Abstract
The continuous outbreaks of foodborne diseases have drawn public attentions to food safety. Early screening of foodborne pathogens is crucial to prevent and control of foodborne diseases. In this study, a novel electrochemical aptasensor was developed for rapid and sensitive detection of E. coli O157:H7 using the coaxial capillary with immune magnetic nanoparticles (MNPs) for specific separation of the target bacteria, the urease with urea for amplification of the impedance signals, and the PCB gold electrode for measurement of the impedance change. The streptavidin modified MNPs were conjugated with the biotinylated polyclonal antibodies (PAbs) to form the immune MNPs, and captured in the coaxial capillary with the line-up high gradient magnetic fields to separate the bacteria from the large volume of sample. Then, the gold nanoparticles (GNPs) were modified with the aptamers against E. coli and the urease, and injected into the capillary to react with the bacteria and form the MNP-PAb-bacteria-aptamer-GNP-urease complexes. Finally, the urease on the complexes was used to catalyze the hydrolysis of urea into ammonium ions and carbonate ions in the capillary, leading to the decrease in the impedance of the catalysate, which was measured by the gold plating PCB electrode. The impedance change of the catalysate and the concentration of the bacteria had a good linear relationship. This aptasensor was able to detect E. coli as low as 101 CFU/mL in 3 h, and the mean recovery of E. coli in the spiked pasteurized milk was ~99%. This proposed aptasensor has the potential for practical applications of foodborne pathogen detection due to its short detection time, high sensitivity and low cost.
Keywords: Electrochemical impedance aptasensor, coaxial capillary, PCB electrode, urease catalysis, Escherichia coli O157:H7.
Introduction
Food safety has attracted worldwide attentions, as a result of the continuous outbreaks of foodborne diseases in recent years. Foodborne pathogens have not only caused tremendous economic losses, but also posed a considerable threaten to public health. It was estimated by the Centers for Disease Control and Prevention that 1 in 6 Americans got sick from contaminated foods or beverages and 3,000 died each year 1. In China, 5,926 people suffered from food poisoning in 2015 and bacteria including Salmonella, Vibrio Parahemolyticus, Bacillus Cereus, Staphylococcus Aureus and Escherichia Coli, etc., were the major causes for illnesses, accounting for 53.7% 2. Escherichia coli O157:H7 is a common foodborne pathogen and has been found in beef, raw milk, chicken, vegetables, and fruits, etc. It can survive at the temperatures as low as 4℃ and at the pH as low as 2.5. It has a strong toxicity and an infection dose as low as 10 to 100 CFU/mL. The infected people might suffer from watery diarrhoea and haemolytic uraemic syndrome, etc., and even die. Thus, early screening of E. coli O157:H7 is essential for food safety.
Currently available methods for detection of E. coli O157:H7 mainly include the gold standard culture plating (Culture), polymerase chain reaction (PCR) and enzyme-linked immune-sorbent assay (ELISA). Culture has the highest sensitivity and accuracy, but needs long time to get final results. PCR is fast and sensitive, but needs complex DNA extraction procedures. ELISA is fast, but lacks sufficient sensitivity and has relatively high false positives. In recent two years, various biosensors, such as optical 3-8, electrochemical 9-14, magnetic 15-16, quartz crystal microbalance 17-18, acoustic 19-20 and surface plasmon resonance 21-22, etc., have been reported for the detection of E. coli O157:H7 and other bacteria. Most biosensors are still in lab research now, and have not been ready for practical use yet. Hence, simple, rapid and sensitive methods are needed for detection of E. coli.
Impedance biosensor is one type of electrochemical biosensors and often featured with compact design, rapid detection, relatively low cost and easy integration, which generally relies on the electrochemical impedance change on the interface of an electrode under an alternating-current potential with a direct-current bias. The bacteria detection strategy based on electrochemical impedance biosensors often includes immunomagnetic separation for isolating the target bacterial cells from the food sample and biosensor detection for determining the concentration of the bacteria. The conventional magnetic separation is based on antigen-antibody reaction and has been widely used for specific separation of various biological and chemical targets 23. Compared to magnetic microparticles, magnetic nanoparticles (MNPs) with higher surface-to-volume ratio, less steric hindrance and more uniform distribution are often modified with the antibodies against the target bacteria and used to capture the bacteria, followed by magnetic separation by a magnetic field and enrichment in a small volume of buffer solution. So far, immunomagnetic separation are often performed by manual operation and not suitable for separation of the target bacteria from a large volume (10 mL or larger) to increase the sensitivity. Besides, interdigitated array microelectrodes with low ohmic drop, short detection time, high signal-to-noise ratio and compact size are often used as the transducer in electrochemical detection. However, they are often fabricated using expensive facilities in super clean conditions by well-trained technicians, resulting in high cost, and this has greatly limited their practical applications.
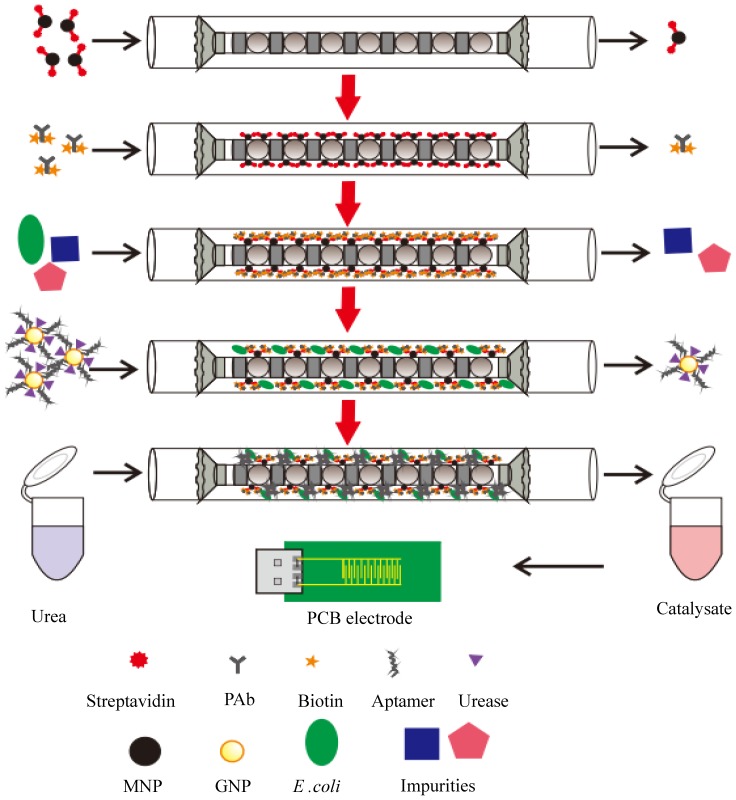
In this study, we intended to develop an electrochemical aptasensor using coaxial capillary with magnetic nanoparticle, urease catalysis and PCB electrode for rapid and sensitive detection of E. coli O157:H7. As shown in Fig. 1, after the streptavidin modified MNPs were injected and captured uniformly in the coaxial capillary that was filled with the line-up high-gradient magnetic fields, the biotinylated polyclonal antibodies (PAbs) were injected and incubated to form the immune MNPs through streptavidin-biotin binding. Then, the bacteria sample was injected into the capillary and the target bacteria would be captured by the MNPs to form the MNP-PAb-bacteria complexes (magnetic bacteria), while the impurities flowed through the capillary. The gold nanoparticles (GNPs), which were modified with the aptamers against E. coli and the urease, were injected into the capillary to react with the bacteria to form the MNP-PAb-bacteria-aptamer-GNP-urease complexes (enzymatic bacteria). Finally, urea was injected to fill the capillary and hydrolyzed into ammonium ions and carbonate ions due to the catalysis of the urease on the enzymatic bacteria, and the decrease in the impedance of the catalysate was measured by the gold plating PCB electrode to determine the concentration of the bacteria.
Figure 1.
Schematic of the electrochemical aptasensor for rapid and sensitive detection of E. coli O157:H7
Materials and Methods
Materials
The biotinylated polyclonal antibodies against E. coli O157:H7 from Thermo Scientific (Waltham, MA, USA) were used to specifically react with the E. coli O157:H7 cells. The aptamers against E. coli O157:H7 were synthesized by the Sangon Biotech (Shanghai, China) according to the previous report 24. The sequence of the aptamers were 5′-biotin-CCGGACGCTTATGCCTTGCCATCTACAGAGCAGGTGTGACGG-3′. The magnetic nanoparticles with the diameter of 150 nm from Ocean Nanotech (SV0152, Fe content: 1 mg/mL, number of particle in 1 mL: 5 x 1010, binding capacity for 1 mg: ~20 µg biotinylated proteins, San Diego, CA, USA) were conjugated with the PAbs for magnetic separation of the target bacteria. Gold chloride tri-hydrate (HAuCl4·3H2O) from Sigma Aldrich (St. Louis, MO, USA) was used to prepare the gold nanoparticles. Urease (E.C.3.5.1.5, Type III, 15,000-50,000 units/g solid), urea (NH2CONH2), ammonium carbonate ((NH4)2CO3), 1-(3-Dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC·HCl), 2-(N-morpholino) ethanesulfonic acid (MES), bovine serum albumin (BSA) and phosphate buffered saline solution (P5493, PBS, diluted 10 times by the deionized water to 10 mM, pH 7.4) were also purchased from Sigma Aldrich. Tween-20 from Amresco (Solon, OH) was prepared in the deionized water for lubricating. The deionized water was produced by Millipore Advantage 10 (18.2 MΩ∙cm, Billerica, MA, USA) and used to prepare all the solutions.
Fabrication of the coaxial capillary
The coaxial capillary were composed of an inner quartz capillary with the inner diameter of 1.1 mm, the outer diameter of 1.3 mm and the length of 150 mm, and an outer quartz one with the inner diameter of 1.8 mm, the outer diameter of 2.0 mm and the length of 300 mm. The inner quartz capillary was filled with small cylinder NdFeB magnets (Grade: N35; diameter: 1 mm; height: 1 mm) and small iron balls (diameter: ~1.0 mm). The adjacent two magnets were set up in mutual repelling layout and separated by one ball. Besides, both ends of the inner capillary were connected with two cylinder aligners fabricated by the 3D printer (Object 24, Stratasys, Eden Prairie, MN, USA) to be concentric with the outer one. The distribution of the magnetic field in the coaxial capillary for capturing the immune MNPs was simulated by the Finite Element Method Magnetics (FEMM) Software.
Design of the fluidic PCB electrode
The fluidic PCB electrode was used for the impedance measurement in this study and consisted of the gold plating PCB electrode and the fluidic channel. The PCB electrode, containing 10 pairs of interdigitated fingers with the length of 6 mm, the width of 200 μm and the spacing of 200 μm, was made using the conventional printed circuit board fabrication technique with a layer (70 µm thick) of copper on the substrate of the FR-4 glass epoxy, and plated with a thick layer (100 nm) of gold for the measurement on the impedance of the catalysate. The electrode has the length of 40 mm, the width of 20 mm and the height of 1 mm.
The fluidic channel with the length of 20 mm, the width of 2 mm and the height of 200 µm was fabricated using the Object24 3D printer to make the mold of the channel and the silicone elastomer kit (Sylgard 184, Dow Corning, Auburn, MI, USA) to cast the poly (dimethoxy) silane (PDMS) channel. The effective working area of the PCB electrode in the channel is about 8 mm2. The holder for housing the electrode was also fabricated by the 3D printer with an inlet, an outlet and a USB port to connect the PCB electrode with the impedance analyzer for impedance measurement.
Preparation of the bacterial cultures
Escherichia coli O157:H7 (ATCC43888) and Salmonella typhimurium (ATCC14028) were used as the target and non-target bacteria, respectively. They were stored with 15% glycerol at -20℃ and revived by streaking on Luria-Bertani (LB) agar plates. Prior to use, they were cultured in the LB medium (Aoboxing Biotech, Beijing, China) at 37℃ with shaking at 180 rpm, respectively. After incubation for 12-16 h, the bacterial cultures were 10-fold diluted with the sterile PBS to obtain the bacterial samples at the concentrations from 101 to 105 CFU/mL, respectively.
For bacterial enumeration, the bacterial samples were serially 10-fold diluted with the sterile PBS, and 100 μL of the diluents were surface plated on the LB agar plates. After incubation at 37℃ for 22-24 h, the colonies were counted to determine the concentration of the bacteria.
Magnetic separation of the bacteria in the coaxial capillary
Prior to use, the coaxial capillary were thoroughly washed successively with alcohol and deionized water, and then blocked with 1% BSA for 30 min, followed by washing with deionized water.
For the magnetic separation of the target bacteria in the coaxial capillary, 50 µg of the streptavidin modified MNPs in 1 mL of PBS were injected into the coaxial capillary and recycled three times at a flow rate of 1.2 mL/min to ensure that almost all the MNPs were captured by the high-gradient magnetic fields and distributed uniformly in the coaxial capillary. Then, 100 μL of the biotinylated polyclonal antibodies (0.1 mg/mL) were injected and filled with the coaxial capillary, followed by incubation for 30 min to allow the forming of the immune MNPs through streptavidin-biotin binding. After washing with PBS, 10 mL of the bacteria sample was injected into the capillary, and the target bacteria were captured by the MNPs through bacteria diffusion and immune reaction, and the capillary was washed with 5 mL of deionized water at a flow rate of 0.36 mL/min to remove the residuals.
Forming of the enzymatic bacteria in the coaxial capillary
The gold nanoparticles (GNPs) with the size of ~20 nm were synthesized in advance according to our previously reported protocol 25-26, and were modified with the aptamers against E. coli and the urease according to our previously reported protocol with slight modifications. Briefly, after the pH for 1 mL of GNPs was adjusted to 7.0 by K2CO3, 2 μL of streptavidin (1 mg/mL) was added drop-by-drop and incubated with gentle stirring for 1 h. Then, 12 μL of the biotinylated aptamers (10 μM) were added drop-by-drop and incubated with gentle stirring for 5 min to conjugate with the GNPs through streptavidin-biotin binding. Finally, 10 μL of the urease (10 mg/mL, in 5 mM MES) diluted with 40 μL of the deionized water were added drop-by-drop and incubated with gentle stirring for 1 h to conjugate with the GNPs, followed by blocking with 1% PEG 20,000 and 10% BSA for 30 min, respectively, and centrifugation at 10,000 rpm for 15 min to remove the excessive aptamers and urease. The aptamer and urease modified GNPs were finally dissolved in 100 μL of the deionized water and stored at 4℃ for further use.
For the forming of the enzymatic bacteria in the coaxial capillary, after the target bacteria were captured by the immune MNPs in the capillary, 10 μL of the aptamers and urease modified GNPs diluted with 90 μL of PBS was used to fill the coaxial capillary and incubated for 30 min. The GNPs were conjugated with the bacteria to form the enzymatic bacteria, followed by successively washing with 5 mL of deionized water with 0.05% Tween-20 and 5 mL of deionized water at the flow rate of 0.36 mL/min to remove the excessive GNPs and the residual conductive ions.
Measurement of the electrochemical impedance change
After the enzymatic bacteria in the capillary were washed, 100 μL of 100 μM urea was injected and incubated for 15 min to allow the urease on the enzymatic bacteria to catalyze the hydrolysis of urea into ammonium ions and carbonate ions. Then, the catalysate was injected into the fluidic PCB electrode for impedance measurement. The electrochemical impedance measurements were conducted on the IM6 electrochemical workstation (ZAHNER, Kronach, Bavaria, Germany) with the Thales analysis software applying a sinusoidal alternating potential with the amplitude of 5 mV, the direct-current bias of 0 V and the frequency range of 1 Hz - 5 MHz on the PCB electrode. After each measurement, the PCB electrode was rinsed with the deionized water to remove the residual ions until its impedance returned to the original level, i.e., the impedance of the deionized water.
Detection of E. coli in milk
Prior to test, the pasteurized milk from the local supermarket was 10 times diluted with the sterile PBS. Different concentrations of the bacterial cultures were prepared and enumerated by gold standard culture method, and then added into the diluted milk to obtain the spiked milk with the bacterial concentrations from 101 to 105 CFU/mL. The optimal amount of the streptavidin modified MNPs were first injected and uniformly distributed in the capillary, followed by conjugation with the biotinylated PAbs for 30 min. Then, 10 mL of the spiked milk at different bacterial concentrations of 101 -105 CFU/mL were injected at the optimal flow rate. After washing with the deionized water, 10 μL of the immune GNPs diluted by 90 μL of PBS were injected and incubated for 30 min. The capillary was washed by the deionized water with Tween-20 and the deionized water, respectively, to remove the unbound aptamer and urease modified AuNPs and the residual conductive ions, and then 100 μL of 100 μM urea were injected to fill the capillary. After incubation for the optimal time, the catalysate was collected and injected into the fluidic PCB electrode for impedance measurement. The impedance change at the characteristic frequency of 10 kHz was calculated as the difference between the impedance of the catalysate and that of deionized water to determine the amount of the E. coli cells.
Results and Discussion
Simulation of the magnetic field in the coaxial capillary
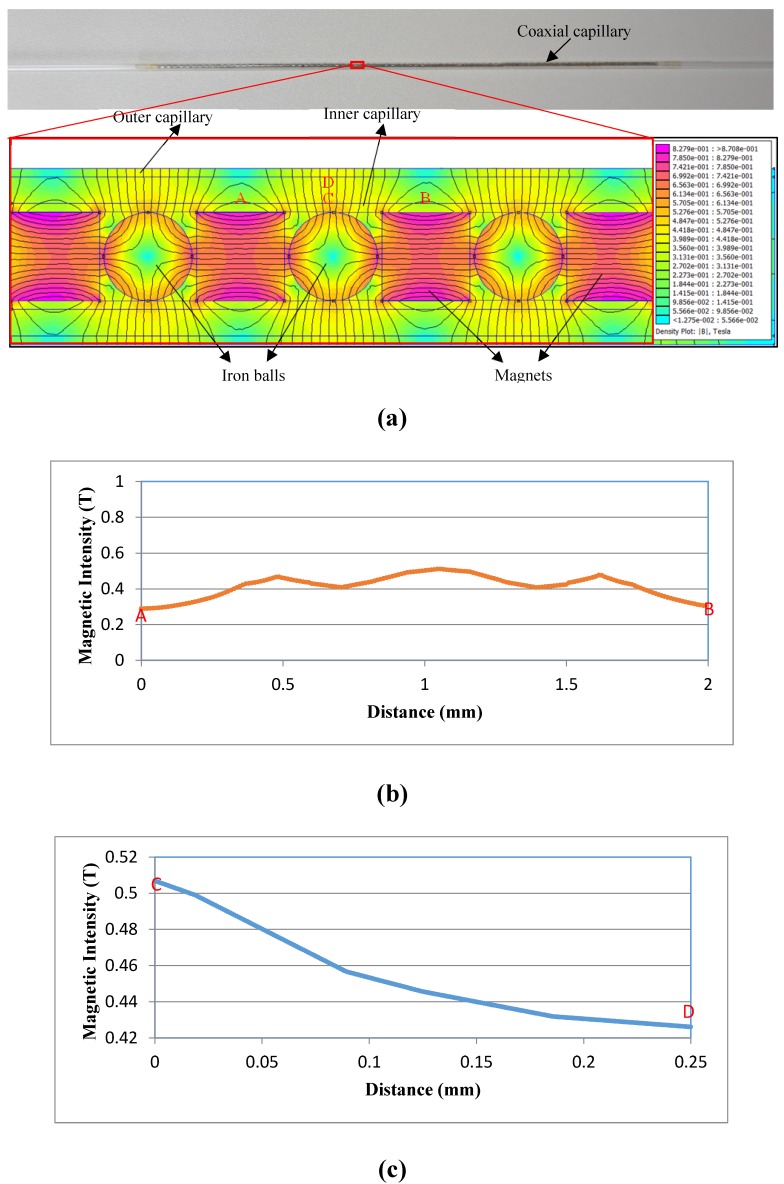
The distribution of the magnetic field can indicate where the immune MNPs are captured in the coaxial capillary for separating the target bacteria from the sample. The simulation of the magnetic field was conducted using the FEMM software and the result was shown in Fig. 2. The adjacent magnets in mutual repelling mode with one iron ball in-between them generated a strong high-gradient magnetic field around the ball. The magnetic field had a mean magnetic intensity of ~0.45 T and a mean gradient of ~350 T/m in the capillary. More importantly, the field had a relatively larger working area, i.e., around 75 1mm-long circular rings around the balls in the coaxial capillary, indicating the MNPs would be captured and distributed uniformly in a larger area. This could increase the opportunity for the antibodies on the MNPs to react with the target bacteria, resulting in an improved separation efficiency of the bacteria.
Figure 2.
(a)The simulation on the distribution of the magnetic field in the coaxial capillary; (b) The horizontal distribution of the magnetic field in the capillary; (c) The vertical distribution of the magnetic field in the capillary.
Optimization of the bacteria separation procedure
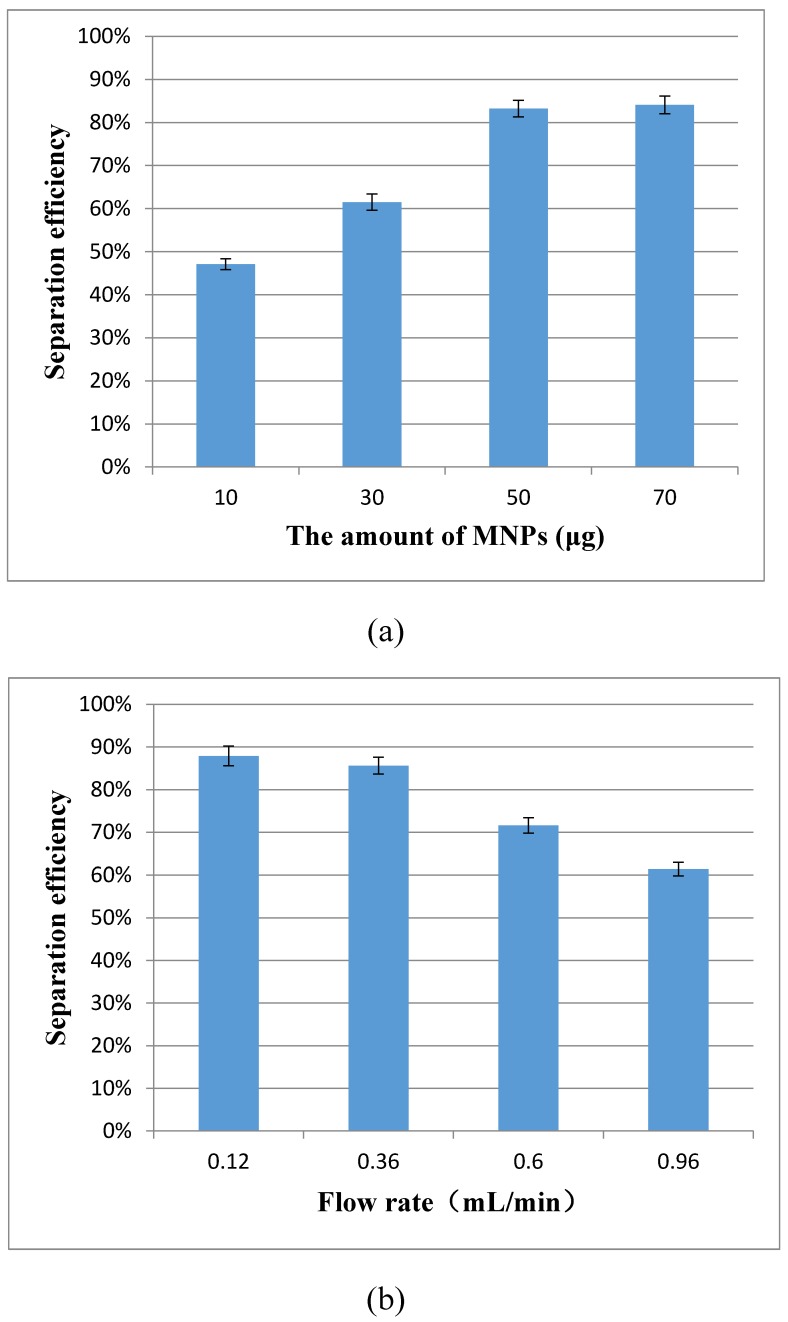
The magnetic separation of the target bacteria in the coaxial capillary is very important to this proposed aptasensor. Two key factors, the amount of the MNPs and the flow rate of the sample, have great impact on the bacteria separation, and were optimized using the separation efficiency to evaluate the performance. The separation efficiency was calculated as the ratio of the concentration of the separated bacteria to that of the original bacteria in this study.
For the optimization of the amount of the immune MNPs for each test, different amounts (10 μg, 30 μg, 50 μg and 70 μg) of the MNPs were injected into the capillary at the flow rate of 1.2 mL/min for separating the target bacteria at the concentration of 1.2×103 CFU/mL. As shown in Fig. 3(a), the separation efficiency increased from 47.1% to 83.2% when the amount of MNPs changed from 10 μg to 50 μg, and the separation efficiency did not increase significantly (84.1%) while 70 μg of the MNPs were used. This indicated that the amount of 50 μg for the MNPs was sufficient to capture the bacteria. Therefore, the optimal amount of 50 μg was used in this study.
Figure 3.
(a) Optimization of the amount of the immune MNPs (N=3); (b) Optimization of the flow rate of the sample (N=3).
For the optimization of the flow rate, different flow rates (0.12, 0.36, 0.60 and 0.96 mL/min) of the sample were used to recycle the target bacteria at the concentration of 1.4×103 CFU/mL to flow through the capillary. As shown in the Fig. 3(b), the separation efficiency increased from 61.4% to 85.6 %, while the flow rates decreased from 0.96 mL/min to 0.36 mL/min, and the separation efficiency remained at ~85% when the flow rate decreased to 0.12 mL/min. Therefore, the optimal flow rate of 0.36 mL/min was used in this study.
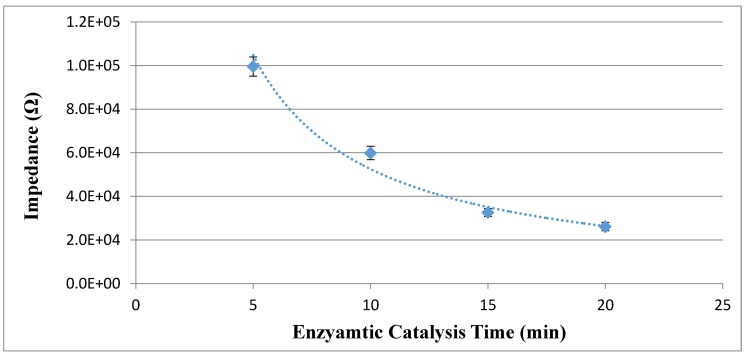
Optimization of the enzymatic catalysis procedure
The signal amplification in this study is based on the enzymatic catalysis, which can improve the sensitivity of the proposed electrochemical aptasensor. Different enzymatic catalysis time ranging from 5 min to 20 min was used to catalyze the hydrolysis of urea into the catalysate (ammonium ions and carbonate ions) by the urease on the enzymatic bacteria at the concentration of 1.0 x 105 CFU/mL. The impedance of the catalysate was measured by the PCB electrode to evaluate the amount of the catalysate. As shown in Fig. 4, when the catalysis time increased from 5 min to 15 min, the impedance of the catalysate decreased from 99.5 kΩ to 32.6 kΩ, indicating that more conductive ions were produced. When the time increased to 20 min, the impedance of 20 min decreased to 26.1 kΩ and did not changed significantly. Therefore, the optimal enzymatic catalysis time of 15 min was used in this study.
Figure 4.
Optimization of the enzymatic catalysis time (N=3).
Evaluation on the PCB electrode for impedance measurement
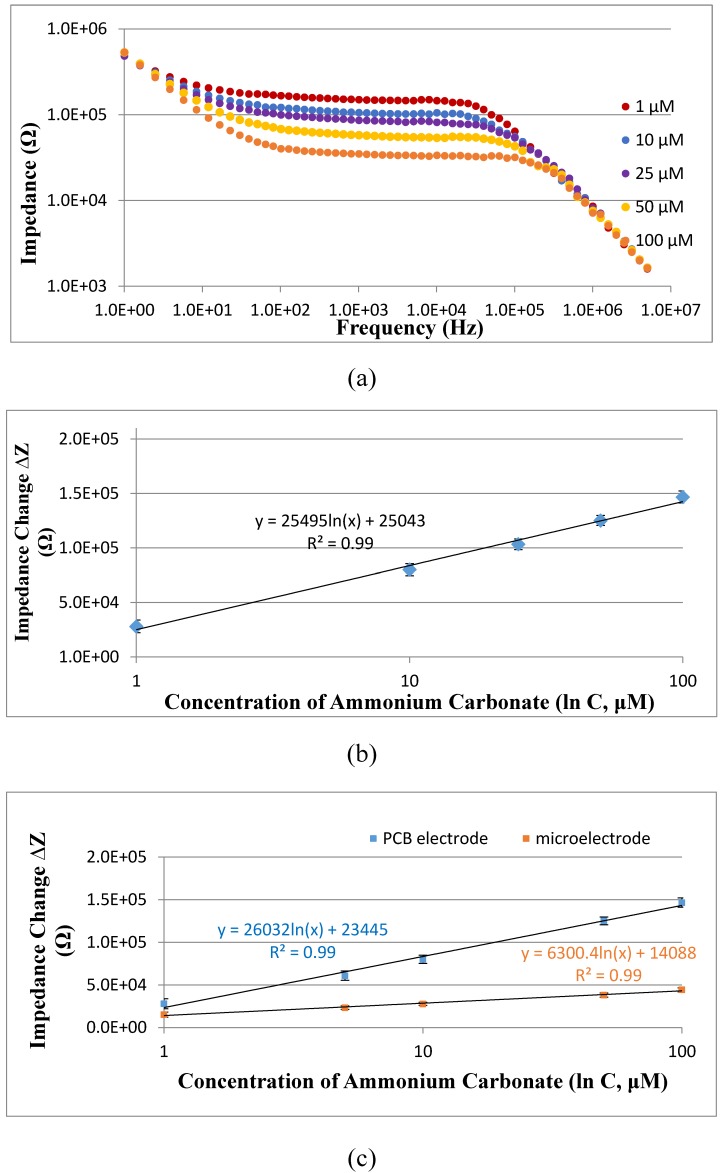
The new PCB electrode was first verified to be able to measure the impedance of different concentrations of ammonium carbonate at the presence of urea. Three parallel tests on the mixtures of different concentrations (1-100 μM) of ammonium carbonate and 100 μM of urea were carried out using the PCB electrode on the IM6 workstation. As shown in Fig. 5(a), when the concentration of ammonium carbonate increases, the impedance of the mixture decreases due to the presence of more conductive ions. To further understand the relationship between the concentration and the impedance change, which was calculated as the difference between the impedance of ammonium carbonate and that of urea or deionized water, the impedance change (∆Z) measured at the characteristic frequency of 10 kHz were plotted with the concentration (C) of ammonium carbonate, and a good linear relationship was found and could be expressed as ∆Z=25495 ln(C) + 25043 (R2=0.99). This indicated that this new gold plating PCB electrode was able to quantitatively detect the concentration of ammonium carbonate, i.e., the catalysate in this study.
Figure 5.
(a) Electrochemical impedance spectra of ammonium carbonate with the concentrations of 1 μM to 100 μM at the presence of 100 μM urea; (b) Linear relationship between the impedance change of ammonium carbonate at the characteristic frequency of 10 kHz measured by the PCB electrode and the concentration of ammonium carbonate ranging from 1 μM to 100 μM (N=3); (c) Comparison on the sensitivity of the interdigitated array microelectrode and the PCB electrode for impedance detection of different concentrations of ammonium carbonate ranging from 1 μM to 100 μM (N=3).
The sensitivity of the PCB electrode is a key factor to the sensitivity of this proposed electrochemical aptasensor. To further evaluate the sensitivity of the PCB electrode, three parallel tests on different concentrations of ammonium carbonate (1-100 μM) were conducted using both our previously reported interdigitated array microelectrode and the PCB electrode, respectively. As shown in Fig. 5(c), the calibration curve for the PCB electrode can be expressed as ∆Z=26032 ln(C) + 23445 with the slope of 26032, and that for the microelectrode can be expressed as ∆Z=6300.4 ln(C) + 14088 with the slope of 6300.4, indicating that the PCB electrode is more sensitive than the microelectrode for impedance detection of ammonium carbonate. Besides, the gold plating PCB electrode was fabricated at a very low cost (~3 US dollars), less than 1/10 of the gold microelectrode, and could be afforded for single use. However, the stability and the biochemical compatibility of the PCB electrode was a little poorer than the microelectrode, since the accuracy and the substrate of the PCB electrode are inferior to that of the microelectrode. To trade off the cost and the performance, the PCB electrode is more suitable for rapid screening of E. coli O157:H7.
Specificity of this proposed electrochemical aptasensor
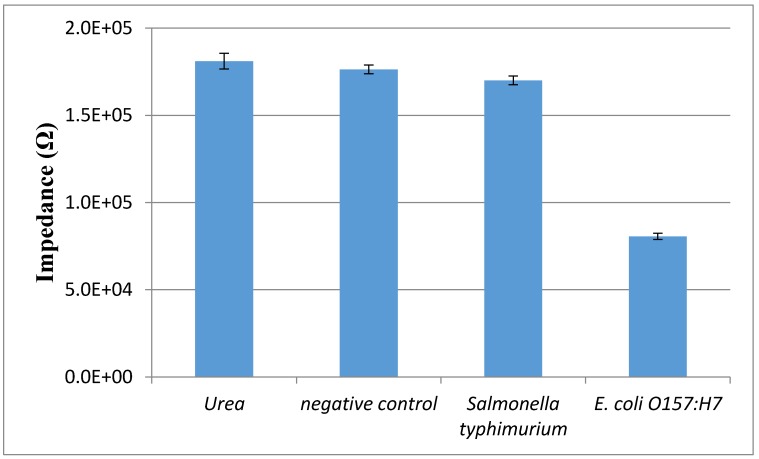
The specificity of this proposed aptasensor is mainly dependent on the polyclonal antibodies and the aptamers against the E. coli O157:H7 cells. Salmonella typhimurium at the concentration of 103 CFU/mL was used as the non-target bacteria to test the specificity of this aptasensor. As shown in Fig. 6, the impedance of urea, negative control (PBS) and Salmonella is 181 kΩ, 176 kΩ and 170 kΩ, respectively, indicating that very few conductive ions were found in their catalysate. The slight impedance difference between Salmonella and negative control might be due to little cross reaction between the PAbs/the aptamers and the Salmonella cells, resulting in a small amount of the catalysate. The impedance of E. coli at the same concentrations with Salmonella is much lower than that of Salmonella due to the forming of the enzymatic bacteria resulting in the hydrolysis of urea to produce more conductive ammonium ions and carbonate ions. This indicated that the proposed aptasensor had a good specificity.
Figure 6.
The specificity of the electrochemical aptasensor. Salmonella typhimurium and E. coli O157:H7 at the same concentration of 103 CFU/mL, urea and negative control were tested using this proposed biosensor (N=3).
Detection of E. coli O157:H7 in the pure cultures and the spiked pasteurized milk
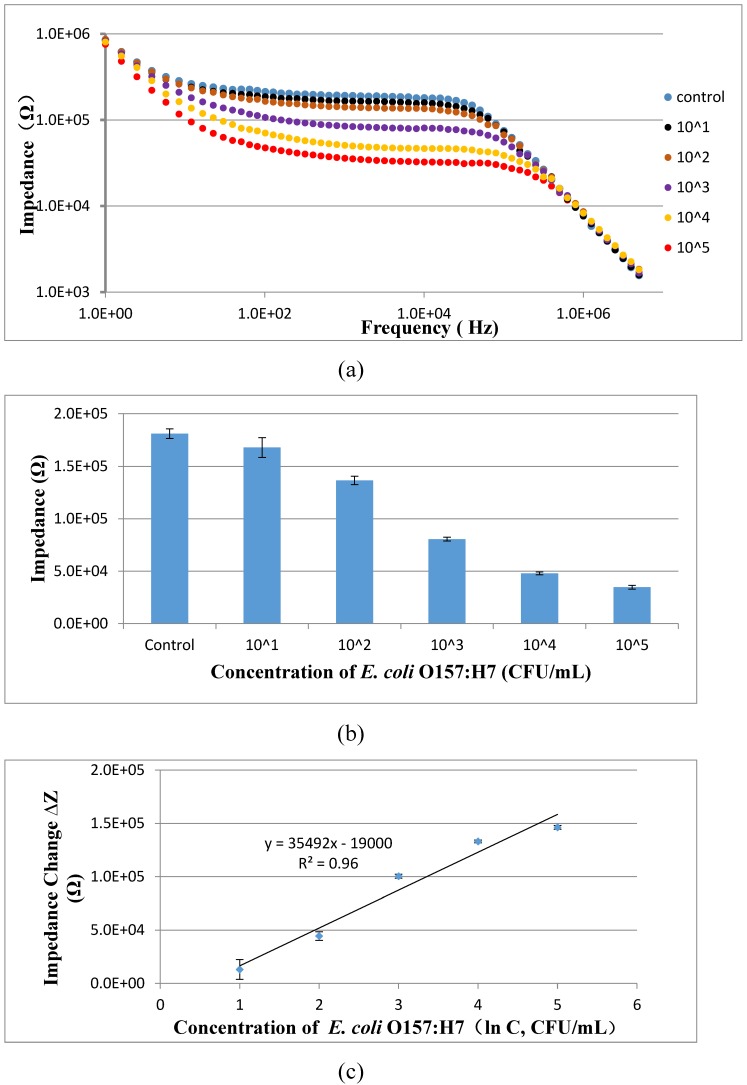
To evaluate the feasibility of the proposed aptasensor, three parallel tests of E. coli O157:H7 with different concentrations in the pure cultures were carried out. As shown in Fig. 7, the impedance of the pure cultures decreased while the concentration of bacteria increased. A good linear relationship between the impedance change at the characteristic frequency of 10 kHz and the concentration of the bacteria in the pure cultures was found and could be expressed as ∆Z = 35492 ln(C) - 19000 (R2=0.96). The low detection limit of this proposed aptasensor was determined to be 101 CFU/mL based on three times of signal/noise ratio. The low LDL is mainly attributed to (1) the larger volume (10 mL) of the bacteria sample was used instead of the conventional volume of 0.5-1 mL; (2) the coaxial capillary was used to obtain the high separation efficiency of the target bacteria; (3) the urease was used to amplify the electrochemical impedance signal.
Figure 7.
(a) Electrochemical impedance spectra of E. coli O157:H7 at the concentrations of 101-105 CFU/mL in the pure cultures; (b) The impedance at the characteristic frequency of 10 kHz for different concentrations of E. coli O157:H7 ranging from 101 CFU/mL to 105 CFU/mL in the pure cultures (N=3); (c) Linear relationship between the impedance change and the concentration of E. coli ranging from 101 to 105 CFU/mL (N=3).
To further evaluate the applicability of the proposed aptasensor, three parallel tests of E. coli O157:H7 with different concentrations in the spiked milk were carried out. As shown in Table 1, the recoveries for E. coli O157:H7 at the concentrations of 1.2×101 CFU/mL, 1.2×102 CFU/mL, 1.2×103 CFU/mL, 1.2×104 CFU/mL and 1.2×105 CFU/mL were 101.2%, 108.8%, 92.9%, 93.1% and 102.5%, respectively, and the mean recovery was 99.7%, indicating this proposed electrochemical aptasensor could be used for detection of E. coli O157:H7 in the real samples.
Table 1.
The recovery of E. coli O157 : H7 at the concentrations of 1.2×101-1.2×105 CFU/mL (N=3)
| E. coli concentration (CFU/mL) | Impedance for the spiked milk (Ω) | Impedance for the pure culture (Ω) | Recovery |
|---|---|---|---|
| 1.2×101 | 1.69×105 | 1.68×105 | 101.2% |
| 1.2×102 | 1.39×105 | 1.36×105 | 108.8% |
| 1.2×103 | 8.96×104 | 8.05×104 | 92.9% |
| 1.2×104 | 5.18×104 | 4.79×104 | 93.1% |
| 1.2×105 | 3.13×104 | 3.47×104 | 102.5% |
Besides, the bacterial sample with different concentrations ranging from 101 to 104 CFU/mL were detected using both the proposed aptasensor and the gold standard culture method for comparison. As shown in Table 2, the relative standard deviation of this aptasensor ranges from 1.4% to 4.3%, indicating this aptasensor has a good accuracy.
Table 2.
The comparison of this aptasensor with the standard culture method for detection of E. coli O157 : H7 at the concentrations of 101-104 CFU/mL (N=3)
| E. coli concentration detected by culture (C1, CFU/mL) | E. coli concentration detected by this aptasensor (C2, CFU/mL) | Relative standard deviation* (RSD, %) |
|---|---|---|
| 7.6×101 | 6.3×101 | 4.3% |
| 2.7×102 | 2.5×102 | 1.4% |
| 2.3×103 | 1.8×103 | 3.2% |
| 5.4×104 | 6.9×104 | 2.2% |
*: RSD= |lg(C1)-lg(C2)|/lg(C1) x 100%
Conclusions
In summary, a novel electrochemical aptasensor based on the coaxial capillary with the magnetic nanoparticles, the urease and the gold plating PCB electrode was developed for rapid, low-cost and sensitive detection of E. coli O157:H7. The proposed aptasensor was able to detect E. coli O157:H7 as low as 101 CFU/mL within 3 h. Besides, this proposed aptasensor could be used for the detection of bacteria in a large volume up to 10 mL. The coaxial capillary based magnetic separation and urease catalysis has greatly improved the sensitivity of this aptasensor. The proposed aptasensor has the potential for the development of an automatic device for sensitive detection of foodborne pathogens in practical application.
Supplementary Material
Supplementary figures.
Acknowledgments
This study was supported by the National Key Research and Development of China (2016YFD0201201) and Walmart Foundation (SA1703161).
References
- 1.Centers for Disease Control and Prevention. https://www.cdc.gov/foodsafety/cdc-and-food-safety.html.
- 2.National Health and Family Planning Commission of China. http://www.nhfpc.gov.cn/yjb/s7859/201604/8d34e4c442c54d33909319954c43311c.shtml.
- 3.Posnicek T, Ettenauer J, Zuser K, Kellner K, Brandl M. A fluorescence based sensor system for automated detection of E. coli in water. Procedia Engineering. 2016;168:574–577. [Google Scholar]
- 4.Khang J, Kim D, Chung KW, Lee JH. Chemiluminescent aptasensor capable of rapidly quantifying Escherichia Coli O157:H7. Talanta. 2016;147:177–183. doi: 10.1016/j.talanta.2015.09.055. [DOI] [PubMed] [Google Scholar]
- 5.Suaifan GA, Alhogail S, Zourob M. Paper-based magnetic nanoparticle-peptide probe for rapid and quantitative colorimetric detection of Escherichia coli O157:H7. Biosensors and Bioelectronics. 2017;92:702–708. doi: 10.1016/j.bios.2016.10.023. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Yan CH, Yang H, Yu JP, Wei HP. Rapid and selective detection of E. coli O157:H7 combining phagomagnetic separation with enzymatic colorimetry. Food Chemistry. 2017;234:332–338. doi: 10.1016/j.foodchem.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 7.Balakrishnan B, Barizuddin S, Wuliji T, El-Dweik M. A rapid and highly specific immunofluorescence method to detect Escherichia coli O157:H7 in infected meat samples. International Journal of Food Microbiology. 2016;231:54–62. doi: 10.1016/j.ijfoodmicro.2016.05.017. [DOI] [PubMed] [Google Scholar]
- 8.Ren W, Liu WJ, Irudayaraj J. A net fishing enrichment strategy for colorimetric detection of E. coli O157:H7. Sensors and Actuators B: Chemical. 2017;247:923–929. [Google Scholar]
- 9.Guner A, Cevik E, Senel M, Alpsoy L. An electrochemical immunosensor for sensitive detection of Escherichia coli O157:H7 by using chitosan, MWCNT, polypyrrole with gold nanoparticles hybrid sensing platform. Food Chemistry. 2017;229:358–365. doi: 10.1016/j.foodchem.2017.02.083. [DOI] [PubMed] [Google Scholar]
- 10.Xu M, Wang RH, Li YB. Electrochemical biosensors for rapid detection of Escherichia coli O157:H7. Talanta. 2017;162:511–22. doi: 10.1016/j.talanta.2016.10.050. [DOI] [PubMed] [Google Scholar]
- 11.Ozkan-Ariksoysal D, Kayran YU, Yilmaz FF, Ciucu AA, David IG, David V, Hosgor-Limoncu M, Ozsoz M. DNA-wrapped multi-walled carbon nanotube modified electrochemical biosensor for the detection of Escherichia coli from real samples. Talanta. 2017;166:27–35. doi: 10.1016/j.talanta.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Wang RH, Lum J, Callaway Z, Lin JH, Bottje W, Li YB. A Label-Free Impedance Immunosensor Using Screen-Printed Interdigitated Electrodes and Magnetic Nanobeads for the Detection of E. coli O157:H7. Biosensors (Basel) 2015;5:791–803. doi: 10.3390/bios5040791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu M, Wang RH, Li YB. Rapid detection of Escherichia coli O157:H7 and Salmonella Typhimurium in foods using an electrochemical immunosensor based on screen-printed interdigitated microelectrode and immunomagnetic separation. Talanta. 2016;148:200–208. doi: 10.1016/j.talanta.2015.10.082. [DOI] [PubMed] [Google Scholar]
- 14.Liu JT, Settu K, Tsai JZ, Chen CJ. Impedance sensor for rapid enumeration of E. coli in milk samples. Electrochimica Acta. 2015;182:89–95. [Google Scholar]
- 15.Guo L, Zhi S, Sun X, Lei C, Zhou Y. Ultrasensitive detection of bioanalytes based on signal amplification of coil-integrated giant magnetoimpedance biosystems. Sensors and Actuators B: Chemical. 2017;247:1–10. [Google Scholar]
- 16.Sun XC, Lei C, Guo L, Zhou Y. Separable detecting of Escherichia coli O157H:H7 by a giant magneto-resistance-based bio-sensing system. Sensors and Actuators B: Chemical. 2016;234:485–492. [Google Scholar]
- 17.Masdor NA, Altintas Z, Tothill IE. Sensitive detection of Campylobacter jejuni using nanoparticles enhanced QCM sensor. Biosensors and Bioelectronics. 2016;78:328–336. doi: 10.1016/j.bios.2015.11.033. [DOI] [PubMed] [Google Scholar]
- 18.Wang LJ, Wang RH, Chen F, Jiang TS, Wang H, Slavik M, Wei H, Li YB. QCM-based aptamer selection and detection of Salmonella typhimurium. Food Chemistry. 2017;221:776–82. doi: 10.1016/j.foodchem.2016.11.104. [DOI] [PubMed] [Google Scholar]
- 19.Ten ST, Hashim U, Gopinath SCB, Liu WW, Poo KL, Sam ST, Rahman SFA, Voon CH, Nordin AN. Highly sensitive Escherichia coli shear horizontal surface acoustic wave biosensor with silicon dioxide nanostructures. Biosensors and Bioelectronics. 2017;93:146–154. doi: 10.1016/j.bios.2016.09.035. [DOI] [PubMed] [Google Scholar]
- 20.Antonis Kordas, George Papadakis, Dimitra Milioni, Jerome Champ, Stephanie Descroix, Electra Gizeli. Rapid Salmonella detection using an acoustic wave device combined with the RCA isothermal DNA amplification method. Sensing and Bio-Sensing Research. 2016;11(2):121–127. [Google Scholar]
- 21.Liu X, Hu YX, Zheng S, Liu Y, He Z, Luo F. Surface plasmon resonance immunosensor for fast, highly sensitive, and in situ detection of the magnetic nanoparticles-enriched Salmonella enteritidis. Sensors and Actuators B: Chemical. 2016;230:191–198. [Google Scholar]
- 22.Nguyen TT, Trinh KTL, Yoon WJ, Lee NY, Ju H. Integration of a microfluidic polymerase chain reaction device and surface plasmon resonance fiber sensor into an inline all-in-one platform for pathogenic bacteria detection. Sensors and Actuators B: Chemical. 2017;242:1–8. [Google Scholar]
- 23.Neethirajan S, Ahmed SR, Chand R, Buozis J, Nagy É. Recent Advances in Biosensor Development for Foodborne Virus Detection. Nanotheranostics. 2017;1:272–95. doi: 10.7150/ntno.20301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu WH, Zhang J, Zheng MQ, Zhong YH, Yang J, Zhao YH, Wu WP, Ye W, Wen J, Wang Q, Lu JX. An aptamer-based biosensor for colorimetric detection of Escherichia coli O157:H7. PLoS One. 2012;7:e48999. doi: 10.1371/journal.pone.0048999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Q, Lin JH, Gan CQ, Wang YH, Wang D, Xiong YH, Lai WH, Li YT, Wang MH. A sensitive impedance biosensor based on immunomagnetic separation and urease catalysis for rapid detection of Listeria monocytogenes using an immobilization-free interdigitated array microelectrode. Biosensors and Bioelectronics. 2015;74:504–11. doi: 10.1016/j.bios.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 26.Wang D, Chen Q, Huo HL, Bai SS, Cai GZ, Xiong YH, Lin JH. Efficient separation and quantitative detection of Listeria monocytogenes based on screen-printed interdigitated electrode, urease and magnetic nanoparticles. Food control. 2017;73:555–561. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.