Abstract
Maintaining the balance of the gut microbiota and its metabolic functions is vital for human health, however, this balance can be disrupted by various external factors including food additives. A range of food and beverages are sweetened by saccharin, which is generally considered to be safe despite controversial debates. However, recent studies indicated that saccharin perturbed the gut microbiota. Inflammation is frequently associated with disruptions of the gut microbiota. The aim of this study is to investigate the relationship between host inflammation and perturbed gut microbiome by saccharin. C57BL/6J male mice were treated with saccharin in drinking water for six months. Q-PCR was used to detect inflammatory markers in mouse liver, while 16S rRNA gene sequencing and metabolomics were used to reveal changes of the gut microbiota and its metabolomic profiles. Elevated expression of pro-inflammatory iNOS and TNF-α in liver indicated that saccharin induced inflammation in mice. The altered gut bacterial genera, enriched orthologs of pathogen-associated molecular patterns, such as LPS and bacterial toxins, in concert with increased pro-inflammatory metabolites suggested that the saccharin-induced liver inflammation could be associated with the perturbation of the gut microbiota and its metabolic functions.
Keywords: Saccharin, Artificial sweetener, Gut microbiota, Inflammation, Metabolite
Introduction
Association between the gut microbiota and host health has raised growing public attention in recent years (P. J. Turnbaugh et al., 2007), as the gut microbiota plays an essential role in host-gut interactions, such as energy metabolism, immune response and epithelial homeostasis (Holmes, Li, Athanasiou, Ashrafian, & Nicholson, 2011). Hence, perturbations on the gut microbiota induced by environmental factors would affect its functional role and contribute to gut microbiota-related human diseases, including immune dysfunction, obesity, diabetes and cardiovascular disease, inflammatory bowel diseases and colorectal cancer (Kau, Ahern, Griffin, Goodman, & Gordon, 2011; Kinross, Roon, Holmes, Darzi, & Nicholson, 2008; Tremaroli & Backhed, 2012). It is well documented that the perturbation of relative abundance of gut microbes as well as gut microbial metabolites can lead to adverse health outcomes. For example, the ratio of Firmicutes and Bacteroidetes was observed to be associated with obesity in mice and human (P. J. Turnbaugh et al., 2006). Likewise, trimethylamine, generated by gut bacteria from dietary choline, can be further metabolized into trimethylamine N-oxide in liver, and the later serves as a predictive maker for cardiovascular disease (Jonsson & Backhed, 2015). Therefore, xenobiotics can potentially lead to diseases by shifting gut microbial compositions or altering the microbial metabolism. Thus, it is noteworthy and imperative to investigate the functional link between exposure to xenobiotics and imbalanced gut microbiota.
Artificial sweetners are ubiquitously used food additives, and the US Food and Drug Administration (FDA) has approved the usage of saccharin, sucralose, aspartame, neotame, acesulfame potassium and advantame to date (Spencer et al., 2016). The effects of artificial sweetners on human health are controversial, and particularly, their role in shaping the gut microbiota is barely understood. As the oldest artificial sweetener, saccharin was discovered and used since 1879 (Shankar, Ahuja, & Sriram, 2013), and the FDA approved accepted daily intake (ADI) in human is 15mg/kg body weight/day. Saccharin is generally considered to be safe, partially because of the fact that it is barely metabolized by human body (Spencer et al., 2016). Saccharin goes through the gastrointestinal (GI) tract un-metabolized, and is absorbed slowly from the intestine but eliminated rapidly in the urine (Renwick, 1985). It has been reported that dietary saccharin could change gut bacterial metabolism in rats by affecting the activity of several bacterial enzymes (Mallett, Rowland, & Bearne, 1985). And saccharin consumption was discovered to be associated with altered amino acid metabolism by gut bacteria (Lawrie, Renwick, & Sims, 1985). Most recently, a study found that saccharin was able to alter the mouse gut microbiota and this alteration led to increased glucose intolerence (Suez et al., 2014). The impact of saccharin on the gut microbiota and its metabolic profiles leads to a new but underappreciated mechanism of the potential toxicity of saccharin, which represents a significant knowledge gap in artificial sweetener toxicity research.
Inflammation is one of the most common physical conditions and associated with a number of human diseases, such as inflammatory bowel diseases, obesity, diabetes, atherosclerosis and cancer (Coussens & Werb, 2002; Furet et al., 2010; Holmes et al., 2011; Libby, Ridker, & Maseri., 2002; Uronis et al., 2009; Xavier & Podolsky, 2007). In previous studies, saccharin was recognized to enrich the biosynthesis pathway of lipopolysaccharide (LPS) of the mouse gut microbiota (Suez et al., 2014), which is a common trigger of inflammation. The gut microbiota and related metabolites are pivotal for host immune response (Kau et al., 2011). For example, the gut microbiota is involved in the regulation of host inflammatory responses through dietary fermentation products, such as short-chain fatty acids (Maslowski et al., 2009). It is of significance to probe the inflammation-inducing effects of saccharin in view of gut microbial and metabolic changes. In the present study, we used a mouse model to investigate its functional impact on the gut microbiota, particularly the microbiota-related alterations associated with inflammation.
We hypothesized that intake of saccharin is able to induce perturbations of the gut microbiota, regarding bacterial abundance and metabolic profiles, which can potentially elicit inflammation in the host via host-gut microbiota interactions. To test this hypothesis, we exposed C57BL/6J male mice to saccharin at the human ADI level for six months. An integrated approach combining high-throughput 16S rRNA gene sequencing and liquid chromatography-mass spectrometry (LC-MS) metabolomics were applied to investigate the alterations of the gut microbiota and its metabolic profiles induced by saccharin. Results of quantitative real-time polymerase chain reaction (qPCR) revealed increased expression of the inflammation mediators, iNOS and TNF-α, in mouse liver after six-month saccharin exposure. More importantly, both 16S rRNA sequencing and metabolomics profiling revealed a pronounced impact of saccharin on the mouse gut microbiota and metabolites, with many changes linked to pro-inflammatory effects.
Materials and Methods
Animals and exposure
We used C57BL/6J mice (male, ~23g, approximately 8 weeks old) purchased from the Jackson Laboratories (Bar Harbor, ME) for this study. Twenty mice were housed upon arrival in the cages under the following environmental conditions (temperature 22°C, 40–70% humidity, and a 12:12 hr light:dark cycle) with standard pelleted rodent diet and water ad libitum for a week before the study. Then, the mice were randomly assigned to the control and treatment group, consisting ten mice in each group. The treatment group received saccharin (Sigma-Aldrich, MO) for six months. Saccharin was dissolved in drinking water to 0.3mg/ml, which was equivalent to the FDA approved ADI in human. Saccharin solution was made every week. Control mice received tap water only. Following the treatment, body weight and water consumption were monitored for both groups. The fecal pellets from individual mouse were collected at baseline, three- and six-month and all samples were frozen at −80 °C for further analysis. This study was carried out in the University of Georgia animal facility. All experiments were approved by the University of Georgia Institutional Animal Care and Use Committee. The mice were treated humanely and with regard for alleviation of sufferings.
RNA extraction, cDNA preparation, and Quantitative real-time polymerase chain reaction (qPCR)
Total RNA from liver treated with RNAlater (Thermo Fisher Scientific) was extracted using the RNeasy mini kit (Qiagen, Valencia, CA) according to the manufacturer’s instruction. The resultant RNA was treated with DNase (DNA-free™ DNA Removal Kit, Thermo Fisher Scientific) to remove genomic DNA contamination. RNA quality was determined using an Agilent TapeStation (Agilent Technologies). Then, 1μg of total RNA was used for cDNA synthesis using iScript™ Reverse Transcription Supermix for RT-qPCR (Bio-Rad Laboratories, CA). The sequences of the primers used for quantitative PCR were listed in Table S1 (Alkhouri et al., 2010; Grivennikov et al., 2012; Song et al., 2015; Wang et al., 2006; Xu et al., 2005; Zhang et al., 2015). All results were normalized to the housekeeping gene β-actin as an endogenous control. Each reaction was prepared as instructed in the SsoAdvanced™ Universal SYBR® Green Supermix (Bio-Rad) manual. Reactions were run on a Bio-Rad CFX96 Touch™ Real-Time PCR Detection System. The qPCR was performed using the following protocol: 95 °C for 10 min, 40 cycles of 15 s at 95 °C, 30 s at 60 °C, 30 s at 72 °C followed by 65 °C to 95 °C increment by 0.5°C for 0.05s. DNase digestion and no-RT control were used to control the potential genomic DNA contamination, and no-template control was used to control technical contamination. Results were analyzed using ΔΔCT method by CFX manager software (Bio-Rad).
16S rRNA gene sequencing of the gut microbiota and data processing
We used 16S rRNA gene sequencing to investigate the gut microbiota in fecal samples at different time points. Fecal DNA from individual mouse was extracted using a PowerSoil DNA Isolation Kit (MO BIO Laboratories) according to the manufacturer’s instructions. The resultant DNA was quantified and stored in −80 °C prior to analysis. For each sample, 1ng of the purified fecal DNA was used as template for the PCR amplification and the barcoded bacterial universal primers of 515 (5′-GTGCCAGCMGCCGCGGTAA) and 806 (5′-GGACTACHVGGGTWTCTAAT) were applied to target variable region 4 of the 16S rRNA gene. Then, the barcoded amplicons from all samples were normalized, pooled and sequenced by Illumina Miseq at the Georgia Genomics Facility. A depth of at least 25,000 reads per sample was prepared by generating pair-end 250 × 250 (PE250, v2 kit) reads. The raw mate-paired files of 16S rRNA gene sequences were first trimmed to dispose bases with high error probability (> 0.01) and merged using Geneious 8.1.5 (Biomatters, Auckland, New Zealand). The pre-processed 16S rRNA gene sequences data were then analyzed using QIIME (Quantitative Insights into Microbial Ecology, version 1.9.1) software package (Caporaso et al., 2010). The operational taxonomic units (OTUs) with 97% sequence similarity against Greengenes database 13.8 were obtained using UCLUST. All OTUs were classified at five different levels: phylum, class, order, family and genus. Mothur software was used to assess the difference in individual gut bacterial component between the controls and treated mice over time.
Functional capability analysis
Tax4Fun, an open-source R package, uses 16S rRNA sequencing dataset to survey the functional profiles of microbial communities (Asshauer, Wemheuer, Daniel, & Meinicke, 2015). It provides functional repertoire with a high correlation coefficient to the corresponding metagenome sequence data based on 16S rRNA datasets (Asshauer et al., 2015). It was used to analyze the enrichment of functional genes of the microbiome in each group to investigate functional responses of bacterial communities to saccharin. QIIME with a SILVA database extension (SILVA 119) was used to pre-process raw data for Tax4Fun as described previously. Further statistical analysis was investigated using Statistical Analysis of Metagenomic Profiles (STAMP) (version 2.1.3) for results obtained from Tax4Fun (Parks, Tyson, Hugenholtz, & Beiko, 2014).
Fecal metabolite analysis
Extraction of metabolic compounds in fecal samples, collected after exposure to saccharin for six months, was conducted using methanol and water as previously described (Lu et al., 2014). The resultant extracts were suspended in 20% Acetonitrile for MS analysis. A HPLC-Q-TOF (Quadrupole-time-of flight (Q-TOF) 6520 mass spectrometer (Agilent Technologies, Santa Clara, CA) with an electrospray ionization source interfaced with Agilent 1200 HPLC) system was used to conduct metabolomic profiling. The daily calibration of Q-TOF with standard tuning solution (Agilent Technologies) was carried out to ensure a mass accuracy of < 5ppm. Metabolites were separated on an YMC Hydrosphere C18 column and a mass range of 30 to 2000 m/z was employed to capture molecular features in a positive mode.
Metabolomics data processing and metabolite identification
Data obtained from the HPLC-Q-TOF system was processed and analyzed as described previously (Lu, Knutson, Wishnok, Fox, & Tannenbaum, 2012). In brief, the MassHunter Workstation software (Agilent) was first used to convert the raw .d data to .mz, and signals with intensity higher than 1000 counts were included for the subsequent analysis. The XCMS Online tools were used to perform peak alignment, intensity calculation and comparison between the control and treatment group. Significantly altered molecular features were profiled and searched against the Human Metabolome Database (HMDB) (http://www.hmdb.ca) and METLIN (http://metlin.scripps.edu). Matched molecular features with database were examined by the product ion scan using a MS/MS mode in the Q-TOF 6520 mass spectrometer for each molecular feature, and the spectra were searched against the HMDB and METLIN MS/MS database for tentative identifications.
Statistical analysis of the data
A two-tailed Student’s t-test was used to determine the statistical significance of pro-inflammatory gene expression between the controls and saccharin-treated mice. The difference in individual gut bacterial component between the controls and sucralose-treated mice at different time points was assessed using a nonparametric test with Metastats (White, Nagarajan, & Pop, 2009). A two-tailed Welch’s t-test was used to compare the difference of functional genes and metabolites between the controls and sucralose-treated mice. Also, partial least squares discriminant analysis (PLS-DA) were performed to visualize metabolomics difference in different groups. A p-value of 0.05 or less was considered statistically significant.
Results
Effect of saccharin on the inflammation mediators in mouse liver
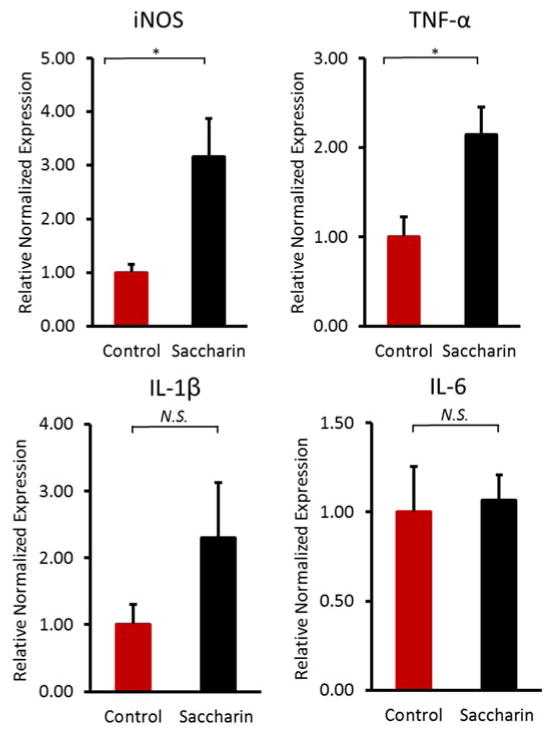
To explore the effect of saccharin consumption on chronic inflammation in mice, we examined the pro-inflammatory gene expression on mRNA level. Notably, we found two key genes, inducible nitric-oxide synthase (iNOS) and tumor necrosis factor alpha (TNF-α), were significantly (p<0.05) elevated in liver of saccharin-treated mice (Figure 1), which suggested a strong link between saccharin consumption and host inflammatory response.
Figure 1.
Comparisons of pro-inflammatory gene expression on mRNA level in liver of saccharin-treated mice. Inducible nitric-oxide synthase (iNOS) and tumor necrosis factor alpha (TNF-α) significantly elevated in liver of saccharin-treated mice. (*p<0.05)
Impact of saccharin on the dynamics of gut bacterial development
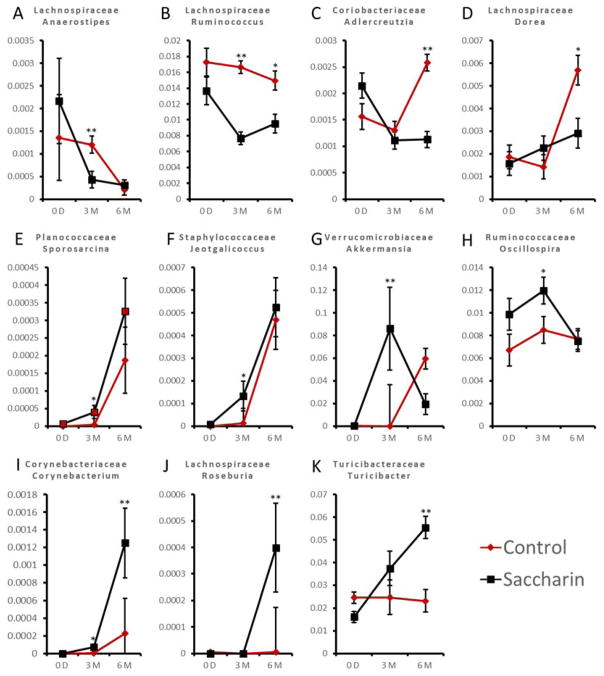
Saccharin induced significant changes to the mouse gut microbiota, manifested by the alterations of gut bacteria. As shown in Figure 2, the relative abundance of bacteria showed no significant difference between the treatment and control group at baseline, however displayed distinction either at three-month or six-month, or both. Eleven genera were significantly changed after three- and six-month treatment, indicating the effect of saccharin on disrupting the dynamics of gut microbiome development. Specifically, Sporosarcina, Jeotgalicoccus, Akkermansia, Oscillospira and Corynebacterium were significantly increased after three-month consumption; Corynebacterium, Roseburia and Turicibacter were increased after six-month consumption. Anaerostipes and Ruminococcus were significantly decreased after three-month consumption; Ruminococcus, Adlercreutzia and Dorea were decreased after six-month consumption. This result indicates that saccharin consumption could perturb the gut microbiota, which is consistent with a previous report (Suez et al., 2014). Of interest, several bacterial genera are demonstrated to be involved in inflammation. For instance, Corynebacterium, Turicibacter, Anaerostipes, Dorea, Roseburia and Ruminococcus (Bajaj et al., 2012; Chamulitrat et al., 1995; Collins et al., 2014; Fernández et al., 2016; Ng et al., 2013) were related to inflammation, suggesting that saccharin-induced gut microbiota changes may be partially responsible for the pro-inflammatory effects of saccharin.
Figure 2.
Saccharin consumption altered the dynamics of gut microbiome development in male C57BL/6J mice. Anaerostipes (A) and Ruminococcus (B) were significantly decreased after three-month consumption; Ruminococcus (B), Adlercreutzia (C) and Dorea (D) were decreased after six-month consumption. Sporosarcina (E), Jeotgalicoccus (F), Akkermansia (G), Oscillospira (H) and Corynebacterium (I) were significantly increased after three-month consumption; Corynebacterium (I), Roseburia (J) and Turicibacter (K) were increased after six-month consumption. (*p<0.05, **p<0.01, ***p<0.001)
Enrichment of bacterial functional genes of pro-inflammatory mediators
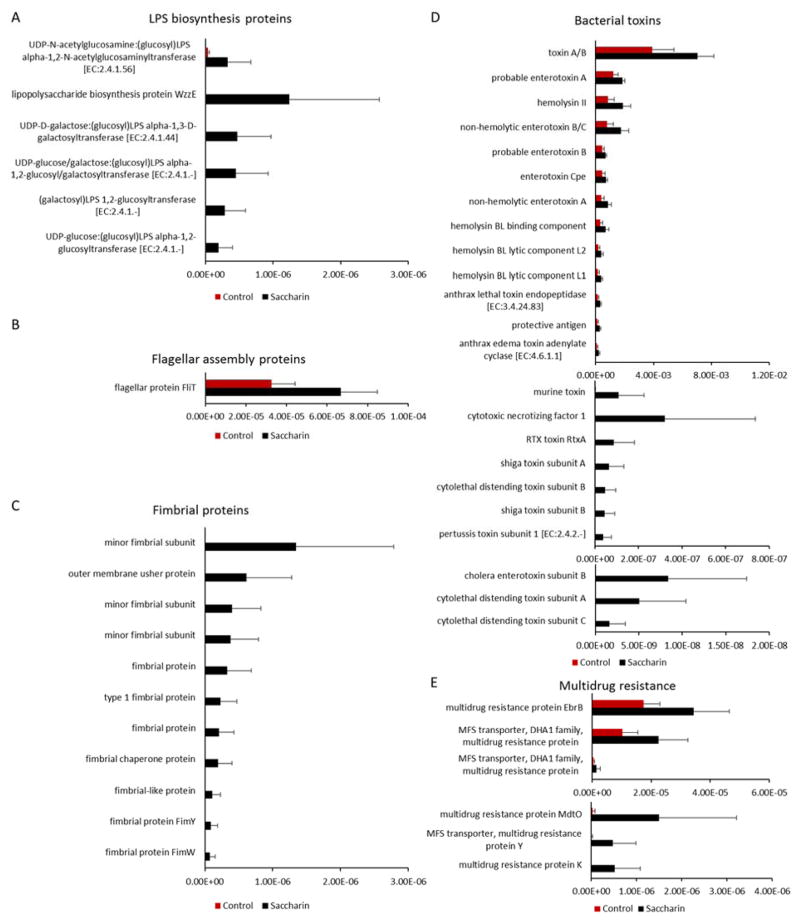
Altered gut microbiota generally functions differently, which can be unveiled by bacterial functional gene profiling. Therefore, we performed the functional enrichment analysis of the mouse gut microbiota based on 16S rRNA sequencing data. The functional comparison of the gut microbiota between the saccharin and control group revealed pronounced difference in term of bacterial inflammation-related pathways. We found that orthologs involved in pro-inflammatory mediators were highly elevated in saccharin-treated mice, as shown in Figure 3. At six-month consumption, 6 Lipopolysaccharide (LPS) biosynthesis orthologs, one Flagellar assembly ortholog, eleven fimbrial orthologs, twenty-three bacterial toxin orthologs, and six multidrug resistance orthologs were significantly increased in saccharin-treated mice. Some of the elevated genes were only observed in saccharin-treated mice. These data suggested that perturbation of the gut microbiota by saccharin increased the abundance of bacterial genes that could increase the risk of inflammation in the host, as these pro-inflammatory mediators can translocate into the host circulation to elicit inflammatory response.
Figure 3.
Significantly altered orthologs of the gut microbiota based on functional enrichment analysis. Genes of lipopolysaccharide (LPS) biosynthesis (A), flagellar assembly (B), fimbria (C), bacterial toxins (D), and multidrug resistance (E) were significantly increased in saccharin-treated mice (p<0.05).
Saccharin altered the gut microbial metabolic profiles
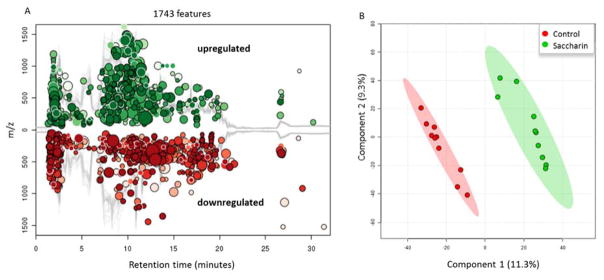
Large amounts of metabolites inside the intestine play crucial roles in the communication between the gut microbiota and host, which helps keep a healthy gut environment. Given the alteration of fecal microbiota by saccharin and the fact that the gut metabolites are co-produced by gut microbes and the host, we further explored the effects of saccharin on gut microbial metabolic profiles. In concert with the perturbation on bacterial abundance, comparative metabolomics analysis indicated that saccharin altered fecal metabolic profiles. Figure 4 shows that saccharin altered the gut metabolome with 1743 significantly changed molecular features (Figure 4A); and the PLS-DA plot reveals a clear separation between controls and saccharin-treated mice (Figure 4B). These results indicated that six-month saccharin consumption altered fecal metabolome.
Figure 4.
(A). Saccharin consumption changed the fecal metabolome in male C57BL/6J mice. 1743 metabolic features were significantly (p<0.05 and fold change>1.5) changed compared to controls. (B). Fecal metabolic profiles of controls were separated from those of saccharin-treated mice by PLS-DA.
Significantly changed functional metabolites by saccharin
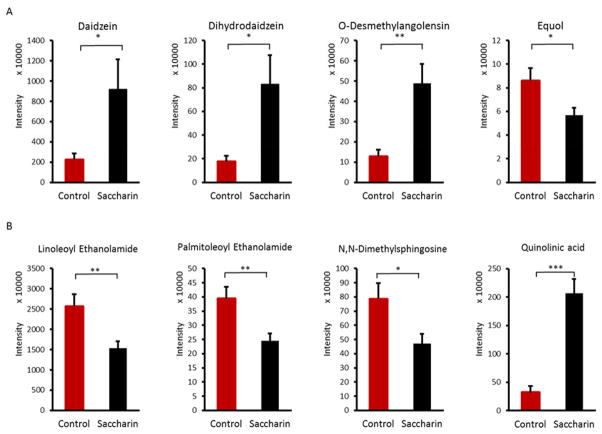
To further investigate the effects of saccharin on chronic inflammation in the host, we identified significantly altered molecular features and obtained a list of changed metabolites (Table S2). Notably, some inflammation-related metabolites were identified. For example, we found that equol, a bacterial metabolite of daidzein, was decreased by saccharin, as illustrated in Figure 5A. Meanwhile, daidzein and its other major bacterial metabolites, dihydrodaidzein and O-desmethylangolensin, were significantly increased in saccharin-treated mice. Increased daidzein and decreased equol suggest a reduced growth or decreased enzymatic activity of metabolizing bacteria. Likewise, linoleoyl ethanolamide, palmitoleoyl ethanolamide, N,N-Dimethylsphingosine and quinolinic acid (Figure 5B), were significantly changed in saccharin-treated mice. The effects of these compounds in mediating inflammation have been demonstrated in previous studies (Ishida et al., 2013; Nishiuma et al., 2008).
Figure 5.
Saccharin consumption significantly altered key fecal metabolites in male C57BL/6J mice, as exemplified by equol, a gut microbiome-catalyzed metabolite of daidzein (A) and metabolites involved in mediating inflammation (B). (*p<0.05, **p<0.01, ***p<0.001)
Discussion
In this study, we applied qPCR to examine the inflammatory markers in mouse liver and two key pro-inflammatory gene expression were found elevated. Also, we used 16S rRNA gene sequencing and metabolomics to explore the effects of saccharin consumption on the gut microbiota and its metabolic functions. We have demonstrated that saccharin consumption altered the fecal microbiota and metabolome, indicating a functional impact of saccharin on the gut microbiota and host. Notably, saccharin consumption increased the abundance of some bacterial genes of pro-inflammatory mediators and decreased the level of anti-inflammatory metabolites, which could increase the risk of developing inflammation in the host. This study provides novel insights into saccharin-induced functional changes in the gut environment and a potential link between gut microbiome perturbations and chronic inflammation in the host.
Previous studies have indicated that saccharin may induce inflammation in animals (Suez et al., 2014), and in our study, we found elevated gene expression of iNOS and TNF-α in the liver of saccharin-treated mice. Inducible NOS (iNOS), present in hepatocytes, endothelium and other immune cells, can synthesize high concentration of nitric oxide (NO), and lead to tissue damage in inflammatory processes. Expression of iNOS is highly regulated at the transcriptional level, through cellular receptors, such as CD14, via the activation of NFκB pathway and the expression can be triggered by LPS through CD14 (Aktan, 2004). iNOS plays an important role in the inflammation in multiple liver diseases, such as liver tumors and liver fibrosis from chronic viral infection (La Mura et al., 2014; Sass, Koerber, Bang, Guehring, & Tiegs, 2001). TNF-α, a key cytokine in inflammation, is mainly produced by activated macrophages, induced by pathogen-associated molecular patterns (PAMPs), such as LPS, through toll-like receptors (TLRs) (Wu & Zhou, 2010). Expression of TNF-α can activate NFκB pathways and induce damage to cells. Previous studies showed that TNF- α could mediate liver damage, which will lead to inflammation in liver, and anti-TNF antibody treatment could improve steatosis in ob/ob mice (Garcia-Ruiz et al., 2006). The elevated expression of iNOS and TNF-α herein suggested that saccharin consumption leads to liver inflammation in mice.
Inflammation triggers are common in the GI tract, for instance, living microbes that behave as pathogens or opportunistic pathogens, PAMPs such as LPS, flagellin, bacterial DNA and RNA, and microbial metabolites such as toxins and secondary bile acids. Perturbation of the gut microbiota and metabolites can potentially increase the level of inflammation triggers and lead to inflammation in the host. In this study, we observed changes in the abundances of various bacteria at the genus level between the saccharin and control group. Among changed gut bacteria, several genera were reported to be related to inflammation in the host. Corynebacterium in the family of Corynebacteriaceae, which were found increased in our study, contains some opportunistic pathogenic species. For example, Corynebacterium parvum could induce chronic inflammation through the over-production of NO in mouse liver, and lead to hepatic necrosis and death if followed by LPS injection (Chamulitrat et al., 1995). Anaerostipes, Dorea and Ruminococcus in family Lachnospiraceae were found decreased in saccharin-treated mice in this study, and the decrease of these three genera were associated with inflammation in previous studies: Anaerostipes, decreased in a biliary inflammation mouse model, Dorea decreased in subjects of irritable bowel syndrome, and the loss of Ruminococcus was associated with colitis induced by dextran sodium sulfate (Fernández et al., 2016; Ng et al., 2013). Taken together, these bacterial changes may partially contribute to elevated expression of inflammatory genes in mouse liver. In addition, the gut microbiome is highly involved in energy metabolism of the host. For example, different gut microbiome community structures are associated with obesity (P.J. Turnbaugh & Gordon, 2009; P.J. Turnbaugh et al., 2006). Saccharin was reported to alter body weight gain in rodents (Parlee et al., 2014; Swithers, Martin, Clark, Laboy, & Davidson, 2010), however, in this study, no significant difference was found in body weight gain between the controls and saccharin-treated mice (10.36±1.41g and 10.30±2.33 g for the control and saccharin group, respectively) after saccharin consumption for 6 months at the dose equivalent to the FDA approved ADI in human (Figure S1).
Consistently, the functional enrichment analysis of the gut microbes also displayed a huge distinction in bacterial functional repertoire between the saccharin and control group. Notably, we found that orthologs related to bacterial pro-inflammatory mediators were significantly increased after saccharin consumption (Fig. 3). LPS, flagella and fimbriae, known PAMPs that can trigger pathological inflammation in the host, were increased in saccharin-treated mice. LPS is an endotoxin from the outer membrane of Gram-negative bacteria, and it can initiate secretion of pro-inflammatory cytokines like interleukin-6 or tumor necrosis factor (TNF)-α via toll-like receptor 4 (TLR4) (de La Serre et al., 2010). Flagella and fimbriae are important bacterial components that are involved in the host inflammatory response modulation (Madianos, Y. A. Bobetsis, & Kinane., 2005). High levels of flagella is associated with gut mucosal barrier breakdown and inflammation (Cullender et al., 2013), while fimbriae play an important role in adhesion and invasion into epithelial cells (Nakagawa, 2002). Likewise, increased bacterial genes involved in multiple bacterial toxins that are also strongly associated with inflammation were found in saccharin-treated mice (Madianos et al., 2005). The enrichment of inflammation-associated orthologs in mouse gut bacterial may contribute to tissue inflammation. In addition, after six-month saccharin treatment, the gut microbiota exhibited a higher frequency and abundance of bacterial genes of multi-drug resistance. For instance, the frequency of several major facilitator superfamily transporters was higher in mice with saccharin treatment. It is well-documented that multi-drug resistance efflux pumps in bacteria could play an important role in resistance to antibiotics as well as the extrusion of host-produced natural substances and xenobiotics (Marshall, Dorothy J. Ochieng, & Levy, 2009; Piddock, 2006). These results strongly suggested a potential effect of saccharin to induce elevated inflammatory responses through altering the gut microbiota.
Saccharin significantly changed bacterial functional metabolites, which may also affect the host. For example, bacterial metabolism of daidzein was impaired by saccharin. Daidzein, an isoflavone compound, works as a weak estrogen in the body, and can be metabolized into dihydrodaidzein, O-desmethylangolensin (O-DMA) and equol by gut bacteria (Decroos, Vanhemmens, Cattoir, Boon, & Verstraete, 2005). Interestingly, the metabolism pattern varies depending on the composition of the gut microbiota, and only about one third of the human population possess the gut microbiota that can produce equol and about 80%–90% of the population are capable to produce O-DMA (Atkinson, Cara L. Frankenfeld, & Lampe, 2005). Equol or O-DMA is more biologically active than daidzein as an estrogen. Recently, equol was found to have the ability to inhibit LPS-induced oxidative stress in macrophages, inhibit superoxide production in cell culture, protect neurons from neuro-inflammatory injury and suppress inflammatory response in mice and, in particular, the antioxidant activity of equol is greater than its parent compound daidzein (Gou, Jiang, Zheng, Tian, & Lin, 2015; Hwang, Wang, Morazzoni, Hodis, & Sevanian, 2003; Lin, Yamashita, Murata, Kumazoe, & Tachibana, 2016; Subedi et al., 2017). Therefore, the capacity of equol production from daidzein by gut bacteria can increase its antioxidative and anti-inflammatory ability in the host. In our study, saccharin consumption significantly decreased the production of equol and increased the level of its parent compound daidzein and other metabolites, O-DMA and dihydrodaidzein (Figure 5), which not only demonstrates the impact of saccharin on the metabolic functions of the gut microbiome, but also may decreases the ability of the gut microbiome to protect the host from inflammatory challenges. Consistent with the decreased level of equol, we found decreased abundance of Adlercreutzia in the gut microbiota (Figure 2), which contains equol-producing bacteria in this genus (Maruo, Sakamoto, Ito, Toda, & Benno, 2008). In addition, several other anti-inflammatory compounds, palmitoleoyl ethanolamide (PEA), linoleoyl ethanolamide (LEA) and N,N-Dimethylsphingosine, were decreased in saccharin-treated mice. As a fatty acid ethanolamide, PEA was found to inhibit inflammation in human adipocytes and peripheral tissues through the regulation of pro-inflammatory proteins, nitric oxide, and neutrophils (Ezzili, Otrubova, & Boger, 2010). Likewise, LEA was reported to reduce LPS-induced inflammation in macrophages (Ishida et al., 2013). N,N-Dimethylsphingosine was shown to attenuate airway inflammation in a mouse model (Nishiuma et al., 2008). The reduction of these compounds can impact chemical signaling between the gut microbiome and host, which may contribute to the development of tissue inflammation in the host. Moreover, we found increased quinolinic acid, a metabolite of tryptophan, which served as a pro-inflammatory compound in a previous study (Keszthelyi, Troost, & Masclee, 2009). Chemical signaling of gut microbiome-host interaction likely involves a vast number of functional metabolites, as exemplified by only few compounds briefly described here. Nevertheless, the change of these metabolites indicates that saccharin consumption may increase the risk of host inflammation through altering metabolites produced or regulated by the gut microbiome.
Conclusions
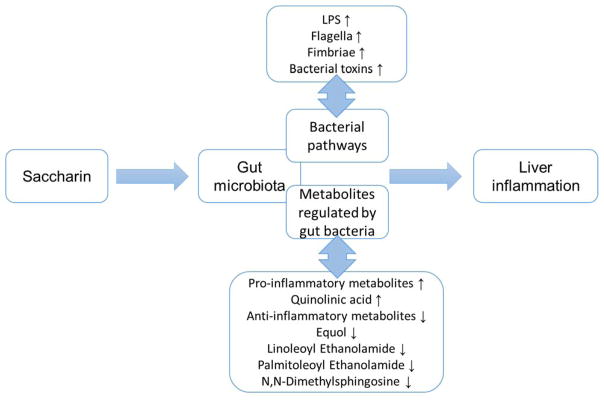
Our results revealed that 6-month saccharin administration in drinking water induced elevated inflammation in mouse liver, which could be functionally associated with saccharin-induced gut microbiome perturbations, exemplified by the alteration of inflammation-related bacterial pathways and metabolites (Figure 6). Elevated expression of iNOS and TNF-α in liver supported that saccharin consumption could increase inflammation in mice, and this change may be the consequence of perturbations of the gut microbiota and metabolites arising from saccharin consumption. Our results highlight the role of disrupted gut microbiome in eliciting systemic adverse response in the host. Consequently, this study provides novel insights regarding the toxicity assessment of food additives, such as artificial sweeteners, in view of their effects on gut microbial homeostasis.
Figure 6.
The functional link between saccharin-induced gut microbiome perturbations and host inflammation.
Supplementary Material
Figure S1. Body weight data of the controls and mice administrated to saccharin for 6 months.
Table S1. Primers used in qPCR.
Table S2. Significantly altered fecal metabolites in saccharin mice compared to control.
Acknowledgments
The authors thank the University of Georgia, University of North Carolina and NIH/NIEHS for partial financial support (R01ES024950).
References
- Aktan F. iNOS-mediated nitric oxide production and its regulation. Life Sci. 2004;75(6):639–653. doi: 10.1016/j.lfs.2003.10.042. [DOI] [PubMed] [Google Scholar]
- Alkhouri N, Gornicka A, Berk MP, Thapaliya S, Dixon LJ, Kashyap S, … Feldstein AE. Adipocyte apoptosis, a link between obesity, insulin resistance, and hepatic steatosis. J Biol Chem. 2010;285(5):3428–3438. doi: 10.1074/jbc.M109.074252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asshauer KP, Wemheuer B, Daniel R, Meinicke P. Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics. 2015;31(17):2882–2884. doi: 10.1093/bioinformatics/btv287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson C, Frankenfeld Cara L, Lampe JW. Gut bacterial metabolism of the soy isoflavone daidzein: exploring the relevance to human health. Experimental biology and medicine. 2005;230(3):155–170. doi: 10.1177/153537020523000302. [DOI] [PubMed] [Google Scholar]
- Bajaj JS, Hylemon PB, Ridlon JM, Heuman DM, Daita K, White MB, … Gillevet PM. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol Gastrointest Liver Physiol. 2012;303(6):G675–685. doi: 10.1152/ajpgi.00152.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, … Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamulitrat W, Jordan Sandra J, Mason Ronald P, Litton Amy L, Wilson Joan G, Wood Edgar R, … Vedia LMY. Targets of nitric oxide in a mouse model of liver inflammation by Corynebacterium parvum. Archives of biochemistry and biophysics. 1995;316(1):30–37. doi: 10.1006/abbi.1995.1006. [DOI] [PubMed] [Google Scholar]
- Collins JW, Chervaux C, Raymond B, Derrien M, Brazeilles R, Kosta A, … Frankel G. Fermented dairy products modulate Citrobacter rodentium-induced colonic hyperplasia. J Infect Dis. 2014;210(7):1029–1041. doi: 10.1093/infdis/jiu205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullender TC, Chassaing B, Janzon A, Kumar K, Muller CE, Werner JJ, … Ley RE. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe. 2013;14(5):571–581. doi: 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de La Serre CB, Ellis CL, Lee J, Hartman AL, Rutledge JC, Raybould HE. Propensity to high-fat diet-induced obesity in rats is associated with changes in the gut microbiota and gut inflammation. Am J Physiol Gastrointest Liver Physiol. 2010;299(2):G440–448. doi: 10.1152/ajpgi.00098.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroos K, Vanhemmens S, Cattoir S, Boon N, Verstraete W. Isolation and characterisation of an equol-producing mixed microbial culture from a human faecal sample and its activity under gastrointestinal conditions. Arch Microbiol. 2005;183(1):45–55. doi: 10.1007/s00203-004-0747-4. [DOI] [PubMed] [Google Scholar]
- Ezzili C, Otrubova K, Boger DL. Fatty acid amide signaling molecules. Bioorg Med Chem Lett. 2010;20(20):5959–5968. doi: 10.1016/j.bmcl.2010.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández J, Redondo-Blanco S, Gutiérrez-del-Río I, Miguélez EM, Villar CJ, Lombó F. Colon microbiota fermentation of dietary prebiotics towards short-chain fatty acids and their roles as anti-inflammatory and antitumour agents: A review. Journal of Functional Foods. 2016;25:511–522. doi: 10.1016/j.jff.2016.06.032. [DOI] [Google Scholar]
- Furet JP, Kong LC, Tap J, Poitou C, Basdevant A, Bouillot JL, … Clement K. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes. 2010;59(12):3049–3057. doi: 10.2337/db10-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ruiz I, Rodriguez-Juan C, Diaz-Sanjuan T, del Hoyo P, Colina F, Munoz-Yague T, Solis-Herruzo JA. Uric acid and anti-TNF antibody improve mitochondrial dysfunction in ob/ob mice. Hepatology. 2006;44(3):581–591. doi: 10.1002/hep.21313. [DOI] [PubMed] [Google Scholar]
- Gou Z, Jiang S, Zheng C, Tian Z, Lin X. Equol Inhibits LPS-Induced Oxidative Stress and Enhances the Immune Response in Chicken HD11 Macrophages. Cell Physiol Biochem. 2015;36(2):611–621. doi: 10.1159/000430124. [DOI] [PubMed] [Google Scholar]
- Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, … Karin M. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012;491(7423):254–258. doi: 10.1038/nature11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E, Li JV, Athanasiou T, Ashrafian H, Nicholson JK. Understanding the role of gut microbiome-host metabolic signal disruption in health and disease. Trends Microbiol. 2011;19(7):349–359. doi: 10.1016/j.tim.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Hwang J, Wang J, Morazzoni P, Hodis HN, Sevanian A. The phytoestrogen equol increases nitric oxide availability by inhibiting superoxide production: an antioxidant mechanism for cell-mediated LDL modification. Free Radical Biology and Medicine. 2003;34(10):1271–1282. doi: 10.1016/s0891-5849(03)00104-7. [DOI] [PubMed] [Google Scholar]
- Ishida T, Nishiumi S, Tanahashi T, Yamasaki A, Yamazaki A, Akashi T, … Mizuno S. Linoleoyl ethanolamide reduces lipopolysaccharide-induced inflammation in macrophages and ameliorates 2,4-dinitrofluorobenzene-induced contact dermatitis in mice. Eur J Pharmacol. 2013;699(1–3):6–13. doi: 10.1016/j.ejphar.2012.11.030. [DOI] [PubMed] [Google Scholar]
- Jonsson AL, Backhed F. Drug the Bug! Cell. 2015;163(7):1565–1566. doi: 10.1016/j.cell.2015.12.005. [DOI] [PubMed] [Google Scholar]
- Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474(7351):327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keszthelyi D, Troost FJ, Masclee AA. Understanding the role of tryptophan and serotonin metabolism in gastrointestinal function. Neurogastroenterol Motil. 2009;21(12):1239–1249. doi: 10.1111/j.1365-2982.2009.01370.x. [DOI] [PubMed] [Google Scholar]
- Kinross JM, Roon ACv, Holmes E, Darzi A, Nicholson JK. The human gut microbiome: implications for future health care. Current gastroenterology reports. 2008;10(4):396–403. doi: 10.1007/s11894-008-0075-y. [DOI] [PubMed] [Google Scholar]
- La Mura V, Pasarin M, Rodriguez-Vilarrupla A, Garcia-Pagan JC, Bosch J, Abraldes JG. Liver sinusoidal endothelial dysfunction after LPS administration: a role for inducible-nitric oxide synthase. J Hepatol. 2014;61(6):1321–1327. doi: 10.1016/j.jhep.2014.07.014. [DOI] [PubMed] [Google Scholar]
- Lawrie CA, Renwick AG, Sims J. The urinary excretion of bacterial amino-acid metabolites by rats fed saccharin in the diet. Food and chemical toxicology. 1985;23(4–5):445–450. doi: 10.1016/0278-6915(85)90138-3. [DOI] [PubMed] [Google Scholar]
- Libby P, Ridker PM, Maseri A. Inflammation and Atherosclerosis. Circulation. 2002;105(9):1135–1143. doi: 10.1161/hc0902.104353. [DOI] [PubMed] [Google Scholar]
- Lin IC, Yamashita S, Murata M, Kumazoe M, Tachibana H. Equol suppresses inflammatory response and bone erosion due to rheumatoid arthritis in mice. J Nutr Biochem. 2016;32:101–106. doi: 10.1016/j.jnutbio.2016.02.012. [DOI] [PubMed] [Google Scholar]
- Lu K, Abo RP, Schlieper KA, Graffam ME, Levine S, Wishnok JS, … Fox JG. Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: an integrated metagenomics and metabolomics analysis. Environ Health Perspect. 2014;122(3):284–291. doi: 10.1289/ehp.1307429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Knutson CG, Wishnok JS, Fox JG, Tannenbaum SR. Serum metabolomics in a Helicobacter hepaticus mouse model of inflammatory bowel disease reveal important changes in the microbiome, serum peptides, and intermediary metabolism. J Proteome Res. 2012;11(10):4916–4926. doi: 10.1021/pr300429x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madianos PN, Bobetsis YA, Kinane DF. Generation of inflammatory stimuli: how bacteria set up inflammatory responses in the gingiva. Journal of clinical periodontology. 2005;32(6):57–71. doi: 10.1111/j.1600-051X.2005.00821.x. [DOI] [PubMed] [Google Scholar]
- Mallett AK, Rowland IR, Bearne CA. Modification of rat caecal microbial biotransformation activities by dietary saccharin. Toxicology. 1985;36(2–3):253–262. doi: 10.1016/0300-483x(85)90058-7. [DOI] [PubMed] [Google Scholar]
- Marshall BM, Ochieng Dorothy J, Levy SB. Commensals: underappreciated reservoir of antibiotic resistance. Microbe. 2009;4(5):231–238. [Google Scholar]
- Maruo T, Sakamoto M, Ito C, Toda T, Benno Y. Adlercreutzia equolifaciens gen. nov., sp. nov., an equol-producing bacterium isolated from human faeces, and emended description of the genus Eggerthella. Int J Syst Evol Microbiol. 2008;58(Pt 5):1221–1227. doi: 10.1099/ijs.0.65404-0. [DOI] [PubMed] [Google Scholar]
- Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, … Mackay CR. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461(7268):1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa I. Functional Differences among FimA Variants of Porphyromonas gingivalis and Their Effects on Adhesion to and Invasion of Human Epithelial Cells. Infection and immunity. 2002;70(1):277–285. doi: 10.1128/iai.70.1.277-285.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SC, Lam EF, Lam TT, Chan Y, Law W, Tse PC, … Wu JC. Effect of probiotic bacteria on the intestinal microbiota in irritable bowel syndrome. J Gastroenterol Hepatol. 2013;28(10):1624–1631. doi: 10.1111/jgh.12306. [DOI] [PubMed] [Google Scholar]
- Nishiuma T, Nishimura Y, Okada T, Kuramoto E, Kotani Y, Jahangeer S, Nakamura S. Inhalation of sphingosine kinase inhibitor attenuates airway inflammation in asthmatic mouse model. Am J Physiol Lung Cell Mol Physiol. 2008;294(6):L1085–1093. doi: 10.1152/ajplung.00445.2007. [DOI] [PubMed] [Google Scholar]
- Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30(21):3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlee SD, Simon BR, Scheller EL, Alejandro EU, Learman BS, Krishnan V, … MacDougald OA. Administration of saccharin to neonatal mice influences body composition of adult males and reduces body weight of females. Endocrinology. 2014;155(4):1313–1326. doi: 10.1210/en.2013-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock LJ. Multidrug-resistance efflux pumps? not just for resistance. Nature Reviews Microbiology. 2006;4(8):629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- Renwick AG. The disposition of saccharin in animals and man—a review. Food and chemical toxicology. 1985;23(4–5):429–435. doi: 10.1016/0278-6915(85)90136-x. [DOI] [PubMed] [Google Scholar]
- Sass G, Koerber K, Bang R, Guehring H, Tiegs G. Inducible nitric oxide synthase is critical for immune-mediated liver injury in mice. J Clin Invest. 2001;107(4):439–447. doi: 10.1172/JCI10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar P, Ahuja S, Sriram K. Non-nutritive sweeteners: review and update. Nutrition. 2013;29(11–12):1293–1299. doi: 10.1016/j.nut.2013.03.024. [DOI] [PubMed] [Google Scholar]
- Song JM, Qian X, Molla K, Teferi F, Upadhyaya P, GOS, … Kassie F. Combinations of indole-3-carbinol and silibinin suppress inflammation-driven mouse lung tumorigenesis by modulating critical cell cycle regulators. Carcinogenesis. 2015;36(6):666–675. doi: 10.1093/carcin/bgv054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer M, Gupta A, Dam LV, Shannon C, Menees S, Chey WD. Artificial Sweeteners: A Systematic Review and Primer for Gastroenterologists. J Neurogastroenterol Motil. 2016;22(2):168–180. doi: 10.5056/jnm15206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subedi L, Ji E, Shin D, Jin J, Yeo JH, Kim SY. Equol, a Dietary Daidzein Gut Metabolite Attenuates Microglial Activation and Potentiates Neuroprotection In Vitro. Nutrients. 2017;9(3) doi: 10.3390/nu9030207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suez J, Korem T, Zeevi D, Zilberman-Schapira G, Thaiss CA, Maza O, … Elinav E. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature. 2014;514(7521):181–186. doi: 10.1038/nature13793. [DOI] [PubMed] [Google Scholar]
- Swithers SE, Martin AA, Clark KM, Laboy AF, Davidson TL. Body weight gain in rats consuming sweetened liquids. Effects of caffeine and diet composition. Appetite. 2010;55(3):528–533. doi: 10.1016/j.appet.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489(7415):242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587(Pt 17):4153–4158. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Uronis JM, Muhlbauer M, Herfarth HH, Rubinas TC, Jones GS, Jobin C. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS One. 2009;4(6):e6026. doi: 10.1371/journal.pone.0006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang ZG, Zhang RL, Gregg SR, Hozeska-Solgot A, LeTourneau Y, … Chopp M. Matrix metalloproteinase 2 (MMP2) and MMP9 secreted by erythropoietin-activated endothelial cells promote neural progenitor cell migration. J Neurosci. 2006;26(22):5996–6003. doi: 10.1523/JNEUROSCI.5380-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol. 2009;5(4):e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhou BP. TNF-alpha/NF-kappaB/Snail pathway in cancer cell migration and invasion. Br J Cancer. 2010;102(4):639–644. doi: 10.1038/sj.bjc.6605530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448(7152):427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- Xu L, Peng H, Wu D, Hu K, Goldring MB, Olsen BR, Li Y. Activation of the discoidin domain receptor 2 induces expression of matrix metalloproteinase 13 associated with osteoarthritis in mice. J Biol Chem. 2005;280(1):548–555. doi: 10.1074/jbc.M411036200. [DOI] [PubMed] [Google Scholar]
- Zhang L, Nichols RG, Correll J, Murray IA, Tanaka N, Smith PB, … Patterson AD. Persistent Organic Pollutants Modify Gut Microbiota-Host Metabolic Homeostasis in Mice Through Aryl Hydrocarbon Receptor Activation. Environ Health Perspect. 2015;123(7):679–688. doi: 10.1289/ehp.1409055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Body weight data of the controls and mice administrated to saccharin for 6 months.
Table S1. Primers used in qPCR.
Table S2. Significantly altered fecal metabolites in saccharin mice compared to control.