Abstract
Confined mixtures of a polymer and nonspecifically binding particles (condensers) are studied as models for viruses containing double-stranded DNA (polymer) and condensing proteins (particles). We explore a model in which all interactions between the packed content (polymer and particles) and its confinement are purely repulsive, with only a short-range attraction between the condensers and polymer to simulate binding. In the range of physical parameters applicable to viruses, the model predicts reduction of pressure in the system effected by the condensers, despite the reduction in free volume. Condensers are found to be interspersed throughout the spherical confinement and only partially wrapped in the polymer, which acts as an effective medium for the condenser interactions. Crowding of the viral interior influences the DNA and protein organization, producing a picture inconsistent with a chromatin-like, beads-on-a-string structure. The model predicts an organization of the confined interior compatible with experimental data on unperturbed adenoviruses and polyomaviruses, at the same time providing insight into the role of condensing proteins in the viral infectious cycles of related viral families.
Introduction
Virus genomes are confined inside a capsid assembled from proteins. In viruses with a double-stranded DNA (dsDNA) genome, confinement of a highly charged DNA molecule may result in huge internal pressures acting on the capsid, depending on the DNA length (25–100 atm in some phages (1, 2)). Such pressures can be reduced through the action of multivalent ions (3) and basic proteins like histones (4). Basic (condensing) proteins have been found accompanying the genome in particles of polyoma- (5, 6) and papillomaviruses (7), as well as in adenovirus (8), baculovirus (9), poxvirus (10, 11, 12), chlorovirus (13), African swine fever virus (14), mimivirus (15), and marseillivirus (16). Most of these condensing proteins have unresolved structures, leaving open questions regarding their DNA-binding mechanism and mode of action, except in polyoma- and papillomaviruses, which have been shown to borrow cellular histones to pack their DNA into a “minichromosome” (7, 17).
The presence of proteins within the capsid may seem a nuisance: they take up the volume that would otherwise be available to viral DNA. Obviously, not just any protein will serve for compacting purposes—only those that bind DNA sufficiently strongly while using up the smallest volume possible. Understanding the role of condensing proteins in packing of genomes inside viral capsids is important in the context of the whole viral infection process and the host cell response (18) and may also provide an evolutionary context, as some of the viruses using them, such as adenovirus and mimivirus, seem to be evolutionarily related (19). Condensing proteins may have evolved in parallel with new viral genes, as a physical support for compacting the longer genome, or their genes may have been hijacked from the host and adapted. Understanding the role of condensing particles (proteins and nanoparticles) is also important in the context of targeted cargo delivery with viruses or other nanocages (20) and gene therapy (21, 22). Packing and ejection of dsDNA in bacteriophages (23) has been intensively studied using various numerical methods (24, 25) that helped elucidate physical aspects of the bacteriophage infectious cycle (26). Although the compaction of eukaryotic and bacterial DNA with condensing proteins is also a well-known problem (4), the mode of action of condensing proteins present within the confines of viral capsids is still a mystery.
Most of the evidence on nucleoprotein structures in viruses with DNA condensing proteins is based on the study of disrupted particles. Data on the structure of the core in intact viruses are scarce due to lack of order or symmetry in the particle contents. Disrupted viral cores show cluster, fiber-like, and bead-on-a-string structures reminiscent of those found in cell chromatin (4) as observed by electron microscopy (EM) after disruption of capsids of SV40 (27), human and bovine papillomavirus (7), and adenovirus (28, 29, 30). Nucleoprotein filaments have also been found using atomic force microscopy in mimiviruses (15), vaccinia (31), and chloroviruses (13). Cryoelectron tomography showed dense areas of presumed DNA-protein complexes in disordered vaccinia capsid interiors (11). Standard methods used to reach high resolution in studies of virus structure rely on averaging data from many particles and enforcing high symmetry, but they give little insight into the core structures, which may be neither ordered nor identical in all particles. Icosahedrally averaged cryo-electron microscopy (cryo-EM) results of studies on adenovirus capsids (28, 32, 33) indicate a more or less flat density profile that lacks the DNA shelling typical of viruses packing naked dsDNA (34, 35, 36). Similar to adenovirus, the polyomavirus SV40 cores have a flat density profile seen by both cryo-EM (37) and small-angle x-ray scattering (5), with no pronounced DNA ordering. A recent cryo-electron tomography study on undisturbed adenovirus virions, where no averaging or symmetry enforcement was used, showed a disordered virus interior containing ∼200 objects that are assumed to consist of condensing proteins binding or wrapping the DNA (38). There are no structural data for any of the adenovirus core proteins (called V, VII, and μ) (33). Two of these have been proposed to contribute to DNA condensation via two universal mechanisms (39): bridging two dsDNA strands (protein μ) or wrapping the DNA (protein VII) (40). EM images of disrupted adenovirus cores showed a bead-on-a-string architecture, where beads would be mainly formed by protein VII bound to the DNA (29, 30). The beads (also called “adenosomes” by analogy with cellular nucleosomes) had an estimated diameter between 2 and 10 nm, and might correspond to the 200 objects observed by cryo-electron tomography inside the intact particle (38). Distributions of adenosome locations extracted from the three-dimensional maps of individual (nonaveraged) particles were explained using a model that treated adenosomes as particles with mutual interactions mediated by the (effectively smeared out) background DNA medium. Elaborations of the model that included the “backbone” between the adenosomes, representing them as beads on a DNA string, were found to produce worse agreement with experiments and adenosome distributions incompatible with the experiments. In a somewhat similar model proposed for SV40, the core was described as a “molten droplet” containing ∼20 nucleosome-like entities bound together by the effective backbone of DNA (5). The SV40 and the adenovirus cases may not be comparable, however. The histones in SV40 are evolved to bend and bind the DNA in chromosomes, and they are likely to perform a similar function within SV40. Much less is known about condensing proteins in adenovirus and they may be quite different from histones.
The role of condensers in virus morphogenesis is also unknown. Two assembly pathways are possible: 1) a sequential pathway, where the DNA is injected into a preformed capsid using a motor complex, or 2) a concurrent pathway, where the capsid is assembled around the genome. The sequential pathway is well established for viruses packing naked DNA, such as bacteriophages (41), but the mode of assembly is much less clear for viruses containing protein condensers. It is in general accepted that polyomavirus assembly follows a concurrent pathway (42). For adenovirus, it has long been assumed that assembly and packaging would occur sequentially, but evidence supporting a concurrent pathway has recently been reported (43). Intriguingly, one of the adenovirus condensing proteins is dispensable for assembly but required for proper uncoating (44). Proteins VII and μ are cleaved by the adenovirus maturation protease, and this cleavage may change their interactions with DNA: after maturation, their condensing action is decreased, resulting in an increase of internal pressure involved in metastabilization of the viral particle for successful initiation of uncoating when entering the host cell (28, 40, 45, 46).
In short, the exact mechanism of DNA binding by the condensing proteins in viruses is unknown; it is not clear whether dsDNA packed together with condensing proteins exhibits some kind of organization and whether the condensing proteins promote it or disrupt it, and there is considerable uncertainty regarding the precise role these proteins play in the virus cycle. All these uncertainties call for a theoretical approach able to encompass general features of a confined and crowded mixture of DNA with condensing particles. Our aim is to study the most basic features of the problem in a simplified model, with the hope of explaining features observed experimentally and providing the basis for future construction of more detailed models.
In previous studies of DNA and condensing proteins inside viruses, the DNA was treated implicitly, either as a tethering bond between proteins or as an effective medium renormalizing the inter-protein interactions (5, 38). In this work, we propose and evaluate a model that explicitly treats the condensing proteins and the DNA polymer in confinement. The explicit treatment of the DNA allows for the examination of DNA-protein complexation without assuming it from the start. It should also shed additional light on the ordering of the core, as DNA appears with its degrees of configurational freedom and not only as a background or linker/spring. The model we use is necessarily simplified, because the DNA, the proteins, and the virus interior are too complex to be treated in all their molecular detail. Still, the same model framework has successfully been applied to naked DNA packing in viral capsids (25, 47), as well as to systems where condensing particles are added to a dilute “solution” of DNA (48). The additional reason for the simplification of the model is to identify the robust aspects of the system. Our approach does not take into account the exact protein shape and amino acid composition, and thus excludes any specificity in binding to the genome. The model should thus be viewed not merely as a description of present day, evolved viruses, but rather as a deliberately simplified variant constructed to elaborate on the possible evolutionary purpose and mechanisms behind compacting proteins. It should serve to pinpoint situations where condensing proteins may be required to compact and pack the DNA, as well as situations where their presence is a tolerable disturbance.
Methods
The system we study comprises two particle types: 1) spheres representing condensing agents and 2) polymer beads on a string, representing DNA, all confined inside a sphere of radius Rc. The model used for condensing particles and polymer is based on previous studies in the literature (25, 47, 48). Notice that in the terminology used here, one condensing particle does not necessarily correspond to a single protein molecule, since homo- or hetero-oligomers may exist and our model does not make any assumption in this respect. To explore the possible configurations of our system, molecular dynamics simulations were performed using the Langevin thermostat as implemented in LAMMPS (49, 50). All particle and particle-confinement interactions are exclusively repulsive, except for condenser-polymer interactions, which have a short-range attractive interaction. DNA connectivity is imposed by finitely extensible nonlinear elastic (FENE) springs acting between neighboring beads (47). The polymer resistance to bending is introduced via the Kratky-Porod potential, which depends on the angle between two neighboring bonds (springs) in the polymer reproducing the polymer persistence length, (51). The number of polymer beads, , and their radius, , determine the volume fraction, , they occupy in confinement, such that , where is a correction due to small overlap of neighboring beads. Condensing particles (proteins and condensers) are represented as spheres of radius (in units of ) interacting with each other via (repulsive only) excluded-volume interactions. Their interaction with polymer beads is represented by a short-range, almost-contact, nonspecific attractive potential, such that the energy gained in a “bond” is . The attractive part of the potential is constant and acts only in a spherical shell around the condenser particle, with interior and exterior radii of and , respectively. There are condensers in the system with volume fraction , where . The properties of such a system are studied by Langevin dynamics simulations as explained in detail in Supporting Material, Section S1. The configurations for different random starting conditions of the system are sampled after sufficient time has passed, so that neither the internal energy nor any of the studied indicators change with time. The initial states of the system represent a random configuration of DNA and polymers within the capsid confinement. We have also tested how a change of protocol for generating starting configurations (e.g., starting from a looser confinement to simulate the situation before genome packaging, Supporting Material, Section S2) or the addition of screened electrostatic interactions extended to include many neighbors (Supporting Material, Section S3) would change the studied indicators, and we have found no significant influence.
To probe the internal organization of the packed DNA and condensing particles, we use the density distribution of particles inside the confinement and correlations between particle positions, along with how the DNA wraps around the condensing particles. The procedures used are explained in detail in the next subsections.
Structural indicators
The radially symmetric (angle-averaged) density, , was calculated by determining the probability of finding a particle in a spherical shell, . The obtained density distributions were normalized to represent the probability density, , so that . We opt to present the probability density, and not the particle density, as it allows direct comparisons between different condenser sizes and different particle numbers in the system.
To determine the correlations between positions of condensing proteins, we use the normalized (reduced) radial distribution function (RDF), (52). The normalized RDF is obtained by calculating the RDF, , and normalizing it to account for the finite size and shape of the “sample” in question. The RDF, , is defined as
| (1) |
where N is the number of particles and the average particle density, and the sum is averaged over all particle pairs at a mutual distance, , in the configuration. The particle density has the volume, V, which does not necessarily correspond to the whole volume of the confinement, . In some cases, condensers might not access the whole interior due to being bound to the polymer and preferring to be away from the repulsive confinement. The function obtained on a finite sample, in our case a spherical cluster of particles, decays to 0 for , as the largest distance between two particles in such a cluster is about two times the cluster’s maximal radius. To compare correlation functions to bulk samples, or two samples of different size, one needs to renormalize the size (and shape) effects. The normalized RDF is then defined as , where is the shape factor. The shape factor has the property and decays to zero at sufficiently large d. The shape factors can be obtained in analytical form for most basic shapes with homogeneous density, but in our case, although the shape is spherical, we do not have a homogeneous particle density. The shape factor normalizes the radial density-density distribution function (52)
| (2) |
where f(d) represents the probability of finding two units of density at a mutual separation of d. is a normalization constant ensuring that .
Our normalized RDFs, , are obtained by first calculating the shape factor from the corresponding particle density function, . We then use the sum rule property of the shape factor (52)
| (3) |
which gives us the volume of the sample. This sum rule allows us to recheck the effective radius of confinement in our simulation runs, and we find that the confinement radius is always within 1% of the specified value. The RDF, , is then calculated with the obtained true particle volume, V, for each configuration, and then averaged over all configurations obtained in simulations. After that, we use the shape factor to obtain the normalized RDF, .
Wrapping indicator
To study the polymer wrapping around a condenser, we need to define what constitutes a bead bound to a condenser. A polymer bead at and condenser at are considered bound if their center-to-center distance, , is such that the bead-condenser interaction is attractive, i.e., . This corresponds to the bead being located in the attractive part of the condenser-DNA interaction potential.
The wrapping number, w, of a particular condenser is defined as the longest length of polymer, in number of beads, that is bound to it. For each snapshot, we make a probability distribution function, , that any condenser in the current configuration has the longest continuous length of polymer consisting of w beads. The total probability of such a function is normalized to unity . The value thus corresponds to the probability that a condenser has no polymer beads bound to it, whereas corresponds to the probability that a condenser has the longest number of continuously wound beads equal to w. Note that does not give any information on the number and distribution of shorter bound segments , or whether there are multiple segments of length w. As such, w is an indicator of the maximal achieved wrapping length on the condenser.
Results
In the following text, all units in the model are reduced so that and (Boltzmann energy). The correspondence between our model and DNA is obtained by taking nm leading to three basepairs per polymer bead (47) and pressure units of atm. To make parallels to real systems, we use a capsid radius of matching the internal mid-radius of adenoviruses ( nm) (28) and comparable to that of SV40 virions (18 nm) (53). Although the short-range part of the electrostatic repulsion is accounted for by the model, we do not include long-range electrostatic repulsion. Coarse-grained modeling of SV40 cores showed that steric interactions are sufficient to explain the internal structure at such high crowding, as the electrostatic interactions provide a minor correction (5). The crowdedness of the system is determined by volume fractions of the polymer and the spheres. Viruses that pack DNA using condensing proteins have smaller DNA densities than typical bacteriophages (T7, ϕ 29, T4, or λ), which have genome volume fractions (at least at dense packing) in the capsid several times larger than SV40 (23) or adenoviruses (38). We estimate the volume occupied by DNA of an effective diameter of 2.5 nm to be for adenovirus (38), for vaccinia (12), for SV40 (5), for chloroviruses (13), and for mimivirus (15). These viruses, all of which are thought to use proteins to compact the DNA, should have sufficient free space in their cores to permit the addition of condensing proteins. In the following, we fix the polymer (DNA) volume fraction at , consistent with the viruses of interest and representative of adenovirus, and we study how a change of other parameters modifies the organization of DNA and condensers in the core.
Pressure of the confined mixture
An important indicator of the state of the viral core is the pressure it exerts on the capsid, which depends on the amount of packaged DNA (54). The resulting stress on the capsid is different for various capsid structures (55) and is known to cause capsids to burst (56). A comparison of pressures for different condenser-DNA binding strengths indicates the existence of two compaction regimes (see Fig. 1 a) determined by the magnitude of binding energy. In a weak (strong) binding regime, is smaller (larger) than the thermal energy. For comparison, histones are assumed to have a binding strength of per binding point to DNA in physiological conditions (4), which would correspond to a strong binding regime in our case. The strong binding enables reduction of pressure on the capsid to below the pressure produced by the polymer packed without condensers, at least for a range of condensed volume fractions. In the weak binding regime, the pressure on the capsid always increases, irrespective of the condenser volume fraction. This means that the condensers’ entropic contribution to the pressure dominates over the binding energy they introduce to the system. Weak binding and/or large condensers do not help the compaction of the genome, yet this does not exclude the existence of such particles in the virus, as their biological role may be different (e.g., facilitation of other aspects of the infection process).
Figure 1.
Pressure on the capsid from a confined mixture of DNA and condensing particles. (a) Pressure versus condenser-DNA interaction energy, , for a DNA volume fraction of and condenser volume fraction of . Data are shown for three different condensing particle radii, . The horizontal dashed line shows the pressure value without condensers . (b) Pressure versus volume fraction, , of condensing particles for a DNA (polymer) volume fraction of and attractive interaction energy between condensers and polymer of . Data are shown for three different condensing protein radii, . (c) Pressure as a function of DNA (polymer) volume fraction, , for three different cases: strongly binding condensing particles with sizes and and no condensing particles, . The shaded area corresponds to typical ranges of volume fractions found in viruses discussed in the text. To see this figure in color, go online.
Fig. 1 b shows the pressure exerted on the capsid by the mixture of polymer and condensers as a function of the volume fraction of condensing particles, ϕs, for different condensing-particle radii, , and (strong binding regime; see below). When only a polymer is confined , the pressure exerted on the capsid is a growing function of polymer volume fraction, (Fig. 1 c). The addition (increase of ) of the strongly binding condensers reduces the pressure until a minimum is reached for a certain, optimal . Further addition of condensers increases the pressure of the mixture, since at high volume fractions, steric repulsion becomes the dominant force and the pressure increases irrespective of the size or binding strength of the condensers. The pressure reduction depends on the size of the condensers, smaller condensers being more efficient. Sufficiently large condensers (depending on ) become inefficient and always increase the pressure of the mixture. The minimal volume fraction of a condensing protein in adenoviruses can be calculated from the hard-core radius of adenosomes estimated from experimental data in (38). For 230 adenosomes of radius nm this gives . Alternative estimates that utilize the information on the total protein content presumed to reside in adenosomes and using typical densities of globular proteins (57) would produce a volume fraction two to three times larger. In the following analysis, we take as the value representing the volume fraction of the adenosome hard cores found in our previous work. Note that the value extracted from the experimental data on adenovirus corresponds well with the interval of values that minimize the internal pressure (from ∼0.06 to ∼0.08, see Fig. 1 b). One could thus speculate that the number of condensing proteins in a virus is tuned to minimize its internal pressure.
We find that condensers efficiently reduce the pressure (by almost an order of magnitude, depending on their size) only below about (Fig. 1 c). This corresponds to the range of DNA volume fractions in viruses that contain proteins in the core (, as estimated before), which points to their role in genome compaction and pressure reduction. The efficiency of pressure reduction depends on the size of the condensers. The maximal number of binding sites that can be realized per condenser is proportional to its surface area, , whereas the number of condensers, , is proportional to . This means that the free energy that can be realized from binding scales as . Thus, for the same volume fraction, the smaller condensers have a larger exposed surface, which enables them to permeate the crowded DNA structure better and realize more bonds with the DNA they compact—making larger condensers less efficient.
Structure of the confined condenser-polymer mixture
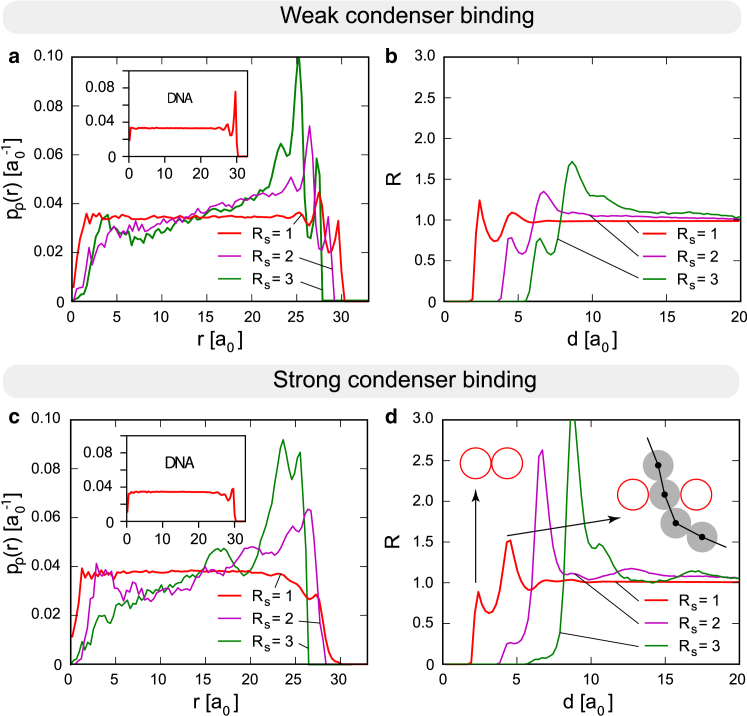
To gain insight into the structure of the condenser-polymer mixture, we examined the particle density distributions and the correlations between condenser particle positions (Fig. 2). The angle-averaged probability density of condenser particles, , based on the particle density, (see Methods), represents the probability of finding a particle at a radial distance, r, from the capsid center normalized so that the total probability is unity. depends on both the condensing interaction energy, , and the size of the condensing particles, . In the weak binding regime , the density of the condensers is either constant (small condensers, ), or increases upon approaching the confinement interior surface (larger condensers, ). In all cases, layering of condensers is present on the interior surface of the confining sphere (capsid) despite there being no attractive interactions with the confining surface. Surface layering is a well-known phenomenon in models that contain either only spherical particles (58) or only DNA polymers (59) and originates in the confinement wall inducing “ordering.” Here, we observe it in the case of a sphere-polymer mixture. Interestingly, we observe layering both in the density of the condensers and in the polymer density (see Fig. 2 a, inset). A similar layering of the total particle density is seen in BK polyomavirus, whose interior density exhibits several discernible layers near the interior capsid surface (6). The particle layering near the capsid surface has been previously detected not only in density but also in the orientation of elongated condensing proteins in the coarse-grained model of SV40 (5). Layering in is weaker in the strong binding regime (Fig. 2 c), especially when the condensers are small . Density for sufficiently small condensers in the strong binding regime shows good agreement with experimentally determined densities. In both cases, a flat density profile in the core and a gradual decay of the density near the interior surface of the confinement (a depletion region) are observed (38). The total density as detected by EM would be a weighted sum of DNA (Fig. 2, a and c, insets) and protein densities. The DNA density shows little layering, and the weighted sum of the two densities corresponds well to density profiles measured in viral cores, which is also mostly constant near the center of the core and features a depletion region near the capsid interior surface (5, 38). Larger condensers tend to occupy regions closer to the confining surface with greater probability, so that their density depletes from the core center and accumulates near the core surface. A somewhat similar phenomenon is found in vaccinia, where an increase in density is seen at the inner-core rim (11). For comparison, protein units that most likely perform the condensing role in vaccinia and mimivirus have rough diameters of nm (12) and 6 nm (15), respectively, comparable to the nm diameter of adenosomes in adenoviruses (38). With a better knowledge of condensing protein structure, as well as interactions of DNA and condensers with the capsid, it would be possible to make a more rigorous prediction of the interior structure based on protein size and interactions.
Figure 2.
Internal structure of confined mixtures in the weak and strong binding regime. (a) Radial probability density, , for finding a condenser in a spherical shell centered at r, for different condenser radii, , with binding parameter (weak binding). (b) RDF for condensers as a function of the interparticle distance, d, for different condenser radii, , in the weak binding regime . (c) The same as in (a), but with binding parameter (strong binding). The inset shows the probability density for DNA polymer beads. (d) The same as (b) but in the strong binding regime . Two schematic representations of direct contact between two condensing proteins and indirect (condenser-bead-condenser) contact are shown with arrows pointing from the corresponding correlation peak in the case. All plots (a–d) show data for , corresponding to polymer beads, , and stiffness To see this figure in color, go online.
The structure and order of the mixture is further characterized by the RDF of the system. The variant used here is normalized to account for finite size effects so that it saturates at 1 at large inter-particle distances, d, i.e., it can be readily compared to RDFs characteristic of bulk systems (see Methods). Two characteristic peaks are observed in the RDFs in Fig. 2, b and d. The first is at the position of direct contact between two condensers, , whereas the second is at and corresponds to the configuration in which the two condensers have a polymer bead between them. The first peak shows that although there are no attractive interactions between two condensers, they are still forced to be in direct contact due to crowding and polymer topological constraints. The second peak is more prominent for larger condensers, i.e., in such systems, it becomes less likely for the two condensers to touch each other, whereas the probability of a single-bead-mediated contact increases. For sufficiently large condensers, the first peak vanishes and all interactions between the condensers become effectively mediated by a polymer layer around them—condensers are better able to realize bonds with the polymer despite the polymer backbone imposing topological constraints. A suppression of direct contacts between condensers and the confinement is seen also in as the binding strength is increased. It should be emphasized that the RDF peaks represent positional correlations between condensers due to a high confinement density, and that they do not necessarily signify tethering of the condensers by the DNA. Whether the polymer wraps around condensers, connecting them to form a beads-on-a-string structure, cannot be answered with the RDF.
In our previous work (38), we suggested DNA wrapping around the condenser as a possibility, but without sequence-specific binding and with DNA easily sliding around the protein. As our new model explicitly accounts for the polymer (DNA), we can examine whether wrapping of the condensers occurs in the equilibrium state of the core. We introduce a wrapping number, w, as the largest number of monomers (beads) in a continuous polymer segment directly attached/bound to the same condenser. Thus, represents a single contact between the condenser and the bead, whereas corresponds to the situation where w successive polymer beads bind to the condenser. It should be remembered that w is an indicator of the maximal achieved wrapping length on a particular condenser—it does not contain information on the length distribution of all bound polymer segments. Fig. 3, a and b, shows the probability distribution for w averaged over all the condensers. For small condensers , we see that the most probable wrapping number is 2 in both weak and strong binding regimes, leading to a very incomplete wrapping. For larger condensers (e.g., ), the probability that is ∼20% in the strong coupling regime. In the ideal case of all four polymer beads being in a plane, this corresponds to only about a quarter of a full circle. The probability of half-circle and more complete wrappings is practically negligible (see Fig. 3), in line with the lack of specific interactions promoting wrapping. An increase of condenser radius should enable a larger w on average, due to geometry only. The obtained growth of mean w with is linear in and, somewhat surprisingly, shows no major dependence on the interaction strength. This indicates that the condenser-polymer complexation is mostly influenced by the geometric constraints effected by the confinement, with the strength of the attractive interaction playing a lesser role.
Figure 3.
Connectivity of DNA and condensing particles in confinement. (a) Probability distribution for the wrapping number, w, averaged over all representative configurations of the system in the weak binding regime. (b) Same as in (a), but for strong binding . The cartoon shows a schematic of the wrapping geometry for a polymer with , corresponding to or about one-fourth of a full turn of DNA around the condenser. (c) Equilibrium state of the system in the strong binding regime. (d) Same as in (a), but after removal of the confinement. (e) Same as in (b), but after removal of the confinement. (f) Equilibrium state of the system in the strong binding regime and after removal of the confinement. Dashed lines in (d) and (e) represent simulations done when starting from a random configuration of DNA and condensers in free space (i.e., configurations without a history of confinement). To see this figure in color, go online.
To better understand the degree to which confinement determines the structure, we study how its removal influences the mixture. After the capsid is removed and a sufficient time has passed for the system to equilibrate, we observe that the mixture relaxes, but it still retains the overall shape of a cluster (Fig. 3 f). When the condensers are small and weakly binding , almost all of the condensers leave the cluster (Fig. 3 d), which is left to unravel after sufficient time. The wrapping numbers of the mixtures without spherical confinement are shown in Fig. 3, d and e. In the weak binding regime, the majority of small condensers do not bind the polymer at all, as they escape the cluster—this is seen as a large probability of . As our simulation starts from a preformed DNA and protein cluster, it can be argued that after sufficient time, the structure will unravel and loose all its condensing proteins. To test this possibility, we performed a simulation of the same DNA length and number of proteins in a simulation box of the same size as the case with a history of confinement but starting from a random configuration without confinement. In Fig. 3, d and e, we can see that for weak binding, almost all of the condensing particles can be lost, whereas with strong binding, >60% of condensers are bound to the DNA. Thus, one would expect the DNA cluster to unravel after sufficient time in the weak binding case, whereas in the strong binding case (Fig. 3 e), a compact structure will remain. This is relevant for capsid assembly, but also for core disassembly, as weak binding cannot support compact structures that might be required for capsid formation, whereas strong binding retains compact structures, thus limiting the release of the genome in the later stages of uncoating. In all cases, the wrapping number decreases when the confinement is removed, indicating a relaxation of the cluster. Confinement promotes wrapping of the polymer on condensing particles, despite the lack of a preference for such behavior in the interactions.
Discussion
Molecular dynamics simulations of mixtures composed of a polymer and the particles that condense it in spherical confinement were performed with the aim of relating the results to viruses that use condensing proteins to pack dsDNA. We found two binding regimes that depend on the condenser-polymer binding strength and that differ with respect to whether condensers can reduce the pressure induced by the packed polymer (strong binding) or not (weak binding).
The density of condensers inside the confinement, and their RDFs, provide a picture of a disordered organization. The density profiles are consistent with the lack of symmetry and order observed in adenoviruses (33, 38), polyomaviruses (5, 6), mimivirus (15), and vaccinia (11) and in stark contrast to the ordered dense packing of DNA in bacteriophages (36). The obtained profiles, although based on a simplified model, are able to cover a range of behaviors seen in experiments, from flat densities, decaying close to the capsid (SV40 and adenovirus) (5, 6, 28, 38), to an increase in the probability of finding condensing particles near the capsid surface in vaccinia (11) and BK (6). However, the RDFs of the condensers indicate some degree of positional correlation. Comparison of the RDFs calculated here with those obtained for condensing proteins in adenoviruses (38) reveals that our model can reproduce two previous findings rooted in experiments. First, there exists a regime in which condensers interact only through a polymer medium, consistent with our previous observations in adenoviruses (38). A tightly bound entity (“quasi-particle”) of a condensing particle and its polymer “coat” is found in which DNA acts as a medium for condenser-condenser interactions. Second, our model does not reduce to proteins tethered by wrapped-around DNA, which could be modeled as an effective spring between them. Each condenser is only partially wrapped by the DNA-like polymer and no positional correlation is seen in condensers beyond the polymer-mediated contact of quasi-particles. It was found in earlier studies that after opening of adenovirus or SV40 capsids, the core assumes a beads-on-a-string configuration (17, 29, 30). However, it has also been shown that the state of the SV40 minichromosome depends on the experimental conditions (60) and, more generally, that a picture of a uniform and regular chromatin structure in vivo is being challenged (61, 62). Thus, care must be taken when extrapolating experiments in diluted solutions to crowded environments like the viral capsid. The model also successfully reproduces the decay of with r, i.e., it features a depletion layer seen also in adenoviruses, so it seems that it can successfully explain the most salient features of the internal structure in adenoviruses, despite its simplicity.
This model explains features observed in adenovirus maturation, which are critical for successful propagation of the virus. As explained above, during maturation, the condenser adenovirus proteins VII and μ are cleaved by the virus protease (63). The observed effects of these cleavages are an increase in internal pressure (40) and a decompaction of the core, such that when the capsid is open, the immature core still forms a tightly knitted sphere, whereas the mature core easily spreads out of the open shell (28, 45, 46). Both aspects (pressure increase and core decompaction) are consistent with our model prediction on the effects of a change of regime from a strong polymer-condenser interaction (before maturation) to a weaker one (after). On the other hand, a weakening of DNA-condenser interactions is necessary for successful genome uncoating. The stability of the genome-protein condensate without confinement is also important for the assembly pathway of virus particles. Adenoviruses may follow a concurrent pathway (43) where capsids assemble around a preformed DNA-protein condensate. Our simulations without confinement support this mode of assembly, with strong binding producing stable DNA-protein structures that could provide a base for stable capsid assembly (64).
Based only on modeling effective particles of adenosomes, we previously estimated a minimal internal pressure of ∼0.1 atm in mature adenovirus (38). Assuming nm, in the model presented here (with and and between 1 and 2), adenovirus would have a pressure of at least ∼0.8 atm in the strong binding regime and ∼2 atm in the weak binding regime, depending on the true size of the condensing particles, with a more precise estimate requiring additional repulsive electrostatic interactions. This estimation agrees with the pressure increase upon maturation previously reported, although it is still smaller than the ∼30 atm value estimated from atomic force microscopy indentations interpreted using continuous elasticity and linear unbranched polymer models (40). As more experimental knowledge on these viruses becomes available, it is expected that more refined models can be built. Also, in icosahedrally averaged maps of immature adenovirus, a weak sign of layering is observed (a slightly higher density beneath the capsid shell over the general flat profile of the core) that disappears upon maturation (28). This effect would be consistent with our model prediction for a decrease in condenser size upon protein cleavage. However, a decrease in condenser size would also be expected to produce a pressure decrease, which is not what is experimentally observed. A possible explanation for this discrepancy is that the apparent weak layering in the immature adenovirus maps comes from the presence of a packaging protein (65). Alternatively, the change in interaction regime between condensers and DNA may predominate over the change in condenser size.
Although condensing proteins seem to help in reducing internal pressure, they are not used by all dsDNA viruses. Other dsDNA viruses, such as tailed bacteriophages, are known to have extreme internal pressures with no condensing proteins present. The different strategy in dsDNA packing and pressure control may be related to differences in the virus infectious cycle. Viruses with medical relevance, such as polyoma- and papillomaviruses, pack histone-bound dsDNA genomes (7, 17). Virus-encoded condensers seem to be most common in members of the PRD1-adenovirus structural lineage, an extended group of viruses infecting hosts throughout the tree of life, from bacteria to humans. Members of the PRD1-adenovirus lineage use β-barrels orthogonal to the capsid surface to assemble icosahedral capsids with diameters ranging from 0.06 to 1 μm (19). The largest, most complex members of the lineage form the recently proposed Megavirales order (66), which includes viruses of ecological (amphibian ranaviruses), economical (fish iridoviruses and poxviruses and African swine fever virus), and biomedical (human poxviruses) importance. There is a strong indication from gene sequencing, as well as molecular and structural analyses, that the presence of DNA-condensing proteins is likely to be a common trait in this growing virus family (8, 9, 10, 11, 12, 13, 14, 15, 16, 67). Assembly of most of these viruses is poorly characterized, and even less work has been done to understand DNA packing within their capsids. Intriguingly, no genes for condensing proteins have been reported for the smallest members of the PRD1-adenovirus lineage (those infecting bacteria), suggesting that this trait is linked to some kind of gene interchange with eukaryotic hosts. Indeed, it has been observed that a particular kind of eukaryotes, phylum Dinoflagellata, lack histones but possess a positively charged, DNA-binding protein whose only known sequence similarity outside the phylum is to a megavirus protein (68). Cryo-EM maps of the PRD1-adenovirus lineage members lacking condensing proteins show internal layered densities similar to those of tailed bacteriophages (69, 70), revealing how the presence of condensers alters DNA packing within the capsid. Although tailed bacteriophages and the PRD1 bacteriophage package their genomes using an ATP-driven motor in a sequential pathway (41, 71), there are experimental indications that viruses with condensing proteins assemble their capsids around the compacting genome (42, 43, 72) in a concurrent pathway. In adenoviruses, proteolytic maturation of the condensing proteins results in physical changes required to turn the particle metastable for successful triggering of sequential uncoating (28, 40, 45, 46). Stepwise dismantling is required for infection of eukaryotic cells, but not so for bacteriophages, which inject their genome through the cell wall, leaving the intact capsid behind. Some of the viruses presumed to use condensing proteins are among the largest known (notably the giant mimivirus). The capacity for a viral capsid to survive internal pressure before bursting drops as the size of the capsid is increased (55). Additionally, no significant change in genome length is required to encode proteins forming larger capsids, and the extra length required to encode the condensing proteins, which also needs to be packed, is more than compensated for by their existence. Both differences in the infectious cycle and the effect of capsid size could explain the lack of condensing proteins in smaller viruses.
Conclusions
Our results clarify the role of condensing proteins in virus particle stability and internal pressure reduction. It is intriguing that a rather generic model, without a specific and detailed account of binding interaction or condenser shape, indicates a potential benefit of condensing proteins in compacting DNA and reducing the pressure in the packed state. We find that the benefits are particularly pronounced in viruses that are not particularly densely packed, i.e., that contain the DNA with volume fractions up to ∼0.3, of which adenovirus is a prime example. Our model, combined with previous experimental observations on adenovirus maturation, indicates how changes in the condenser-binding regime may be critical to ensure the assembly/disassembly interplay during the infectious cycle. The dependence of internal pressure on the condenser size, binding strength, and concentration, which we document and explain here, will contribute to the current understanding of stability modulation of complex virus capsids, advance their development as nanocontainers for controlled cargo delivery in therapeutics, and ultimately illustrate the different mechanisms used for DNA compaction throughout nature.
Author Contributions
S.M. and A.Š. designed the study. S.M. carried out the simulations and analyzed the data. S.M., C.S.M., and A.Š. provided important suggestions for the simulations, discussed the results, and wrote the manuscript.
Acknowledgments
S.M. thanks T. Ivek and K. Salamon for useful discussions. C.S.M. acknowledges support via grant BFU2016-74868-P from the Spanish Agencia Estatal de Investigación, cofunded by the European Regional Development Fund, as well as BFU2013-41249-P and the Spanish Adenovirus Network (AdenoNet, BIO2015-68990-REDT) from the Ministerio de Economía y Competitividad of Spain.
Editor: Andrew Spakowitz.
Footnotes
Supporting Materials and Methods and four figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30930-X.
Supporting Citations
References (73, 74) appear in the Supporting Material.
Supporting Material
References
- 1.Smith D.E., Tans S.J., Bustamante C. The bacteriophage straight ϕ29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 2.Šiber A., Božič A.L., Podgornik R. Energies and pressures in viruses: contribution of nonspecific electrostatic interactions. Phys. Chem. Chem. Phys. 2012;14:3746–3765. doi: 10.1039/c1cp22756d. [DOI] [PubMed] [Google Scholar]
- 3.Bloomfield V.A. DNA condensation by multivalent cations. Biopolymers. 1997;44:269–282. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 4.Schiessel H. The physics of chromatin. J. Phys. Condens. Matter. 2003;15:R699–R774. doi: 10.1088/0953-8984/27/6/060301. [DOI] [PubMed] [Google Scholar]
- 5.Saper G., Kler S., Harries D. Effect of capsid confinement on the chromatin organization of the SV40 minichromosome. Nucleic Acids Res. 2013;41:1569–1580. doi: 10.1093/nar/gks1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurdiss D.L., Morgan E.L., Ranson N.A. New structural insights into the genome and minor capsid proteins of BK polyomavirus using cryo-electron microscopy. Structure. 2016;24:528–536. doi: 10.1016/j.str.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Favre M., Breitburd F., Orth G. Chromatin-like structures obtained after alkaline disruption of bovine and human papillomaviruses. J. Virol. 1977;21:1205–1209. doi: 10.1128/jvi.21.3.1205-1209.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giberson A.N., Davidson A.R., Parks R.J. Chromatin structure of adenovirus DNA throughout infection. Nucleic Acids Res. 2012;40:2369–2376. doi: 10.1093/nar/gkr1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tweeten K.A., Bulla L.A., Consigli R.A. Characterization of an extremely basic protein derived from granulosis virus nucleocapsids. J. Virol. 1980;33:866–876. doi: 10.1128/jvi.33.2.866-876.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tseng M., Palaniyar N., Evans D.H. DNA binding and aggregation properties of the vaccinia virus I3L gene product. J. Biol. Chem. 1999;274:21637–21644. doi: 10.1074/jbc.274.31.21637. [DOI] [PubMed] [Google Scholar]
- 11.Cyrklaff M., Risco C., Carrascosa J.L. Cryo-electron tomography of vaccinia virus. Proc. Natl. Acad. Sci. USA. 2005;102:2772–2777. doi: 10.1073/pnas.0409825102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Condit R.C., Moussatche N., Traktman P. In a nutshell: structure and assembly of the vaccinia virion. Adv. Virus Res. 2006;66:31–124. doi: 10.1016/S0065-3527(06)66002-8. [DOI] [PubMed] [Google Scholar]
- 13.Wulfmeyer T., Polzer C., Meckel T. Structural organization of DNA in chlorella viruses. PLoS One. 2012;7:e30133. doi: 10.1371/journal.pone.0030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borca M.V., Irusta P.M., Rock D.L. A structural DNA binding protein of African swine fever virus with similarity to bacterial histone-like proteins. Arch. Virol. 1996;141:301–313. doi: 10.1007/BF01718401. [DOI] [PubMed] [Google Scholar]
- 15.Xiao C., Kuznetso Y.G., Rossmann M.G. Structural studies of the giant Mimivirus. PLoS Biol. 2009;7:e1000092. doi: 10.1371/journal.pbio.1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okamoto K., Miyazaki N., Svenda M. Cryo-EM of a marseilleviridae virus particle reveals a large internal microassembly. bioRxiv. 2017 doi: 10.1016/j.virol.2018.01.021. [DOI] [PubMed] [Google Scholar]
- 17.Germond J.E., Hirt B., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc. Natl. Acad. Sci. USA. 1975;72:1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fields B.N., Knipe D.M., Howley P.M. Wolters Kluwer Health/Lippincott Williams & Wilkins; Philadelphia, PA: 2007. Fields Virology, Sixth Edition. [Google Scholar]
- 19.Abrescia N.G.A., Bamford D.H., Stuart D.I. Structure unifies the viral universe. Annu. Rev. Biochem. 2012;81:795–822. doi: 10.1146/annurev-biochem-060910-095130. [DOI] [PubMed] [Google Scholar]
- 20.Schoonen L., van Hest J.C.M. Functionalization of protein-based nanocages for drug delivery applications. Nanoscale. 2014;6:7124–7141. doi: 10.1039/c4nr00915k. [DOI] [PubMed] [Google Scholar]
- 21.Amalfitano A., Parks R.J. Separating fact from fiction: assessing the potential of modified adenovirus vectors for use in human gene therapy. Curr. Gene Ther. 2002;2:111–133. doi: 10.2174/1566523024605618. [DOI] [PubMed] [Google Scholar]
- 22.Thomas C.E., Ehrhardt A., Kay M.A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 23.Purohit P.K., Inamdar M.M., Phillips R. Forces during bacteriophage DNA packaging and ejection. Biophys. J. 2005;88:851–866. doi: 10.1529/biophysj.104.047134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forrey C., Muthukumar M. Langevin dynamics simulations of genome packing in bacteriophage. Biophys. J. 2006;91:25–41. doi: 10.1529/biophysj.105.073429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Locker C.R., Fuller S.D., Harvey S.C. DNA organization and thermodynamics during viral packing. Biophys. J. 2007;93:2861–2869. doi: 10.1529/biophysj.106.094771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molineux I.J., Panja D. Popping the cork: mechanisms of phage genome ejection. Nat. Rev. Microbiol. 2013;11:194–204. doi: 10.1038/nrmicro2988. [DOI] [PubMed] [Google Scholar]
- 27.Christiansen G., Landers T., Berg P. Characterization of components released by alkali disruption of simian virus 40. J. Virol. 1977;21:1079–1084. doi: 10.1128/jvi.21.3.1079-1084.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pérez-Berná A.J., Marabini R., San Martín C. Structure and uncoating of immature adenovirus. J. Mol. Biol. 2009;392:547–557. doi: 10.1016/j.jmb.2009.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vayda M.E., Rogers A.E., Flint S.J. The structure of nucleoprotein cores released from adenovirions. Nucleic Acids Res. 1983;11:441–460. doi: 10.1093/nar/11.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mirza M.A., Weber J. Structure of adenovirus chromatin. Biochim. Biophys. Acta. 1982;696:76–86. doi: 10.1016/0167-4781(82)90012-4. [DOI] [PubMed] [Google Scholar]
- 31.Kuznetsov Y., Gershon P.D., McPherson A. Atomic force microscopy investigation of vaccinia virus structure. J. Virol. 2008;82:7551–7566. doi: 10.1128/JVI.00016-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silvestry M., Lindert S., Stewart P.L. Cryo-electron microscopy structure of adenovirus type 2 temperature-sensitive mutant 1 reveals insight into the cell entry defect. J. Virol. 2009;83:7375–7383. doi: 10.1128/JVI.00331-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.San Martín C. Latest insights on adenovirus structure and assembly. Viruses. 2012;4:847–877. doi: 10.3390/v4050847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Booy F.P., Newcomb W.W., Steven A.C. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell. 1991;64:1007–1015. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.San Martín C., Burnett R.M., Bamford D.H. Combined EM/x-ray imaging yields a quasi-atomic model of the adenovirus-related bacteriophage PRD1 and shows key capsid and membrane interactions. Structure. 2001;9:917–930. doi: 10.1016/s0969-2126(01)00642-6. [DOI] [PubMed] [Google Scholar]
- 36.De Frutos M., Leforestier A., Livolant F. Relationship between the genome packing in the bacteriophage capsid and the kinetics of DNA ejection. Biophys. Rev. Lett. 2014;9:81–104. [Google Scholar]
- 37.Shen P.S., Enderlein D., Belnap D.M. The structure of avian polyomavirus reveals variably sized capsids, non-conserved inter-capsomere interactions, and a possible location of the minor capsid protein VP4. Virology. 2011;411:142–152. doi: 10.1016/j.virol.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pérez-Berná A.J., Marion S., San Martín C. Distribution of DNA-condensing protein complexes in the adenovirus core. Nucleic Acids Res. 2015;43:4274–4283. doi: 10.1093/nar/gkv187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luijsterburg M.S., White M.F., Dame R.T. The major architects of chromatin: architectural proteins in bacteria, archaea and eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2008;43:393–418. doi: 10.1080/10409230802528488. [DOI] [PubMed] [Google Scholar]
- 40.Ortega-Esteban A., Condezo G.N., de Pablo P.J. Mechanics of viral chromatin reveals the pressurization of human adenovirus. ACS Nano. 2015;9:10826–10833. doi: 10.1021/acsnano.5b03417. [DOI] [PubMed] [Google Scholar]
- 41.Cuervo A., Daudén M.I., Carrascosa J.L. Nucleic acid packaging in viruses. Subcell. Biochem. 2013;68:361–394. doi: 10.1007/978-94-007-6552-8_12. [DOI] [PubMed] [Google Scholar]
- 42.Gordon-Shaag A., Ben-Nun-Shaul O., Oppenheim A. Cellular transcription factor Sp1 recruits simian virus 40 capsid proteins to the viral packaging signal, ses. J. Virol. 2002;76:5915–5924. doi: 10.1128/JVI.76.12.5915-5924.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Condezo G.N., San Martín C. Localization of adenovirus morphogenesis players, together with visualization of assembly intermediates and failed products, favor a model where assembly and packaging occur concurrently at the periphery of the replication center. PLoS Pathog. 2017;13:e1006320. doi: 10.1371/journal.ppat.1006320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ostapchuk P., Suomalainen M., Hearing P. The adenovirus major core protein VII is dispensable for virion assembly but is essential for lytic infection. PLoS Pathog. 2017;13:e1006455. doi: 10.1371/journal.ppat.1006455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pérez-Berná A.J., Ortega-Esteban A., San Martín C. The role of capsid maturation on adenovirus priming for sequential uncoating. J. Biol. Chem. 2012;287:31582–31595. doi: 10.1074/jbc.M112.389957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ortega-Esteban A., Pérez-Berná A.J., de Pablo P.J. Monitoring dynamics of human adenovirus disassembly induced by mechanical fatigue. Sci. Rep. 2013;3:1434. doi: 10.1038/srep01434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Locker C., Harvey S. A model for viral genome packing. Multiscale Model. Simul. 2006;5:1264–1279. [Google Scholar]
- 48.Le Treut G., Képès F., Orland H. Phase behavior of DNA in the presence of DNA-binding proteins. Biophys. J. 2016;110:51–62. doi: 10.1016/j.bpj.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider T., Stoll E. Molecular-dynamics study of a three-dimensional one-component model for distortive phase transitions. Phys. Rev. B. 1978;17:1302–1322. [Google Scholar]
- 50.Plimpton S. Fast parallel algorithms for short-range molecular dynamics. J. Comput. Phys. 1995;117:1–19. [Google Scholar]
- 51.Rubinstein M., Colby R.H. Oxford University Press; Oxford, United Kingdom: 2003. Polymer Physics. [Google Scholar]
- 52.Lei M., de Graff A., Sartbaeva A. Uncovering the intrinsic geometry from the atomic pair distribution function of nanomaterials. Phys. Rev. B. 2009;80:024118. [Google Scholar]
- 53.Liddington R.C., Yan Y., Harrison S.C. Structure of simian virus 40 at 3.8-Å resolution. Nature. 1991;354:278–284. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- 54.Cordova A., Deserno M., Ben-Shaul A. Osmotic shock and the strength of viral capsids. Biophys. J. 2003;85:70–74. doi: 10.1016/S0006-3495(03)74455-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zandi R., Reguera D. Mechanical properties of viral capsids. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2005;72:021917. doi: 10.1103/PhysRevE.72.021917. [DOI] [PubMed] [Google Scholar]
- 56.Anderson T.F., Rappaport C., Muscatine N.A. On the structure and osmotic properties of phage particles. Ann. Inst. Pasteur (Paris) 1953;84:5–14. [PubMed] [Google Scholar]
- 57.Fischer H., Polikarpov I., Craievich A.F. Average protein density is a molecular-weight-dependent function. Protein Sci. 2004;13:2825–2828. doi: 10.1110/ps.04688204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Desmond K.W., Weeks E.R. Random close packing of disks and spheres in confined geometries. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2009;80:051305. doi: 10.1103/PhysRevE.80.051305. [DOI] [PubMed] [Google Scholar]
- 59.Spakowitz A.J., Wang Z.-G. DNA packaging in bacteriophage: is twist important? Biophys. J. 2005;88:3912–3923. doi: 10.1529/biophysj.104.052738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schlick T., Hayes J., Grigoryev S. Toward convergence of experimental studies and theoretical modeling of the chromatin fiber. J. Biol. Chem. 2012;287:5183–5191. doi: 10.1074/jbc.R111.305763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maeshima K., Hihara S., Eltsov M. Chromatin structure: does the 30-nm fibre exist in vivo? Curr. Opin. Cell Biol. 2010;22:291–297. doi: 10.1016/j.ceb.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 62.Luger K., Dechassa M.L., Tremethick D.J. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat. Rev. Mol. Cell Biol. 2012;13:436–447. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mangel W.F., San Martín C. Structure, function and dynamics in adenovirus maturation. Viruses. 2014;6:4536–4570. doi: 10.3390/v6114536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perlmutter J.D., Mohajerani F., Hagan M.F. Many-molecule encapsulation by an icosahedral shell. eLife. 2016;5:e14078. doi: 10.7554/eLife.14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Condezo G.N., Marabini R., San Martín C. Structures of adenovirus incomplete particles clarify capsid architecture and show maturation changes of packaging protein L1 52/55k. J. Virol. 2015;89:9653–9664. doi: 10.1128/JVI.01453-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Colson P., De Lamballerie X., Raoult D. “Megavirales”, a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch. Virol. 2013;158:2517–2521. doi: 10.1007/s00705-013-1768-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frouco G., Freitas F.B., Ferreira F. DNA-binding properties of African swine fever virus pA104R, a histone-like protein involved in viral replication and transcription. J. Virol. 2017;91 doi: 10.1128/JVI.02498-16. e02498–e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gornik S.G., Ford K.L., Waller R.F. Loss of nucleosomal DNA condensation coincides with appearance of a novel nuclear protein in dinoflagellates. Curr. Biol. 2012;22:2303–2312. doi: 10.1016/j.cub.2012.10.036. [DOI] [PubMed] [Google Scholar]
- 69.San Martín C., Huiskonen J.T., Burnett R.M. Minor proteins, mobile arms and membrane-capsid interactions in the bacteriophage PRD1 capsid. Nat. Struct. Biol. 2002;9:756–763. doi: 10.1038/nsb837. [DOI] [PubMed] [Google Scholar]
- 70.Huiskonen J.T., Kivelä H.M., Butcher S.J. The PM2 virion has a novel organization with an internal membrane and pentameric receptor binding spikes. Nat. Struct. Mol. Biol. 2004;11:850–856. doi: 10.1038/nsmb807. [DOI] [PubMed] [Google Scholar]
- 71.Hong C., Oksanen H.M., Chiu W. A structural model of the genome packaging process in a membrane-containing double stranded DNA virus. PLoS Biol. 2014;12:e1002024. doi: 10.1371/journal.pbio.1002024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mutsafi Y., Shimoni E., Minsky A. Membrane assembly during the infection cycle of the giant Mimivirus. PLoS Pathog. 2013;9:e1003367. doi: 10.1371/journal.ppat.1003367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Andelman D. Introduction to electrostatics in soft and biological matter. In: Poon W.C.K., Andelman D., editors. Soft Condensed Matter Physics in Molecular and Cell Biology. Taylor and Francis; 2006. pp. 97–122. [Google Scholar]
- 74.Naji A., Kanduč M., Podgornik R. Exotic electrostatics: unusual features of electrostatic interactions between macroions. In: Hu W., Shi A.-C., editors. Understanding Soft Condensed Matter via Modeling and Computation. World Scientific; 2010. pp. 265–295. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.