e., DiI) within the membrane. Additionally, it is feasible that the local concentration of GM1 was increased sufficiently to drive the local lipid environment into a more ordered state, and cause an increase in the effective membrane viscosity, as would be expected for more ordered lipid environment (39, 40, 41). It is not likely that the 99.4 mol % POPC bilayer would have significant phase separation, but CTxB-encouraged GM1 accumulations are possible. Further, a change in the lipid headgroups dynamics and orientation in the vicinity of the toxin may also contribute to changes in the lipid diffusion rates (42).
Finally, it is feasible that the membrane bending itself affects the local effective lipid viscosity and the free diffusion of lipids and proteins through the membrane buds. This effect was observed when membrane curvature was engineered by draping SLBs over nanoparticles and the lipid diffusion was measured with SPT (25, 43) but not when measured with fluorescence recovery after photobleaching (FRAP) (44), as expected (34). In comparing the diffusion coefficient measured by SPT to that measured by FRAP or fluorescence correlation spectroscopy, it is critical to consider the differences in sensitivity to detecting mobile versus immobile diffusers, length- and timescale-dependent processes, and subpopulations of diffusers (33, 34). The SPT results presented here are consistent with prior SPT results and, as expected, report a slower diffusion rate than FRAP or fluorescence correlation spectroscopy measurements (43, 45). If the effective membrane viscosity changes upon membrane bending, then the diffusion of the DiI molecules in both leaflets could be affected, regardless of the supporting substrate or local CTxB concentrations.
Bud formation and molecular sorting did not require lipid phase separation
SLBs of >99% POPC or DPhyPC were used for these studies due to their disordered acyl tails and the minimal possibility of phase separation with 0.3% GM1 and 0.3% DiI included. The liquid-to-gel transition temperatures for POPC and DPhyPC are −2°C and <−120°C, respectively.
A localization density rate of (1.3 ± 0.1) × 10−6 localizations/nm2/frame was desired to reconstruct the super-resolution images, such rate being obtained by utilizing a 0.3% membrane tracer (DiI, DiO, or DiD). A lower DiI composition would have prohibited the reconstruction of the pPLM images at the desired time points with a low localization density. Increasing the DiI composition is possible in future experiments for faster processes that require pPLM images reconstruction at earlier times with higher localization densities.
To minimize the possibility of a spontaneous GM1 clustering in the bilayers, a low GM1 concentration was used here (0.3 mol %); however, GM1-rich gel phases have been observed in otherwise fluid bilayers (46). For a binary system of POPC and GM1, a GM1-rich gel phase is expected to form at low temperatures and high GM1 concentrations (>1% GM1). Prior studies that reported sorting of CTxB to the neck of membrane buds required a ternary mixture of cholesterol, sphingomyelin, and POPC to observe CTxB sorting (23). CTxB sorting, in that case, might have been driven by nanoscale lipid phase separation or by the existing membrane curvature. However, curvature in these prior studies was of a significantly larger radius of curvature (micrometer scale) such that the curvature-dependent sorting was presumably weaker than that observed here (nanometer scale). Even though lipid phase separation is not likely in these experiments, it is feasible that nanodomains of GM1 were formed and were stabilized by the multivalent CTxB binding, as further discussed below.
Bud formation is energetically feasible
The spontaneous bud nucleation and tubulation are controlled by the membrane bending rigidity, density of CTxB, and the adhesion of the SLB with the substrate. For bud formation to be spontaneous, the energy released by CTxB-GM1 binding must be greater than the energy put into bending the membrane and separating the membrane from the glass. We have estimated the energy required to bend the membrane (EBend) by the Helfrich energy model (47), the energy required to release the SLB from the substrate (EAdhesion), and the energy released by the binding of the CTxB to the SLB (EBind), as described in detail in the Supporting Material. In comparing these three energetic components, we found | EBind | > | EBend + EAdhesion |. There is sufficient energy from CTxB binding to drive nanoscale bud formation spontaneously. However, further discussion is warranted to consider how much of this energy of CTxB binding is expected to drive membrane bending and the preferred radius of curvature for a membrane under CTxB.
The forces that drive budding
A complex interplay of factors could contribute to the budding process such as CTxB steric crowding (48), CTxB insertion into the membrane (49), CTxB-GM1 cross-linking, the positive intrinsic curvature of GM1 (50), GM1 clustering via lipid phase separation (51), the extended long acyl chain of GM1 causing wedging in the opposing bilayer leaflet (16), and the asymmetric GM1 concentrations in the bilayer leaflets. The steric pressure between the crowded CTxB within a nanoscale area could inherently encourage membrane bending if there was an attractive force on the GM1 to counter the steric repulsion between CTxB and provide a local membrane torque (48). An attraction between GM1 is plausible considering the strong liquid-ordered phase preference of GM1 in ternary lipid mixtures, the affinity of GM1 to self-cluster (46, 49, 51), and the possibility of GM1 unbound to CTxB further accumulating around the clusters of CTxB-GM1. A leaflet asymmetry in GM1-GM1 clustering and CTxB binding could encourage a mismatch of composition between the leaflets and encourage curvature.
Further, the molecular shape of CTxB itself is likely to encourage a negative membrane curvature. The molecular shape of CTxB and Shiga toxin both have glycolipid binding pockets that are elevated above from the bottom of the folded protein. The toxin-lipid binding encourages a penetration of the protein into the bilayer and/or a local wrapping of the membrane around the protein, as has been most explicitly shown for Shiga toxin (49). As the membrane is pulled to wrap around the toxin, a bending force would be created by the toxin with its peripheral GM1 binding pockets. A wrapping of the membrane around the toxin would encourage a negative membrane curvature. In a suspended membrane or a plasma membrane, this typically manifests as the formation of a membrane invagination, where CTxB is located on the inner leaflet with both principal planes being negatively curved. On an SLB, where vesiculation of the CTxB is inhibited by the substrate, the local wrapping of the membrane around the toxin was sufficiently preferable to drive the membrane to bud away from the coverslip. In this case, CTxB primarily partitioned on the bud neck with one principal plane of negative curvature. Although this budding toward the CTxB is not observed on plasma membranes, these experiments demonstrate the nanoscale effects of CTxB on SLBs, and the curvature induction capabilities of CTxB, broadly. The importance of the spontaneous curvature generation by CTxB is likely essential for its trafficking through the cell. Similarly, the complex curvature-dependent sorting profile of CTxB is likely impactful in complex organelle membrane topographies.
Finally, the multivalent binding of CTxB may be critical for the preference of CTxB to bind to negatively curved membranes, such as regions of negative Gaussian curvature over planar membranes. CTxB is typically saturated by binding to five GM1 molecules simultaneously; however, the number of bound GM1 molecules directly affects the orientation of CTxB on the membrane (42, 52). CTxB is parallel to the membrane when bound to five GM1, but tilted when bound to only three GM1 (42). The rotational asymmetry of the occupied GM1 binding pockets on the CTxB may result in a shifted CTxB preference to bind to one dimension of negative curvature and have a minimal preference for the membrane curvature in the other principal curvature dimension. Accordingly, if introduced upon a negatively curved membrane (i.e., inside of a vesicle), a CTxB bound to three GM1 would be stable at any rotation. However, if introduced upon a negative Gaussian curvature membrane (i.e., bud neck), the CTxB bound to three GM1s would require it to be rotated. This rotation allows the GM1s to be spaced along the dimension of negative curvature, limiting the degrees of freedom for CTxB orientation while maintaining membrane wrapping around toxin (40, 42, 52). The number of GM1 bound to the CTxB might not only affect its curvature preference, but also its diffusion rate. This might be the reason for the observed variation in the diffusion rates reported here on the flat membrane, and the importance of valency in lipid phase separation (53). Unfortunately, these explanations for the curvature induction mechanisms by CTxB are highly speculative at this time. Further experimentation to clarify the relationship between CTxB-induced membrane curvature and CTxB valency is warranted.
Membrane curvature-induced CTxB sorting
Studies regarding CTxB location at membrane curvature gradients have shown that CTxB preferentially localizes at negatively curved membrane regions (3), at the neck of micronscale engineered membrane buds (23), on tubules (37), and occasionally onto membrane ridges (Fig. S6). Here, CTxBs were observed to initially cluster and spontaneously form small membrane buds (Fig. 1 A, black arrows). As the buds grew in size, the accumulation of CTxB around the perimeter of larger membrane protrusions became increasingly apparent (Fig. 1 A, white arrow). CTxB also localized at similar membrane geometry on bilayer draped over a 70 nm-radius nanoparticle (Figs. 7 and S2). Thus, the ringlike shape of localizations obtained for CTxB demonstrates the preferential location of CTxB at the neck of the curvature. This observation falls into agreement with previous reports that observed the accumulation of CTxB at the neck of lithographically patterned spherical membrane protrusions with 25 μm diameter that was dependent on coexisting liquid lipid phases (23). By observing 20× smaller curvatures here, the curvature-based sorting forces were presumably larger and not requiring assistance by lipid phase separation for CTxB sorting to occur.
PLM detects nanoscale membrane bending
PLM enabled the detection of the membrane budding process induced by CTxB and resolved the sizes of the membrane buds at the various budding stages. PLM is a single-molecule, super-resolution, imaging technique that requires no alteration of the emission path and no sacrifice in the signal. The feasibility of performing PLM on aqueous samples under physical conditions makes it suitable for live cell imaging. Detecting the live budding process induced by CTxB reflects the ability to study dynamic processes and structures in living systems via PLM. Future improvements to PLM will include the following: a faster frame rate, a higher localization density, and a lower localization imprecision of individual fluorophores. Further details are reported in a companion article in this journal (25).
Conclusions
PLM has enabled the direct observation of CTxB effects on membrane dynamics to reveal CTxB-induced membrane budding on SLBs. In this article, PLM was used to reveal simultaneous multicolor super-resolution images of CTxB and the induced bud growth on a supported, quasi-one-component lipid bilayer. Our data provide context to prior studies with CTxB that observed time-dependent diffusion rates and diverse internalization mechanisms. We demonstrated that the molecular mobility of CTxB and DiI is affected by the nanoscale membrane structures induced by CTxB. On planar SLBs, the diffusion rates of DiI and CTxB are in agreement with previous reports on cells and synthetic membranes. However, the diffusion coefficients at the center of the induced buds are (82 ± 4) and (81 ± 9)% less than that on the planar membranes for DiI and CTxB, respectively. DiI and CTxB underwent transient confinement in regions that later appeared to be nanoscale protrusions as small as a 30 nm radius. Our studies demonstrated the budding process was reversible and dependent on CTxB concentration. PLM will aid in providing new information for previously untestable nanoscale processes coupled with changes in membrane topography. We propose a mechanism of CTxB trafficking in cells dependent on the spontaneous membrane-curvature induction and curvature-based sorting by CTxB. In this case, CTxB sorts to negative Gaussian curvature over planar membranes. In further articles, we will explore the effects of changing the GM1 structure and membrane composition on the budding process, as well as using mutant, monovalent CTxB that binds to one GM1 (53). Biological functions of the cell are dictated by the sorting, mobility, and organization of its constituents, which affect the structure of the cell membrane to facilitate diverse essential membrane processes.
Author Contributions
A.M.K. and C.V.K. designed the experiments, analyzed the data, and prepared the manuscript. A.M.K. performed the experiments.
Acknowledgments
The authors thank Xinxin Woodward, Dipanwita De, Rebecca Meerschaert, and Eric Stimpson for valuable discussions.
A.M.K. was funded by the Thomas C. Rumble Fellowship Award. Financial support was provided by Wayne State University laboratory start-up funds and Richard J. Barber. This material is based upon work supported by the National Science Foundation under grant no. DMR-1652316.
Editor: Tommy Nylander.
Footnotes
Supporting Materials and Methods and six figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30925-6.
Supporting Material
References
- 1.Heinrich M., Tian A., Baumgart T. Dynamic sorting of lipids and proteins in membrane tubes with a moving phase boundary. Proc. Natl. Acad. Sci. USA. 2010;107:7208–7213. doi: 10.1073/pnas.0913997107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tian A., Baumgart T. Sorting of lipids and proteins in membrane curvature gradients. Biophys. J. 2009;96:2676–2688. doi: 10.1016/j.bpj.2008.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsieh W.-T., Hsu C.-J., Baumgart T. Curvature sorting of peripheral proteins on solid-supported wavy membranes. Langmuir. 2012;28:12838–12843. doi: 10.1021/la302205b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mikhalyov I., Samsonov A. Lipid raft detecting in membranes of live erythrocytes. Biochim. Biophys. Acta. 2011;1808:1930–1939. doi: 10.1016/j.bbamem.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Baumgart T., Hammond A.T., Webb W.W. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc. Natl. Acad. Sci. USA. 2007;104:3165–3170. doi: 10.1073/pnas.0611357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Day C.A., Kenworthy A.K. Functions of cholera toxin B-subunit as a raft cross-linker. Essays Biochem. 2015;57:135–145. doi: 10.1042/bse0570135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammond A.T., Heberle F.A., Feigenson G.W. Crosslinking a lipid raft component triggers liquid ordered-liquid disordered phase separation in model plasma membranes. Proc. Natl. Acad. Sci. USA. 2005;102:6320–6325. doi: 10.1073/pnas.0405654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Windschiegl B., Orth A., Steinem C. Lipid reorganization induced by Shiga toxin clustering on planar membranes. PLoS One. 2009;4:e6238. doi: 10.1371/journal.pone.0006238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chinnapen D.J.-F., Hsieh W.-T., Lencer W.I. Lipid sorting by ceramide structure from plasma membrane to ER for the cholera toxin receptor ganglioside GM1. Dev. Cell. 2012;23:573–586. doi: 10.1016/j.devcel.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ewers H., Römer W., Johannes L. GM1 structure determines SV40-induced membrane invagination and infection. Nat. Cell Biol. 2010;12:11–18. doi: 10.1038/ncb1999. 1–12. [DOI] [PubMed] [Google Scholar]
- 11.Yanagisawa K. GM1 ganglioside and the seeding of amyloid in Alzheimer’s disease: endogenous seed for Alzheimer amyloid. Neuroscientist. 2005;11:250–260. doi: 10.1177/1073858405275177. [DOI] [PubMed] [Google Scholar]
- 12.Yanagisawa K., Odaka A., Ihara Y. GM1 ganglioside-bound amyloid beta-protein (A β): a possible form of preamyloid in Alzheimer’s disease. Nat. Med. 1995;1:1062–1066. doi: 10.1038/nm1095-1062. [DOI] [PubMed] [Google Scholar]
- 13.Wolf A.A., Fujinaga Y., Lencer W.I. Uncoupling of the cholera toxin-G(M1) ganglioside receptor complex from endocytosis, retrograde Golgi trafficking, and downstream signal transduction by depletion of membrane cholesterol. J. Biol. Chem. 2002;277:16249–16256. doi: 10.1074/jbc.M109834200. [DOI] [PubMed] [Google Scholar]
- 14.Stoddart A., Jackson A.P., Brodsky F.M. Plasticity of B cell receptor internalization upon conditional depletion of clathrin. Mol. Biol. Cell. 2005;16:2339–2348. doi: 10.1091/mbc.E05-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moens P.D.J., Digman M.A., Gratton E. Modes of diffusion of cholera toxin bound to GM1 on live cell membrane by image mean square displacement analysis. Biophys. J. 2015;108:1448–1458. doi: 10.1016/j.bpj.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spillane K.M., Ortega-Arroyo J., Kukura P. High-speed single-particle tracking of GM1 in model membranes reveals anomalous diffusion due to interleaflet coupling and molecular pinning. Nano Lett. 2014;14:5390–5397. doi: 10.1021/nl502536u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsieh C.-L., Spindler S., Sandoghdar V. Tracking single particles on supported lipid membranes: multimobility diffusion and nanoscopic confinement. J. Phys. Chem. B. 2014;118:1545–1554. doi: 10.1021/jp412203t. [DOI] [PubMed] [Google Scholar]
- 18.Kirkham M., Fujita A., Parton R.G. Ultrastructural identification of uncoated caveolin-independent early endocytic vehicles. J. Cell Biol. 2005;168:465–476. doi: 10.1083/jcb.200407078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lajoie P., Kojic L.D., Nabi I.R. Caveolin-1 regulation of dynamin-dependent, raft-mediated endocytosis of cholera toxin-B sub-unit occurs independently of caveolae. J. Cell. Mol. Med. 2009;13(9B):3218–3225. doi: 10.1111/j.1582-4934.2009.00732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montesano R., Roth J., Orci L. Non-coated membrane invaginations are involved in binding and internalization of cholera and tetanus toxins. Nature. 1982;296:651–653. doi: 10.1038/296651a0. [DOI] [PubMed] [Google Scholar]
- 21.Römer W., Berland L., Johannes L. Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature. 2007;450:670–675. doi: 10.1038/nature05996. [DOI] [PubMed] [Google Scholar]
- 22.Day C.A., Baetz N.W., Kenworthy A.K. Microtubule motors power plasma membrane tubulation in clathrin-independent endocytosis. Traffic. 2015;16:572–590. doi: 10.1111/tra.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryu Y.-S., Lee I.-H., Lee S.-D. Reconstituting ring-rafts in bud-mimicking topography of model membranes. Nat. Commun. 2014;5:4507. doi: 10.1038/ncomms5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pezeshkian W., Hansen A.G., Ipsen J.H. Membrane invagination induced by Shiga toxin B-subunit: from molecular structure to tube formation. Soft Matter. 2016;12:5164–5171. doi: 10.1039/c6sm00464d. [DOI] [PubMed] [Google Scholar]
- 25.Kabbani A.M., Kelly C.V. The detection of nanoscale membrane bending with polarized localization microscopy. Biophys. J. 2017;113:1782–1794. doi: 10.1016/j.bpj.2017.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dempsey G.T., Bates M., Zhuang X. Photoswitching mechanism of cyanine dyes. J. Am. Chem. Soc. 2009;131:18192–18193. doi: 10.1021/ja904588g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sund S.E., Swanson J.A., Axelrod D. Cell membrane orientation visualized by polarized total internal reflection fluorescence. Biophys. J. 1999;77:2266–2283. doi: 10.1016/S0006-3495(99)77066-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anantharam A., Axelrod D., Holz R.W. Polarized TIRFM reveals changes in plasma membrane topology before and during granule fusion. Cell. Mol. Neurobiol. 2010;30:1343–1349. doi: 10.1007/s10571-010-9590-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anantharam A., Axelrod D., Holz R.W. Real-time imaging of plasma membrane deformations reveals pre-fusion membrane curvature changes and a role for dynamin in the regulation of fusion pore expansion. J. Neurochem. 2012;122:661–671. doi: 10.1111/j.1471-4159.2012.07816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van de Linde S., Löschberger A., Sauer M. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 2011;6:991–1009. doi: 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]
- 31.Veatch S. Electro-formation and fluorescence microscopy of giant vesicles with coexisting liquid phases. In: McIntosh T., editor. Lipid Rafts. Humana Press; New York, NY: 2007. pp. 59–72. [DOI] [PubMed] [Google Scholar]
- 32.Ovesný M., Křížek P., Hagen G.M. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30:2389–2390. doi: 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagerholm B.C., Andrade D.M., Eggeling C. Convergence of lateral dynamic measurements in the plasma membrane of live cells from single particle tracking and STED-FCS. J. Phys. D Appl. Phys. 2017;50:063001. doi: 10.1088/1361-6463/aa519e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kabbani, A. M., X. Woodward, and C. V. Kelly. 2017. Revealing the effects of nanoscale membrane curvature on lipid mobility. https://arXiv.org/abs/1706.00087. [DOI] [PMC free article] [PubMed]
- 35.Lew M.D., Backlund M.P., Moerner W.E. Rotational mobility of single molecules affects localization accuracy in super-resolution fluorescence microscopy. Nano Lett. 2013;13:3967–3972. doi: 10.1021/nl304359p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lobovkina T., Gözen I., Orwar O. Protrusive growth and periodic contractile motion in surface-adhered vesicles induced by Ca2+-gradients. Soft Matter. 2010;6:268–272. [Google Scholar]
- 37.Day C.A., Kenworthy A.K. Mechanisms underlying the confined diffusion of cholera toxin B-subunit in intact cell membranes. PLoS One. 2012;7:e34923. doi: 10.1371/journal.pone.0034923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Zanten T.S., Gómez J., Garcia-Parajo M.F. Direct mapping of nanoscale compositional connectivity on intact cell membranes. Proc. Natl. Acad. Sci. USA. 2010;107:15437–15442. doi: 10.1073/pnas.1003876107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Štefl M., Šachl R., Hof M. Dynamics and size of cross-linking-induced lipid nanodomains in model membranes. Biophys. J. 2012;102:2104–2113. doi: 10.1016/j.bpj.2012.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watkins E.B., Miller C.E., Kuhl T.L. Membrane texture induced by specific protein binding and receptor clustering: active roles for lipids in cellular function. Proc. Natl. Acad. Sci. USA. 2011;108:6975–6980. doi: 10.1073/pnas.1014579108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johannes L., Mayor S. Induced domain formation in endocytic invagination, lipid sorting, and scission. Cell. 2010;142:507–510. doi: 10.1016/j.cell.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 42.Basu I., Mukhopadhyay C. Insights into binding of cholera toxin to GM1 containing membrane. Langmuir. 2014;30:15244–15252. doi: 10.1021/la5036618. [DOI] [PubMed] [Google Scholar]
- 43.Cheney P.P., Weisgerber A.W., Knowles M.K. Single lipid molecule dynamics on supported lipid bilayers with membrane curvature. Membranes (Basel) 2017;7:15. doi: 10.3390/membranes7010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Black J.C., Cheney P.P., Knowles M.K. Membrane curvature based lipid sorting using a nanoparticle patterned substrate. Soft Matter. 2014;10:2016–2023. doi: 10.1039/c3sm52522h. [DOI] [PubMed] [Google Scholar]
- 45.Bag N., Yap D.H.X., Wohland T. Temperature dependence of diffusion in model and live cell membranes characterized by imaging fluorescence correlation spectroscopy. Biochim. Biophys. Acta. 2014;1838:802–813. [PubMed] [Google Scholar]
- 46.Becucci L., Vizza F., Guidelli R. The GM1 ganglioside forms GM1-rich gel phase microdomains within lipid rafts. Coatings. 2014;4:450–464. [Google Scholar]
- 47.Helfrich W. Elastic properties of lipid bilayers: theory and possible experiments. Z. Naturforschung C. 1973;28:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- 48.Stachowiak J.C., Schmid E.M., Hayden C.C. Membrane bending by protein-protein crowding. Nat. Cell Biol. 2012;14:944–949. doi: 10.1038/ncb2561. [DOI] [PubMed] [Google Scholar]
- 49.Sun H., Chen L., Fang W. Nanodomain formation of ganglioside GM1 in lipid membrane: effects of cholera toxin-mediated cross-linking. Langmuir. 2015;31:9105–9114. doi: 10.1021/acs.langmuir.5b01866. [DOI] [PubMed] [Google Scholar]
- 50.Kabaso D., Lokar M., Iglič A. Temperature and cholera toxin B are factors that influence formation of membrane nanotubes in RT4 and T24 urothelial cancer cell lines. Int. J. Nanomedicine. 2011;6:495–509. doi: 10.2147/IJN.S16982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi J., Yang T., Cremer P.S. GM1 clustering inhibits cholera toxin binding in supported phospholipid membranes. J. Am. Chem. Soc. 2007;129:5954–5961. doi: 10.1021/ja069375w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sridhar A., Kumar A., Dasmahapatra A.K. Multi-scale molecular dynamics study of cholera pentamer binding to a GM1-phospholipid membrane. J. Mol. Graph. Model. 2016;68:236–251. doi: 10.1016/j.jmgm.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 53.Raghunathan K., Wong T.H., Kenworthy A.K. Glycolipid crosslinking is required for cholera toxin to partition into and stabilize ordered domains. Biophys. J. 2016;111:2547–2550. doi: 10.1016/j.bpj.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.