Abstract
Artificial intelligence (AI), a computer system aiming to mimic human intelligence, is gaining increasing interest and is being incorporated into many fields, including medicine. Stroke medicine is one such area of application of AI, for improving the accuracy of diagnosis and the quality of patient care. For stroke management, adequate analysis of stroke imaging is crucial. Recently, AI techniques have been applied to decipher the data from stroke imaging and have demonstrated some promising results. In the very near future, such AI techniques may play a pivotal role in determining the therapeutic methods and predicting the prognosis for stroke patients in an individualized manner. In this review, we offer a glimpse at the use of AI in stroke imaging, specifically focusing on its technical principles, clinical application, and future perspectives.
Keywords: Artificial intelligence, Machine learning, Stroke
Introduction
Artificial intelligence (AI) is machine intelligence that is meant to imitate human brain processes [1-3]. With rapid advances in computer science, AI has recently been applied to many areas by large information technology companies. The recent defeat of a human Go champion by Google DeepMind AlphaGo surprised the public worldwide and demonstrated that AI may even be superior to the human brain in some decision-making processes [4].
Precision medicine, defined as tailored diagnosis and therapy for an individual patient, is an important emerging concept in future medicine [5]. The recent explosion of clinical, biological (genomic and immunological), and imaging data is creating a path for such personalized medicine. Integration of these multiple streams of information and appropriate interpretation thereof are important, and AI techniques may be useful in this respect.
Stroke medicine may be a suitable application for precision medicine and AI techniques because of the vast amount of data and multidisciplinary approaches used in making clinical decisions [1,6]. In particular, brain imaging, which is the key factor in stroke management and forms the basis for numerous complex go/no-go decisions, is an attractive subject for AI techniques. In this review article, we will briefly review AI techniques in stroke imaging and discuss their potential clinical applications and future roles.
AI
Machine learning is a technique of AI that is widely used in interpreting medical images. It recognizes patterns of imaging information and renders medical diagnoses [3]. Supervised and unsupervised learning are widely used typical machine learning types. Deep learning is a more recently developed technique, which mimics the human brain, using multiple layers of artificial neuronal networks.
Machine learning
Machine learning is typically classified into supervised and unsupervised learning [1-3]. Supervised machine learning uses a training dataset labeled by humans to define the desired or known answers. It may expedite classification or regression processes with large datasets and would be useful for predicting or discriminating clinical outcomes. However, it requires a human labeling process, which is often cumbersome and time-consuming. Examples of a supervised learning method include the support vector machine, decision tree, linear regression, logistic regression, naive Bayes, and random forest methods. In contrast, unsupervised machine learning does not use human-defined answers; instead, it seeks to identify hidden patterns, on its own, in large datasets, which are usually invisible to humans. Therefore, unsupervised learning may be useful in seeking novel disease mechanisms, genotypes, and phenotypes. Examples of unsupervised learning include K-means, mean shift, affinity propagation, hierarchical clustering, and Gaussian mixture modeling.
These two learning methods can be easily distinguished by whether or not they use human feedback. If a machine is to be developed that could distinguish between ischemic and hemorrhagic stroke images, machines with a supervised algorithm will be trained with human pre-labeled correct answers, while those with an unsupervised algorithm will learn to categorize images into two or three groups according to certain patterns, which they would identify during learning processes, by themselves.
In machine learning, variables used as input data are generally referred to as features; these can be numerical or nominal values. Because the performance of machines is variable according to the features inputted, it is important to select and extract features from the data appropriately. The input features are usually determined by researchers and data scientists. Nowadays, various feature selection methods have been developed to enhance the selection process and establish machine models with high accuracy. Using these features, machine learning algorithms determine the optimal decision boundary—selecting features and developing a model—to conduct a set task. For imaging data, various image features, such as the size, location, shape, and signal intensities of the lesion, can be used for machine learning. Machines can distinguish and make use of additional imaging features, such as texture information, e.g., signal intensity gradient, and skewness, which are not discernible by humans [3,7].
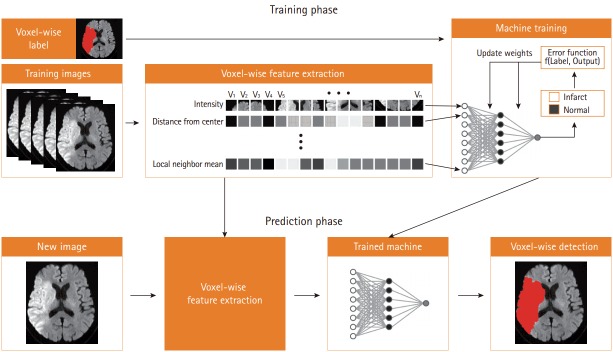
Figure 1 demonstrates an example of automated segmentation of infarct lesions using supervised machine learning techniques. In the training phase, machines learn how to detect infarct lesions using various voxel-wise imaging features under the supervision of human-labeled answers. In this phase, the machine attempts to develop an optimal model to perform the task with selective input variables. In this case, the machine determines several image features and devises a model to automatically draw stroke lesion margins. In the prediction phase, the machine encounters a new image, and applies its determined mechanism to the selected image.
Figure 1.
Schematic workflow of supervised machine learning.
There are two representative machine learning algorithms, the support vector machine (SVM) and the artificial neural network (ANN).
SVM
SVM is a supervised machine learning method, which is useful for developing a model to allocate an object to one category or the other. Therefore, SVM is widely used in clinical imaging analysis, which categorizes or classifies a diagnosis. SVM constructs a hyper-plane in a high-dimensional space as the decision surface. To accomplish better performance, the margin of separation between classes needs to be maximized (Figure 2) [8]. For a non-linear classification, SVM uses the kernel technique, which implicitly converts the input features into high-dimensional feature spaces. Therefore, selection of the kernel should be appropriate, to avoid increases in error rates.
Figure 2.
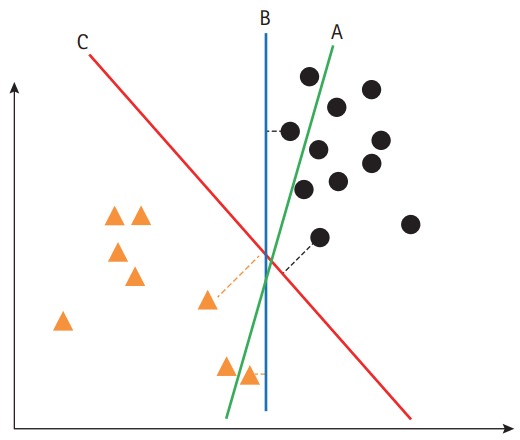
Hyper-plane of support vector machine. (A) Hyper-plane fails to distinguish the groups. (B) Hyper-plane distinguishes the groups but with a small margin. (C) Hyper-plane distinguishes the groups with the maximum margin.
ANN
ANN is inspired by biological neural networks. An artificial neuron receives inputs from other neurons, integrates the inputs with weights, and activates when a defined condition is satisfied (Figure 3). ANN consists of input, output, and hidden layers. The input layer receives observed values, while the output layer represents the target (a value or class). The layers between input and output layers are called hidden layers. Typically, nodes within each layer are all connected to each other between the layers. ANN is very flexible in terms of application to various data types, and can be applied to conditions in which other statistical analyses are unsuccessful. However, ANN has limitations in that it is susceptible to data-overfitting, and tends to require a long computation time.
Figure 3.
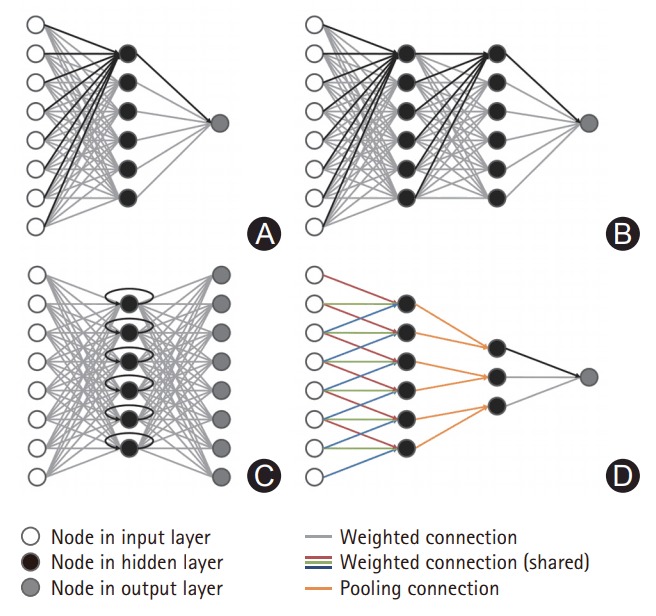
Schematic representation of neural network. (A) Artificial neural network with a single hidden layer. All nodes are fully connected between layers. (B) Deep neural network with two hidden layers. Deep learning has multiple hidden layers. (C) Recurrent neural network with a single hidden layer architecture. Nodes in the hidden layer have a directed cycle. (D) Convolutional neural network. Weighted connections are indicated with the same color in convolutional hidden layers.
Deep learning
Deep learning is a more recently developed technique of machine learning, which mimics the human brain using multiple layers of ANN. Although there are no explicit criteria on the threshold of depth to discriminate between shallow and deep learning, the latter is conventionally defined as having multiple hidden layers.
Recurrent neural network (RNN) is a subtype of ANN that uses connections between nodes forming a cycle with a one-way direction (Figure 3). RNN can use the temporal memory of networks, which is in contrast to other typical neural networks with feed-forward structures. Therefore, RNN is specialized for the processing of temporal data, which include recognition of natural language, handwriting, and speech.
Convolutional neural network (CNN) also uses a neural network but assumes that the CNN input signal has geometric information, such as the receptive field in the visual cortex, individual cortical neurons here belong to the receptive field in the visual cortex, not to CNN, therefore we think that current form would be better [9]. CNN nodes connect to each other in a geometrical structure, not showing all-to-all connections (Figure 3). In imaging analysis, nodes in the input layer are arranged to produce a convolution of a small part of the image (kernel) [3]; this kernel then moves around the image to produce an output value. Remarkably, the CNN algorithms do not require a computing process as a first step, minimizing human effort in selecting and designing features.
Consideration
There are several issues to be addressed before machine and deep learning techniques can be introduced into clinical practice. A machine learns from various imaging features by itself, including those even unrecognizable to humans. Its technical mechanisms and logic are mostly impossible to understand. This “black box” nature may be against the current concept of evidence-based medicine. Moreover, a lack of understanding of the working mechanism may raise legal and ethical issues in using the machine model in real-world practice. Overfitting problems also need to be addressed. Overfitting refers to a model that functions too well in the training dataset but is less applicable to other datasets. Essentially, the performance of the machine depends strongly on the quality of the data that are used in the training session. It can be problematic if a model learns to use unique features, such as details and noise, which are only present in the training data. Moreover, the “black box” characteristics of the machine may augment this problem. Therefore, it is mandatory to develop standardized methods to test the function of various machine models, which may be in common use in the near future, thoroughly and precisely.
Clinical applications
AI techniques have been applied to stroke imaging in the following two respects: 1) automatic or accurate diagnosis and 2) prediction of prognosis (Table 1).
Table 1.
Machine-learning studies on stroke imaging
| Application | Setting | Imaging tool | Performance |
|---|---|---|---|
| Diagnosis | |||
| Automatic lesion segmentation (ischemic stroke) [17] | Subacute stroke (> 24 hours and < 2 weeks) | MRI | Inferior to human segmentation |
| Automatic lesion segmentation (ischemic stroke) [18] | Chronic stroke | MRI (T1-weighted) | Comparable to manual segmentation |
| Automatic lesion segmentation (ischemic stroke) [19] | Acute stroke | DWI | Comparable to manual segmentation |
| Determination of ASPECTS (e-ASPECTS) [20,21] | Acute stroke | CT | Non-inferior to human reading |
| Automatic diagnosis of MCA dot sign [22] | Acute stroke (< 24 hours) | CT | Sensitivity 97.5% |
| Estimation of CSF volume for infarct edema [23] | Acute stroke | CT | Better than conventional method |
| Automatic lesion segmentation (hemorrhagic stroke) [25] | Acute stroke | CT | Comparable to manual segmentation |
| Prognosis | |||
| Symptomatic ICH after thrombolysis [27] | Acute stroke | CT | Improved the prognostic prediction |
| Improvement of visual function in PCA infarcts [28] | Subacute stroke (within 7 days) | MRI | Improved the prognostic prediction |
| Long-term mortality of AVM [34] | After endovascular treatment | CT, MRI | Accuracy of 97.5% to predict outcome |
| Impairment in multiple behavioral domains [35] | Subacute stroke (within 2 weeks) | MRI, fMRI | Enabled the prognostic prediction |
| Motor impairment [36] | Chronic stroke (≥ 3 months) | MRI, fMRI | Enabled the prognostic prediction |
MRI, magnetic resonance imaging; DWI, diffusion weighted imaging; ASPECTS, Alberta Stroke Program Early Computed Tomography Score; CT, computed tomography; MCA, middle cerebral artery; CSF, cerebrospinal fluid; ICH, intracerebral hemorrhage; PCA, posterior cerebral artery; AVM, arteriovenous malformation; fMRI, functional magnetic resonance imaging.
Diagnosis
Automatic lesion identification or segmentation is one of the most important elements in precision medicine dealing with huge datasets of brain imaging because manual lesion segmentation is cumbersome and inconsistent across raters [10,11]. In addition, automatic lesion identification may expedite accurate diagnoses by non-neurologists. However, automatic segmentation in magnetic resonance imaging (MRI) is not an easy task. The lesion shapes and locations vary according to time-from-symptom onset, vessel occlusion site, and collateral status [12]. Noise related to lesion signals, such as leukoaraiosis, T2 shine-through effects, and tissue defects, may further hamper lesion segmentation. Accordingly, a number of attempts have been made to delineate stroke lesions automatically using machine learning techniques [13-17] but have failed to demonstrate superiority over human manual segmentation [15-17].
Machine learning techniques have only recently begun to show some promising results, comparable to manual segmentation, in chronic stroke [18]. In that study, the authors proposed a supervised lesion segmentation algorithm in T1-weighted imaging. They built a machine with a dataset of 60 left-hemispheric chronic stroke patients (post-stroke interval 2.6±2 years), and tested their model in an independent dataset of an additional 45 patients. The correlations between the predicted manual lesion volume and the predicted lesion volume were r=0.961 for the former and r=0.957 for the latter datasets, respectively.
More recently, the CNN deep learning technique has been applied to lesion segmentation of acute ischemic stroke with diffusion-weighted imaging (DWI) [19]. Two already available CNNs were used together to develop their model: one CNN was an ensemble of two DeconvNets (EDD Net) and the other CNN was a multi-scale convolutional label evaluation net (MUSCLE Net), which aimed to evaluate the results from the EDD Net, removing potential false positives. DWIs from a total of 380 patients were used to train and validate CNNs, and another 361 DWIs were used for testing the model. The mean accuracy of the Dice coefficient (the quotient of pixel-overlap between the candidate segmentation and the manual segmentation; range: 0−1) was 0.67, which was higher than any previous results from the other groups [19], while the detection rate (the quotient of overlap between the number of subjects with any true-positive lesion detections and the number of all subjects) was 0.94. This study was notable because it did not use any manual editing method during training or developing of the machine (unsupervised learning).
Machine learning-based diagnosis by means of computed tomography (CT) has also been attempted. The Alberta Stroke Program Early Computed Tomography Score (ASPECTS), a topographic scoring system for acute ischemic damage to the brain, was automatically assessed using the e-ASPECTS software [20,21]. This software is a standardized, fully automated, CE (Conformité Européene, European Conformity)-mark approved, commercial ASPECTS scoring tool (Brainomix Ltd., Oxford, UK). Remarkably, e-ASPECTS showed a non-inferior performance in reading a total of 2,640 regions (132 patients×20 regions per patient) to that of stroke experts in the assessment of brain CT using ASPECTS.
An attempt was also made to detect the hyperdense middle cerebral artery (MCA) dot sign on CT, which represents a thromboembolus in acute MCA infarcts, using machine learning [22]. To accomplish this, the authors isolated the Sylvian fissure region, extracting several features, and classified the candidates using a SVM classifier. A total of 297 CT images (from 7 patients) were initially considered; 109 CT images with extracted regions of the Sylvian fissure regions were eventually used, which contained 40 islands of the MCA dot sign. The performance was examined using a leave-one-case-out method (cross-validation tool). This system achieved a sensitivity of 97.5% (39/40) for detection of the MCA dot sign at a false-positive rate of 0.5 per image (54 false-positive MCA dot signals/109 CT images) on the cerebral hemisphere suspected of acute stroke.
In patients with a malignant hemispheric infarct, automated quantification of cerebral edema has been attempted [23]. The authors of that article had previously proposed that a reduction in cerebrospinal fluid (CSF) volume on serial CT scans constituted an early and sensitive biomarker of cerebral edema; changes in CSF volumes from baseline to follow-up scans (ΔCSF) was strongly correlated with a midline shift at peak edema [24]. Because manual segmentation of hemispheric CSF on serial CT scans is time-consuming and clinically impractical, the authors developed and validated an automated technique for CSF segmentation via integration of random forest-based machine learning with geodesic active contour segmentation [23]. The algorithm demonstrated excellent results in measuring ΔCSF, which were well-correlated with those from manual segmentation (Pearson coefficient r=0.879, P<10-6). In terms of intracranial hemorrhage (ICH), automatic segmentation of CT lesions has also been successfully achieved [25]. This automated method was more accurate in estimating ICH volume than the conventional ABC/2 method.
Prognosis
Knowing stroke prognosis assists in making clinical decisions [26]. Prediction of treatment complications may be useful for screening a high-risk group receiving acute treatment, such as thrombolysis [27], whereas prediction of neurological long-term outcomes may guide stroke management [28].
Despite its rarity, symptomatic ICH remains a feared complication of acute thrombolysis [29]. Several prognostic scoring systems, such as the Hemorrhage After Thrombolysis (HAT) and Sugar, Early infarct signs, Dense cerebral artery sign, Age, and National Institute of Health Stroke Scale (SEDAN) scores, have been developed for estimating the risk of symptomatic ICH [30,31]; however, external validation of these scores failed to demonstrate convincing results [32,33]. Recently, machine learning of CT images has been attempted for predicting the symptomatic ICH risk with a small cohort of acute ischemic stroke patients [27]. The authors retrospectively used CT brain scan images of 116 acute ischemic stroke patients who were treated with intravenous thrombolysis (including 16 who developed symptomatic ICH), and split these into training (n=106) and test sets (n=10), repeatedly, involving 1,760 different combinations. These CT brain scan images were used as inputs into an SVM, along with clinical severity, to predict the risk of symptomatic ICH. Remarkably, the predictive performance of this SVM-based model (area under the receiver operating characteristic curve [AUC] 0.744, 95% confidence intervals [CIs] 0.738−0.748) was superior to those of established prognostication tools, such as the HAT (AUC 0.629, 95% CIs 0.628−0.630) and SEDAN (AUC 0.626, 95% CIs 0.625−0.627) scores.
Imaging findings are not always a target of machine learning, but can be used as an input feature in machines, for predicting stroke prognosis. In patients (n=35) with posterior cerebral artery infarcts, an SVM-based model, which integrates the regional extent of ischemic lesions and other clinical variables, has successfully predicted the improvement in visual field defects at 3 months [28]. In addition, in patients with brain arteriovenous malformation, a machine learning model using neural networks showed superior accuracy (97.5%) for predicting fatal outcome after endovascular treatment over a mean follow-up of 5 years, while a conventional regression model demonstrated an accuracy of 43% [34]. Recently, findings in both functional and structural MRI have also been used as input features in machine learning to predict post-stroke outcomes, such as motor dysfunction [35] and impairment in multiple behavioral domains [36].
Future
AI techniques in stroke imaging could markedly change the milieu of stroke diagnosis and management in the near future. Automated diagnosis of stroke may be popular in an era where fast thrombolysis, and even prehospital thrombolysis, is recommended [37-40]. The machine-based diagnosis would be particularly helpful for medical staff who are not accustomed to stroke imaging, such as general practitioners or paramedics; the decision to give thrombolysis may be thus be markedly faster. Prediction of prognosis with AI techniques will also be widely used in stroke management. A precise risk stratification for stroke treatment, such as acute thrombolysis or intervention, would be made possible. In addition, foreseeing the degree of post-stroke recovery in advance and informing patients/family members of the prognosis may enhance the treatment rapport and rehabilitation processes.
Perspectives
Establishing a well-constructed, large imaging database is a prerequisite for grafting AI techniques successfully into stroke-imaging analysis. This imaging database should be integrated and interpreted along with other large databases, containing clinical and biological data.
Construction of a “big” database for stroke imaging
Imaging the brain is a sine qua non in stroke medicine, and is obtained at every stage of stroke diagnosis and management [41-46]. Moreover, even a single brain imaging scan contains a number of MRI/CT sequences, which consist of numerous brain slices. In clinical practice, an enormous amount of imaging data is thus rapidly accumulated. Generally, however, such imaging data are not appropriately stored in many hospitals and are rather abandoned after a limited time period. Because AI techniques require a large database to function efficiently, it is imperative to collect imaging data in a structured and systematic way. Cooperative efforts across multiple hospitals and centers are indispensable. On the basis of these necessities, there has recently been a move to gather stroke imaging data worldwide [47,48]. The aim is to build a Million Brains Initiative of real world data for precision medicine in stroke. Besides the global cooperation, there are also many pre-conditions that need to be addressed, such as standardization of imaging protocols, and development of a user-friendly image-uploading server, as well as a cloud system with huge storage capacity. In addition, it is also necessary to build a stroke imaging database of high quality. The accuracy of the dataset, and particularly of the training dataset, is vital for machines to perform reliably. This high-quality database may also facilitate the process of developing novel important imaging parameters and reliable quantification methods. Moreover, it would be challenging to determine who should lead the data collection and analysis processes. Therefore, continuous and enthusiastic efforts across continents and countries are mandatory if a successful imaging database is to be established.
Imaging as a block of “bigger” data
Stroke patients present a large amount of varied data, which include clinical, biological (genetic, immunological, and serological markers), in addition to imaging information. Although these datasets are huge, ever-changing over time, difficult to collect systemically, and often lack value in terms of clinical meaning, they are evidently important for defining stroke characteristics and for predicting the prognosis of stroke [1,6]. Currently, novel histological information derived from proteomic analysis of clots retrieved during thrombectomy [49] or cerebrovascular endothelial cells captured from stent retrievers has also emerged. Understanding these multidimensional large datasets is essential in precision medicine, for choosing optimal management in an individual patient, and is the way of the future (issues related to such precision medicine have been thoroughly reviewed by Hinman et al.) [6]. Because not only imaging, but also clinical and biological data, are important in terms of stroke characteristics, stroke imaging data should be interpreted along with the layers of other dimensional data to analyze stroke prognosis and weigh the treatment options. Remarkably, AI-based analysis would also be mandatory in these interpretations. Therefore, efforts to decipher huge multidimensional datasets should run parallel with application of AI techniques in the analysis of stroke imaging.
Conclusions
Although AI techniques in medicine are in their infancy, they have been enthusiastically applied to stroke imaging analysis, showing some encouraging results. The future role of AI techniques in stroke medicine may be promising, because large databases of imaging and other parameters are exponentially being accumulated. Global cooperation and efforts are mandatory to expedite this process.
Acknowledgments
This research was supported by National Research Foundation of Korea Grant NRF 2014R1A2A1A11051280 funded by the Korea government, and the Korea Health Technology R&D Project, Ministry for Health & Welfare, Republic of Korea Grants HI12C1847 and HI14C1983.
Footnotes
The authors have no financial conflicts of interest.
References
- 1.Krittanawong C, Zhang H, Wang Z, Aydar M, Kitai T. Artificial intelligence in precision cardiovascular medicine. J Am Coll Cardiol. 2017;69:2657–2664. doi: 10.1016/j.jacc.2017.03.571. [DOI] [PubMed] [Google Scholar]
- 2.Lee JG, Jun S, Cho YW, Lee H, Kim GB, Seo JB, et al. Deep learning in medical imaging: general overview. Korean J Radiol. 2017;18:570–584. doi: 10.3348/kjr.2017.18.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erickson BJ, Korfiatis P, Akkus Z, Kline TL. Machine learning for medical imaging. Radiographics. 2017;37:505–515. doi: 10.1148/rg.2017160130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silver D, Huang A, Maddison CJ, Guez A, Sifre L, van den Driessche G, et al. Mastering the game of go with deep neural networks and tree search. Nature. 2016;529:484–489. doi: 10.1038/nature16961. [DOI] [PubMed] [Google Scholar]
- 5.Hamburg MA, Collins FS. The path to personalized medicine. N Engl J Med. 2010;363:301–304. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 6.Hinman JD, Rost NS, Leung TW, Montaner J, Muir KW, Brown S, et al. Principles of precision medicine in stroke. J Neurol Neurosurg Psychiatry. 2017;88:54–61. doi: 10.1136/jnnp-2016-314587. [DOI] [PubMed] [Google Scholar]
- 7.Nixon MS, Aguado AS. Feature extraction & image processing for computer vision. 3rd ed. Elsevier: Academic Press; 2012. [Google Scholar]
- 8.Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–297. [Google Scholar]
- 9.Matsugu M, Mori K, Mitari Y, Kaneda Y. Subject independent facial expression recognition with robust face detection using a convolutional neural network. Neural Netw. 2003;16:555–559. doi: 10.1016/S0893-6080(03)00115-1. [DOI] [PubMed] [Google Scholar]
- 10.Ashton EA, Takahashi C, Berg MJ, Goodman A, Totterman S, Ekholm S. Accuracy and reproducibility of manual and semiautomated quantification of MS lesions by MRI. J Magn Reson Imaging. 2003;17:300–308. doi: 10.1002/jmri.10258. [DOI] [PubMed] [Google Scholar]
- 11.Fiez JA, Damasio H, Grabowski TJ. Lesion segmentation and manual warping to a reference brain: intra- and interobserver reliability. Hum Brain Mapp. 2000;9:192–211. doi: 10.1002/(SICI)1097-0193(200004)9:4<192::AID-HBM2>3.0.CO;2-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaesemann P, Thomalla G, Cheng B, Treszl A, Fiehler J, Forkert ND. Impact of severe extracranial ICA stenosis on MRI perfusion and diffusion parameters in acute ischemic stroke. Front Neurol. 2014;5:254. doi: 10.3389/fneur.2014.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seghier ML, Ramlackhansingh A, Crinion J, Leff AP, Price CJ. Lesion identification using unified segmentation-normalisation models and fuzzy clustering. Neuroimage. 2008;41:1253–1266. doi: 10.1016/j.neuroimage.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maier O, Wilms M, von der Gablentz J, Krämer UM, Münte TF, Handels H. Extra tree forests for sub-acute ischemic stroke lesion segmentation in MR sequences. J Neurosci Methods. 2015;240:89–100. doi: 10.1016/j.jneumeth.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 15.Wilke M, de Haan B, Juenger H, Karnath HO. Manual, semi-automated, and automated delineation of chronic brain lesions: a comparison of methods. Neuroimage. 2011;56:2038–2046. doi: 10.1016/j.neuroimage.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 16.Mitra J, Bourgeat P, Fripp J, Ghose S, Rose S, Salvado O, et al. Lesion segmentation from multimodal MRI using random forest following ischemic stroke. Neuroimage. 2014;98:324–335. doi: 10.1016/j.neuroimage.2014.04.056. [DOI] [PubMed] [Google Scholar]
- 17.Maier O, Schröder C, Forkert ND, Martinetz T, Handels H. Classifiers for ischemic stroke lesion segmentation: a comparison study. PLoS One. 2015;10:e0145118. doi: 10.1371/journal.pone.0145118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pustina D, Coslett HB, Turkeltaub PE, Tustison N, Schwartz MF, Avants B. Automated segmentation of chronic stroke lesions using LINDA: lesion identification with neighborhood data analysis. Hum Brain Mapp. 2016;37:1405–1421. doi: 10.1002/hbm.23110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Bentley P, Rueckert D. Fully automatic acute ischemic lesion segmentation in DWI using convolutional neural networks. Neuroimage Clin. 2017;15:633–643. doi: 10.1016/j.nicl.2017.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herweh C, Ringleb PA, Rauch G, Gerry S, Behrens L, Möhlenbruch M, et al. Performance of e-ASPECTS software in comparison to that of stroke physicians on assessing CT scans of acute ischemic stroke patients. Int J Stroke. 2016;11:438–445. doi: 10.1177/1747493016632244. [DOI] [PubMed] [Google Scholar]
- 21.Nagel S, Sinha D, Day D, Reith W, Chapot R, Papanagiotou P, et al. e-ASPECTS software is non-inferior to neuroradiologists in applying the ASPECT score to computed tomography scans of acute ischemic stroke patients. Int J Stroke. 2017;12:615–622. doi: 10.1177/1747493016681020. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi N, Lee Y, Tsai DY, Matsuyama E, Kinoshita T, Ishii K. An automated detection method for the MCA dot sign of acute stroke in unenhanced CT. Radiol Phys Technol. 2014;7:79–88. doi: 10.1007/s12194-013-0234-1. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Dhar R, Heitsch L, Ford A, Fernandez-Cadenas I, Carrera C, et al. Automated quantification of cerebral edema following hemispheric infarction: application of a machine-learning algorithm to evaluate CSF shifts on serial head CTs. Neuroimage Clin. 2016;12:673–680. doi: 10.1016/j.nicl.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dhar R, Yuan K, Kulik T, Chen Y, Heitsch L, An H, et al. CSF volumetric analysis for quantification of cerebral edema after hemispheric infarction. Neurocrit Care. 2016;24:420–427. doi: 10.1007/s12028-015-0204-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scherer M, Cordes J, Younsi A, Sahin YA, Götz M, Möhlenbruch M, et al. Development and validation of an automatic segmentation algorithm for quantification of intracerebral hemorrhage. Stroke. 2016;47:2776–2782. doi: 10.1161/STROKEAHA.116.013779. [DOI] [PubMed] [Google Scholar]
- 26.Holloway RG, Benesch CG, Burgin WS, Zentner JB. Prognosis and decision making in severe stroke. JAMA. 2005;294:725–733. doi: 10.1001/jama.294.6.725. [DOI] [PubMed] [Google Scholar]
- 27.Bentley P, Ganesalingam J, Carlton Jones AL, Mahady K, Epton S, Rinne P, et al. Prediction of stroke thrombolysis outcome using CT brain machine learning. Neuroimage Clin. 2014;4:635–640. doi: 10.1016/j.nicl.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim BJ, Kim YH, Kim N, Kwon SU, Kim SJ, Kim JS, et al. Lesion location-based prediction of visual field improvement after cerebral infarction. PLoS One. 2015;10:e0143882. doi: 10.1371/journal.pone.0143882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wardlaw JM, Murray V, Berge E, del Zoppo G, Sandercock P, Lindley RL, et al. Recombinant tissue plasminogen activator for acute ischaemic stroke: an updated systematic review and meta-analysis. Lancet. 2012;379:2364–2372. doi: 10.1016/S0140-6736(12)60738-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lou M, Safdar A, Mehdiratta M, Kumar S, Schlaug G, Caplan L, et al. The HAT Score: a simple grading scale for predicting hemorrhage after thrombolysis. Neurology. 2008;71:1417–1423. doi: 10.1212/01.wnl.0000330297.58334.dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strbian D, Engelter S, Michel P, Meretoja A, Sekoranja L, Ahlhelm FJ, et al. Symptomatic intracranial hemorrhage after stroke thrombolysis: the SEDAN score. Ann Neurol. 2012;71:634–641. doi: 10.1002/ana.23546. [DOI] [PubMed] [Google Scholar]
- 32.Mazya MV, Bovi P, Castillo J, Jatuzis D, Kobayashi A, Wahlgren N, et al. External validation of the SEDAN score for prediction of intracerebral hemorrhage in stroke thrombolysis. Stroke. 2013;44:1595–1600. doi: 10.1161/STROKEAHA.113.000794. [DOI] [PubMed] [Google Scholar]
- 33.Sung SF, Chen SC, Lin HJ, Chen YW, Tseng MC, Chen CH. Comparison of risk-scoring systems in predicting symptomatic intracerebral hemorrhage after intravenous thrombolysis. Stroke. 2013;44:1561–1566. doi: 10.1161/STROKEAHA.111.000651. [DOI] [PubMed] [Google Scholar]
- 34.Asadi H, Kok HK, Looby S, Brennan P, O’Hare A, Thornton J. Outcomes and complications after endovascular treatment of brain arteriovenous malformations: a prognostication attempt using artificial intelligence. World Neurosurg. 2016;96:562–569. doi: 10.1016/j.wneu.2016.09.086. e1. [DOI] [PubMed] [Google Scholar]
- 35.Siegel JS, Ramsey LE, Snyder AZ, Metcalf NV, Chacko RV, Weinberger K, et al. Disruptions of network connectivity predict impairment in multiple behavioral domains after stroke. Proc Natl Acad Sci U S A. 2016;113:E4367–E4376. doi: 10.1073/pnas.1521083113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rondina JM, Filippone M, Girolami M, Ward NS. Decoding post-stroke motor function from structural brain imaging. Neuroimage Clin. 2016;12:372–380. doi: 10.1016/j.nicl.2016.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber JE, Ebinger M, Rozanski M, Waldschmidt C, Wendt M, Winter B, et al. Prehospital thrombolysis in acute stroke: results of the PHANTOM-S pilot study. Neurology. 2013;80:163–168. doi: 10.1212/WNL.0b013e31827b90e5. [DOI] [PubMed] [Google Scholar]
- 38.Gierhake D, Weber JE, Villringer K, Ebinger M, Audebert HJ, Fiebach JB. Mobile CT: technical aspects of prehospital stroke imaging before intravenous thrombolysis. RoFo. 2013;185:55–59. doi: 10.1055/s-0032-1325399. [DOI] [PubMed] [Google Scholar]
- 39.Nam HS, Park E, Heo JH. Facilitating stroke management using modern information technology. J Stroke. 2013;15:135–143. doi: 10.5853/jos.2013.15.3.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeon SB, Ryoo SM, Lee DH, Kwon SU, Jang S, Lee EJ, et al. Multidisciplinary approach to decrease in-hospital delay for stroke thrombolysis. J Stroke. 2017;19:196–204. doi: 10.5853/jos.2016.01802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim BJ, Kang HG, Kim HJ, Ahn SH, Kim NY, Warach S, et al. Magnetic resonance imaging in acute ischemic stroke treatment. J Stroke. 2014;16:131–145. doi: 10.5853/jos.2014.16.3.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merino JG, Warach S. Imaging of acute stroke. Nat Rev Neurol. 2010;6:560–571. doi: 10.1038/nrneurol.2010.129. [DOI] [PubMed] [Google Scholar]
- 43.Muir KW, Buchan A, von Kummer R, Rother J, Baron JC. Imaging of acute stroke. Lancet Neurol. 2006;5:755–768. doi: 10.1016/S1474-4422(06)70545-2. [DOI] [PubMed] [Google Scholar]
- 44.Menon BK, Puetz V, Kochar P, Demchuk AM. ASPECTS and other neuroimaging scores in the triage and prediction of outcome in acute stroke patients. Neuroimaging Clin N Am. 2011;21:407–423. doi: 10.1016/j.nic.2011.01.007. xii. [DOI] [PubMed] [Google Scholar]
- 45.Lee EJ, Kang DW, Warach S. Silent new brain lesions: innocent bystander or guilty party? J Stroke. 2016;18:38–49. doi: 10.5853/jos.2015.01410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Munuera J, Blasco G, Hernández-Pérez M, Daunis-I-Estadella P, Dávalos A, Liebeskind DS, et al. Venous imaging-based biomarkers in acute ischaemic stroke. J Neurol Neurosurg Psychiatry. 2017;88:62–69. doi: 10.1136/jnnp-2016-314814. [DOI] [PubMed] [Google Scholar]
- 47.Liebeskind DS, Malhotra K, Hinman JD. Imaging as the nidus of precision cerebrovascular health: a million brains initiative. JAMA Neurol. 2017;74:257–258. doi: 10.1001/jamaneurol.2016.4896. [DOI] [PubMed] [Google Scholar]
- 48.Liebeskind DS, Woolf GW, Shuaib A, Collaterals 2016 Consortium. Collaterals 2016: translating the collaterome around the globe. Int J Stroke. 2017;12:338–342. doi: 10.1177/1747493017701942. [DOI] [PubMed] [Google Scholar]
- 49.Hinman JD, Rao NM, Yallapragada A, Capri J, Souda P, Whitelegge J, et al. Drip, ship, and grip, then slice and dice: Comprehensive Stroke Center management of cervical and intracranial emboli. Front Neurol. 2013;4:104. doi: 10.3389/fneur.2013.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]