Abstract
KIF3C is one subunit of the functional microtubule-based kinesin-2 KIF3AC motor, an anterograde cargo transporter in neurons. However, KIF3C has also been implicated as an injury-specific kinesin that is a key regulator of axonal growth and regeneration by promoting microtubule dynamics for reorganization at the neuronal growth cone. To test its potential role as a modulator of microtubule dynamics in vitro, an engineered homodimeric KIF3CC was incorporated into a dynamic microtubule assay and examined by total internal reflection fluorescence microscopy. The results reveal that KIF3CC is targeted to the microtubule plus-end, acts as a potent catastrophe factor through an increase in microtubule catastrophe frequency, and does so by elimination of the dependence of the catastrophe rate on microtubule lifetime. Moreover, KIF3CC accelerates the catastrophe rate without altering the microtubule growth rate. Therefore, the ATP-promoted KIF3CC mechanism of catastrophe is different from the well-described catastrophe factors kinesin-13 MCAK and kinesin-8 Kip3/KIF18A. The properties of KIF3CC were not shared by heterodimeric KIF3AC and required the unique KIF3C-specific sequence extension in loop L11 at the microtubule interface. At the microtubule plus-end, the presence of KIF3CC resulted in modulation of the tapered structure typically seen in growing dynamic microtubules to microtubule blunt plus-ends. Overall our results implicate homodimeric KIF3CC as a unique promoter of microtubule catastrophe and substantiate its physiological role in cytoskeletal remodeling.
Introduction
The microtubule (MT) cytoskeleton is an organized array of rodlike tubulin polymers that supports long-range motor-driven cargo transport. Yet at the same time these MTs can be highly dynamic and exhibit significant rearrangements that drive cytoskeleton reorganization. In neurons, these dynamic properties have fundamental roles in cell polarity specification, axon differentiation, and growth. Although KIF3C has largely been regarded as the partner of KIF3A to generate the heterodimeric KIF3AC for cargo transport, a number of studies have alluded to a developmentally regulated subpopulation of homodimeric KIF3C with proposed roles in the regulation of MT dynamics, particularly in axon regeneration after injury and in neuronal growth cone remodeling (1, 2, 3).
A recent study by Gumy et al. (2) showed that in response to injury, KIF3C was locally translated in rat embryonic sensory axons and growth cones yet was transported from the cell body to growth cones in adult neurons in a manner dependent upon EB3. Depletion of KIF3C in adult neurons resulted in an increase in stable, overgrown, and looped MTs indicating that KIF3C functioned as a MT-destabilizing factor. The authors also reported that in the absence of KIF3C either by RNA interference or KIF3C gene knock-out in adult neurons, axonal outgrowth was impaired in vitro with regeneration after injury delayed both in vitro and in vivo. Yet a report by Yang et al. (4) showed that KIF3C knock-out mice were viable, apparently developed normally, and were able to reproduce.
Although these earlier studies provide evidence that KIF3C promotes MT dynamics, questions remain. Can KIF3C truly function independently from KIF3A as a homodimer (KIF3CC)? If so, does its function require ATP turnover as expected of a kinesin motor? What is its mechanism of action to remodel the MT cytoskeleton at the site of injury? For example, is KIF3C a catastrophe factor functioning similarly to kinesin-8 (Kip3/KIF18A) and kinesin-13 (MCAK)? Moreover, KIF3C unlike other kinesins, contains a signature motif conserved in mammalian species, a 25-residue insert in loop L11 of the catalytic motor domain (5, 6, 7). Is this loop L11 motif involved in the promotion of MT dynamics by homodimeric KIF3CC? To address these questions directly and quantitatively, we examined the capability of homodimeric KIF3CC to promote MT dynamics in vitro.
Each MT is polarized in such a way that a relatively stable MT minus-end acts as a seed for extension from the dynamic MT plus-end (8). MT plus-ends undergo a stochastic cycle termed “dynamic instability” in which they alternate between phases of steady growth and rapid disassembly, termed “catastrophe”. Whereas growth phases generate outward forces to drive cell migration or mitosis, catastrophe is critical to MT length regulation for rearrangement of the MT cytoskeleton in response to cellular cues (9).
The transition between growth and catastrophe is determined by the nucleotide state of the tubulin subunits (10, 11) and involves a process termed “MT aging”. Growth from the MT plus-end occurs by the longitudinal incorporation of GTP-bound tubulin subunits to each of the 13 protofilaments that make up a MT polymer. As the MT continues to grow, incorporated subunits gradually hydrolyze their GTP to GDP as new GTP-tubulin subunits are added to the tip. The GTP cap that forms at the MT plus-end protects the growing MT and supports the continuation of the growth phase. However, the integrity of the GTP cap is compromised as it ages over time and becomes gradually less capable of sustaining MT growth. Various mechanisms for MT aging have been proposed, but it is generally considered a multistep process that involves the accumulation of errors. These errors can include premature GTP hydrolysis within the GTP cap or loss of a GTP-tubulin subunit, either of which can destabilize the polymer and inevitably lead to catastrophe at the MT plus-end (12, 13, 14, 15, 16).
The rate of accumulation of MT aging errors can be altered through additional proteins termed “catastrophe factors” that influence the frequency of MT catastrophe. Some members of the kinesin superfamily of MT motors including kinesin-8 (Kip3/KIF18A) and kinesin-13 (MCAK) have been identified as catastrophe factors. The mechanisms they employ and their downstream cellular outcomes differ greatly: MCAK eliminates the aging process to impact mitotic spindle length, and Kip3/KIF18A accelerates aging to influence spindle positioning and chromosome alignment (17, 18, 19, 20, 21, 22, 23).
Previously, we reported that an engineered KIF3CC homodimer in single-molecule quantum-dot (Qdot) assays operated as a slow processive motor capable of traveling only 0.5 μm on average along a MT at an exceedingly slow ATP turnover rate of 1 s−1 for each 8 nm step (5). This behavior was in stark contrast to heterodimeric KIF3AB and KIF3AC motors that traveled on average 1.6 and 1.2 μm, respectively, toward the MT plus-end, and at much faster rates, as expected for cargo transporters. In these experiments, KIF3CC-promoted MT dynamics were not observed because the MTs were stabilized with paclitaxel, which masked the MT-destabilizing properties of KIF3CC. Thus, the question remains of whether KIF3CC can influence MT dynamics.
To test the hypothesis that homodimeric KIF3CC can promote MT dynamics, total internal reflection fluorescence (TIRF) microscopy experiments were performed using the engineered KIF3CC previously characterized (5). The results show that KIF3CC is targeted to the MT plus-end, acts as a MT catastrophe factor through an increase of MT catastrophe frequency, and does so while also eliminating the MT aging process. These properties were not evident for the heterodimeric counterpart, KIF3AC, and are also dependent upon the KIF3C-specific extension of loop L11. The impact of KIF3CC on MT catastrophe involves at least in part a regulation of MT plus-end architecture, where in the presence of KIF3CC the MT tips are blunt rather than tapered. Furthermore, the localization of KIF3CC-Qdot motors suggests that multiple KIF3CC dimers are likely working concurrently at the MT plus-end to spur catastrophe. These results support the existence of a cellular subpopulation of KIF3CC that is expressed in response to injury to remodel the MT cytoskeleton.
Materials and Methods
Mus musculus KIF3 proteins
The KIF3CC, KIF3AC, and KIF3ACΔL11 proteins used herein were designed, expressed, and purified as previously described (5, 24). See also the Supporting Material and Methods. Briefly, the KIF3A and KIF3C contain the native sequence of the N-terminal motor domain, neck linker, and helix α7 that are then fused with an in-register dimerization motif to stabilize the native coiled-coil and C-terminal purification tags (His8 for homodimers; His8 and StrepII for heterodimers). Before experiments, all motors were clarified with a 10-min high speed centrifugation spin at 4°C (Optima TLX Ultracentrifuge TLA-100 rotor at 313,000 × g; Beckman Coulter, Brea, CA). All motor concentrations reported for these assays are for the dimer concentrations.
Dynamic MTs and extension assay
To generate GMPCPP-stabilized MT seeds, unlabeled bovine tubulin and lyophilized X-rhodamine tubulin (Cytoskeleton, Denver, CO) were diluted to 20 μM in PME80 buffer (80 mM PIPES, pH 6.9 with KOH, 5 mM MgCl2, and 1 mM EGTA). Both tubulin stocks were then mixed in a 1:1 ratio in the presence of 1 mM GMPCPP and incubated in the dark for 15 min at room temperature, followed by further elongation at 37°C for 6 min. The seeds were then sheared with a 30G × 1/2'' needle to generate a stock of seeds for microscopy that were stored in the dark at room temperature. Perfusion chambers were made from an acid-washed, silanized coverslip (22 × 22 mm) mounted on an acid-washed nonsilanized slide (22 × 60 mm) separated by strips of double-sided tape to generate a 10 μL flow cell. Each perfusion chamber was first treated with 0.4% rat anti-α-tubulin (ABD Serotec, Raleigh, NC) for 5 min followed by blocking the chamber with 5% Pluronic F-127 (Sigma-Aldrich, St. Louis, MO) for 5 min. GMPCPP-seeds were then diluted fresh for each slide in PME80 with 10 mM DTT to ∼60 nM (1:300, optimized for seed distribution density on the imaging surface) and flowed into the chamber for an 8-min incubation in the dark at room temperature. The perfusion chamber was then transferred to an incubated microscope stage to equilibrate to 37°C for an additional 2 min. Unbound GMPCPP seeds were washed out with prewarmed Blocking Mix (PME80, 0.5 mg/mL casein, 2 mM MgATP, 10 mM 2-mercaptoethanol, 0.3 mg/mL glucose oxidase, 0.2 mg/mL catalase, and 25 mM glucose). Seeds were then briefly primed for extension by flowing in 2× chamber volume of prewarmed Priming Mix (PME80, 10 μM unlabeled tubulin, 0.4 mg/mL casein, 2 mM MgATP, 1 mM GTP, 20 mM DTT, 0.3 mg/mL glucose oxidase, 0.2 mg/mL catalase, and 25 mM glucose). MT extension was then initiated with prewarmed Extension Mix (Priming Mix that contained 10 μM tubulin consisting of a mixture of a 1:8.5 ratio of X-rhodamine labeled to unlabeled tubulin). KIF3 motors were introduced to the chamber as a component of the Extension Mix. Chambers were sealed and imaged with TIRF microscopy using an Inverted Axio Observer Z1 MOT microscope with a 100× oil 1.46 N.A. Plan-Apochromat objective (Carl Zeiss Microscopy, Jena, Germany). MTs were imaged at 561 nm (2% laser power, 80 ms exposure) every 2 s for 10 min.
For the AMPPNP control, 2 mM MgATP in the assay mix was replaced accordingly with 2 mM MgAMPPNP. For the no nucleotide control, apyrase treatment could not be used due to the requirement of MgGTP for MT dynamics. Instead, for this control, MgATP in the assay was replaced with PME80 buffer.
Data analysis
Seeds were selected for analysis that were 1–3 μm long, that were separated from other seeds on the chamber surface, and whose extensions did not cross any other MTs during the time course of the video. MT lifetime data were collected through kymograph analysis of MT seeds (MultipleKymograph plugin for ImageJ; National Institutes of Health, Bethesda, MD). These data were corroborated with an additional analysis using the TipTracker algorithm for the software MATLAB (The MathWorks, Natick, MA), as further described below (25, 26, 27).
Gamma fit of MT lifetime data
Cumulative distribution plots and probability density function (PDF) plots (17) were evaluated using the Distribution Fitting application within the Statistics Toolbox of MATLAB (R2016a v. 9.0.0) as described (17) to determine the step parameter values and aging rate for all experimental conditions (Figs. 1 and 2).
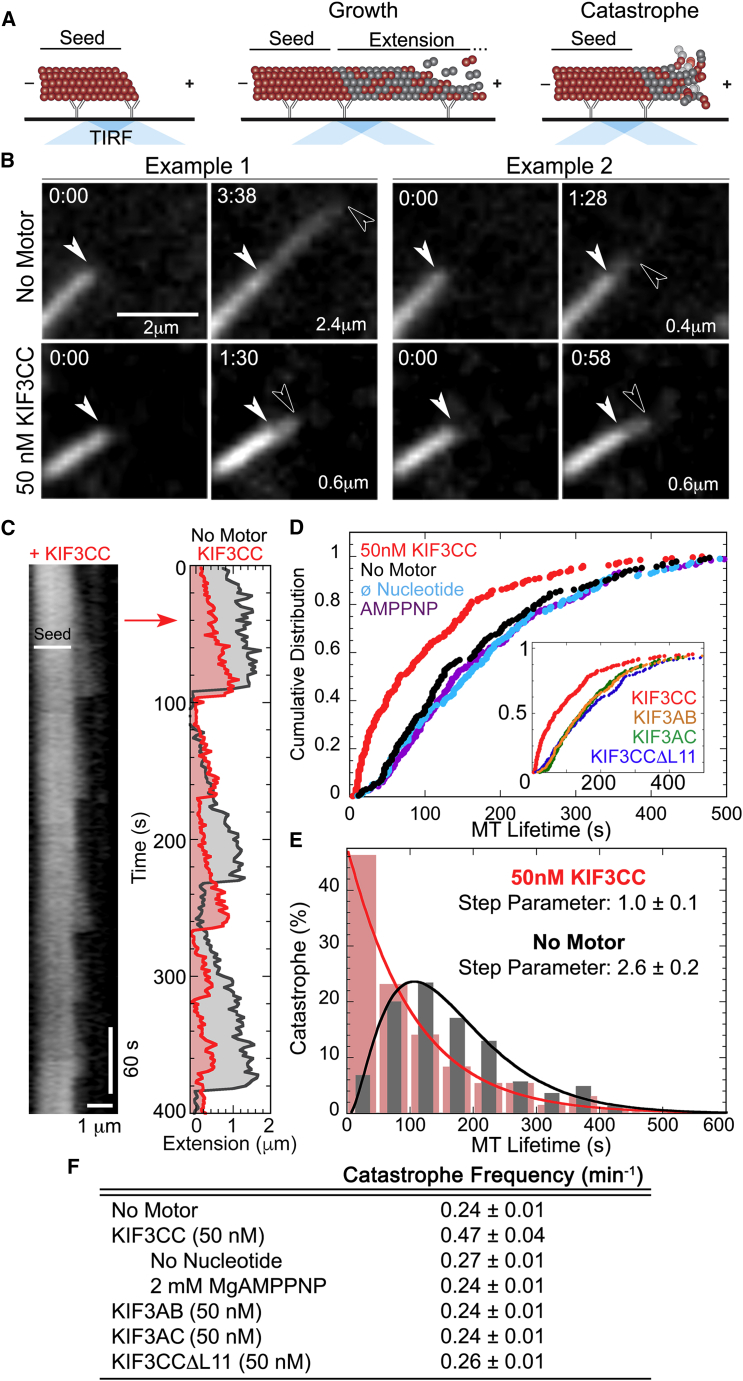
Figure 1.
KIF3CC enhances MT dynamics and shortens MT lifetimes. (A) Illustration of MT dynamics as examined with TIRF microscopy. A brightly labeled GMPCPP seed acts as the initiation point for a dim plus-end extension grown in the presence of 1 mM MgGTP. The seed remains stable throughout the assay. Both growth and catastrophe events occur from the MT plus-end and retract back to the MT seed. (B) Two representative examples of dynamic MTs from each of the no-motor and +50 nM KIF3CC conditions. Extension phases begin at 0 s with the initiation of extension and end with the timestamp immediately before a catastrophe event. Extension lengths are also provided in the lower right of each panel. White arrowheads denote the end of the GMPCPP seed and outlined arrowheads show the location of the tip of the extension. See also Table 1. Scale bar represents 2 μm. (C) Kymograph of MT dynamics (left) from a representative MT seed in the presence of 50 nM KIF3CC, shown over the first 400 s of data acquisition. The location of the MT plus-tip was determined by the MATLAB TipTracker algorithm for the same seed (right). An additional TipTracker analysis from a representative seed of the no-motor control is provided for comparison (gray). For the TipTracker plot, the x axis provides only the length of the MT extension, with 0 μm representing the plus-end of the GMPCPP seed. (D) Plot of cumulative distribution as a function of MT lifetime in the presence of 50 nM KIF3CC (N = 252) and 2 mM MgATP compared with control conditions: no motor, no nucleotide (∅; MgATP was left out of the reaction mix), and with 2 mM MgAMPPNP. Further comparison of KIF3CC with other kinesin-2 motors, KIF3AB and KIF3AC and KIF3CCΔL11 each at 50 nM dimer and 2 mM MgATP, are provided as insets. For all comparisons, the analysis included between 177 and 230 extensions. See also Fig. S1 for further examination of KIF3AC. (E) MT lifetime datasets from 50 nM KIF3CC and the no-motor control (N = 230) represented with a PDF plot. The gamma fit of the data provided the step parameter that gives a quantification of the number of aging steps that are accumulated before MT catastrophe in each condition. (F) Table of values for the population catastrophe frequency for each dataset calculated as the number of catastrophe events that occur per min in each condition. See Table 1 for N values for each dataset. Values reported are mean ± SE. To see this figure in color, go online.
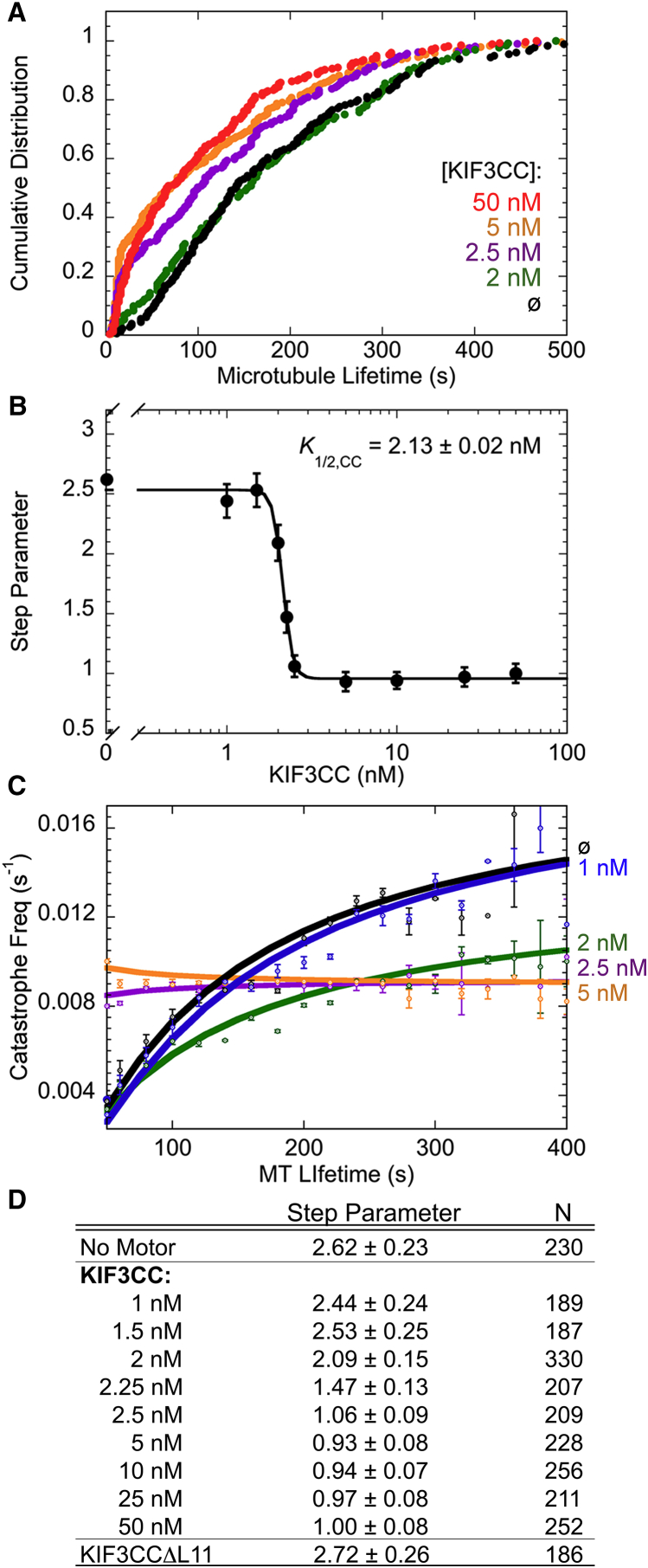
Figure 2.
Low concentrations of KIF3CC alter MT dynamics. (A) Datasets of cumulative distribution as a function of MT lifetime at decreasing concentrations of KIF3CC. The 50 nM KIF3CC trace is repeated from Fig. 1. ∅, 0 nM KIF3CC in the reaction mixture. (B) Step parameter plot as a function of KIF3CC concentration. The step parameters were determined from the gamma function of PDF plots from each MT lifetime dataset. The Hill slope fit of the step parameter data provides a K1/2,KIF3CC of 2.13 ± 0.03 nM, reported as mean ± SE. (C) Time-dependent MT catastrophe frequency plotted as a function of MT lifetime for a subset of the titration data with KIF3CC from 0 nM (∅) to 5 nM. Fit lines represent a single exponential equation using the nonlinear least-squares method, with error bars representing mean ± SE. (D) Table of compiled step parameters from the KIF3CC titration shown in (B). See also Table S1. To see this figure in color, go online.
First, each unbinned dataset was imported as a vector into the MATLAB Distribution Fitting application for analysis. Within the fitting tool, a gamma fit was applied to each dataset. The relationship of the PDF plot of the dataset (dF(t)/dt) to a gamma function (Γ(n)) is given by
| (1) |
where r is the rate of accumulation of MT aging errors and n is the number of aging steps before catastrophe. From this equation, the Distribution Fitting application provided the maximum likelihood estimates for each of the gamma-fit parameters (step and rate). Additionally, each fit was accompanied by quantification of mean ± SE as well as confidence intervals for the fit (Figs. 1 and 2).
To analyze the step parameter as a function of KIF3CC concentration, each datapoint was plotted with a Hill slope fit to the data according to
| (2) |
where d is the minimum step parameter, a represents the maximum step parameter value, c is the K1/2,KIF3CC, and b is the Hill slope (Fig. 2).
Catastrophe frequency
Population catastrophe frequency values were calculated as the number of catastrophe events that occur per minute in each condition, as determined by kymograph analysis of dynamic MT assays (Fig. 1 F).
Time-dependent catastrophe frequency, f(t), kinetically examines the age-dependence of MT catastrophe by calculating a ratio of the population of MTs that undergoes catastrophe by a certain MT lifetime to the population of MTs that reach that specific lifetime (17) (Fig. 2 C). This parameter was calculated by
| (3) |
where F(t) is the cumulative distribution for each MT lifetime, t. First, cumulative distribution plots of each dataset were compiled, and the values along each plot were then used to calculate catastrophe frequency according to the equation above. The calculated catastrophe frequency values were then plotted as a function of MT lifetime for each condition tested. Subsequently, fits to the data were performed within the Curve Fitting tool of MATLAB using a single exponential,
| (4) |
where A is amplitude of the plot, k is the rate constant, and C is the limit of the catastrophe frequency as the plot approaches infinity. The best fit from this equation was determined with Levenberg-Marquardt nonlinear least-squared analysis. The individual datapoints that are shown along the plot, as well as the corresponding error bars (mean ± SE), represent values calculated from 20 s bins of the f(t) data (Fig. 2 C).
TipTracker for MT tip analysis
For analysis of tip structure of growing MT extensions, the TipTracker algorithm (v. 3) for MATLAB was used with no modifications made to the original script and analysis conducted as previously described (26, 27). However, the use of TipTracker for analysis of the data presented herein required a higher signal-to-noise ratio at the MT tip than could be achieved with the previously described MT labeling method. Therefore, the labeling ratios used for the MT seed and extension mix were reversed. Rather than the bright seeds and dim extensions reported elsewhere in this text, the experiments conducted for analysis with TipTracker were performed with dim MT seeds (1:8.5 ratio of X-rhodamine labeled to unlabeled tubulin) and more highly fluorescent extensions (1:2 ratio of X-rhodamine labeled to unlabeled tubulin). These labeling ratios were then incorporated into the polymerization of GMPCPP seeds and for the extension mix previously described. From the acquired videos, regions of interest that contained a single MT seed were blindly selected and scaled to 32 nm/pixel for characterization with TipTracker (25). During subsequent evaluation, any region of interest that contained a dynamic MT that crossed other MT seeds or extensions during the time of the experiment were eliminated from further analysis due to possible interference on MT growth and catastrophe from such interactions. In total, TipTracker was used to analyze 25 regions of interest for KIF3CC and 36 regions of interest for the no-motor control (see Fig. 4).
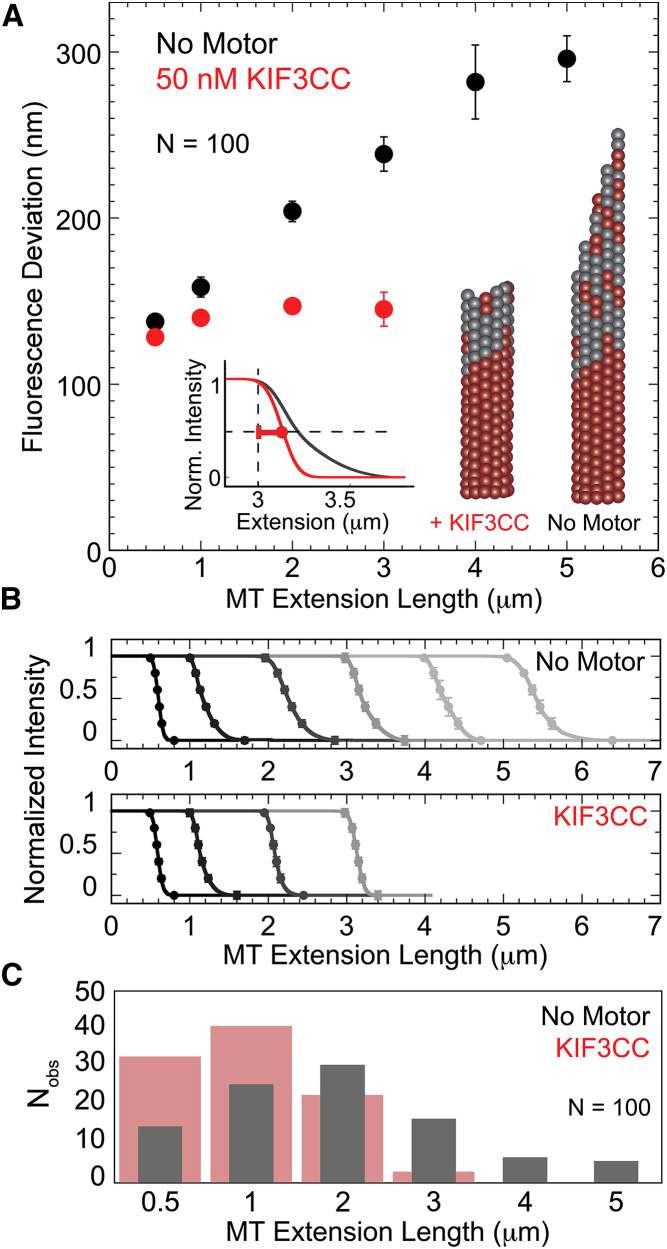
Figure 4.
KIF3CC modulates the taper of extending MT tips. (A) Compiled data for tip fluorescence deviation measurements for the no-motor control and 50 nM KIF3CC as a function of extension length. Data were collected using the TipTracker algorithm for MATLAB on inversely labeled MTs (bright extensions at a ratio of 1:2 of X-Rhodamine labeled to unlabeled tubulin; dim seeds with a ratio of 1:8.5 of X-Rhodamine labeled to unlabeled tubulin). Fluorescence deviation was defined as half of the distance over which the fluorescence signal depletes at the MT tip from maximum intensity to background, as shown by the red bar in the overlay of representative 3-μm MT extension profiles from the KIF3CC (red) and no motor (gray) conditions (inset). As illustrated, the presence of KIF3CC results in protofilaments with a lower fluorescence deviation than the no-motor control. (B) Average MT extension profiles for increasing extension lengths, illustrated by a progressive grayscale value with short MT extensions (0.5 μm) in black and the longest extensions (up to 5 μm) in light gray. Normalized intensity values are plotted as a function of MT extension length in the no-motor control and in the presence of 50 nM KIF3CC. Datapoints along each trace for the no-motor control as well as the 0.5–2 μm extension traces for KIF3CC represent the average of six extensions. The 3-μm trace for KIF3CC was limited to N = 3 due to the limited population that reach this range. Error bars represent mean ± SE of the normalized intensity values from the averaged traces. (C) Distribution of the population of MT extension lengths in the absence and presence of 50 nM KIF3CC. This bar graph represents the dataset used to estimate fluorescence deviations in (A). To see this figure in color, go online.
The tip structure estimation in TipTracker fits a Gaussian survival function to the decrease in fluorescence intensity from maximum signal along the MT extension to background (25, 26, 27). Along with this profile a quantification of the standard deviation of the taper profile is determined, which is the result of two contributing factors: the fluorescence signal of the MT taper itself, and the point spread function of the microscope. The data points on the plot of fluorescence deviation versus MT extension length reported here are corrected standard deviation values that remove the contribution from point spread function and are comparable to the tip standard deviation previously reported (25, 27). Tip structures that are more blunt have low fluorescence deviation values that result from little variation in protofilament lengths and therefore a steep drop in fluorescence signal at the MT plus-end. Tip structures that are tapered have higher fluorescence deviation values that result from a larger distribution of protofilament lengths at the MT plus-end and therefore a larger distance over which the fluorescence signal gradually drops (see Fig. 4 A, inset; and Fig. 4 B).
The length of MT extensions immediately before catastrophe was determined from the length of the MT in the video frame immediately before the start of the catastrophe event, measured from the end of the seed to the MT tip. Representative MT extensions were then binned by length ± 200 nm, such that the 2-μm extension trace represents a bin of MT extensions ranging from 1.8 to 2.2 μm. In the raw data images, this bin size corresponds to 2.5 pixels (160 nm/pixel). Due to slight variations in the illumination of replicate experiments, the fluorescence intensity values of the plots of intensity versus MT extension length were normalized from 0 to 1. The normalized intensity plots from each representative MT within a binned population were aligned based on the datapoint representing 0.96 for normalized fluorescence intensity. This method of alignment allowed for the calculation of an average intensity value along the MT extension length, as well as a calculation of mean ± SE as illustrated by error bars. For both the no-motor control and the 50 nM KIF3CC condition, each binned population of MTs contained an average of six representative extensions, with the exception of the 3 μm bin for KIF3CC for which there were only three representative extensions within the pool of data analyzed (see Fig. 4).
Qdot localization
To visualize individual motors in MT dynamics assays (see Fig. 5), the concentration of motor needed to be far lower than the required saturating concentrations of KIF3CC used for catastrophe stimulation. Therefore, we used spiking experiments in which only a subpopulation (8%) of the KIF3CC present in the reaction mix was labeled with quantum dots (28). Full details of the spiking process are found in the Supporting Materials and Methods. Briefly, 100 nM Qdot•KIF3 complex was supplemented with KIF3 dimer to a final concentration of 1.2 μM dimer. This working stock was incorporated into the Extension Mix to yield a final total chamber concentration of 50 nM KIF3 (including 4 nM Qdot•KIF3 complex). Qdots were visualized at 488 nm (5% laser power, 35 ms exposure) in conjunction with MT imaging as described above.
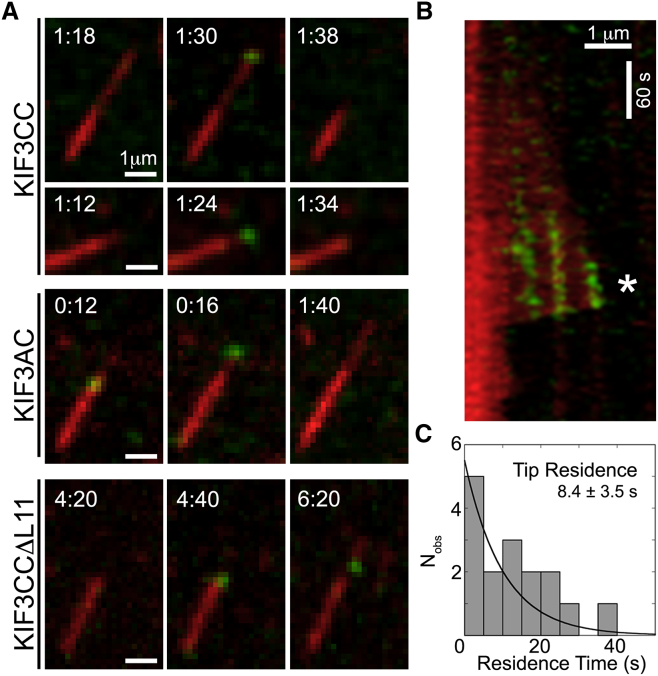
Figure 5.
Catastrophe occurs upon arrival of KIF3CC at the tip and not as a result of lattice binding. (A) Representative dual-channel images of Qdot-motor complexes (green) with dynamic MTs (red; brightly labeled GMPCPP MT seeds with dim MT extensions) at three timepoints. In the presence of 50 nM KIF3CC (top 2 rows), KIF3AC (middle row), or KIF3CCΔL11 (bottom row), the three timepoints for each set correspond to the frame before motor binding, the frame during which the motor binds to the MT, and the first frame immediately after the initiation of MT catastrophe. Timestamps for each image are provided relative to the beginning of the raw data image acquisition. Scale bars represent 1 μm. (B) Representative dual-channel kymograph of two KIF3CC motors bound along the MT lattice and one KIF3CC binding at the tip of a growing MT extension leading to MT catastrophe (∗). See also Fig. S2 for additional lattice loading examples. Scale bars represent 1 μm on x axis and 60 s on the y axis. (C) Histogram of residence time before catastrophe for KIF3CC motors that land on the MT plus-end in the Qdot localization assay. On 34 MT seeds observed over a period of 10 min, 16 events out of a total of 151 catastrophe events (10.6%) exhibited Qdot binding at the MT plus-end that coincided with MT catastrophe. The average residence time mean ± SE is reported from a single exponential fit to the data. To see this figure in color, go online.
Results
KIF3CC enhances MT dynamics
To first test the hypothesis that KIF3CC influences MT behavior, we adapted assays established to study dynamic MTs in vitro using TIRF microscopy (17, 21). MTs were extended from GMPCPP-stabilized seeds in a perfusion chamber (Fig. 1). Upon addition of an Extension Mix containing 10 μM tubulin (labeling ratio of 1:8.5 of X-Rhodamine-labeled to unlabeled-tubulin) and 1 mM MgGTP plus 2 mM MgATP, we observed MT dynamics that consisted of alternating phases of extensive growth followed by catastrophe events that rapidly shortened the extension back to the seed. In two representative examples of extensions of a no-motor control, the growth phase continued for a period of 1.5–3.5 min with extensions ranging from 0.6 to 2.4 μm (Fig. 1, A and B).
When 50 nM KIF3CC was included in the extension mix, the observed dynamics shifted considerably. In lieu of the long extension phases seen in the no-motor control, the majority of the KIF3CC-promoted extensions were limited to 1 min, and their lengths were ∼0.5 μm (Fig. 1 B). A direct comparison of the dynamics of two representative MT seeds is provided in Fig. 1 C (right panel), which shows the location of each MT tip tracked over time. The rates of both MT growth and shortening were consistent in both conditions. However, in the absence of motor, the seed displayed consistent growth phases that approached 2 μm whereas the addition of 50 nM KIF3CC limited the length of growth events (Movies S1 and S2; Table 1). We did not observe rescue events at the conditions used for these assays. Therefore, the presence of KIF3CC seemingly spurred catastrophe, interrupted the growth phase resulting in shorter MT lengths, and ultimately increased the number of dynamic events during the same time period.
Table 1.
Dynamic MT Parameters for Growth and Catastrophe
| Growth |
Shortening |
Catastrophe |
|||||
|---|---|---|---|---|---|---|---|
| N | Rate (nm/s) | Duration (s) | Length (μm) | Rate (nm/s) | Duration (s) | Frequency (min−1) | |
| No motor | 230 | 16.8 ± 0.3 | 165.2 ± 6.8 | 2.5 ± 0.1 | 352.7 ± 10.8 | 7.9 ± 0.3 | 0.24 ± 0.01 |
| KIF3CC (50 nM) | 252 | 24.5 ± 0.8 | 95.2 ± 6.6 | 1.5 ± 0.1 | 513.6 ± 20.7 | 2.9 ± 0.1 | 0.47 ± 0.04 |
| No nucleotide | 194 | 16.7 ± 0.4 | 174.1 ± 7.5 | 2.6 ± 0.1 | 474.0 ± 14.9 | 6.0 ± 0.2 | 0.27 ± 0.01 |
| 2 mM MgAMPPNP | 229 | 19.4 ± 0.4 | 183.6 ± 7.4 | 3.3 ± 0.1 | 709.4 ± 20.0 | 4.9 ± 0.2 | 0.24 ± 0.01 |
| KIF3AB (50 nM) | 201 | 17.3 ± 0.4 | 153.7 ± 6.9 | 2.4 ± 0.1 | 460.6 ± 15.4 | 6.0 ± 0.3 | 0.24 ± 0.01 |
| KIF3AC (50 nM) | 177 | 18.1 ± 0.5 | 146.3 ± 6.7 | 2.3 ± 0.1 | 454.4 ± 14.2 | 5.8 ± 0.2 | 0.24 ± 0.01 |
| KIF3CCΔL11 (50 nM) | 186 | 19.5 ± 0.5 | 162.1 ± 6.9 | 2.8 ± 0.1 | 451.6 ± 14.8 | 7.6 ± 0.3 | 0.26 ± 0.01 |
| KIF3CC | |||||||
| 1 nM | 189 | 20.4 ± 0.4 | 156.6 ± 7.0 | 2.8 ± 0.1 | 375.4 ± 14.8 | 8.9 ± 0.4 | 0.26 ± 0.01 |
| 1.5 nM | 187 | 20.2 ± 0.5 | 150.8 ± 6.5 | 2.6 ± 0.1 | 403.1 ± 11.5 | 7.1 ± 0.2 | 0.27 ± 0.01 |
| 2 nM | 330 | 18.8 ± 0.5 | 179.6 ± 6.0 | 2.8 ± 0.1 | 310.7 ± 7.2 | 9.9 ± 0.3 | 0.29 ± 0.01 |
| 2.25 nM | 207 | 21.8 ± 0.5 | 125.0 ± 7.3 | 2.2 ± 0.1 | 402.9 ± 13.7 | 5.8 ± 0.2 | 0.31 ± 0.02 |
| 2.5 nM | 209 | 16.2 ± 1.1 | 127.7 ± 7.6 | 2.4 ± 0.1 | 320.6 ± 9.0 | 8.6 ± 0.4 | 0.32 ± 0.02 |
| 5 nM | 228 | 17.2 ± 1.1 | 116.2 ± 7.4 | 1.9 ± 0.1 | 258.9 ± 6.3 | 7.8 ± 0.4 | 0.36 ± 0.02 |
| 10 nM | 256 | 16.8 ± 1.0 | 123.1 ± 7.5 | 2.5 ± 0.1 | 353.5 ± 10.1 | 7.4 ± 0.3 | 0.37 ± 0.02 |
| 25 nM | 211 | 15.6 ± 1.1 | 102.3 ± 6.9 | 2.0 ± 0.1 | 356.3 ± 12.6 | 6.4 ± 0.3 | 0.45 ± 0.02 |
| 50 nMa | 252 | 24.5 ± 0.8 | 95.2 ± 6.6 | 1.5 ± 0.1 | 513.6 ± 20.7 | 2.9 ± 0.1 | 0.47 ± 0.04 |
Table of growth and catastrophe parameters from all control and KIF3CC titration assays performed herein. The data were compiled from kymograph analysis of the plus-ends of dynamic MT seeds over a period of 10 min. The number of MT seeds used for each condition ranged from 34 to 39 seeds from three replicate days of experiments. All values are reported mean ± SE.
Data for 50 nM KIF3CC are presented twice on this table.
MT catastrophe occurs at earlier MT ages in the presence of KIF3CC
The difference in MT dynamic behavior in the analysis of KIF3CC could be further examined through analysis of “MT lifetime”, a term for the length of time a MT extension grows before catastrophe. One approach is to plot a cumulative distribution as a function of MT lifetime, whereby each MT lifetime datapoint is represented as the fraction (from 0 to 1) of the MT population that undergoes catastrophe by each time point. Previous work established that cumulative distribution plots are a robust tool for detecting condition-dependent behavior differences in a population of aging MTs (17). Typically, these plots show a lag in the initial phase of MT lifetime where the cumulative distribution remains close to zero because the MT GTP cap is intact. As a function of time, errors accumulate increasing the probability of catastrophe.
Disruption to MT dynamics by catastrophe factors has been reliably detected as an increase in the population of MTs that undergo catastrophe even at the earliest time points of MT extension (17). The cumulative distribution plot of MT dynamics in the presence of 50 nM KIF3CC features a prominent increase in the population of MTs undergoing catastrophe at early MT lifetimes (Fig. 1 D). This is in contrast to MTs in the absence of KIF3CC that exhibit the previously described lag phase at early time points in the MT lifetime curve. These results both qualitatively and quantitatively reveal that MTs in the presence of KIF3CC are susceptible to early catastrophe events.
To test whether ATP turnover was required for KIF3CC to promote MT catastrophe, the ATPase cycle was disrupted with modifications to our assay conditions: one condition to omit ATP from the extension mix, and one to replace ATP with the nonhydrolyzable ATP analog AMPPNP. Both of these conditions resulted in MT dynamics that revert to the behavior of the no-motor control, thus confirming that KIF3CC does in fact require ATP turnover to promote the observed MT dynamics (Fig. 1 D).
We asked whether other members of the kinesin-2 subfamily could stimulate catastrophe. MTs in the presence of either 50 nM KIF3AB or KIF3AC exhibited the same lag phase for catastrophe observed in controls (Fig. 1 D, inset; Movie S3). However, KIF3AC has a lower affinity for MTs (K1/2,MT = 133 nM) than that of KIF3CC at 44 nM (5). Possibly, the concentration of KIF3AC used for these experiments was not sufficiently high for MT binding. To test this hypothesis directly, experiments were performed at 0.5, 1, and 2 μM KIF3AC (Fig. S1 A). Even when the concentration of KIF3AC was increased >10-fold the K1/2,MT, the MT dynamics did not change and the lag phase in the cumulative distribution plot was comparable to the other controls. These results confirm that the catastrophe promotion of MTs by KIF3CC is unique to the homodimer, and heterodimeric KIF3AC and KIF3AB do not promote MT catastrophe.
We also examined whether the KIF3C-specific extension of loop L11 was required for KIF3CC-promoted MT dynamics. The L11 flexible surface loop is extended by 25 residues in KIF3C and is located at the MT-motor interface. In earlier studies, results showed that truncation of this extension to a length characteristic of other kinesins increased the processive run length of both KIF3AC and KIF3CC (5). To test the role of loop L11 in MT catastrophe, we used the KIF3CCΔL11 homodimer in which the L11 sequence of each polypeptide was shortened (Movie S4). In the presence of 50 nM KIF3CCΔL11, the cumulative distribution plots exhibited the lag phase seen in the no-motor control (Fig. 1 D, inset). Thus, the characteristic extension of the KIF3C loop L11 appears critical for KIF3CC-promoted MT catastrophe.
To explore the mechanism by which KIF3CC promotes MT catastrophe, the MT lifetime data were plotted as a PDF. The use of a PDF plot provides the percent of catastrophe events that occur at each MT lifetime. The relationship of the PDF dataset (dF(t)/dt) to a gamma function (Γ(n)) can be used to derive parameters for the shape of a gamma function, a quantification of both the number of aging steps that are accumulated before catastrophe and the rate of their accumulation, according to Eq. 1. From this fit, the no-motor control exhibits 2.6 ± 0.2 aging steps (Fig. 1 E), in agreement with previous reports of MT aging (17). In contrast, MTs in the presence of KIF3CC show a shift to a high likelihood of catastrophe at early MT lifetimes (Fig. 1 E), resulting in a step parameter value of 1.0 ± 0.1 (fit with a 95% confidence interval) (Fig. 1, D and E; Table S1).
This analysis indicates that the presence of homodimeric KIF3CC results in dynamic MTs that exhibit growth phases interrupted by a higher frequency of catastrophe events (Fig. 1 F; Table 1). Furthermore, this increased catastrophe frequency is linked to a prevalence of MT catastrophe events at early MT lifetimes, which is at least in part due to the shortening of a typically multistep MT aging process to a single step in the presence of KIF3CC.
KIF3CC is a potent stimulator of MT catastrophe
The concentration of 50 nM KIF3CC was chosen based on experiments in which all the MTs in the field of view show similar KIF3CC-promoted MT dynamics. At this concentration, every MT has KIF3CC bound with at least 16 motors per MT (Supporting Material). However, the use of this concentration of KIF3CC raised the question of what concentration of KIF3CC was required to stimulate MT catastrophe. Cumulative distribution plots were compiled from a titration of KIF3CC from 1 to 50 nM, and these results confirmed that lower concentrations of the motor also alter MT dynamics (Fig. 2 A). Interestingly even at just 2.5 nM, KIF3CC could stimulate MTs to undergo catastrophe at early MT lifetimes. Note that the data analysis for each KIF3CC concentration included the entire MT population, although not every MT in the field of view showed the same behavior. The step parameter values for each concentration in the titration were determined from the gamma distribution of the PDF plots. The values were then compiled as a function of KIF3CC concentration (Fig. 2 B), and a Hill slope model fit to the data provided the K1/2,KIF3CC at 2.1 nM KIF3CC. These results reveal that KIF3CC is a potent MT catastrophe factor that requires only a small concentration of motor to promote MT catastrophe. However, this experiment does not provide information about the number of KIF3CC dimers at the MT plus-end for catastrophe.
The kinetics of the MT aging process can be examined with a calculation of time-dependent catastrophe frequency. Unlike a population catastrophe frequency parameter (Fig. 1 F; Table 1) that looks at all catastrophe events during the entire time of acquisition, the time-dependent catastrophe frequency examines the MT age-dependence of MT catastrophe. Calculations of the time-dependent catastrophe frequency quantify the population of MTs that undergo a catastrophe by a certain MT lifetime versus the MTs that reach that lifetime without a catastrophe event. A direct examination of the time-dependent catastrophe frequency for each dataset in the KIF3CC titration (Fig. 2 C) shows that in the absence of KIF3CC, the catastrophe rate is zero for short MT lifetimes and asymptotically approaches 0.014 s−1. As the concentration of KIF3CC increases, the time-dependent catastrophe frequency increases at much shorter MT lifetimes. The addition of 2 nM KIF3CC into the assay illustrates a transition in these data where the time-dependent catastrophe frequency curvature dampens, and the maximum rate of catastrophe decreases to 0.010 s−1. With 2.5 nM KIF3CC, the rate drops to 0.008 s−1 and the curve flattens because the catastrophe frequency becomes independent of MT lifetime. These data indicate that even in the presence of low concentrations of KIF3CC, the MT aging process is eliminated as catastrophe shifts from a multistep aging process to elimination of MT aging (Fig. 2 D), which is reminiscent of MT catastrophe in the presence of MCAK (17, 29).
To examine the MT aging process kinetically, the rate of accumulation of MT aging errors was pursued. This rate parameter, along with the step parameter that tells the number of aging errors that accumulate before catastrophe, comes from the gamma fit of the original PDF plots of MT lifetime data. A plot of the rate of error accumulation versus KIF3CC concentration exhibits an increase at low KIF3CC concentrations that reaches a maximum rate of 0.01 s−1 at 20 nM KIF3CC (Fig. 3 A). In contrast, KIF3CC has little impact on the growth rate of polymerizing MTs (Fig. 3 B).
Figure 3.
KIF3CC accelerates the rate parameter for catastrophe but not MT elongation. (A) Plot of the rate parameter for MT catastrophe plotted as a function of KIF3CC concentration from 0 to 50 nM dimer. Each data point reports the rate parameter determined by the gamma fit of the PDF plot for MT lifetime data. The nonlinear least-squared fit is shown, which provides a maximum rate of accumulation of MT aging steps. See also Fig. 2D for N values of each data point. All data are shown mean ± SE. (B) Rate of growth at the MT plus-end (nm/s) in the dynamic MT assay at 37°C for 10 min as determined from kymograph analysis, plotted as a function of KIF3CC dimer concentration. Error bars represent mean ± SE. See also Table 1 for calculated rates and N values for each condition.
When compared with the characteristics of MT catastrophe in the presence of MCAK or Kip3, the combination of KIF3CC-induced characteristics suggests a different mechanism for catastrophe promotion (Table 2). Although Kip3 was shown to accelerate the rate of accumulation of MT aging steps, it does so without altering the number of steps required to induce MT catastrophe (17). The MCAK mechanism involves a decrease in the number of required steps to spur MT catastrophe but without a change in the rate of accumulation of errors (17). The results for the KIF3CC step parameter and catastrophe frequency reported here reveal that KIF3CC promotion of MT catastrophe results from a combination of properties. It decreases the number of MT aging errors that accumulate before MT catastrophe to a single step, and also accelerates the rate parameter of MT catastrophe to eliminate the MT aging process without an impact on the rate of growth at the elongating MT plus-end.
Table 2.
The Mechanism Employed by KIF3CC Is Distinct from Other Catastrophe Factors
| MCAK (Kinesin-13)a | Kip3/KIF18A (Kinesin-8)b | KIF3CC (Kinesin-2) | ||
|---|---|---|---|---|
| MT tip localization | tip recognition | yes | yes | yes |
| motility | bilateral diffusion | processivity | no | |
| plus-end targeting | no | yes | yes | |
| Mechanism of influencing MT dynamics | catastrophe factor | yes | yes | yes |
| structural features | curls protofilaments | detects curved subunits | eliminates tip taper | |
| method of MT disassembly | catastrophe | depolymerization by cooperative removal of subunits | catastrophe | |
| Impact on MT aging | change number of MT aging steps | decreases | no change | decreases |
| rate of catastrophe | no impact | accelerates | accelerates | |
| Rescue events | no | no | no |
MT tips appear blunt in the presence of KIF3CC
Recent work from Coombes et al. (25) suggested that in addition to the accumulation of errors in the protective GTP cap, a structural change at the MT tip precedes catastrophe. For this finding, the authors measured the depletion of the fluorescence signal at the MT tip to detect MT tip structure according to protofilament length. They reported that in short MT extensions, the fluorescence signal exhibited an abrupt drop in intensity at the MT tip. The interpretation of this drop was that during a short growth phase, the protofilaments are approximately similar in length extending from the MT seed and thus the fluorescence signal of each protofilament end is approximately the same distance away from the MT seed. Alternatively, MTs that grow for longer times exhibited a fluorescence signal decrease that gradually depleted, which was interpreted as variability in each of the 13 protofilament lengths due to variability in protofilament elongation. This behavior ultimately leads to a tapered structure at the tips of long MT extensions (Fig. 4 A, illustration). The authors subsequently applied this method to probe MT tip structure in the presence of MCAK (25). We pursued this experimental approach to assess the impact of KIF3CC on the MT tip structure.
The depletion of the X-rhodamine-tubulin fluorescence signal for MTs in both the absence and presence of KIF3CC was calculated as the fluorescence deviation, which reports half of the distance over which the fluorescence signal decreases from maximum fluorescence intensity to background. For the shortest MT extensions, both the absence and presence of KIF3CC have fluorescence deviation values at ∼125 nm (Fig. 4 A). Comparable to previous reports, MTs in the absence of motor show an increase in the length of fluorescence deviation at increasing extension lengths because of high variability in length for each of the 13 protofilaments (25, 30). In contrast, the presence of 50 nM KIF3CC resulted in a steep drop in fluorescence at the MT plus-end with fluorescence deviations that remain around 125–140 nm even at extensions up to 3 μm.
The difference in the depletion of the fluorescence signal in these two conditions is made evident by a plot of the MT tip fluorescence intensity. A direct comparison of the profiles from representative 3-μm extensions in both the presence and absence of KIF3CC illustrate the structural impact of KIF3CC that alters the MT tip taper despite significant MT extension (Fig. 4 A, inset). In further comparisons of profiles from different length extensions, the evolution of tip structure as a function of MT elongation becomes evident (Fig. 4 B). The gradual loss of fluorescence signal in the MT tips from long (4–5 μm) extensions in the no-motor control contrasts starkly with the steep drop in fluorescence signal of MT tips in the presence of KIF3CC. This steep drop in fluorescence signal is sustained at all lengths of MT extensions that were observed in the presence of KIF3CC. Furthermore, the population of MTs that is capable of reaching long extension lengths differs greatly in the absence and in the presence of KIF3CC, as seen by the distribution of extension lengths observed in each condition (Fig. 4 C). Notably, the extension lengths of MTs in the presence of KIF3CC are limited such that only a handful of extensions are capable of reaching 3 μm.
One interpretation of these data could be that the correlation between blunt MT tips and KIF3CC is an indication that KIF3CC identifies blunt MT plus-ends for catastrophe. However, the fluorescence deviation measurements in combination with the shift in catastrophe frequency to early MT lifetimes (Fig. 1) reveals KIF3CC as a potent catastrophe factor that limits the extent of MT growth before catastrophe, and thereby hinders the development of a characteristically tapered tip structure. Therefore, the data indicate that the KIF3CC-induced MT catastrophe occurs at least in part through restrictions on both the length that MT extensions can reach before they undergo catastrophe and on the tip structure of the protofilaments immediately before catastrophe.
Catastrophe events correlate with brief KIF3CC MT tip-localization events
Tip localization is a likely target for the KIF3CC mechanism if tip structure is altered. We adapted a single molecule assay using Qdots to visualize the KIF3CC in relation to the dynamic MTs in our TIRF assay. For continuation of our dataset, these experiments contained a final concentration of 50 nM KIF3CC. However, for visualization in the single molecule regime, the pool of motor was spiked with a subpopulation (4 nM; 8% of the total motor concentration) of Qdot-labeled KIF3CC (28).
KIF3CC was found at the plus-end of dynamic MTs, and its association was followed on average ∼8 s later by MT catastrophe (Fig. 5 A, top section; Fig. 5 C). A quantification of these Qdot tip localizations determined that 10.6 ± 0.1% of all catastrophe events was preceded by KIF3CC landing at the MT plus-tip, in close agreement with our 8% labeling ratio. Based on these results, we conclude that it is highly likely that each catastrophe event in saturating KIF3CC coincides with localization of the motor at the MT plus-end. However, this experimental design cannot determine whether a single KIF3CC at the MT plus-tip is responsible for each catastrophe event or multiple KIF3CC motors. Rather, the labeling ratio leads us to hypothesize that although we see only a single Qdot at the MT tip before catastrophe in saturating motor conditions, there is likely a larger number of additional unlabeled motors interacting simultaneously at the tip to promote MT disassembly.
KIF3CC-Qdots also associated along the MT lattice where they were observed to move processively, albeit slowly, toward the dynamic MT plus-end. These lattice-bound motors allowed us to determine the average velocity of KIF3CC on dynamic MTs at 7.7 ± 0.3 nm/s (Fig. S2 A). This value is in agreement with our previously published 8 nm/s velocity for KIF3CC on paclitaxel-stabilized MTs (5). On average, each extending MT had ∼1 lattice-bound Qdot (Fig. S2 B). However, the presence of Qdots along the lattice did not seem to affect catastrophe events at the MT plus-ends, either on MTs with a single lattice-bound Qdot or in rare instances where multiple Qdots were seen bound along a MT lattice (Fig. 5 B; Fig. S2 C). Moreover, KIF3CC-Qdots were not localized at MT minus-ends, and MT dynamics at the MT minus-ends were not observed.
Homodimerization and loop L11 are central to KIF3CC catastrophe promotion
Because KIF3C can heterodimerize with KIF3A, we examined whether KIF3AC can localize to MT plus tips and promote MT catastrophe (Fig. 5 A, middle; Fig. S1). KIF3AC motors were seen landing along the MT lattice where they moved rapidly and processively toward the dynamic MT plus-end. Neither the movement of KIF3AC along the lattice nor its interaction with the MT tip correlated with catastrophe. Rather, MT extension continued both for substantial lengths and times even after the KIF3AC-Qdot had run the full length of the MT. This result further establishes that the heterodimeric KIF3AC does not promote MT catastrophe and moreover that KIF3C in a heterodimeric association is not sufficient for catastrophe promotion.
KIF3CCΔL11, like KIF3CC, landed both on the MT lattice and at the dynamic MT plus-end. When tip binding occurred, only 2.2 ± 0.4% of these events were immediately followed by catastrophe. In the remaining tip binding events, KIF3CCΔL11 seemingly posed no barrier to continued MT extension (Fig. 5 A, bottom). Along with cumulative distribution plots (Fig. 1 D), these imaging data provide further proof that the loop L11 extension unique to KIF3C is a structural motif required for KIF3CC-induced MT catastrophe and promotion of MT dynamics.
Discussion
Kinesin-2 KIF3CC is a novel catastrophe factor
Our previous work explored the single molecule motility and the presteady-state kinetics of KIF3CC to gain a better understanding of the motile properties of KIF3C in the context of the KIF3AC heterodimer (5, 31). The analysis presented here expands our understanding of one member of the kinesin-2 subfamily, revealing that homodimeric KIF3CC acts as a catastrophe factor, substantiating its proposed physiological role for regulation of MT dynamics in neuronal growth cones. Moreover, this analysis provides further insight into the mechanochemical diversity that can be achieved within the kinesin-superfamily. The homodimeric kinesin-2 KIF17 was previously shown to regulate MT length through protection of the MT plus-end to dampen MT dynamics leading to long MT growth phases and inhibition of depolymerization (32). However, to the best of our knowledge, the results presented here are the first direct evidence of a kinesin-2 operating in the opposite manner, as a catastrophe factor that stimulates and accelerates MT catastrophe.
As examined herein, the mechanism used by KIF3CC to enhance the frequency of MT catastrophe involves a combination of increasing the rate of the catastrophe parameter (Fig. 3 A) and eliminating the stepwise aging process that normally leads to MT catastrophe (Fig. 1), without altering the rate of growth at the MT plus-end (Fig. 3 B). Among the kinesins classified as catastrophe factors, KIF3CC appears to use a mechanism that is altogether distinct from the mechanisms employed by either kinesin-8 (Kip3/KIF18A) or kinesin-13 (MCAK) for promotion of MT catastrophe (Table 2).
A recently published study of Kip3/kinesin-8 identified its loop L11 as a critical component to Kip3 MT depolymerization activity (33). Although the data reported herein also identify L11 as being critical for MT catastrophe promotion by KIF3CC, there are notable differences. The sequences of loop L11 of Kip3 and KIF3C are entirely different, with Kip3 featuring a string of basic residues in contrast to the significantly extended length of KIF3C’s loop L11 that is highly enriched in glycines, serines, and prolines (5, 33). Specifically, there are no notable features or motifs within the two loop L11 sequences that are shared or that would suggest a shared mechanism for their influence on MTs.
Our previous work on loop L11 in KIF3C showed that shortening the extended sequence to the length seen in other kinesins resulted in an increase in MT affinity. Compared to the other heterodimeric and engineered homodimeric KIF3 motors, KIF3CC itself has a high MT affinity (K1/2,MT = 44 nM (5)) that was further increased by shortening loop L11. Both Kip3 and MCAK have high MT affinities for certain attributes of their function: the Kip3 affinity is pertinent, as the motors move processively toward the MT plus-end and exhibit preferential binding for curved tubulin to cooperatively remove one tubulin dimer at a time, whereas MCAK has a high affinity for a binding site at the MT plus-tip where it applies torque to curl protofilaments away from the polymer structure (28, 33, 34, 35, 36). Although not known at this time, the high affinity of KIF3CC and its extended loop L11 may play a role in targeting KIF3CC to the plus-end tip of MTs. The data reported here imply that tip recognition and plus-end localization are likely critical elements for the KIF3CC mechanism.
It is unlikely, however, that KIF3CC utilizes its processive movement for such plus-end localization. Although the slow processivity of KIF3CC agrees with the single molecule data on paclitaxel-stabilized MTs (5), the motility of KIF3CC is not fast enough to deliver this catastrophe factor to the MT plus-end to drive MT dynamics where the rate of the growth is ∼20 nm/s (Table 1). This KIF3CC characteristic itself is highly inconsistent with the mechanism employed by Kip3, whereby the velocity of the motors exceeds the MT growth rate and thus lattice loading of the motor leads to MT plus-end localization over time (28, 37).
MT tip localization by MCAK also differs from KIF3CC. Although they are similar in their elimination of the MT aging process, the rapid random diffusive search used by MCAK to reach either the MT-plus or minus-end to promote catastrophe (38, 39, 40) contrasts with KIF3CC. KIF3CC-promoted catastrophe was not observed at the MT minus-end nor were KIF3CC motors localized to the MT minus-end.
With such a slow processive motor and with no apparent impact of lattice-bound KIF3CC on MT catastrophe (Fig. S2), how can KIF3CC efficiently arrive at the MT plus-end to stimulate MT catastrophe? One possibility is through association with a MT-end binding protein. The MT depolymerizing kinesin-8 Kif18B, for example, directly associates with EB1 to establish its association with the MT plus-end (22). The tip localization of KIF3C itself has been reported to be in part driven through an association to the MT plus-end binding protein EB3 in neurons (2). However, our data imply that KIF3CC may have some inherent ability to detect the MT plus-end because no EB3 is present. Furthermore, our motor constructs encode a C-terminal truncation that eliminates the EB3-binding site on KIF3C. Despite these considerations, KIF3CC can still readily detect the MT tip and initiate MT catastrophe in our minimal component in vitro system.
KIF3CC as a potent stimulator of MT dynamics in cells
The data reported herein show that once KIF3CC arrives at the MT plus-end, it acts as a robust stimulator of MT catastrophe with a unique mechanism of catastrophe promotion. The work published by Gumy et al. (2) showed that catastrophe frequency increased as a result of KIF3C gene overexpression. Our results substantiate that report and also point out that only a modest concentration of the KIF3C gene product would need to homodimerize to exert this effect. This mechanism bolsters KIF3CC’s proposed role at the growth cone where rapid MT restructuring and remodeling are required.
We expect, however, that the complexity of the neuronal milieu likely requires additional targeting and regulatory mechanisms beyond the inherent properties of the KIF3C motor to ensure efficient delivery of the motor to the MT plus-end. Future work will need to confirm the cellular presence of homodimeric KIF3CC and directly explore its impact on the dynamics of an intact MT cytoskeleton.
Author Contributions
S.G.-L. and S.P.G. designed research. S.G.-L. performed research and analyzed data. S.G.-L., I.R., and S.P.G. wrote and revised the manuscript.
Acknowledgments
We thank Melissa Gardner for assistance in adapting the TipTracker program for these experiments. We also acknowledge Alexey Khodjakov, Scott Forth, and Lee Ligon for thoughtful discussions, and input from members of the S.P.G. lab.
This work was supported by National Institutes of Health (NIH) grant R37-GM054141 (to S.P.G.).
Editor: David Odde.
Footnotes
Supporting Materials and Methods, two figures, one table, and four movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)31027-5.
Supporting Material
The average rate of elongation is 16.8 ± 0.3 nm/s with an average length of 2.5 ± 0.1 μm. Video playback speed is at 120× real-time.
The average rate of elongation is 24.5 ± 0.8 nm/s with an average length of 1.5 ± 0.1 μm. Video playback speed is at 120× real-time. Two MTs are shown.
The average rate of elongation is 18.1 ± 0.5 nm/s with an average length of 2.3 ± 0.1 μm. Video playback speed is at 120× real-time.
The average rate of elongation is 19.5 ± 0.5 nm/s, with an average length of 2.8 ± 0.1 μm. Video playback speed is at 120× real-time.
References
- 1.Davidovic L., Jaglin X.H., Khandjian E.W. The fragile X mental retardation protein is a molecular adaptor between the neurospecific KIF3C kinesin and dendritic RNA granules. Hum. Mol. Genet. 2007;16:3047–3058. doi: 10.1093/hmg/ddm263. [DOI] [PubMed] [Google Scholar]
- 2.Gumy L.F., Chew D.J., Fawcett J.W. The kinesin-2 family member KIF3C regulates microtubule dynamics and is required for axon growth and regeneration. J. Neurosci. 2013;33:11329–11345. doi: 10.1523/JNEUROSCI.5221-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muresan V., Abramson T., Schnapp B.J. KIF3C and KIF3A form a novel neuronal heteromeric kinesin that associates with membrane vesicles. Mol. Biol. Cell. 1998;9:637–652. doi: 10.1091/mbc.9.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Z., Roberts E.A., Goldstein L.S. Functional analysis of mouse kinesin motor Kif3C. Mol. Cell. Biol. 2001;21:5306–5311. doi: 10.1128/MCB.21.16.5306-5311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guzik-Lendrum S., Rank K.C., Gilbert S.P. Kinesin-2 KIF3AC and KIF3AB can drive long-range transport along microtubules. Biophys. J. 2015;109:1472–1482. doi: 10.1016/j.bpj.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang C.F., Banker G. The translocation selectivity of the kinesins that mediate neuronal organelle transport. Traffic. 2012;13:549–564. doi: 10.1111/j.1600-0854.2011.01325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sardella M., Navone F., Cabibbo A. KIF3C, a novel member of the kinesin superfamily: sequence, expression, and mapping to human chromosome 2 at 2p23. Genomics. 1998;47:405–408. doi: 10.1006/geno.1997.5123. [DOI] [PubMed] [Google Scholar]
- 8.Mitchison T., Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 9.Walker R.A., O’Brien E.T., Salmon E.D. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J. Cell Biol. 1988;107:1437–1448. doi: 10.1083/jcb.107.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyman A.A., Salser S., Mitchison T.J. Role of GTP hydrolysis in microtubule dynamics: information from a slowly hydrolysable analogue, GMPCPP. Mol. Biol. Cell. 1992;3:1155–1167. doi: 10.1091/mbc.3.10.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandelkow E.M., Mandelkow E., Milligan R.A. Microtubule dynamics and microtubule caps: a time-resolved cryo-electron microscopy study. J. Cell Biol. 1991;114:977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowne-Anderson H., Zanic M., Howard J. Microtubule dynamic instability: a new model with coupled GTP hydrolysis and multistep catastrophe. BioEssays. 2013;35:452–461. doi: 10.1002/bies.201200131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zakharov P., Gudimchuk N., Grishchuk E.L. Molecular and mechanical causes of microtubule catastrophe and aging. Biophys. J. 2015;109:2574–2591. doi: 10.1016/j.bpj.2015.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.VanBuren V., Cassimeris L., Odde D.J. Mechanochemical model of microtubule structure and self-assembly kinetics. Biophys. J. 2005;89:2911–2926. doi: 10.1529/biophysj.105.060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohi R., Zanic M. Ahead of the curve: new insights into microtubule dynamics. F1000 Res. 2016;5:314. doi: 10.12688/f1000research.7439.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brouhard G.J. Dynamic instability 30 years later: complexities in microtubule growth and catastrophe. Mol. Biol. Cell. 2015;26:1207–1210. doi: 10.1091/mbc.E13-10-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardner M.K., Zanic M., Howard J. Depolymerizing kinesins Kip3 and MCAK shape cellular microtubule architecture by differential control of catastrophe. Cell. 2011;147:1092–1103. doi: 10.1016/j.cell.2011.10.037. [DOI] [PubMed] [Google Scholar]
- 18.Maney T., Hunter A.W., Wordeman L. Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J. Cell Biol. 1998;142:787–801. doi: 10.1083/jcb.142.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayr M.I., Hümmer S., Mayer T.U. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr. Biol. 2007;17:488–498. doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 20.Stumpff J., von Dassow G., Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev. Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varga V., Helenius J., Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat. Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- 22.Stout J.R., Yount A.L., Walczak C.E. Kif18B interacts with EB1 and controls astral microtubule length during mitosis. Mol. Biol. Cell. 2011;22:3070–3080. doi: 10.1091/mbc.E11-04-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su X., Arellano-Santoyo H., Pellman D. Microtubule-sliding activity of a kinesin-8 promotes spindle assembly and spindle-length control. Nat. Cell Biol. 2013;15:948–957. doi: 10.1038/ncb2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albracht C.D., Rank K.C., Gilbert S.P. Kinesin-2 KIF3AB exhibits novel ATPase characteristics. J. Biol. Chem. 2014;289:27836–27848. doi: 10.1074/jbc.M114.583914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coombes C.E., Yamamoto A., Gardner M.K. Evolving tip structures can explain age-dependent microtubule catastrophe. Curr. Biol. 2013;23:1342–1348. doi: 10.1016/j.cub.2013.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prahl L.S., Castle B.T., Odde D.J. Quantitative analysis of microtubule self-assembly kinetics and tip structure. Methods Enzymol. 2014;540:35–52. doi: 10.1016/B978-0-12-397924-7.00003-0. [DOI] [PubMed] [Google Scholar]
- 27.Demchouk A.O., Gardner M.K., Odde D.J. Microtubule tip tracking and tip structures at the nanometer scale using digital fluorescence microscopy. Cell. Mol. Bioeng. 2011;4:192–204. doi: 10.1007/s12195-010-0155-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varga V., Leduc C., Howard J. Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell. 2009;138:1174–1183. doi: 10.1016/j.cell.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 29.Odde D.J., Cassimeris L., Buettner H.M. Kinetics of microtubule catastrophe assessed by probabilistic analysis. Biophys. J. 1995;69:796–802. doi: 10.1016/S0006-3495(95)79953-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chrétien D., Fuller S.D., Karsenti E. Structure of growing microtubule ends: two-dimensional sheets close into tubes at variable rates. J. Cell Biol. 1995;129:1311–1328. doi: 10.1083/jcb.129.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang P., Rayment I., Gilbert S.P. Fast or slow, either head can start the processive run of Kinesin-2 KIF3AC. J. Biol. Chem. 2016;291:4407–4416. doi: 10.1074/jbc.M115.705970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Acharya B.R., Espenel C., Kreitzer G. Direct regulation of microtubule dynamics by KIF17 motor and tail domains. J. Biol. Chem. 2013;288:32302–32313. doi: 10.1074/jbc.M113.494989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arellano-Santoyo H., Geyer E.A., Pellman D. A tubulin binding switch underlies Kip3/Kinesin-8 depolymerase activity. Dev. Cell. 2017;42:37–51. doi: 10.1016/j.devcel.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moores C.A., Yu M., Milligan R.A. A mechanism for microtubule depolymerization by KinI kinesins. Mol. Cell. 2002;9:903–909. doi: 10.1016/s1097-2765(02)00503-8. [DOI] [PubMed] [Google Scholar]
- 35.Moores C.A., Hekmat-Nejad M., Milligan R.A. Regulation of KinI kinesin ATPase activity by binding to the microtubule lattice. J. Cell Biol. 2003;163:963–971. doi: 10.1083/jcb.200304034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan D., Rice W.J., Sosa H. Structure of the kinesin13-microtubule ring complex. Structure. 2008;16:1732–1739. doi: 10.1016/j.str.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta M.L., Jr., Carvalho P., Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat. Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- 38.Helenius J., Brouhard G., Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- 39.Hunter A.W., Caplow M., Howard J. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol. Cell. 2003;11:445–457. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desai A., Verma S., Walczak C.E. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The average rate of elongation is 16.8 ± 0.3 nm/s with an average length of 2.5 ± 0.1 μm. Video playback speed is at 120× real-time.
The average rate of elongation is 24.5 ± 0.8 nm/s with an average length of 1.5 ± 0.1 μm. Video playback speed is at 120× real-time. Two MTs are shown.
The average rate of elongation is 18.1 ± 0.5 nm/s with an average length of 2.3 ± 0.1 μm. Video playback speed is at 120× real-time.
The average rate of elongation is 19.5 ± 0.5 nm/s, with an average length of 2.8 ± 0.1 μm. Video playback speed is at 120× real-time.