SUMMARY
The nervous system—in particular, the brain and its cognitive abilities—is among humans’ most distinctive and impressive attributes. How the nervous system has changed in the human lineage and how it differs from that of closely related primates is not well understood. Here, we consider recent comparative analyses of extant species that are uncovering new evidence for evolutionary changes in the size and the number of neurons in the human nervous system, as well as the cellular and molecular reorganization of its neural circuits. We also discuss the developmental mechanisms and underlying genetic and molecular changes that generate these structural and functional differences. As relevant new information and tools materialize at an unprecedented pace, the field is now ripe for systematic and functionally relevant studies of the development and evolution of human nervous system specializations.
The question of what makes us human has fascinated humankind throughout modern history. Today, we view the brain as the core component of human identity, and an understanding of this organ is consequently essential for answering why we as a species are what we are. The remarkable abilities of the human brain are among the most obvious features that set us apart from our taxonomically closest living relatives, the other great apes of Africa. However, our understanding of how the nervous system—in particular, the brain—has changed in the human lineage remains incomplete (Bae et al., 2015; Enard, 2016; Franchini and Pollard, 2015; Gazzaniga, 2009; Geschwind and Rakic, 2013; Herculano-Houzel, 2016; Hrvoj-Mihic et al., 2013; Lieberman, 2016; Lui et al., 2011; Pääbo, 2014; Passingham, 2008; Penn et al., 2008; Preuss, 2012; Silbereis et al., 2016; Sherwood et al., 2012; Van Schaik, 2016).
Although humans share most of their genetic, molecular, and cellular features with other non-human primates (NHPs), there are compelling differences in cognitive and behavioral capacities between humans and NHPs. Syntactical-grammatical language, symbolic thought, self-reflection, long-term planning ability, autobiographical memory, the theory of mind, and the capacity to create art are among the most distinctively human aspects of cognition and behavior (Gazzaniga, 2009; Lieberman, 2016; Passingham, 2008; Penn et al., 2008). Whether these differences represent incremental, quantitative advances in cognitive ability rather than a novel or qualitative leap is an open question. Historically, some cognitive faculties present in humans were thought to represent such a leap (Lashley, 1949; Penn et al., 2008), but there is increasingly persuasive evidence that even potentially ‘‘special’’ mental abilities are not restricted to humans, but rather exist in some rudimentary form in other apes (Call and Tomasello, 2008; Premack, 2007). For example, great apes have a more sophisticated natural behavioral repertoire than previously thought, and some behavioral skills are transmitted culturally, similar to how human infants learn from observing adults (Horner et al., 2006; Whiten et al., 1999; Watson et al., 2017). This implies that human mental abilities have gradually evolved from ancestral forms and functions, with culture in addition to genomic information playing a critical role in the emergence and transmission of complex behavioral skills. These observations also suggest that a greater understanding of the evolutionary differences separating humans and NHPs will yield new insights into human neurodevelopment, function, and disease. Importantly, as more and better-designed studies are conducted, it is likely that further behavioral and cognitive homologies and differences between humans and NHPs will also be identified and provide insights into how the underlying structure and function of neural circuits have changed in the human lineage.
Comparative studies of the human and NHP brain have historically been difficult to accomplish in systematic and functionally relevant manners. Most of the procedures used in experimental biology are not, due to ethical and legal limitations, applicable to human and NHP brains. Therefore, it is not surprising that much of our understanding of the molecular and cellular mechanisms governing the development and function of the primate brain is derived from experimental studies of a handful of relatively distantly related model organisms. While the use of these model systems has revolutionized the biomedical sciences, extrapolating from them can be problematic; the prolonged development of the human brain, its remarkable cellular complexity and connectivity, and the reliance of the brain on experience for the shaping and refinement of synaptic connections all conspire to make translating data across experimental contexts difficult. Furthermore, the type of studies that can be accomplished with the human or NHP brain is also hampered by experimental limitations, particularly those typical of in vitro assays, imaging modalities, and tissue collected postmortem, often only available to researchers after substantial delays. As a result, characterizing the genome-to-structure-to-function relationships in the nervous system and comparing these relationships between humans and NHPs is especially complicated (Enard, 2016; Franchini and Pollard, 2015; Reilly and Noonan, 2016; Silver, 2016).
Fortunately, recent advances in methods and tools are easing many of these limitations. Increased tissue banking, multi-institutional and multi-disciplinary consortia, and the development of improved protocols for tissue processing and preservation allow greater access to high-quality developmental and adult postmortem human and NHP tissues with reduced postmortem intervals. The development of genomic methods and in vitro systems, including induced pluripotent stem cells (iPSCs) and primary human neural cells, have also expanded the range of studies possible. Non-invasive technologies, such as diffusion tensor imaging, that can be applied to living human and NHP subjects are similarly allowing new avenues of research. Together, these advances have enabled comparative studies of cognition, behavior, neuroanatomy, neurophysiology, proteome, transcriptome, and genome that have provided the necessary foundation to begin a comprehensive exploration of the mechanisms underlying human cognitive capacities.
In this Review, we summarize current advances in our understanding of potentially distinguishing features of the function, organization, and development of the human central nervous system (CNS). Emphasis is given to studies on evolutionary changes in the size and number of neurons of the human brain and the organization of neural circuits—in particular, the long-distance projection systems associated with language and digital dexterity. We also highlight the importance of conducting comparative studies throughout development, as some of the most distinctively human aspects of cognition and behavior are apparent as early as infancy and toddlerhood. Next, we consider the search for the changes in genomic content and molecular processes that make these features possible. We also provide a perspective on the emerging experimental investigations linking the development of human evolutionary specializations to genetic changes and related molecular and cellular mechanisms. Throughout this review, studies of the cerebral neocortex will be highlighted because of its importance in higher-order cognition and complex behaviors and because it has been the focus of many comparative and developmental studies. We concentrate on comparative studies of extant primates and direct readers to other recent reviews on the insights into evolution based on the fossil and archaeological record (Falk, 2014; Neubauer, 2014; Pearce et al., 2013; Wood and Baker, 2011; Zollikofer and De León, 2013). Lastly, we direct readers to other recent reviews for in-depth information on the cognitive, behavioral, and ecological aspects of human nervous system evolution (Defelipe, 2011; MacLean, 2016; Mars et al., 2014; Passingham, 2008; Penn et al., 2008; Preuss, 2012; Rilling, 2014; Schmahmann and Pandya, 2006; Sherwood et al., 2012; Van Schaik, 2016).
Evolutionary Perspective on Human Nervous System Structure
Humans Have the Largest Brain among Extant Primates
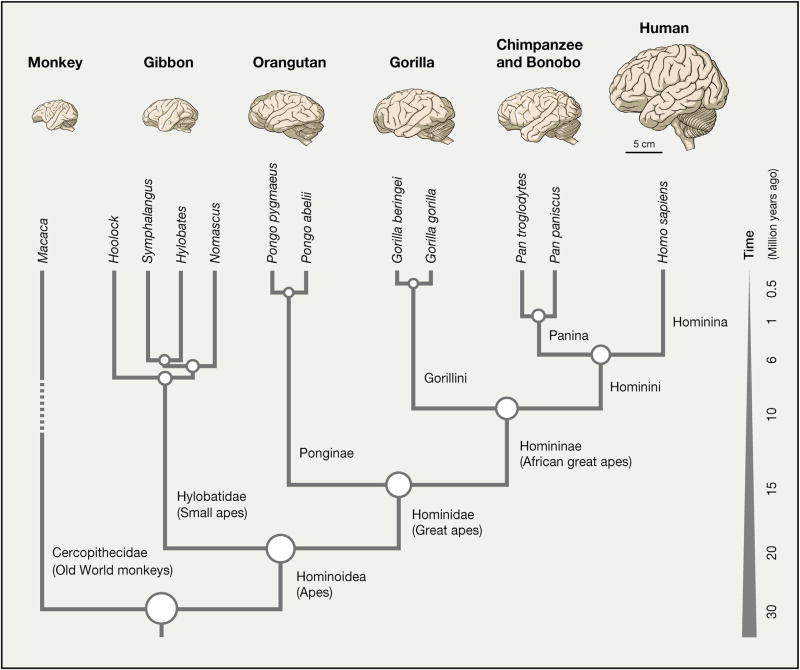
The overall size of the CNS has been correlated with general intelligence and other indicators of cognitive capacities (Jerison, 1973; Williams and Herrup, 1988), but this relationship is neither robust nor mechanistically understood. The size of the human brain tends to support such a relationship, as at roughly 1,400g (Dekaban, 1978) and with only 30 g belonging to the spinal cord (MacLarnon, 1996), the human CNS is about three times larger than that of chimpanzees (Figure 1; Blinkov and Glezer, 1968; Stephan et al., 1981). Similarly, absolute brain size is a good predictor of cognitive capabilities across primates (Deaner et al., 2007; Reader and Laland, 2002). However, several large mammals such as the sperm whale (~7.8 kg; Physeter macrocephalus; Kojima, 1951), long-finned pilot whale (~3.6 kg; Globicephala melas; Mortensen et al., 2014), and elephant (~4.6 kg; Loxodonta africana; Herculano-Houzel et al., 2015) have substantially larger brains than humans do. This raises a question: how can brain size or similar metrics sometimes, but not always, be related to cognitive capacity?
Figure 1. Humans Are Part of the African Great Ape Clade and Have the Largest Brain among Extant Primates.
The images depict adult brains of the species indicated at the leaf node. Chimpanzee and bonobo are represented with identical schematics due to their close similarity. The estimated divergence times with respect to humans among great apes (chimpanzee [4.5–7 mya], gorilla [7–10 mya], and orangutan [12–16 mya]), a small ape (gibbon [18–20 mya]), and an Old World monkey (Rhesus macaque [25–33 mya]) are based on unique gap-free sequence identity (adapted from Locke et al. (2011), Yu et al. (2003), and Chen and Li [2001]) and recent analyses of the fossil record (Katoh et al., 2016; Almécija et al., 2013). For simplicity, only one old-world monkey is depicted (Rhesus macaque, the most commonly studied non-human primate); the dashed line represents the approximate split time from the other extant monkey species. ~1 million years ago (mya) implies approximately 1 mya or less.
One possibility is that larger animals have bigger brains simply as a consequence of isometric scaling. Several efforts have been made to correct for such scaling. The most simple utilizes a ratio between brain mass and body size, and while brain mass is larger relative to body mass for humans than for other great apes, this simple relative measurement is not a reliable predictor of cognitive capacities, as smaller animals like mice have ratios similar to that of humans (Jerison, 1973). A more complex measurement, the encephalization quotient, was formulated to measure how big the brain is relative to the brains of other similarly sized animals (Jerison, 1973; Roth and Dicke, 2005), and by this measure, humans surpass all other primates and most, but not all, mammals that have been assessed.
A second possibility is that the relative sizes of discrete territories in the brain differ across species. Allometric considerations, therefore, may be essential for explaining the cognitive capacities of different species. For example, the cerebral white matter, which contains not only axons but around 2 billion neurons and a large, but yet unknown, number of glia in humans (Herculano-Houzel et al., 2015; Herculano-Houzel, 2016; Sigaard et al., 2016; Silbereis et al., 2016), seems to increase disproportionately compared to gray matter as brain size scales across species (Hofman, 2012; Rilling and Insel, 1999; Zhang and Sejnowski, 2000). Not surprisingly, the cerebral neocortex and cerebellum, which contain the largest amounts of white matter, tend to make up greater proportions of larger mammalian brains (Barton and Venditti, 2014; Finlay and Darlington, 1995; Stephan et al., 1981). Of particular interest for cognitive research are association areas of the cerebral neocortex, which are thought to be essential for higher-order cognition. However, when compared to other primates, the relative size of human association areas falls within the expected range (Sherwood et al., 2008). Likewise, the overall size of the human frontal cortex, a region enriched for association areas, seems no larger than expected as compared to the rest of the brain or the cortex (Bush and Allman, 2004; Semendeferi et al., 2002). Additional studies have provided evidence that the prefrontal cortex or a portion of the prefrontal cortex may be expanded in humans relative to NHPs (Schoenemann et al., 2005; Semendeferi et al., 2011), but these results are not universally accepted for various reasons (Sherwood et al., 2008; Holloway, 2002). While still a matter of considerable discussion and study, recent evidence suggests that certain features of the human brain anatomy scale as expected for a primate (Herculano-Houzel, 2016).
As the absolute size of the brain increases across taxa, some features of the brain do not scale uniformly and should not be expected to do so. Structural and functional differences associated with scaling the brain, including neuronal populations at the root of functional connectivity, may therefore be among the substrates susceptible to evolutionary and developmental variation and consequently associated with the emergence of human cognition. Consistent with this, changes in the numbers, as well as the structural and functional properties, of both neurons and glia have been observed in humans compared to NHPs (for example, Bianchi et al., 2013; Elston et al., 2011; Herculano-Houzel, 2016; Kwan et al., 2012; Oberheim et al., 2012; Sherwood et al., 2004). However, many of these and other characteristics have not yet been explicitly and systematically assayed across different NHPs or other mammalian species.
Humans Have the Most Cortical Neurons among Extant Primates
Several attempts have been made to estimate the number and distribution of neurons in the human CNS. The human CNS has approximately 86 billion neurons, with a roughly equal number of glial cells, and 99.9% of these neurons are located in the brain (i.e., the forebrain, brain stem, and cerebellum; Herculano-Houzel et al., 2015; Silbereis et al., 2016; Williams and Herrup, 1988). While the cerebral cortex, including white matter, represents ~82% of the mass of the CNS, only ~20% of all neurons (around 16 billion) reside within this structure (Herculano-Houzel et al., 2015; Herculano-Houzel, 2016; Sigaard et al., 2016), with published estimates of the number of neocortical neurons varying by as much as a factor of 2 (between 14.7–32.0 billion) (Herculano-Houzel et al., 2015; Herculano-Houzel, 2016; Pakkenberg and Gundersen, 1997; Pakkenberg et al., 2003; Sigaard et al., 2016). In contrast, the cerebellum represents ~10% of the mass of the CNS but has ~80% of the neurons (69 billion), most notably granular neurons. An additional 700 million neurons (< 1%) are present in the rest of the CNS, including as few as 20 million neurons in the spinal cord (Herculano-Houzel et al., 2015). For comparison, there are around 400–600 million neurons in the human enteric nervous system, the largest part of the peripheral nervous system (Furness, 2006). Crucially, recent comparative studies suggest that both the numbers of neurons and glia and the ratio of these cell types to one another in the human cerebral cortex and cerebellum likely follow general scaling rules common to non-great ape primates (Herculano-Houzel, 2016); as human brains are larger than those of other primates, this again suggests a positive relationship between neuron number and cognitive capacities.
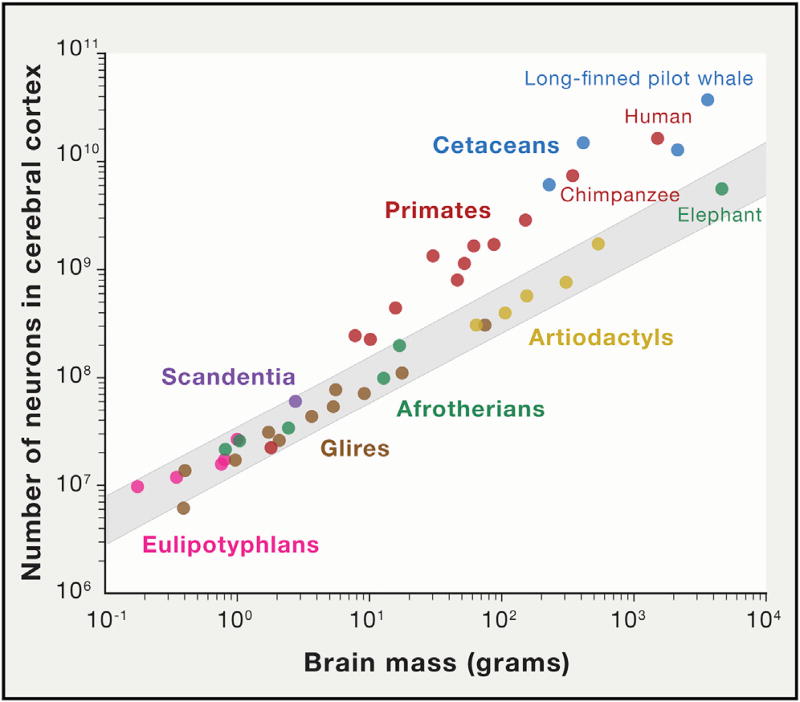
However, while the human brain has more neurons than any extant NHP several mammalian species possess more neurons in both the CNS and the cerebral cortex, including the long-finned pilot whale, a species of dolphin with more neurons (37.29 billion) in the neocortex than any mammal studied to date and almost twice as many as humans (Figure 2; Mortensen et al., 2014). Moreover, humans affected by either congenital or acquired conditions where portions of the brain are severely underdeveloped or missing can have normal or near-normal intelligence and cognitive skills, with examples including some severe forms of microcephaly (Haslam and Smith, 1979; Kozma et al., 1996); childhood hemispherectomy (i.e., disconnection or removal of an entire cerebral hemisphere; Borgstein and Grootendorst, 2002; Devlin et al., 2003; Liégeois et al., 2010; Pulsifer et al., 2004); a patient born with only one hemisphere (Muckli et al., 2009); craniopagus malformation (Lansdell, 1999; Stone and Goodrich, 2006; Todorov et al., 1974), including a pair of craniopagus twins that share a brain and mind (Squair, 2012); individuals with almost complete absence of the cerebellum (Glickstein, 1994; Yu et al., 2015); and a case of severe hydrocephaly (Feuillet et al., 2007). Nevertheless, how a simple change in brain size or neuron and/or glia number could lead to differences in cognitive capabilities is not, at a mechanistic level, well understood.
Figure 2. Variation in the Number of Cerebral Cortical Neurons across Mammals.
The shaded region represents the 95% prediction interval for the data points excluding primate and cetacean points, highlighting that the observed number of cortical neurons in primates does not follow the same scaling rules. What scaling rule cetaceans adhere to is an open question that will be resolved as more data become available. Data for this figure were taken from several sources that used different methodologies and data reporting methods. The numbers on cetaceans were compiled from cerebral neocortical samples by Eriksen and Pakkenberg (2007), Walløe et al. (2010), and Mortensen et al. (2014). Information for the chimpanzee data point was obtained from neocortex by Collins et al. (2016). All other data were retrieved from the dataset generated by Herculano-Houzel et al. (2015) from the entire cerebral cortex.
Therefore, the key to our brain’s unique capacities may not be simply its absolute or relative size, or even its number of neurons and glia, but instead more nuanced components such as increased diversity of neural cell types, molecular changes, and expanded or more complex patterns of neuronal connectivity. One possibility is that increased cognitive abilities emerged following the expansion of the human brain because this expansion untethered large portions of association cortices from previously strong constraints imposed by molecular gradients and neuronal activity patterns, thereby allowing new sets of cortico-cortical synaptic projections, the re-wiring of ancestral circuits, and the development of new behavioral, cognitive, and phenotypic outcomes (Buckner and Krienen, 2013). Such a process may have also occurred in some whales, elephants, and other species with large brains and high numbers of neurons, but any specific changes that occurred would be independent and consequently likely unique to a single lineage. Nevertheless, increasing our knowledge of distant species remains important to distinguish lineage-specific brain features from those that can be predicted solely on the basis of increased brain size. Relationships between brain size and neuron number with cognitive ability might then be largely constant, though not strictly causal, within taxa but less robust across more divergent species. Given that the human brain appears to respect the same general scaling properties observed in NHPs and that brain size and neuron (and likely glia) number are generally predictive of cognitive capacity among primates, any pursuit seeking to determine unique features of the human brain must therefore consider the context of primate CNS structure, physiology, and development.
Humans Have Specialized Neuronal Connections
Myriad neuronal cell types and their specific synaptic connections comprise the core components of neural circuits and networks, which are collectively referred to as the connectome (van den Heuvel et al., 2016). While the immense complexity of the human connectome has become increasingly evident over the past decade, largely due to advances in imaging technologies, our understanding of its organization and function at the level of long range projections, local synaptic circuits, and intracellular signaling remains highly incomplete. Recent estimates suggest there may be between several hundred trillion and well over a quadrillion synapses in the human CNS (see Silbereis et al., 2016), with an average of 164 trillion synapses in the neocortex of young adult males (Tang et al., 2001). Moreover, the cerebral white matter of young adults contains ~149,000–176,000 km of myelinated axons (Marner et al., 2003), which may form hundreds of thousands of distinct long-range projection systems (Irimia et al., 2012). The topology and fidelity of the connectome are central to the establishment of the dynamic activity patterns that underlie species-specific cognition and behavior (Markov et al., 2013; Mesulam, 2000; van den Heuvel et al., 2016). In principle, small changes in the wiring of the connectome can lead to profound and specific functional changes. In keeping with the hypothesis presented in the previous section, multiple lines of evidence suggest that such changes in neuronal connectivity have occurred in the human lineage.
The allometric expansion of the human brain, in particular the cerebral cortex, was likely accompanied by the structural reorganization of the connectome, as evidenced in part by the prominent changes in gyral anatomy (Hofman, 2012; Rogers et al., 2010). However, while changes in gyrification are obvious, it is not clear if humans have any brain structures, cell types, or neural circuits that are not present in other primates. Most of the effort in identifying human-specific changes in cellular diversity and connectome wiring has been dedicated to comparative studies of the neocortex. Despite the expansion of the human neocortex, there is no compelling evidence that any neocortical area is unique to humans. This includes human perisylvian areas involved in speech and language, which appear to have homologs in at least some NHPs (Grefkes and Fink, 2005; Petrides et al., 2005). Likewise, histological comparisons have not identified human-specific changes in structure, with the exception of changes within layer 4a in the human primary visual cortex, which is thought to contribute to differences in the cortical representation of the magnocellular visual pathway (Preuss and Coleman, 2002).
In contrast, several studies support human-specific changes in neuronal organization and connectivity patterns. Even though no neural cell types unique to the human brain have been identified to date, there are changes in the morphology and abundance of both excitatory and inhibitory neurons, as well as glia between humans and NHPs (Bianchi et al., 2013; Elston et al., 2011; Herculano-Houzel, 2016; Kwan et al., 2012; Oberheim et al., 2012; Sherwood et al., 2004). For example, human excitatory projection neurons, also known as pyramidal cells, are larger, have more complex dendritic arborization, and have a higher density of spines compared to excitatory projection neurons of NHPs (Elston et al., 2011; Sherwood et al., 2003), suggesting that they may have an increased potential for integrative connectivity. A notable example of this centers upon a subgroup of modified pyramidal neurons, known as spindle or von Economo neurons, characterized by a large spindle-shaped soma mainly found in layer 5b of the fronto-insular and anterior cingulate cortex of several primates, elephants, and cetaceans (Nimchinsky et al., 1999; Seeley et al., 2012), which have been shown to be larger and more numerous in humans than in other apes (Allman et al., 2010). Along similar lines, another group of modified pyramidal neurons, fork neurons, which feature a von Economo neuron-related but nevertheless distinct morphology, are intermingled with von Economo neurons in the human fronto-insular cortex and are scattered in chimpanzees and orangutans, but not cats (see Seeley et al., 2012 for references). However, the extent of species differences for von Economo and fork neurons requires further investigation before conclusions about their phylogenies can be determined. There are also several lines of evidence for species-specific differences in the regional localization and abundance of certain subclasses of inhibitory neurons and axons from serotoninergic, dopaminergic, and cholinergic neuromodulatory systems in the neocortex of humans and NHPs (Defelipe, 2011; Raghanti et al., 2016; Raghanti et al., 2009; Raghanti et al., 2008a, b; Sherwood et al., 2004). Human-specific changes at the brain tissue level, likely reflecting alterations in the molecular and biochemical properties of neurons and glia, have also been observed (Khaitovich et al., 2004; Oldham et al., 2006; Somel et al., 2009; Somel et al., 2013; Zhu et al., 2014). For example, differences in gene expression, signaling pathways, and increased complexity in astrocyte morphology and function have been reported between human and non-human neocortical glial cells (Han et al., 2013; Oberheim et al., 2012; Spocter et al., 2012; Zhang and Barres, 2013). Overall, it is necessary to appreciate that these species differences may reflect an expansion of the functional role of glia in synaptic modulation and perhaps cognition in humans.
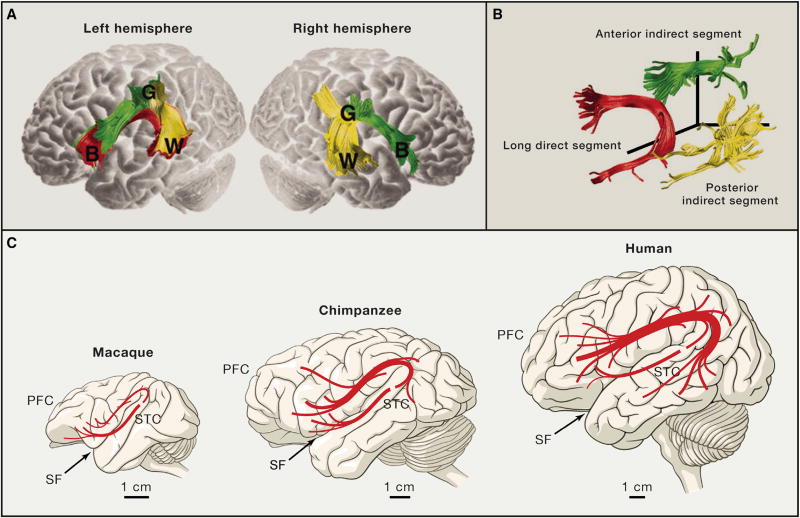
There are also compelling examples of species differences in the organization of long-distance projection systems between primate species. One example is the arcuate fasciculus, a tract that connects perisylvian temporo-parietal and frontal neocortical areas (Figures 3A and 3B). Alterations of the arcuate fasciculus result in conduction aphasia, a rare audition-based aphasia resulting in speech repetition inabilities, indicating the importance of these fibers for human language (Anderson et al., 1999). Diffusion tensor imaging analysis of the homologous perisylvian axonal tracts in humans, chimpanzees, and macaques revealed that temporal projections present in humans seem to be greatly reduced in chimpanzees and absent in macaques, especially in the left medial and inferior temporal gyri (Figure 3C; Rilling et al., 2008), which may have implications for the evolution of language. In addition, species differences in the organization of the superior longitudinal fasciculus, the primary white matter tract connecting lateral frontal with lateral parietal neocortical areas, have been observed between humans and chimpanzees (Hecht et al., 2015), which may have implications for the evolution of fronto-parietal functions, including spatial attention to observed actions, social learning, and tool use. In addition to structurally based studies, a recent comparative functional magnetic resonance imaging study observed two lateralized human fronto-parietal networks in the cortical regions displaying the greatest evolutionary expansion that had neither topological nor functional monkey correspondents (Mantini et al., 2013), suggesting that functions of certain structural networks have diverged during human evolution. There is also evidence for structural and functional reorganization of corticofugal neurons and their long-range axons, such as the corticospinal projection system in primates (Heffner and Masterton, 1983; Herculano-Houzel et al., 2016; Kuypers, 1987; Nakajima et al., 2000), which might be relevant for the corticalization of motor control and the evolution of digital dexterity.
Figure 3. Language-Related Pathways Are Strongly Lateralized and Modified in Humans.
(A and B) Language-related pathways connecting Broca’s (B), Geschwind’s (G), and Wernicke’s (W) neocortical territories, reconstructed based on diffusion tensor imaging data. Both hemispheres share the long direct segment and the posterior indirect segments, which connect Broca’s territory in the frontal cortex with Geschwind’s territory in the parietal cortex and connect Geschwind’s territory with Wernicke’s territory in the temporal cortex, respectively. The direct segment is found exclusively in the left hemisphere and connects Broca’s and Wernicke’s territories.
(C) Comparison of arcuate fasciculus projections in humans and non-human primates. In humans, projections extend far into the medial and inferior temporal gyri, whereas projections to the chimpanzee temporal lobe are less extensive into the inferior temporal gyrus. In macaques, the projection into the inferior temporal gyrus appears to be completely absent. PFC, prefrontal cortex; SF, Sylvian fissure; STC, superior temporal cortex. Adapted from Catani and Mesulam (2008); Rilling et al. (2008), and Ghazanfar (2008).
The principles of routing complexity and connectivity scaling (Deacon, 1990; Ringo, 1991; Stevens, 1989) have been used to suggest that there is an increase in the amount of cortical local short-range connections, including short U-shaped fibers, in the human cortex (Catani et al., 2012; Herbert et al., 2003; van den Heuvel et al., 2016). These short-range connections are expected to arise from pyramidal neurons in layers 2 and 3. These cortical layers exhibit increased thickness and neuron number in humans compared to NHPs and other analyzed mammals (Hutsler et al., 2005; Marin-Padilla, 1978; Rockel et al., 1980). Cortical short-range projections connect neighboring areas and are hypothesized to generate more elaborate gyrification (Van Essen, 1997), which is a prominent characteristic in humans as compared to NHPs (Hofman, 2012). Another study (Spocter et al., 2012) quantified the neuropil fraction (used as a proxy for the total connectivity under the assumption that more neuropil indicates more connectivity) of six different neocortical (primary and association) areas of human and chimpanzee brains. Human association areas, especially in the prefrontal cortex, contained a higher fraction of neuropil compared to primary areas, while chimpanzee association and primary areas had similar neuropil levels. In addition, neurons in association areas of the frontal (Schenker et al., 2008) and temporal (Semendeferi et al., 2011) cortices are more separated than in other areas of the brain in humans compared to NHPs.
The human connectome also appears unique in the timing and pattern of neocortical myelination, which may have implications on the conduction velocity along axons (Glasser et al., 2014; Miller et al., 2012; Olmos-Serrano et al., 2016). While its overall myelination maps are comparable to those of chimpanzee and macaque brains, the human brain has a larger total axon surface that is less myelinated, representing mostly the association areas (Glasser et al., 2014). These results reinforce the idea that humans have reorganized long-range corticopetal, intra-cortical, and corticofugal projection systems, especially those associated with the prefrontal and temporal association cortices, which are regions involved in higher-order cognitive functions. Together, these studies suggest that both local circuits and long-range projection systems and networks have undergone structural, molecular, and functional reorganization during human evolution and that these features may have evolved independently of brain enlargement. However, the evidence for human-specific changes in some cases is not conclusive due to sample size, tissue quality, and methods used, indicating the need for further studies and the development and implementation of new methodologies.
Interestingly, the large size and expansive connectivity of the human brain create two problems that may also drive some aspects of human divergence. Generating and maintaining an immensely complex connectome comes at a substantial metabolic cost, as the human brain uses 18% of the body’s oxygen at rest but accounts for only 2.5% of a human’s total body weight (Kety and Schmidt, 1948). As a consequence, molecular adaptations for high levels of neuronal activity, along with changes in energy allocation and diet, occurred during human evolution (Aiello and Wheeler, 1995; Khaitovich et al., 2008; Pontzer et al., 2016; Wrangham et al., 1999). For extended information and discussion on the potential role of diet and energetics in the evolution of brain size, we refer readers to Navarrete et al. (2011) and Isler and Van Schaik (2014). In addition, the development of a large brain and the generation of the human-specific features of the neural connectome necessitate an extended developmental period relative to NHPs and mammals with smaller brains or brains with lower complexity in terms of connectivity.
Developmental Mechanisms Underlying the Evolution of Human Nervous System
We have thus far reviewed phenotypic similarities and differences between gross brain features, as well as smaller structures and cell types in human and non-human brains. These phenotypic differences between human and NHP CNS most likely arise during the normal brain development of each species. But what governs the developmental occurrences that allow these structures to arise?
Owing to its remarkable complexity, the human brain, and most especially the association areas of the neocortex, develops more slowly than the brains of other primates. It takes over two decades to build a fully mature human brain, a period of time greater than the entire lifespan of some NHPs (Silbereis et al., 2016). As a consequence, in addition to a particularly long gestational time, humans have an extended childhood and adolescence (Bogin, 1994). This prolonged developmental course and period of dependency allows, more so than in other primates, for longer critical periods and a greater effect of environmental factors on the development of cognitive, emotional, and social capacities (Bogin, 1994; Silbereis et al., 2016; Sousa et al., 2015).
Compared to either the macaque, one of the most commonly studied NHPs, or other non-primate mammals whose CNS development has been extensively studied, the developing human CNS possesses divergent and highly derived features. Specifically, humans have expanded proliferative zones and have diverse subtypes of neural stem and progenitor cells with enhanced proliferative capacities that facilitate brain expansion, especially of the neocortex (see recent reviews for details: Bae et al., 2015; Dehay et al., 2015; Florio et al., 2015; Geschwind and Rakic, 2013; Gulden and Šestan, 2014; Lui et al., 2011; Taverna et al., 2014). As the timing and duration of neurogenesis is correlated with the size of the resulting neocortex, and as the duration of neurogenesis in humans is protracted, more progenitor cells can be added to the pool that gives rise to the neocortex (Finlay and Darlington, 1995; Geschwind and Rakic, 2013). This is reflected in the large expansion of the subventricular zone, particularly the outer subventricular zone, in the primate and human developing neocortex (Lui et al., 2011; Smart et al., 2002; Taverna et al., 2014). Similarly, upper-layer neurons, which are the last postmitotic neurons to migrate to their final position in the laminar neocortex and might therefore be most affected by a prolonged neurogenic period, have an extraordinary numeric expansion in primates, especially in humans, when compared to other mammals (Hutsler et al., 2005; Marin-Padilla, 1978; Rockel et al., 1980).
Experimentally investigating the details of primate and human neurogenesis is technically challenging. A study of cell-cycle kinetics during cortical neurogenesis in macaque embryos has revealed longer cell-cycle duration in monkeys compared with rodents (Dehay et al., 2015; Kornack and Rakic, 1998). Using directed differentiation of human, chimpanzee, and macaque iPSCs, Otani et al. (2016) recently recapitulated key features of cortical neurogenesis in vitro. Furthermore, they reported species differences in cell-cycle length and proliferative capacities of neural progenitors, including changes in the timing (heterochrony) of key developmental events and extended proliferation of chimpanzee and human progenitors compared to macaque.
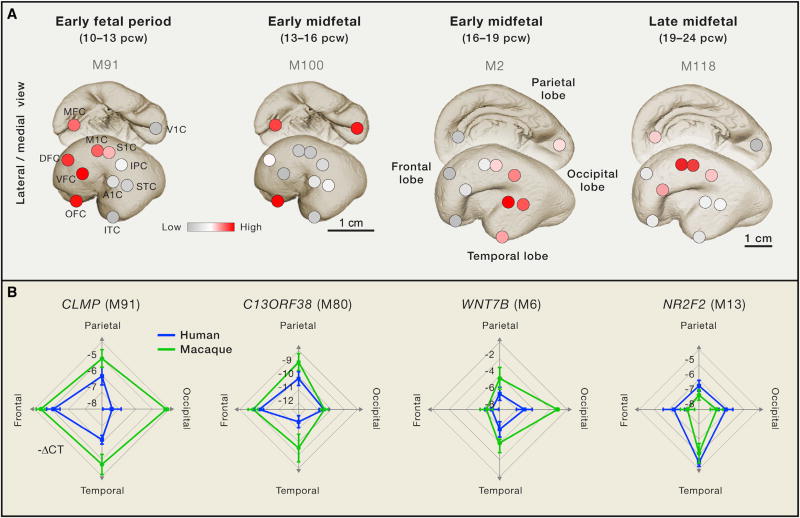
The complexity that defines the structural and functional patterns of neural connectivity is rooted in the diversification of developing neurons and glial cells into functionally distinct subtypes and the formation of proper connections (Bae et al., 2015; Lui et al., 2011; Shibata et al., 2015). While we have made substantial progress in understanding how molecular and cellular changes can lead to variations in the size of the mammalian CNS, in particular the neocortex, we know very little about how developmental changes in neuronal complexity and connectivity arose during human evolution. Notably, a small number of recent comparative studies of postmortem prenatal and early postnatal human and NHP brains have provided some relevant insights by revealing certain region and cell-type-specific developmental changes across those species (Figure 4; Bakken et al., 2016; Florio et al., 2015; Kwan et al., 2012; Pletikos et al., 2014). However, neural connectivity and developmental processes cannot be easily assessed in postmortem human tissue through imaging studies or in existing in vitro neural cell systems. As a consequence, much of the complexity of the human brain and the observed human-specific differences discussed so far in this review, as well as cell-type and phenotypic specializations, can best be studied through the genomic changes that have occurred in the human lineage. In view of the current technological boom in genomics, this is a welcome circumstance for investigators, as human-specific phenotypes must ultimately be linked to the genomic changes giving rise to that innovation.
Figure 4. Comparative Analysis of Neocortical Gene Expression between Human and Macaque Reveal Species Differences during Fetal Development.
(A) Illustration showing gradients of gene co-expression modules (M) during human neocortical development. Four gradients are shown: anterio-posterior (10–13 pcw), ventro-medial (13–16 pcw), temporal (16–19 pcw), and perisylvian (19–24). Genes present within each co-expression module shown here are listed in Pletikos et al. (2014). pcw: postconcepional week; MFC: medial prefrontal cortex; OFC: orbital prefrontal cortex; DFC: dorsolateral prefrontal cortex; VFC: ventrolateral prefrontal cortex; M1C: primary motor cortex; S1C: primary somatosensory cortex; IPC: inferior parietal cortex; STC: superior temporal cortex; ITC: inferior temporal cortex; A1C: primary auditory cortex; V1C: primary visual cortex.
(B) Quantitative real-time PCR of hub genes belonging to the coexpression modules represented in (A) show divergence between human and macaque, particularly in the expression of CLMP and C13ORF38. Adapted from Pletikos et al. (2014).
Genetic Basis and Molecular Mechanisms Underlying Human Brain Evolution
Given a phenotypic difference, identifying underlying genomic changes is a challenging task. Functionally relevant sequence variation between species must be identified from a much larger set of variants, most of which are expected to be neutral. Once a connection between sequence and trait variation is supported, it is even more difficult to gain a satisfactorily mechanistic understanding of the relationship, even when the species of interest are genetically tractable model organisms. The classes of genetic changes behind human-specific traits can range from single-base-pair substitutions to large chromosomal rearrangements, which can lead to a variety of effects ranging from small modifications of gene expression levels to the appearance or destruction of whole genes and regulatory regions. Human and chimpanzee genomes differ by ~35 million nucleotide substitutions (1.2% of base pairs) and 5 million insertions or deletions (indels) (Chimpanzee Sequencing and Analysis Consortium, 2005). In fact, structural variation, including indels, inversions, and duplications, accounts for 3–4 times more sequence divergence than single-base-pair mutations between human and chimpanzee genomes (90 Mb versus 35 Mb from substitutions; Cheng et al., 2005).
In the next section, we describe studies that have begun to characterize genomic variation in the context of brain development and function, with an emphasis on studies that have attempted to functionally investigate variants that occurred in the human lineage after the split with chimpanzees. The degree to which changes in noncoding regulatory regions, particularly cis-regulatory elements, versus changes in protein-coding regions drive phenotypic evolution remains an unresolved question. For discussion of this topic, see reviews arguing in favor of a dominant role for cis-regulatory evolution (Carroll, 2008; Wray, 2007), as well as reviews providing different perspectives (Alonso and Wilkins, 2005; Hoekstra and Coyne, 2007; Wagner and Lynch, 2008). Despite the relative contributions of different mutation types, there are clear examples of each type driving lineage- and species-specific differences, and both types likely contribute to human-specific features of brain development.
Protein-Coding Mutations
While many of the genetic differences between species lie outside of protein-coding regions, focusing on amino-acid substitutions makes it easier to identify changes that likely have a functional consequence (for example, when an amino acid with very different properties is substituted or a mutation leads to a stop codon). Furthermore, comparing the ratio of nonsynonymous to synonymous mutations provides a straightforward method for estimating whether a sequence has evolved at a rate expected under neutral evolution. In the study of human brain specializations, brain-expressed genes that show signs of positive selection are of particular interest. If human-specific features of brain development involved changes in the function of many proteins, a reasonable expectation is for brain-ex-pressed genes to display elevated rates of evolution in the human lineage. Although a few studies reported results that support this idea (Dorus et al., 2004; Yu et al., 2006), other studies found no evidence that brain-expressed genes have evolved faster in humans (Bustamante et al., 2005; Clark et al., 2003; Nielsen et al., 2005; Shi et al., 2006; Wang et al., 2007). While these studies indicate that global rates of amino-acid substitutions in brain-expressed genes are not exceptional in the human lineage, these data provide no direct information about the functional consequences of the mutations that have occurred. As such, it is important that specific genes that appear to be under positive selection are functionally characterized.
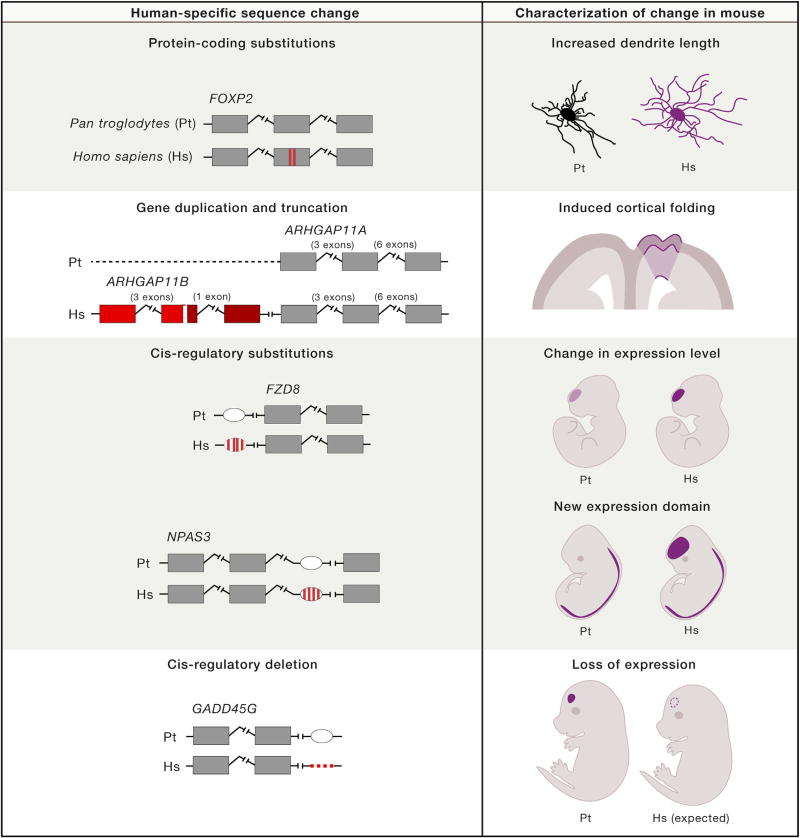
FOXP2 is an example of a gene for which there are multiple lines of evidence, including functional studies, that point to its involvement in human-specific specializations of the brain. FOXP2 is an extremely well-conserved protein, with only three amino-acid substitutions between human and mouse orthologs (Enard et al., 2009; Zhang et al., 2002), and was the first gene found to harbor mutations that cause problems in speech and language development (Lai et al., 2001). Of these substitutions, two occurred in the human FOXP2 after the divergence of humans and chimpanzees, and these mutations appear to be fixed in all human populations, with one of the mutations also arising independently in carnivores and a suborder of bat species (Zhang et al., 2002; Li et al., 2007). The potential functional consequences of these two human-specific substitutions have been investigated by introducing the two mutations into the endogenous mouse Foxp2 (Enard et al., 2009). Mice with the humanized version of Foxp2 showed differences restricted to neural-related phenotypes, specifically in cortico-basal ganglia circuits, including changes in dopamine levels, striatal synaptic plasticity, dendrite morphology, and stimulus-response learning (Figure 5; Enard et al., 2009; Schreiweis et al., 2014). Compared to their wild-type littermates, juvenile mice carrying the two human-specific mutations also showed differences in ultrasonic vocalizations (Enard et al., 2009), but these effects appear to be transient, as they were not observed in adult mice carrying the two human-specific mutations (Hammerschmidt et al., 2015).
Figure 5. Mouse Models of Human-Specific Genomic Features.
Each row presents a study that investigated a different type of human-specific change in a mouse model, with the right column highlighting a specific finding of the study. In order, the studies are Enard et al. (2009), Florio et al. (2015), Boyd et al. (2015), Kamm et al. (2013a), and McLean et al. (2011). The darker shade in ARHGAP11B represents a frameshift and termination that followed a deletion in the fifth exon. Pt, Pan troglodytes, common chimpanzee; Hs, Homo sapiens, human. Note that the findings displayed, particularly for the Enard et al. (2009) and Boyd et al. (2015) studies, are not intended to serve as a summary for all the phenotypes investigated.
Beyond single nucleotide substitutions, studies have begun to explore the contribution of larger structural changes, such as duplications, to phenotypic differences between human and NHP brains. Human-specific duplications of genes with neurodevelopmental functions are of particular interest, as these duplications may have resulted in novel gene function that is restricted to humans. Investigating these events generally involves accurate sequencing of large-insert bacterial artificial chromosome (BAC) libraries to fully resolve the structure and evolution of these highly complex regions. There are more than 30 gene families expanded specifically in humans (Sudmant et al., 2010), including many genes involved in neurodevelopment (Fortna et al.,2004; Goidts et al., 2006; Sudmant et al., 2010). Among human-specific gene duplicates, SRGAP2 has been one of the most extensively studied. After the divergence of human and chimpanzee lineages, SRGAP2 underwent a series of duplications in humans, with one duplication fixed across all human populations studied (Dennis et al., 2012). Interestingly, experiments in mouse suggest that this human-specific paralog interferes with the function of the ancestral copy of SRGAP2, antagonizing its role in synapse regulation, maturation, and density, resulting in protracted synapse maturation, increased synaptic density in neocortical pyramidal neurons, and prolonged spine maturation (Fossati et al., 2016; Charrier et al., 2012).
Another human-specific duplication with evidence for neuro-developmental function, ARHGAP11B, resides in a ~2.5 Mb region on chromosome 15q13.3, known as one of the most unstable loci in the human genome, with structural variants associated with several developmental disorders (Antonacci et al., 2014; El-Hattab et al., 2009; Shinawi et al., 2009). Recently, Florio and collaborators discovered that this gene is highly expressed in isolated radial glial cells, while its expression was undetectable in cortical neurons and cortical plate. A series of experiments in mouse demonstrated that ARHGAP11B, which was also present in Neanderthals and Denisovan (Prüfer et al., 2014), increases basal progenitor mitosis when expressed in the mouse neocortex at E13.5. It appears to do so by promoting apical radial glia cells to undergo symmetric-differentiative divisions producing two basal progenitors in each mitosis (Florio et al., 2015). Notably, when expressed in the mouse brain, ARHGAP11B produced folding of the neocortex in an otherwise lissencephalic brain (Figure 5). Due to their human-specific evolution at the genomic level along with their phenotypic characterization in mouse, SRGAP2 and ARHGAP11B are strong candidates for playing an important role in producing differences in human brain development that occurred after the split of human and chimpanzees.
Among other promising candidate genes under study in the context of evolutionary neuroscience are BOLA2 and DUF1220 repeats, which are both linked to autism spectrum disorder (Davis et al., 2014; Kumar et al., 2008). BOLA2 is contained in a modern-human-specific segmental duplication in a complex region of chromosome 16, showing 2–5 copies in almost all present-day humans analyzed (Prüfer et al., 2014; Sudmant et al., 2013; Nuttle et al., 2016), which is an instance of a quick fixation after a duplication event. The DUF1220 protein domain, extensively studied by Sikela and colleagues, shows the largest expansion of protein-coding sequence in the human genome, with an estimated addition of 28 copies every million years since the split with chimpanzees, resulting in ~270 copies in humans compared to the 90–125 copies in great apes (Fortna et al., 2004; O’Bleness et al., 2012a; O’Bleness et al., 2012b; Popesco et al., 2006). These domains are mostly present in the NBPFgene family and mainly found in chromosome 1 (1q21.1). Almost all DUF1220 regions accumulated in humans since the split with chimpanzee are located within a human-specific pericentromeric inversion. Deletions and duplications in this DUF1220-rich locus have been implicated in microcephaly and macrocephaly, respectively (Brunetti-Pierri et al., 2008). There appears to be a positive correlation between brain size and DUF1220 content in the genome within the human population and between species (Keeney et al., 2014), and recent findings suggest that DUF1220 is expressed in the ventricular zone primarily during cortical neurogenesis in the human fetal brain and promotes proliferation when expressed in vitro in neural stem cells (Keeney et al., 2015).
All of the protein-coding changes discussed above stem from modifications of existing genes. Human-specific protein-coding genes created de novo from noncoding sequence are rare. Estimates on the number of de novo protein-coding genes present only in humans vary widely, with up to 60 genes proposed (Knowles and McLysaght, 2009; McLysaght and Guerzoni, 2015; Guerzoni and McLysaght. 2016; Wu et al., 2011). These genes are largely functionally uncharacterized but show a tendency for increased expression in the brain and testes (Wu et al., 2011; Xie et al., 2012), although a recent analysis of de novo genes (which was not restricted to protein-coding genes) did not find evidence for brain-enriched expression (Ruiz-Orera et al., 2015).
Mutations to Noncoding Regulatory Regions
Many phenotypic differences between humans and NHPs, including those involving the nervous system, may be rooted in regulatory variation. A meta-analysis of positive selection in coding and noncoding regions of the genome has provided support for this idea, as neural genes are enriched for regulatory evolution (Haygood et al., 2010). In addition, comparative genomics has facilitated the identification of putative regulatory regions marked by exceptional change in the human lineage. Collectively, genome-wide scans have revealed more than 2,000 non-coding regions that, while highly conserved in other species, have accumulated a remarkable number of substitutions in humans (Bird et al., 2007; Kim and Pritchard, 2007; Lindblad-Toh et al., 2011; Pollard et al., 2006a; Pollard et al., 2006b; Prabhakar et al., 2006). These studies provide a set of putative regulatory elements that, due to their pattern of sequence evolution, are candidates for producing human-specific gene regulation, which may ultimately lead to human-specific phenotypes, including unique features of human neurodevelopment. Because these human-specific regions are identified from genomic comparisons, additional information is needed to infer the spatiotemporal context in which any given sequence operates. For example, several studies have reported that differentially expressed genes across brain regions disproportionately neighbor conserved noncoding sequences that display human-specific acceleration (Lambert et al., 2011; Johnson et al., 2009; Miller et al., 2014). However, a recent study suggests that this association is confounded by the number of conserved noncoding sequences that a gene neighbors (Meyer et al., 2017).
One of the accelerated regions identified, HAR1, lies within two overlapping RNA genes, HAR1F and HAR1R. Interestingly, HAR1F is expressed in Cajal-Retzius neurons of the developing human cortex (Pollard et al., 2006b), but the functional role of HAR1F and the consequences of the human-specific acceleration within the gene are still unknown. Although HAR1 falls inside RNA genes, many human-accelerated regions likely mark enhancers. A combined analysis of human-accelerated regions considered a diverse set of genomic data (such as sequence composition, transcription factor binding sites, and chromatin state) and predicted that at least 30% of these regions act as developmental enhancers (Capra et al., 2013). Using enhancer assays in transgenic mice, several studies have identified cases where a human-accelerated region, compared to the orthologous region from other species, drives a distinct pattern of expression of a reporter gene in the developing mouse brain (Boyd et al., 2015; Capra et al., 2013; Kamm et al., 2013a; Oksenberg et al., 2013). Another recent study found that some rare human mutations occurring in specific human-accelerated regions were associated with autism spectrum disorder (Doan et al., 2016), further supporting the functional relevance for particular accelerated sequences.
The highest density of human accelerated noncoding regions occurs within the gene NPAS3 (Kamm et al., 2013b), a gene coding for a transcription factor involved in brain development (Brunskill et al., 2005). Of the 14 accelerated regions, all of which reside in introns of NPAS3, transgenic assays in zebrafish provided evidence that 11 were capable of driving reproducible expression of a reporter gene in the CNS. Focusing on one of these regions, Kamm et al. found that the orthologous sequences from mouse and chimpanzee drove LacZ expression in a pattern similar to endogenous NPAS3 expression in the mouse CNS. The human-accelerated sequence, by contrast, drove LacZ expression in an extended region that included the developing anterior telencephalon (Figure 5; Kamm et al., 2013a). This example is among the currently small catalog of human-specific changes in regulatory regions that likely generate human-specific expression patterns of genes that are important in brain development.
In a search for conserved sequences with human-specific deletions instead of an accelerated rate of base-pair substitutions, McLean et al. (2011) identified over 500 noncoding regions that are conserved in other mammals and deleted in humans. As with accelerated sequences, transgenic enhancer assays have provided evidence for the relevance of a human-specific deletion in brain development. The chimpanzee and mouse versions of a sequence that was deleted near GADD45G are able to drive expression of a reporter gene in the subventricular zone of developing mice (Figure 5). This finding raises the possibility that a human-specific deletion of this enhancer contributes to the expansion of the human neocortex.
While expression assays are a useful step in linking genomic variants to phenotypic change, only recently has a study carried out more extensive functional characterization of a human-specific regulatory region. Boyd et al. (2015) identified a human accelerated region that functions as an enhancer in the neocortex during prenatal development. First, they showed that the human enhancer drives the expression of a LacZ reporter more robustly and in an earlier developmental time than the homologous chimpanzee sequence (Figure 5). Then, they determined that the enhancer regulates its nearest gene, FZD8, a member of the frizzled gene family, which produces receptors for the WNT proteins. The authors generated transgenic mice with FZD8 under the control of either the human or chimpanzee enhancer. Mice with the human enhancer had neural progenitors with a faster cell cycle and displayed an increase in brain size.
To date, many genome-wide scans have been performed using a wide range of methods and species to catalog human-specific sequence changes, and there are still changes that remain to be described. For example, a recent analysis of tandem repeats between humans and NHPs found that genes containing repeats show higher expression divergence between species, including in brain samples (Bilgin Sonay et al., 2015). Repeats that are conserved in NHPs but different in human (either fixed across humans or polymorphic) show enrichment for neural-development-related categories. However, while there are still sequence-level differences to explore, particularly as technological advances allow for investigation of complex regions of the genome, more extended experimental work is needed to gain a more comprehensive understanding of the role of human-specific sequences in human brain specializations.
Changes in Patterns of Developmental and Adult Gene Expression
Transcriptome profiling of tissues provides an additional level of information that can be used to prioritize genes for functional studies of human-specific gene regulation during neurodevelopment. Recent comprehensive analyses of gene expression across multiple human brain regions and time points have revealed that gene expression is dynamically regulated across brain regions and time (Colantuoni et al., 2011; Johnson et al., 2009; Kang et al., 2011; Miller et al., 2014; Pletikos et al., 2014), most prominently during early and mid-fetal development (Johnson et al., 2009; Miller et al., 2014; Pletikos et al., 2014), a crucial time in the formation of neural circuits. Within the midfetal neocortex, robust inter-regional and areal transcriptional differences were observed at the level of individual genes, as well as groups of highly co-expressed genes (modules), and included specific patterns of expression demarcating prospective prefrontal and perisylvian areas, which are involved in some of the most distinctly human aspects of cognition and behavior (Figure 4A) (Johnson et al., 2009; Kwan et al., 2012; Miller et al., 2014; Pletikos et al., 2014). Moreover, these studies have also revealed differences among humans, NHPs, and rodents in the expression of certain genes and proteins previously implicated in neurodevelopment (Figure 4B). Together, these results suggest that early and mid fetal periods are key to exploring the development of human brain specializations and that human-specific, and likely transient, changes in the spatiotemporal expression of certain genes during these periods may play a role in the unique features of human neural circuit development.
Most studies comparing gene expression between humans and NHPs have analyzed transcriptional differences in adult samples. This is largely due to the lack of high-quality prenatal and early postnatal tissues, especially from great apes. Given the difficulty of interpreting adult studies in the context of brain development, we will only briefly summarize these adult studies. Some of these comparative analyses have reported that there are more genes upregulated in the human brain compared to NHP brain, but not in other examined tissues (Cáceres et al., 2003; Gu and Gu, 2003; Khaitovich et al., 2004). However, these findings were not corroborated by other studies, which have reported that gene expression in the human brain is not more divergent (Hsieh et al., 2003) and that there is no bias for upregulated expression in the human brain (Uddin et al., 2004). Other works reported several interesting findings on human-specific differences in the expression of genes involved in aerobic metabolism (Babbitt et al., 2011; Uddin et al., 2008) and on genes that are organized into modules of co-expression that are specific to humans (Konopka et al., 2012; Oldham et al., 2006), as well as a surprising conservation of noncoding RNAs among the analyzed species (Babbitt et al., 2010).
Although it is nearly impossible to comprehensively study gene expression variation among developing primates, especially the great apes, several studies have made progress in identifying human-specific differences. These studies highlighted groups of genes with developmental time-shifts and focused on neotenic features (Somel et al., 2009), miRNA regulation (Hu et al., 2011; Somel et al., 2011), RNA editing (Li et al., 2013), and transcription factor regulation (Liu et al., 2012) of groups of co-expressed genes. Inter-species expression differences were found to be more pronounced in the human prefrontal cortex than in the cerebellum. Though intriguing, this finding is difficult to interpret because no other neocortical areas were examined and because the analyses were limited to postnatal development (except Liu et al. [2012], which includes prenatal macaque samples). According to Kang et al. (2011) and Pletikos et al. (2014), human neocortical areas are more similar in terms of expression during postnatal development, suggesting that the difference observed between the cerebellum and prefrontal cortex might not be specific to the prefrontal cortex. Extending transcriptional evolution studies to prenatal stages, when neocortical regions are most distinct from each other in terms of gene expression, will likely be essential for capturing the transcriptional differences that are important for unique features of human neurodevelopment.
Beyond a global characterization of spatiotemporal gene expression differences across species, these studies are useful for selecting promising genes for future functional studies. Alone, it is difficult to interpret gene-expression differences between species for a given gene because these differences may be due to variation in environment or tissue quality, among other confounding variables. Furthermore, even if the difference in expression is biological, it may not have an effect on a phenotypic outcome. However, despite these limitations, genome-wide expression profiling provides a valuable snapshot that can be used to generate hypotheses and steer functional studies. One such candidate is the gene coding the transcription factor MEF2A, which appears to regulate a module of co-expressed genes involved in synaptogenesis (Liu et al., 2012), highlighting the fact that alteration of the expression time course of a single transcription factor may drive differences in crucial developmental processes. Genes in this module are expressed later in human development than in other NHPs, concurrent with the expression of MEF2A. These expression changes match the delay of synaptogenesis observed in humans (Petanjek et al., 2011). Interestingly, the comparison of the human, Neanderthal, chimpanzee, and macaque MEF2A locus has shown that there is an excess of SNPs in the human lineage in an upstream region to the gene, indicating that this region is under positive selection in the human lineage (Liu et al., 2012). Given these findings, MEF2A is an interesting candidate for functional characterization that leverages mouse genetics.
Changes in Patterns of Developmental and Adult Regulatory Marks
In addition to transcriptome data, several contemporary techniques provide valuable information for comparing gene regulation across species. Chromatin immunoprecipitation sequencing (ChIP-seq) enables a genome-wide survey of genetic marks that signal regulatory activity of putative promoter and enhancers (Reilly and Noonan, 2016). Recent studies have compared promoter and/or enhancer activity in the brain of humans and closely related species: one capturing differences during the time of corticogenesis in human, macaque, and mouse brains (Reilly et al., 2015); one examining numerous brain regions from adult human, chimpanzee, and macaque samples (Vermunt et al., 2016); and a third assessing cell-type-specific promoter landscapes in prefrontal cortex of human, chimpanzee, and macaque brains (Shulha et al., 2012). These inter-species comparisons of active regulatory elements in different tissues have so far demonstrated that promoters and enhancers are largely conserved between humans and primates with respect to position in spite of sequence divergence (Cotney et al., 2013; Prescott et al., 2015; Vermunt et al., 2016) and that a subset of positionally conserved marks appear to be either tissue-specific, species-specific, or both in terms of activity, as marks for these enhancers (and to a lesser extent promoters) have been observed in disparate tissues and among different brain areas from one species to another (Cotney et al., 2013; Vermunt et al., 2016). Conserved epigenetic marks can also be cell type specific, as neuronal epigenomes in prefrontal cortex are more similar across human and NHP (chimpanzee and macaque) species than they are to non-neuronal epigenomes within the same species (Shulha et al., 2012). These studies emphasize the need to include a closely related species to identify human-specific gain or loss of enhancer and promoter activity. While ~1400 enhancers and ~90 promoters are specifically enriched in humans when comparing adult human and macaques, only 139 enhancers and 7 promoters remain as human-specific gains when introducing chimpanzee in the comparison (Vermunt et al., 2016). This classification cannot be performed in the embryonic enhancer catalog because data from chimpanzees are absent, and thus, prenatal human-specific enhancer activity is yet to be uncovered. Nevertheless, those embryonic enhancers and promoters either specifically active or absent in the human samples involve genes in co-expression modules associated with neuronal proliferation and differentiation (Reilly et al., 2015). In these studies, human-accelerated sequence overlapped with some human-specific enhancers, but an overall enrichment was not observed. These new catalogs of putative human-specific enhancers active at specific stages of development and in different brain regions open a novel research space for testing hypotheses linking genes to human-acquired brain differences.
Promoters, enhancers, and other regulatory elements are subject to dynamic regulation in part through the methylation of the ~1 billion cytosines in the human genome, most notably the ~28 million CpG sites (Feng et al., 2005; Spiers et al., 2015). This methylation, driven by the differential expression of DNA methyltransferases, can result in species-, tissue-, and cell-type-specific patterns of gene expression (Hernando-Herraez et al., 2015). For example, methylation in the brain is particularly pronounced in adult neurons as compared to non-neuronal populations (Kozlenkov et al., 2014; Lister et al., 2013). Promoter methylation is also associated with reduced gene expression, and comparison of human and chimpanzee liver, heart, and kidney tissues suggests that 12%–18% of the differential gene expression between these two species can be explained by differences in promoter methylation (Pai et al., 2011).
A comparison of methylation at putative regulatory regions of 36 genes found that CpG sites appear to be more methylated in the human brain than in the chimpanzee brain (Enard et al., 2004). On the other hand, promoter regions of several genes were shown to be significantly less methylated in the human brain than in the chimpanzee brain (Zeng et al., 2012). Although no obvious global differences were found, it was reported that humans and NHPs have different methylation states at both DNA (Farcas et al., 2009; Schneider et al., 2014; Schneider et al., 2012) and histone levels (Shulha et al., 2012) for specific genes involved in neuronal function. CNTNAP2, a transcriptional target of FOXP2 implicated in neurological and psychiatric disorders, including language impairments (Graham and Fisher, 2013; Rodenas-Cuadrado et al., 2014), is an example of a neurodevelopmental gene where methylation differences between human and chimpanzees have been studied in detail. Like FOXP2, CNTNAP2 contains strong signals of recent positive selection in various human populations (Ayub et al., 2013). CNTNAP2 was found to be differentially expressed between humans and chimpanzees, largely due to changes in one CNTNAP2 isoform, which were accompanied by widespread gene methylation differences (Schneider et al., 2014).
Functional Modeling of Human Brain Development and Evolution
There are a number of methods available for investigating these findings in experimental systems, many of which have been discussed above. The most time-efficient method is in vitro characterization, but the separation of the cellular system from a realistic biological context, especially when it comes to the modeling of specific neural circuits, makes the results difficult to interpret (for example, see Heissig et al., 2005; Shim et al., 2012). Genome editing and transgenesis in mice, on the other hand, allow for evolutionary hypotheses to be tested in the context of the whole organism using a wide range of genetic tools. Species- and lineage-specific gene variants or regulatory regions can be added or replace endogenous sequences in a model organism (so far mainly mice), allowing for in vivo characterization of the feature (for example, see Boyd et al., 2015; Cotney et al., 2013; Enard et al., 2009; Kwan et al., 2012; Shim et al., 2012).
Developments in iPSC research provide another method for evolutionary and functional analyses (Gallego Romero et al., 2015; Hrvoj-Mihic et al., 2014; Wunderlich et al., 2014). Methods for generating iPSCs and directing their differentiation into specific human neural cell types and cell culture systems, including organoids, have provided much-needed tools for comparative studies of neurodevelopment in humans and NHPs (Gallego Romero et al., 2015; Hrvoj-Mihic et al., 2014; Otani et al., 2016; Prescott et al., 2015; Wunderlich et al., 2014). Recently, Prescott et al. (2015) differentiated cranial neural crest cells from iPSCs derived from human and chimpanzee fibroblasts to study the evolution of human craniofacial morphology. The authors compared genome-wide enhancer and promoter activity and identified a novel motif enriched in regions with species-biased activity. This study provides a prime example of ‘‘cellular anthropology’’ (Prescott et al., 2015) and highlights newfound methods for the identification of putatively relevant genomic changes between species during development.
The functional studies discussed above are encouraging attempts to provide experimental evidence of the molecular mechanisms involved in human brain evolution and underscore the challenges of experimentally testing such evolutionary hypotheses. While mouse models provide the most powerful tools for genomic manipulation in mammals, lineages leading to humans and mice diverged 75 million years ago (Mouse Genome Sequencing Consortium et al., 2002). This difference in genetic background complicates the interpretation of functional studies, as genes evolve in concert with the rest of the genome and an alteration or loss of function can be due the absence of any number of unknown players. Thus, while a positive experimental result provides support for a given interpretation, a negative result is ambiguous. Aside from the difficulties of interpreting the results of mouse models specifically, demonstrating that a mutation has a functional consequence that may have been evolutionarily selected is a necessary step to supporting adaptive evolution, but by itself does not provide strong evidence that the mutation was selected in the context hypothesized (Nielsen, 2009). Therefore, these experimental studies, while far from conclusive, should be viewed as sources of information that serve to either increase or decrease the probability of a given hypothesis.
Conclusions and Future Directions
Multiple lines of evidence show that key aspects of human brain organization and development scale as expected, while cognition does not. Even though the way our brain is built is not exceptional, we differ by a unique combination of mental abilities, combined with higher general intelligence. While higher general intelligence compared to NHPs may likely be the product of increased relative and absolute neuron number, especially in the neocortex, our superiority in specific cognitive abilities is likely the result of mosaic structural rewiring and molecular reorganization of specific neural circuits and cell types. These observations, as well as the study of brain size and organization in extinct primates (Falk, 2014; Neubauer, 2014; Pearce et al., 2013; Wood and Baker, 2011; Zollikofer and De León, 2013), suggest that many independent changes rather than one single defining event have occurred across the nervous system in the human lineage. It is also likely that these changes have affected many, if not all, brain structures and levels of organization.
Advancements in high-throughput molecular biology and biochemistry technologies have enabled unprecedented identification of human-specific features that may contribute to unique aspects of human brain development. For example, several recent studies have mapped gene expression to specific cell types during brain development, using various single-cell sequencing approaches (Johnson et al., 2015; Lui et al., 2014; Onorati et al., 2016; Pollen et al., 2015). The cellular-level resolution of differential expression possible through these approaches, both among neural progenitor cells and subsequent postmitotic populations, allows a better understanding of the myriad fine-tuned processes governing early brain development. High-throughput techniques also permit, to some degree, the systematization of key aspects of the functional characterization of human specific genomic elements. This is the case for the massively parallel reporter assays, which enable simultaneous testing of the regulatory activity of hundreds of thousands of putative regulatory elements (Shlyueva et al., 2014). Continued efforts are both important and critical in many areas, including characterizing the extent of human diversity, expanding high-quality annotations of NHP genomes, and increasing the coverage of gene expression profiles across more tissues and time points. Therefore, within the context of human CNS development and evolution, it will be valuable to profile transcriptional dynamics in developmental NHP samples from an extended number of CNS regions. Most of the recent efforts have focused on a few regions of the forebrain and cerebellum, while almost no data are available on the majority of other regions of the human or NHP CNS.
Furthermore, there is an obvious lack of comparative studies on diverse types of neuronal and glial cells across primates. So far, most of the effort has been largely focused on neocortical neurons. However, astrocytes, oligodendrocytes, and microglia are relevant in many aspects of CNS function and represent half of the cellular composition of the nervous system. It is thus a promising and almost uncharted field that must be explored to generate a more complete and integrated picture of the human CNS specializations.
In addition to NHP comparisons, advances in the acquisition and processing of genomic material from archaic humans (Neanderthals and Denisovans) can improve temporal resolution of human-specific sequence change. Initial comparisons of high-quality modern human and Neanderthal/Denisovan genomes identified ~31,000 single nucleotide changes and ~4,000 indels present and fixed in Homo sapiens but absent from the genomes of the other two human species. These changes gave rise to only 87 proteins containing amino-acid substitutions. Of note, the expression of these proteins was found to be enriched in the ventricular zone during cortical development (Prüfer et al., 2014). In addition, around 3,000 of the changes, including both single nucleotide changes and indels, exclusively found and fixed in Homo sapiens were predicted to have an effect on gene regulation. Although Neanderthals and Denisovans carry the derived state for the great majority of the substitution events within human accelerated regions, around 8% of them retain the ancestral state in the archaic genomes (Burbano et al., 2012; Hubisz and Pollard 2014). Some of them, for instance, are found in an intronic HAR of AUTS2, a gene associated with several neurological phenotypes, in a region also showing strong evidence of a selective sweep that occurred in modern humans after the split with Neanderthals (Green et al., 2010; Oksenberg et al., 2013). Other genes involved in cognition and behavior, such as DYRK1A, NRG3, and CADPS2, were also likely subject to such selective sweeps. Recent studies offer other intriguing observations, such as the fact that genes expressed in developing cortex and adult striatum are significantly depleted in introgressed archaic genetic material (Vernot et al., 2016). One of these regions of introgression deserts includes FOXP2.
Finally, the emerging field of imaging genetics, which allows the identification of genetic loci explaining variation in brain structures and functions among extant humans (Hibar et al., 2015), might offer new candidate genetic elements for generating evolutionary hypotheses about how and when human-specific neurodevelopmental events took place. Furthermore, it will be necessary to develop in vitro cellular systems that are able to model the complexities of human neurodevelopment more accurately. Building on work that has characterized human-specific changes at multiple levels, it will be important for future work to integrate this information and shift focus to detailed experimental studies in order to gain a better understanding of the mechanisms underlying the development and evolutionary specializations of the human nervous system.
Acknowledgments
We thank members of our laboratory for thoughtful discussions and comments on the manuscript and Amanda Gautier for illustrating the primate brains in Figure 1. Also, we would like to thank reviewers for their insightful comments, as these comments led to an improved manuscript. We apologize to all colleagues whose relevant studies were not cited because of space limitations, the broad scope of this review, and the emphasis on recent studies. Work in our laboratory was supported by the NIH, the Kavli Foundation, the James S. McDonnell Foundation, and the Simons Foundation.
References
- Aiello LC, Wheeler P. The expensive-tissue hypothesis: the brain and the digestive system in human and primate evolution. Curr. Anthropol. 1995;36:199–221. [Google Scholar]
- Allman JM, Tetreault NA, Hakeem AY, Manaye KF, Semendeferi K, Erwin JM, Park S, Goubert V, Hof PR. The von Economo neurons in frontoinsular and anterior cingulate cortex in great apes and humans. Brain Struct. Funct. 2010;214:495–517. doi: 10.1007/s00429-010-0254-0. [DOI] [PubMed] [Google Scholar]
- Almécija S, Tallman M, Alba DM, Pina M, Moyá-Solá S, Jungers WL. The femur of Orrorin tugenensis exhibits morphometric affinities with both Miocene apes and later hominins. Nat. Commun. 2013;4:2888. doi: 10.1038/ncomms3888. [DOI] [PubMed] [Google Scholar]
- Alonso CR, Wilkins AS. The molecular elements that underlie developmental evolution. Nat. Rev. Genet. 2005;6:709–715. doi: 10.1038/nrg1676. [DOI] [PubMed] [Google Scholar]
- Anderson JM, Gilmore R, Roper S, Crosson B, Bauer RM, Nadeau S, Beversdorf DQ, Cibula J, Rogish M, 3rd, Kortencamp S, et al. Conduction aphasia and the arcuate fasciculus: A reexamination of the Wernicke-Geschwind model. Brain Lang. 1999;70:1–12. doi: 10.1006/brln.1999.2135. [DOI] [PubMed] [Google Scholar]
- Antonacci F, Dennis MY, Huddleston J, Sudmant PH, Steinberg KM, Rosenfeld JA, Miroballo M, Graves TA, Vives L, Malig M, et al. Palindromic GOLGA8 core duplicons promote chromosome 15q13.3 microdeletion and evolutionary instability. Nat. Genet. 2014;46:1293–1302. doi: 10.1038/ng.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayub Q, Yngvadottir B, Chen Y, Xue Y, Hu M, Vernes SC, Fisher SE, Tyler-Smith C. FOXP2 targets show evidence of positive selection in European populations. Am. J. Hum. Genet. 2013;92:696–706. doi: 10.1016/j.ajhg.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbitt CC, Fedrigo O, Pfefferle AD, Boyle AP, Horvath JE, Furey TS, Wray GA. Both noncoding and protein-coding RNAs contribute to gene expression evolution in the primate brain. Genome Biol. Evol. 2010;2:67–79. doi: 10.1093/gbe/evq002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbitt CC, Warner LR, Fedrigo O, Wall CE, Wray GA. Genomic signatures of diet-related shifts during human origins. Proc. Biol. Sci. 2011;278:961–969. doi: 10.1098/rspb.2010.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae BI, Jayaraman D, Walsh CA. Genetic changes shaping the human brain. Dev. Cell. 2015;32:423–434. doi: 10.1016/j.devcel.2015.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken TE, Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Dalley RA, Royall JJ, Lemon T, et al. A comprehensive transcriptional map of primate brain development. Nature. 2016;535:367–375. doi: 10.1038/nature18637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton RA, Venditti C. Rapid evolution of the cerebellum in humans and other great apes. Curr. Biol. 2014;24:2440–2444. doi: 10.1016/j.cub.2014.08.056. [DOI] [PubMed] [Google Scholar]
- Bianchi S, Stimpson CD, Bauernfeind AL, Schapiro SJ, Baze WB, McArthur MJ, Bronson E, Hopkins WD, Semendeferi K, Jacobs B, et al. Dendritic morphology of pyramidal neurons in the chimpanzee neocortex: regional specializations and comparison to humans. Cereb. Cortex. 2013;23:2429–2436. doi: 10.1093/cercor/bhs239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgin Sonay T, Carvalho T, Robinson MD, Greminger MP, Krützen M, Comas D, Highnam G, Mittelman D, Sharp A, Marques-Bonet T, Wagner A. Tandem repeat variation in human and great ape populations and its impact on gene expression divergence. Genome Res. 2015;25:1591–1599. doi: 10.1101/gr.190868.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird CP, Stranger BE, Liu M, Thomas DJ, Ingle CE, Beazley C, Miller W, Hurles ME, Dermitzakis ET. Fast-evolving noncoding sequences in the human genome. Genome Biol. 2007;8:R118. doi: 10.1186/gb-2007-8-6-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blinkov SMI, Glezer II. The human brain in figures and tables; a quantitative handbook. New York: Basic Books; 1968. [Google Scholar]
- Bogin B. Adolescence in evolutionary perspective. Acta Paediatr. Suppl. 1994;406:29–35. doi: 10.1111/j.1651-2227.1994.tb13418.x. discussion 36. [DOI] [PubMed] [Google Scholar]
- Borgstein J, Grootendorst C. Clinical picture: half a brain. Lancet. 2002;359:473. doi: 10.1016/s0140-6736(02)07676-6. [DOI] [PubMed] [Google Scholar]
- Boyd JL, Skove SL, Rouanet JP, Pilaz L-J, Bepler T, Gordân R, Wray GA, Silver DL. Human-chimpanzee differences in a FZD8 enhancer alter cell-cycle dynamics in the developing neocortex. Curr. Biol. 2015;25:772–779. doi: 10.1016/j.cub.2015.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, Sahoo T, Lalani SR, Graham B, Lee B, Shinawi M, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephalyor macrocephaly and developmental and behavioral abnormalities. Nat. Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill EW, Ehrman LA, Williams MT, Klanke J, Hammer D, Schaefer TL, Sah R, Dorn GW, 2nd, Potter SS, Vorhees CV. Abnormal neurodevelopment, neurosignaling and behaviour in Npas3-deficient mice. Eur. J. Neurosci. 2005;22:1265–1276. doi: 10.1111/j.1460-9568.2005.04291.x. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Krienen FM. The evolution of distributed association networks in the human brain. Trends Cogn. Sci. 2013;17:648–665. doi: 10.1016/j.tics.2013.09.017. [DOI] [PubMed] [Google Scholar]
- Burbano HA, Green RE, Maricic T, Lalueza-Fox C, de la Rasilla M, Rosas A, Kelso J, Pollard KS, Lachmann M, Pääbo S. Analysis of human accelerated DNA regions using archaic hominin genomes. PLoS ONE. 2012;7:e32877. doi: 10.1371/journal.pone.0032877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush EC, Allman JM. The scaling of frontal cortex in primates and carnivores. Proc. Natl. Acad. Sci. USA. 2004;101:3962–3966. doi: 10.1073/pnas.0305760101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Hubisz MT, Glanowski S, Tanenbaum DM, White TJ, Sninsky JJ, Hernandez RD, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- Cáceres M, Lachuer J, Zapala MA, Redmond JC, Kudo L, Geschwind DH, Lockhart DJ, Preuss TM, Barlow C. Elevated gene expression levels distinguish human from non-human primate brains. Proc. Natl. Acad. Sci. USA. 2003;100:13030–13035. doi: 10.1073/pnas.2135499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call J, Tomasello M. Does the chimpanzee have a theory of mind? 30 years later. Trends Cogn. Sci. 2008;12:187–192. doi: 10.1016/j.tics.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Capra JA, Erwin GD, McKinsey G, Rubenstein JLR, Pollard KS. Many human accelerated regions are developmental enhancers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20130025. doi: 10.1098/rstb.2013.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Catani M, Mesulam M. The arcuate fasciculus and the disconnection theme in language and aphasia: history and current state. Cortex. 2008;44:953–961. doi: 10.1016/j.cortex.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catani M, Dell’acqua F, Vergani F, Malik F, Hodge H, Roy P, Valabregue R, Thiebaut de Schotten M. Short frontal lobe connections of the human brain. Cortex. 2012;48:273–291. doi: 10.1016/j.cortex.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Charrier C, Joshi K, Coutinho-Budd J, Kim J-E, Lambert N, de Marchena J, Jin W-L, Vanderhaeghen P, Ghosh A, Sassa T, Polleux F. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell. 2012;149:923–935. doi: 10.1016/j.cell.2012.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FC, Li WH. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 2001;68:444–456. doi: 10.1086/318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Ventura M, She X, Khaitovich P, Graves T, Osoegawa K, Church D, DeJong P, Wilson RK, Pääbo S, et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Clark AG, Glanowski S, Nielsen R, Thomas PD, Kejariwal A, Todd MA, Tanenbaum DM, Civello D, Lu F, Murphy B, et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DR, Kleinman JE. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CE, Turner EC, Sawyer EK, Reed JL, Young NA, Flaherty DK, Kaas JH. Cortical cell and neuron density estimates in one chimpanzee hemisphere. Proc. Natl. Acad. Sci. USA. 2016;113:740–745. doi: 10.1073/pnas.1524208113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J, Leng J, Yin J, Reilly SK, DeMare LE, Emera D, Ayoub AE, Rakic P, Noonan JP. The evolution of lineage-specific regulatory activities in the human embryonic limb. Cell. 2013;154:185–196. doi: 10.1016/j.cell.2013.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JM, Searles VB, Anderson N, Keeney J, Dumas L, Sikela JM. DUF1220 dosage is linearly associated with increasing severity of the three primary symptoms of autism. PLoS Genet. 2014;10:e1004241. doi: 10.1371/journal.pgen.1004241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon TW. Rethinking mammalian brain evolution. Am. Zool. 1990;30:629–705. [Google Scholar]
- Deaner RO, Isler K, Burkart J, van Schaik C. Overall brain size, and not encephalization quotient, best predicts cognitive ability across non-human primates. Brain Behav. Evol. 2007;70:115–124. doi: 10.1159/000102973. [DOI] [PubMed] [Google Scholar]
- Defelipe J. The evolution of the brain, the human nature of cortical circuits, and intellectual creativity. Front. Neuroanat. 2011;5:29. doi: 10.3389/fnana.2011.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay C, Kennedy H, Kosik KS. The outer subventricular zone and primate-specific cortical complexification. Neuron. 2015;85:683–694. doi: 10.1016/j.neuron.2014.12.060. [DOI] [PubMed] [Google Scholar]
- Dekaban AS. Changes in brain weights during the span of human life: relation of brain weights to body heights and body weights. Ann. Neurol. 1978;4:345–356. doi: 10.1002/ana.410040410. [DOI] [PubMed] [Google Scholar]
- Dennis MY, Nuttle X, Sudmant PH, Antonacci F, Graves TA, Nefedov M, Rosenfeld JA, Sajjadian S, Malig M, Kotkiewicz H, et al. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell. 2012;149:912–922. doi: 10.1016/j.cell.2012.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin AM, Cross JH, Harkness W, Chong WK, Harding B, Vargha-Khadem F, Neville BG. Clinical outcomes of hemispherectomy for epilepsy in childhood and adolescence. Brain. 2003;126:556–566. doi: 10.1093/brain/awg052. [DOI] [PubMed] [Google Scholar]
- Doan RN, Bae BI, Cubelos B, Chang C, Hossain AA, Al-Saad S, Mukaddes NM, Oner O, Al-Saffar M, Balkhy S, et al. Homozygosity Mapping Consortium for Autism (2016). Mutations in human accelerated regions disrupt cognition and social behavior. Cell. 167:341–354. doi: 10.1016/j.cell.2016.08.071. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorus S, Vallender EJ, Evans PD, Anderson JR, Gilbert SL, Mahowald M, Wyckoff GJ, Malcom CM, Lahn BT. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell. 2004;119:1027–1040. doi: 10.1016/j.cell.2004.11.040. [DOI] [PubMed] [Google Scholar]
- El-Hattab AW, Smolarek TA, Walker ME, Schorry EK, Immken LL, Patel G, Abbott M-A, Lanpher BC, Ou Z, Kang S-HL, et al. Redefined genomic architecture in 15q24 directed by patient deletion/duplication breakpoint mapping. Hum. Genet. 2009;126:589–602. doi: 10.1007/s00439-009-0706-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston GN, Benavides-Piccione R, Elston A, Manger PR, Defelipe J. Pyramidal cells in prefrontal cortex of primates: marked differences in neuronal structure among species. Front. Neuroanat. 2011;5:2. doi: 10.3389/fnana.2011.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard W. The molecular basis of human brain evolution. Curr. Biol. 2016;26:R1109–R1117. doi: 10.1016/j.cub.2016.09.030. [DOI] [PubMed] [Google Scholar]
- Enard W, Fassbender A, Model F, Adorján P, Pääbo S, Olek A. Differences in DNA methylation patterns between humans and chimpanzees. Curr. Biol. 2004;14:R148–R149. [PubMed] [Google Scholar]
- Enard W, Gehre S, Hammerschmidt K, Hölter SM, Blass T, Somel M, Brückner MK, Schreiweis C, Winter C, Sohr R, et al. A humanized version of Foxp2 affects corticobasal ganglia circuits in mice. Cell. 2009;137:961–971. doi: 10.1016/j.cell.2009.03.041. [DOI] [PubMed] [Google Scholar]
- Eriksen N, Pakkenberg B. Total neocortical cell number in the mysticete brain. Anat. Rec. (Hoboken) 2007;290:83–95. doi: 10.1002/ar.20404. [DOI] [PubMed] [Google Scholar]
- Falk D. Interpreting sulci on hominin endocasts: old hypotheses and new findings. Front. Hum. Neurosci. 2014;8:134. doi: 10.3389/fnhum.2014.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farcas R, Schneider E, Frauenknecht K, Kondova I, Bontrop R, Bohl J, Navarro B, Metzler M, Zischler H, Zechner U, et al. Differences in DNA methylation patterns and expression of the CCRK gene in human and nonhuman primate cortices. Mol. Biol. Evol. 2009;26:1379–1389. doi: 10.1093/molbev/msp046. [DOI] [PubMed] [Google Scholar]
- Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J. Neurosci. Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- Feuillet L, Dufour H, Pelletier J. Brain of a white-collar worker. Lancet. 2007;370:262. doi: 10.1016/S0140-6736(07)61127-1. [DOI] [PubMed] [Google Scholar]
- Finlay BL, Darlington RB. Linked regularities in the development and evolution of mammalian brains. Science. 1995;268:1578–1584. doi: 10.1126/science.7777856. [DOI] [PubMed] [Google Scholar]
- Florio M, Albert M, Taverna E, Namba T, Brandl H, Lewitus E, Haffner C, Sykes A, Wong FK, Peters J, et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 2015;347:1465–1470. doi: 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- Fortna A, Kim Y, MacLaren E, Marshall K, Hahn G, Meltesen L, Brenton M, Hink R, Burgers S, Hernandez-Boussard T, et al. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004;2:E207. doi: 10.1371/journal.pbio.0020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossati M, Pizzarelli R, Schmidt ER, Kupferman JV, Stroebel D, Polleux F, Charrier C. SRGAP2 and its human-specific paralog co-regulate the development of excitatory and inhibitory synapses. Neuron. 2016;91:356–369. doi: 10.1016/j.neuron.2016.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini LF, Pollard KS. Genomic approaches to studying human-specific developmental traits. Development. 2015;142:3100–3112. doi: 10.1242/dev.120048. [DOI] [PubMed] [Google Scholar]
- Furness JB. The enteric nervous system. Malden, Mass: Black-well Pub; 2006. [Google Scholar]
- Gallego Romero I, Pavlovic BJ, Hernando-Herraez I, Zhou X, Ward MC, Banovich NE, Kagan CL, Burnett JE, Huang CH, Mitrano A, et al. A panel of induced pluripotent stem cells from chimpanzees: a resource for comparative functional genomics. eLife. 2015;4:e07103. doi: 10.7554/eLife.07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzaniga MS. Human: The science behind what makes us unique. New York: Harper Collins; 2009. [Google Scholar]
- Geschwind DH, Rakic P. Cortical evolution: judge the brain by its cover. Neuron. 2013;80:633–647. doi: 10.1016/j.neuron.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazanfar AA. Language evolution: neural differences that make a difference. Nat. Neurosci. 2008;11:382–384. doi: 10.1038/nn0408-382. [DOI] [PubMed] [Google Scholar]
- Glasser MF, Goyal MS, Preuss TM, Raichle ME, Van Essen DC. Trends and properties of human cerebral cortex: correlations with cortical myelin content. Neuroimage. 2014;93:165–175. doi: 10.1016/j.neuroimage.2013.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickstein M. Cerebellar agenesis. Brain. 1994;117:1209–1212. doi: 10.1093/brain/117.5.1209. [DOI] [PubMed] [Google Scholar]
- Goidts V, Cooper DN, Armengol L, Schempp W, Conroy J, Estivill X, Nowak N, Hameister H, Kehrer-Sawatzki H. Complex patterns of copy number variation at sites of segmental duplications: an important category of structural variation in the human genome. Hum. Genet. 2006;120:270–284. doi: 10.1007/s00439-006-0217-y. [DOI] [PubMed] [Google Scholar]
- Graham SA, Fisher SE. Decoding the genetics of speech and language. Curr. Opin. Neurobiol. 2013;23:43–51. doi: 10.1016/j.conb.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefkes C, Fink GR. The functional organization of the intraparietal sulcus in humans and monkeys. J. Anat. 2005;207:3–17. doi: 10.1111/j.1469-7580.2005.00426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Gu X. Induced gene expression in human brain after the split from chimpanzee. Trends Genet. 2003;19:63–65. doi: 10.1016/s0168-9525(02)00040-9. [DOI] [PubMed] [Google Scholar]
- Guerzoni D, McLysaght A. De novo genes arise at a slow but steady rate along the primate lineage and have been subject to incomplete lineage sorting. Genome Biol. Evol. 2016;8:1222–1232. doi: 10.1093/gbe/evw074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulden FO, Sestan N. Neurobiology: building a bigger brain. Nature. 2014;515:206–207. doi: 10.1038/515206a. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt K, Schreiweis C, Minge C, Pääbo S, Fischer J, Enard W. A humanized version of Foxp2 does not affect ultrasonic vocalization in adult mice. Genes Brain Behav. 2015;14:583–590. doi: 10.1111/gbb.12237. [DOI] [PubMed] [Google Scholar]
- Han X, Chen M, Wang F, Windrem M, Wang S, Shanz S, Xu Q, Oberheim NA, Bekar L, Betstadt S, et al. Forebrain engraftment by human glial progenitor cells enhances synaptic plasticity and learning in adult mice. Cell Stem Cell. 2013;12:342–353. doi: 10.1016/j.stem.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslam RH, Smith DW. Autosomal dominant microcephaly. J. Pediatr. 1979;95:701–705. doi: 10.1016/s0022-3476(79)80714-3. [DOI] [PubMed] [Google Scholar]
- Haygood R, Babbitt CC, Fedrigo O, Wray GA. Contrasts between adaptive coding and noncoding changes during human evolution. Proc. Natl. Acad. Sci. USA. 2010;107:7853–7857. doi: 10.1073/pnas.0911249107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht EE, Gutman DA, Bradley BA, Preuss TM, Stout D. Virtual dissection and comparative connectivity of the superior longitudinal fasciculus in chimpanzees and humans. Neuroimage. 2015;108:124–137. doi: 10.1016/j.neuroimage.2014.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffner RS, Masterton RB. The role of the corticospinal tract in the evolution of human digital dexterity. Brain Behav. Evol. 1983;23:165–183. doi: 10.1159/000121494. [DOI] [PubMed] [Google Scholar]
- Heissig F, Krause J, Bryk J, Khaitovich P, Enard W, Pääbo S. Functional analysis of human and chimpanzee promoters. Genome Biol. 2005;6:R57. doi: 10.1186/gb-2005-6-7-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert MR, Ziegler DA, Deutsch CK, O’Brien LM, Lange N, Bakardjiev A, Hodgson J, Adrien KT, Steele S, Makris N, et al. Dissociations of cerebral cortex, subcortical and cerebral white matter volumes in autistic boys. Brain. 2003;126:1182–1192. doi: 10.1093/brain/awg110. [DOI] [PubMed] [Google Scholar]
- Herculano-Houzel S. The human advantage: a new understanding of how our brain became remarkable. Cambridge, MA: The MIT Press; 2016. [Google Scholar]
- Herculano-Houzel S, Catania K, Manger PR, Kaas JH. Mammalian brains are made of these: a dataset of the numbers and densities of neuronal and nonneuronal cells in the brain of glires, primates, scandentia, eulipotyphlans, afrotherians and artiodactyls, and their relationship with body mass. Brain Behav. Evol. 2015;86:145–163. doi: 10.1159/000437413. [DOI] [PubMed] [Google Scholar]
- Herculano-Houzel S, Kaas JH, de Oliveira-Souza R. Corticalization of motor control in humans is a consequence of brain scaling in primate evolution. J. Comp. Neurol. 2016;524:448–455. doi: 10.1002/cne.23792. [DOI] [PubMed] [Google Scholar]
- Hernando-Herraez I, Garcia-Perez R, Sharp AJ, Marques-Bonet T. DNA methylation: insights into human evolution. PLoS Genet. 2015;11:e1005661. doi: 10.1371/journal.pgen.1005661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desriviéres S, Jahanshad N, Toro R, Wittfeld K, Abramovic L, Andersson M, et al. Alzheimer’s Disease Neuroimaging Initiative; CHARGE Consortium; EPIGEN; IMAGEN; SYS. Common genetic variants influence human subcortical brain structures. Nature. 2015;520:224–229. doi: 10.1038/nature14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. The locus of evolution: evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- Hofman MA. Design principles of the human brain: an evolutionary perspective. Prog. Brain Res. 2012;195:373–390. doi: 10.1016/B978-0-444-53860-4.00018-0. [DOI] [PubMed] [Google Scholar]
- Hollo-way RL. Brief communication: how much larger is the relative volume of area 10 of the prefrontal cortex in humans? Am. J. Phys. Anthropol. 2002;118:399–401. doi: 10.1002/ajpa.10090. [DOI] [PubMed] [Google Scholar]
- Horner V, Whiten A, Flynn E, de Waal FB. Faithful replication of foraging techniques along cultural transmission chains by chimpanzees and children. Proc. Natl. Acad. Sci. USA. 2006;103:13878–13883. doi: 10.1073/pnas.0606015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrvoj-Mihic B, Bienvenu T, Stefanacci L, Muotri AR, Semendeferi K. Evolution, development, and plasticity of the human brain: from molecules to bones. Front. Hum. Neurosci. 2013;7:707. doi: 10.3389/fnhum.2013.00707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrvoj-Mihic B, Marchetto MCN, Gage FH, Semendeferi K, Muotri AR. Novel tools, classic techniques: evolutionary studies using primate pluripotent stem cells. Biol. Psychiatry. 2014;75:929–935. doi: 10.1016/j.biopsych.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Hsieh W-P, Chu T-M, Wolfinger RD, Gibson G. Mixed-model reanalysis of primate data suggests tissue and species biases in oligonucleotide-based gene expression profiles. Genetics. 2003;165:747–757. doi: 10.1093/genetics/165.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HY, Guo S, Xi J, Yan Z, Fu N, Zhang X, Menzel C, Liang H, Yang H, Zhao M, et al. MicroRNA expression and regulation in human, chimpanzee, and macaque brains. PLoS Genet. 2011;7:e1002327. doi: 10.1371/journal.pgen.1002327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz MJ, Pollard KS. Exploring the genesis and functions of Human Accelerated Regions sheds light on their role in human evolution. Curr. Opin. Genet. Dev. 2014;29:15–21. doi: 10.1016/j.gde.2014.07.005. [DOI] [PubMed] [Google Scholar]
- Hutsler JJ, Lee DG, Porter KK. Comparative analysis of cortical layering and supragranular layer enlargement in rodent carnivore and primate species. Brain Res. 2005;1052:71–81. doi: 10.1016/j.brainres.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Irimia A, Chambers MC, Torgerson CM, Van Horn JD. Circular representation of human cortical networks for subject and population-level connectomic visualization. Neuroimage. 2012;60:1340–1351. doi: 10.1016/j.neuroimage.2012.01.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isler K, Van Schaik CP. How humans evolved large brains: comparative evidence. Evol. Anthropol. 2014;23:65–75. doi: 10.1002/evan.21403. [DOI] [PubMed] [Google Scholar]
- Jerison HJ. Evolution of the brain and intelligence. New York: Academic; 1973. [Google Scholar]
- Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanović D, Geschwind DH, Mane SM, State MW, Sestan N. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Wang PP, Atabay KD, Murphy EA, Doan RN, Hecht JL, Walsh CA. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat. Neurosci. 2015;18:637–646. doi: 10.1038/nn.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamm GB, Lopez-Leal R, Lorenzo JR, Franchini LF. A fast-evolving human NPAS3 enhancer gained reporter expression in the developing forebrain of transgenic mice. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013a;368:20130019. doi: 10.1098/rstb.2013.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamm GB, Pisciottano F, Kliger R, Franchini LF. The developmental brain gene NPAS3 contains the largest number of accelerated regulatory sequences in the human genome. Mol. Biol. Evol. 2013b;30:1088–1102. doi: 10.1093/molbev/mst023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AMM, Pletikos M, Meyer KA, Sedmak G, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh S, Beyene Y, Itaya T, Hyodo H, Hyodo M, Yagi K, Gouzu C, WoldeGabriel G, Hart WK, Ambrose SH, et al. New geological and palaeontological age constraint for the gorilla-human lineage split. Nature. 2016;530:215–218. doi: 10.1038/nature16510. [DOI] [PubMed] [Google Scholar]
- Keeney JG, Dumas L, Sikela JM. The case for DUF1220 domain dosage as a primary contributorto anthropoid brain expansion. Front. Hum. Neurosci. 2014;8:427. doi: 10.3389/fnhum.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney JG, Davis JM, Siegenthaler J, Post MD, Nielsen BS, Hopkins WD, Sikela JM. DUF1220 protein domains drive proliferation in human neural stem cells and are associated with increased cortical volume in anthropoid primates. Brain Struct. Funct. 2015;220:3053–3060. doi: 10.1007/s00429-014-0814-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kety SS, Schmidt CF. The nitrous oxide method for the quantitative determination of cerebral blood flow in man: Theory, procedure and normal values. J. Clin. Invest. 1948;27:476–483. doi: 10.1172/JCI101994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Muetzel B, She X, Lachmann M, Hellmann I, Dietzsch J, Steigele S, Do H-H, Weiss G, Enard W, et al. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 2004;14:1462–1473. doi: 10.1101/gr.2538704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Lockstone HE, Wayland MT, Tsang TM, Jayatilaka SD, Guo AJ, Zhou J, Somel M, Harris LW, Holmes E, et al. Metabolic changes in schizophrenia and human brain evolution. Genome Biol. 2008;9:R124. doi: 10.1186/gb-2008-9-8-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Pritchard JK. Adaptive evolution of conserved noncoding elements in mammals. PLoS Genet. 2007;3:1572–1586. doi: 10.1371/journal.pgen.0030147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles DG, McLysaght A. Recent de novo origin of human protein-coding genes. Genome Res. 2009;19:1752–1759. doi: 10.1101/gr.095026.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima T. On the brain of the sperm whale (Physeter catadon L.) Sci. Rep. Whales Res. Inst. Tokyo. 1951;6:49–72. [Google Scholar]
- Konopka G, Friedrich T, Davis-Turak J, Winden K, Oldham MC, Gao F, Chen L, Wang G-Z, Luo R, Preuss TM, Geschwind DH. Human-specific transcriptional networks in the brain. Neuron. 2012;75:601–617. doi: 10.1016/j.neuron.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornack DR, Rakic P. Changes in cell-cycle kinetics during the development and evolution of primate neocortex. Proc. Natl. Acad. Sci. USA. 1998;95:1242–1246. doi: 10.1073/pnas.95.3.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlenkov A, Roussos P, Timashpolsky A, Barbu M, Rudchenko S, Bibikova M, Klotzle B, Byne W, Lyddon R, Di Narzo AF, et al. Differences in DNA methylation between human neuronal and glial cells are concentrated in enhancers and non-CpG sites. Nucleic Acids Res. 2014;42:109–127. doi: 10.1093/nar/gkt838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozma C, Scribanu N, Gersh E. The microcephaly-lymphoedema syndrome: report of an additional family. Clin. Dysmorphol. 1996;5:49–54. doi: 10.1097/00019605-199601000-00007. [DOI] [PubMed] [Google Scholar]
- Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, Gilliam TC, Nowak NJ, Cook EH, Jr, Dobyns WB, Christian SL. Recurrent 16p11.2 microdeletions in autism. Hum. Mol. Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- Kuypers HG. Some aspects of the organization of the output of the motor cortex. Ciba Found. Symp. 1987;132:63–82. doi: 10.1002/9780470513545.ch5. [DOI] [PubMed] [Google Scholar]
- Kwan KY, Lam MMS, Johnson MB, Dube U, Shim S, Rašin M-R, Sousa AMM, Fertuzinhos S, Chen J-G, Arellano JI, et al. Species-dependent posttranscriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell. 2012;149:899–911. doi: 10.1016/j.cell.2012.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- Lambert N, Lambot M-A, Bilheu A, Albert V, Englert Y, Libert F, Noel JC, Sotiriou C, Holloway AK, Pollard KS, et al. Genes expressed in specific areas of the human fetal cerebral cortex display distinct patterns of evolution. PLoS ONE. 2011;6:e17753. doi: 10.1371/journal.pone.0017753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansdell H. Intelligence test scores from infancy to adulthood for a craniopagus twin pair neurosurgically separated at 4 months of age. Psychol. Rep. 1999;84:209–217. doi: 10.2466/pr0.1999.84.1.209. [DOI] [PubMed] [Google Scholar]
- Lashley KS. Persistent problems in the evolution of mind. Q. Rev. Biol. 1949;24:28–42. doi: 10.1086/396806. [DOI] [PubMed] [Google Scholar]
- Li G, Wang J, Rossiter SJ, Jones G, Zhang S. Accelerated FoxP2 evolution in echolocating bats. PLoS ONE. 2007;2:e900. doi: 10.1371/journal.pone.0000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Bammann H, Li M, Liang H, Yan Z, Phoebe Chen Y-P, Zhao M, Khaitovich P. Evolutionary and ontogenetic changes in RNA edit-ng in human, chimpanzee, and macaque brains. RNA. 2013;19:1693–1702. doi: 10.1261/rna.039206.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman P. The evolution of language and thought. J. Anthropol. Sci. 2016;94:127–146. doi: 10.4436/JASS.94029. [DOI] [PubMed] [Google Scholar]
- Liégeois F, Morgan AT, Stewart LH, Helen Cross J, Vogel AP, Vargha-Khadem F. Speech and oral motor profile after childhood hemispherectomy. Brain Lang. 2010;114:126–134. doi: 10.1016/j.bandl.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. Broad Institute Sequencing Platform and Whole Genome Assembly Team; Baylor College of Medicine Human Genome Sequencing Center Sequencing Team; Genome Institute at Washington University. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Somel M, Tang L, Yan Z, Jiang X, Guo S, Yuan Y, He L, Oleksiak A, Zhang Y, et al. Extension of cortical synaptic development distinguishes humans from chimpanzees and macaques. Genome Res. 2012;22:611–622. doi: 10.1101/gr.127324.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, Yang S-P, Wang Z, Chinwalla AT, Minx P, et al. Comparative and demographic analysis of orangutan genomes. Nature. 2011;469:529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Nowakowski TJ, Pollen AA, Javaherian A, Kriegstein AR, Oldham MC. Radial glia require PDGFD-PDGFRp signalling in human but not mouse neocortex. Nature. 2014;515:264–268. doi: 10.1038/nature13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLarnon A. The scaling of gross dimensions of the spinal cord in primates and other species. J. Hum. Evol. 1996;30:71–87. [Google Scholar]
- MacLean EL. Unraveling the evolution of uniquely human cognition. Proc. Natl. Acad. Sci. USA. 2016;113:6348–6354. doi: 10.1073/pnas.1521270113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantini D, Corbetta M, Romani GL, Orban GA, Vanduffel W. Evolutionary novel functional networks in the human brain? J. Neurosci. 2013;33:3259–3275. doi: 10.1523/JNEUROSCI.4392-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Padilla M. Dual origin of the mammalian neocortex and evolution of the cortical plate. Anat. Embryol. (Berl.) 1978;152:109–126. doi: 10.1007/BF00315920. [DOI] [PubMed] [Google Scholar]
- Markov NT, Ercsey-Ravasz M, Van Essen DC, Knoblauch K, Toroczkai Z, Kennedy H. Cortical high-density counter stream architectures. Science. 2013;342:1238406. doi: 10.1126/science.1238406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marner L, Nyengaard JR, Tang Y, Pakkenberg B. Marked loss of myelinated nerve fibers in the human brain with age. J. Comp. Neurol. 2003;462:144–152. doi: 10.1002/cne.10714. [DOI] [PubMed] [Google Scholar]
- Mars RB, Neubert FX, Verhagen L, Sallet J, Miller KL, Dunbar RI, Barton RA. Primate comparative neuroscience using magnetic resonance imaging: promises and challenges. Front. Neurosci. 2014;8:298. doi: 10.3389/fnins.2014.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Reno PL, Pollen AA, Bassan AI, Capellini TD, Guenther C, Indjeian VB, Lim X, Menke DB, Schaar BT, et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature. 2011;471:216–219. doi: 10.1038/nature09774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLysaght A, Guerzoni D. New genes from non-coding sequence: the role of de novo protein-coding genes in eukaryotic evolutionary innovation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015;370:20140332. doi: 10.1098/rstb.2014.0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesulam M. Brain, mind, and the evolution of connectivity. Brain Cogn. 2000;42:4–6. doi: 10.1006/brcg.1999.1145. [DOI] [PubMed] [Google Scholar]
- Meyer KA, Marques-Bonet T, Sestan N. Differential gene expression in the human brain is associated with conserved, but not accelerated, noncoding sequences. Mol. Biol. Evol. 2017;34:1217–1229. doi: 10.1093/molbev/msx076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DJ, Duka T, Stimpson CD, Schapiro SJ, Baze WB, McArthur MJ, Fobbs AJ, Sousa AM, Sestan N, Wildman DE, et al. Prolonged myelination in human neocortical evolution. Proc. Natl. Acad. Sci. USA. 2012;109:16480–16485. doi: 10.1073/pnas.1117943109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Ding S-L, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen HS, Pakkenberg B, Dam M, Dietz R, Sonne C, Mikkelsen B, Eriksen N. Quantitative relationships in delphinid neocortex. Front. Neuroanat. 2014;8:132. doi: 10.3389/fnana.2014.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckli L, Naumer MJ, Singer W. Bilateral visual field maps in a patient with only one hemisphere. Proc. Natl. Acad. Sci. USA. 2009;106:13034–13039. doi: 10.1073/pnas.0809688106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Maier MA, Kirkwood PA, Lemon RN. Striking differences in transmission of corticospinal excitation to upper limb motoneurons in two primate species. J. Neurophysiol. 2000;84:698–709. doi: 10.1152/jn.2000.84.2.698. [DOI] [PubMed] [Google Scholar]
- Navarrete A, van Schaik CP, Isler K. Energetics and the evolution of human brain size. Nature. 2011;480:91–93. doi: 10.1038/nature10629. [DOI] [PubMed] [Google Scholar]
- Neubauer S. Endocasts: possibilities and limitations for the interpretation of human brain evolution. Brain Behav. Evol. 2014;84:117–134. doi: 10.1159/000365276. [DOI] [PubMed] [Google Scholar]
- Nielsen R. Adaptionism-30 years after Gould and Lewontin. Evolution. 2009;63:2487–2490. doi: 10.1111/j.1558-5646.2009.00799.x. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Bustamante C, Clark AG, Glanowski S, Sackton TB, Hubisz MJ, Fledel-Alon A, Tanenbaum DM, Civello D, White TJ, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchinsky EA, Gilissen E, Allman JM, Perl DP, Erwin JM, Hof PR. A neuronal morphologic type unique to humans and great apes. Proc. Natl. Acad. Sci. USA. 1999;96:5268–5273. doi: 10.1073/pnas.96.9.5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuttle X, Giannuzzi G, Duyzend MH, Schraiber JG, Narvaiza I, Sudmant PH, Penn O, Chiatante G, Malig M, Huddleston J, et al. Emergence of a Homo sapiens-specific gene family and chromosome 16p11.2 CNV susceptibility. Nature. 2016;536:205–209. doi: 10.1038/nature19075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Bleness M, Searles VB, Varki A, Gagneux P, Sikela JM. Evolution of genetic and genomic features unique to the human lineage. Nat. Rev. Genet. 2012a;13:853–866. doi: 10.1038/nrg3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Bleness MS, Dickens CM, Dumas LJ, Kehrer-Sawatzki H, Wyckoff GJ, Sikela JM. Evolutionary history and genome organization of DUF1220 protein domains. G3 (Bethesda) 2012b;2:977–986. doi: 10.1534/g3.112.003061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberheim NA, Goldman SA, Nedergaard M. Heterogeneity of astrocytic form and function. Methods Mol. Biol. 2012;814:23–45. doi: 10.1007/978-1-61779-452-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksenberg N, Stevison L, Wall JD, Ahituv N. Function and regulation of AUTS2, a gene implicated in autism and human evolution. PLoS Genet. 2013;9:e1003221. doi: 10.1371/journal.pgen.1003221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc. Natl. Acad. Sci. USA. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos-Serrano JL, Kang HJ, Tyler WA, Silbereis JC, Cheng F, Zhu Y, Pletikos M, Jankovic-Rapan L, Cramer NP, Galdzicki Z, et al. Down syndrome developmental brain transcriptome reveals defective oligodendrocyte differentiation and myelination. Neuron. 2016;89:1208–1222. doi: 10.1016/j.neuron.2016.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onorati M, Li Z, Liu F, Sousa AM, Nakagawa N, Li M, Dell’Anno MT, Gulden FO, Pochareddy S, Tebbenkamp AT, et al. Zika virus disrupts phospho-TBK1 localization and mitosis in human neuroepithelial stem cells and radial glia. Cell Rep. 2016;16:2576–2592. doi: 10.1016/j.celrep.2016.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani T, Marchetto MC, Gage FH, Simons BD, Livesey FJ. 2D and 3D stem cell models of primate cortical development identify species-specific differences in progenitor behavior contributing to brain size. Cell Stem Cell. 2016;18:467–480. doi: 10.1016/j.stem.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pääbo S. The human condition-a molecular approach. Cell. 2014;157:216–226. doi: 10.1016/j.cell.2013.12.036. [DOI] [PubMed] [Google Scholar]
- Pai AA, Bell JT, Marioni JC, Pritchard JK, Gilad Y. A genome-wide study of DNA methylation patterns and gene expression levels in multiple human and chimpanzee tissues. PLoS Genet. 2011;7:e1001316. doi: 10.1371/journal.pgen.1001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakkenberg B, Gundersen HJ. Neocortical neuron number in humans: effect of sex and age. J. Comp. Neurol. 1997;384:312–320. [PubMed] [Google Scholar]
- Pakkenberg B, Pelvig D, Marner L, Bundgaard MJ, Gundersen HJ, Nyengaard JR, Regeur L. Aging and the human neocortex. Exp. Gerontol. 2003;38:95–99. doi: 10.1016/s0531-5565(02)00151-1. [DOI] [PubMed] [Google Scholar]
- Passingham RE. What is special about the human brain? Oxford, New York: Oxford University Press; 2008. [Google Scholar]
- Pearce E, Stringer C, Dunbar RIM. New insights into differences in brain organization between Neanderthals and anatomically modern humans. Proc. Biol. Sci. 2013;280:20130168. doi: 10.1098/rspb.2013.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn DC, Holyoak KJ, Povinelli DJ. Darwin’s mistake: explaining the discontinuity between human and nonhuman minds. Behav. Brain Sci. 2008;31:109–130. doi: 10.1017/S0140525X08003543. discussion 130–178. [DOI] [PubMed] [Google Scholar]
- Petanjek Z, Judaš M, Šimic G, Rasin MR, Uylings HBM, Rakic P, Kostovic I. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc. Natl. Acad. Sci. USA. 2011;108:13281–13286. doi: 10.1073/pnas.1105108108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrides M, Cadoret G, Mackey S. Orofacial somatomotor responses in the macaque monkey homologue of Broca’s area. Nature. 2005;435:1235–1238. doi: 10.1038/nature03628. [DOI] [PubMed] [Google Scholar]
- Pletikos M, Sousa AM, Sedmak G, Meyer KA, Zhu Y, Cheng F, Li M, Kawasawa YI, Sestan N. Temporal specification and bilaterality of human neocortical topographic gene expression. Neuron. 2014;81:321–332. doi: 10.1016/j.neuron.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, King B, Kern AD, Dreszer T, Katzman S, Siepel A, Pedersen JS, Bejerano G, Baertsch R, et al. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006a;2:e168. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot M-A, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006b;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, Shuga J, Liu SJ, Oldham MC, Diaz A, et al. Molecular identity of human outer radial glia during cortical development. Cell. 2015;163:55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontzer H, Brown MH, Raichlen DA, Dunsworth H, Hare B, Walker K, Luke A, Dugas LR, Durazo-Arvizu R, Schoeller D, et al. Metabolic acceleration and the evolution of human brain size and life history. Nature. 2016;533:390–392. doi: 10.1038/nature17654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popesco MC, Maclaren EJ, Hopkins J, Dumas L, Cox M, Meltesen L, McGavran L, Wyckoff GJ, Sikela JM. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science. 2006;313:1304–1307. doi: 10.1126/science.1127980. [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Noonan JP, Pääbo S, Rubin EM. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- Premack D. Human and animal cognition: continuity and discontinuity. Proc. Natl. Acad. Sci. USA. 2007;104:13861–13867. doi: 10.1073/pnas.0706147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott SL, Srinivasan R, Marchetto MC, Grishina I, Narvaiza I, Selleri L, Gage FH, Swigut T, Wysocka J. Enhancer divergence and cis-regulatory evolution in the human and chimp neural crest. Cell. 2015;163:68–83. doi: 10.1016/j.cell.2015.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss TM. Human brain evolution: from gene discovery to phenotype discovery. Proc. Natl. Acad. Sci. USA. 2012;109(Suppl 1):10709–10716. doi: 10.1073/pnas.1201894109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss TM, Coleman GQ. Human-specific organization of primary visual cortex: alternating compartments of dense Cat-301 and calbindin immunoreactivity in layer 4A. Cereb. Cortex. 2002;12:671–691. doi: 10.1093/cercor/12.7.671. [DOI] [PubMed] [Google Scholar]
- Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulsifer MB, Brandt J, Salorio CF, Vining EP, Carson BS, Freeman JM. The cognitive outcome of hemispherectomy in 71 children. Epilepsia. 2004;45:243–254. doi: 10.1111/j.0013-9580.2004.15303.x. [DOI] [PubMed] [Google Scholar]
- Raghanti MA, Stimpson CD, Marcinkiewicz JL, Erwin JM, Hof PR, Sherwood CC. Cholinergic innervation of the frontal cortex: differences among humans, chimpanzees, and macaque monkeys. J. Comp. Neurol. 2008a;506:409–424. doi: 10.1002/cne.21546. [DOI] [PubMed] [Google Scholar]
- Raghanti MA, Stimpson CD, Marcinkiewicz JL, Erwin JM, Hof PR, Sherwood CC. Differences in cortical serotonergic innervation among humans, chimpanzees, and macaque monkeys: a comparative study. Cereb. Cortex. 2008b;18:584–597. doi: 10.1093/cercor/bhm089. [DOI] [PubMed] [Google Scholar]
- Raghanti MA, Spocter MA, Stimpson CD, Erwin JM, Bonar CJ, All-man JM, Hof PR, Sherwood CC. Species-specific distributions of tyrosine hydroxylase-immunoreactive neurons in the prefrontal cortex of anthropoid primates. NeuroScience. 2009;158:1551–1559. doi: 10.1016/j.neuroscience.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghanti MA, Edler MK, Stephenson AR, Wilson LJ, Hopkins WD, Ely JJ, Erwin JM, Jacobs B, Hof PR, Sherwood CC. Human-specific increase of dopaminergic innervation in a striatal region associated with speech and language: A comparative analysis of the primate basal ganglia. J. Comp. Neurol. 2016;524:2117–2129. doi: 10.1002/cne.23937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reader SM, Laland KN. Social intelligence, innovation, and enhanced brain size in primates. Proc. Natl. Acad. Sci. USA. 2002;99:4436–4441. doi: 10.1073/pnas.062041299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly SK, Noonan JP. Evolution of gene regulation in humans. Annu. Rev. Genomics Hum. Genet. 2016;17:45–67. doi: 10.1146/annurev-genom-090314-045935. [DOI] [PubMed] [Google Scholar]
- Reilly SK, Yin J, Ayoub AE, Emera D, Leng J, Cotney J, Sarro R, Rakic P, Noonan JP. Evolutionary genomics. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science. 2015;347:1155–1159. doi: 10.1126/science.1260943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rilling JK. Comparative primate neuroimaging: insights into human brain evolution. Trends Cogn. Sci. 2014;18:46–55. doi: 10.1016/j.tics.2013.09.013. [DOI] [PubMed] [Google Scholar]
- Rilling JK, Insel TR. The primate neocortex in comparative perspective using magnetic resonance imaging. J. Hum. Evol. 1999;37:191–223. doi: 10.1006/jhev.1999.0313. [DOI] [PubMed] [Google Scholar]
- Rilling JK, Glasser MF, Preuss TM, Ma X, Zhao T, Hu X, Behrens TEJ. The evolution of the arcuate fasciculus revealed with comparative DTI. Nat. Neurosci. 2008;11:426–428. doi: 10.1038/nn2072. [DOI] [PubMed] [Google Scholar]
- Ringo JL. Neuronal interconnection as a function of brain size. Brain Behav. Evol. 1991;38:1–6. doi: 10.1159/000114375. [DOI] [PubMed] [Google Scholar]
- Rockel AJ, Hiorns RW, Powell TP. The basic uniformity in structure of the neocortex. Brain. 1980;103:221–244. doi: 10.1093/brain/103.2.221. [DOI] [PubMed] [Google Scholar]
- Rodenas-Cuadrado P, Ho J, Vernes SC. Shining a light on CNTNAP2: complex functions to complex disorders. Eur. J. Hum. Genet. 2014;22:171–178. doi: 10.1038/ejhg.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J, Kochunov P, Zilles K, Shelledy W, Lancaster J, Thompson P, Duggirala R, Blangero J, Fox PT, Glahn DC. On the genetic architecture of cortical folding and brain volume in primates. Neuroimage. 2010;53:1103–1108. doi: 10.1016/j.neuroimage.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth G, Dicke U. Evolution of the brain and intelligence. Trends Cogn. Sci. 2005;9:250–257. doi: 10.1016/j.tics.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Ruiz-Orera J, Hernandez-Rodriguez J, Chiva C, Sabidó E, Kondova I, Bontrop R, Marqués-Bonet T, Albá MM. Origins of de novo genes in human and chimpanzee. PLoS Genet. 2015;11:e1005721. doi: 10.1371/journal.pgen.1005721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenker NM, Buxhoeveden DP, Blackmon WL, Amunts K, Zilles K, Semendeferi K. A comparative quantitative analysis of cytoarchitecture and minicolumnar organization in Broca’s area in humans and great apes. J. Comp. Neurol. 2008;510:117–128. doi: 10.1002/cne.21792. [DOI] [PubMed] [Google Scholar]
- Schmahmann JD, Pandya DN. Fiber pathways of the Brain. Oxford, New York: Oxford University Press; 2006. [Google Scholar]
- Schneider E, Mayer S, El Hajj N, Jensen LR, Kuss AW, Zischler H, Kondova I, Bontrop RE, Navarro B, Fuchs E, et al. Methylation and expression analyses of the 7q autism susceptibility locus genes MEST, COPG2, and TSGA14 in human and anthropoid primate cortices. Cytogenet. Genome Res. 2012;136:278–287. doi: 10.1159/000337298. [DOI] [PubMed] [Google Scholar]
- Schneider E, El Hajj N, Richter S, Roche-Santiago J, Nanda I, Schempp W, Riederer P, Navarro B, Bontrop RE, Kondova I, et al. Widespread differences in cortex DNA methylation of the “language gene” CNTNAP2 between humans and chimpanzees. EpiGenetics. 2014;9:533–545. doi: 10.4161/epi.27689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenemann PT, Sheehan MJ, Glotzer LD. Prefrontal white matter volume is disproportionately larger in humans than in other primates. Nat. Neurosci. 2005;8:242–252. doi: 10.1038/nn1394. [DOI] [PubMed] [Google Scholar]
- Schreiweis C, Bornschein U, Burguière E, Kerimoglu C, Schreiter S, Dannemann M, Goyal S, Rea E, French CA, Puliyadi R, et al. Humanized Foxp2 accelerates learning by enhancing transitions from declarative to procedural performance. Proc. Natl. Acad. Sci. USA. 2014;111:14253–14258. doi: 10.1073/pnas.1414542111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Merkle FT, Gaus SE, Craig AD, Allman JM, Hof PR. Distinctive neurons of the anterior cingulate and frontoinsular cortex: a historical perspective. Cereb. Cortex. 2012;22:245–250. doi: 10.1093/cercor/bhr005. [DOI] [PubMed] [Google Scholar]
- Semendeferi K, Lu A, Schenker N, Damasio H. Humans and great apes share a large frontal cortex. Nat. Neurosci. 2002;5:272–276. doi: 10.1038/nn814. [DOI] [PubMed] [Google Scholar]
- Semendeferi K, Teffer K, Buxhoeveden DP, Park MS, Bludau S, Amunts K, Travis K, Buckwalter J. Spatial organization of neurons in the frontal pole sets humans apart from great apes. Cereb. Cortex. 2011;21:1485–1497. doi: 10.1093/cercor/bhq191. [DOI] [PubMed] [Google Scholar]
- Sherwood CC, Lee PWH, Rivara C-B, Holloway RL, Gilissen EPE, Simmons RMT, Hakeem A, Allman JM, Erwin JM, Hof PR. Evolution of specialized pyramidal neurons in primate visual and motor cortex. Brain Behav. Evol. 2003;61:28–44. doi: 10.1159/000068879. [DOI] [PubMed] [Google Scholar]
- Sherwood CC, Holloway RL, Erwin JM, Hof PR. Cortical orofacial motor representation in Old World monkeys, great apes, and humans. II. Stereologic analysis of chemoarchitecture. Brain Behav. Evol. 2004;63:82–106. doi: 10.1159/000075673. [DOI] [PubMed] [Google Scholar]
- Sherwood CC, Subiaul F, Zawidzki TW. A natural history of the human mind: tracing evolutionary changes in brain and cognition. J. Anat. 2008;212:426–454. doi: 10.1111/j.1469-7580.2008.00868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood CC, Bauernfeind AL, Bianchi S, Raghanti MA, Hof PR. Human brain evolution writ large and small. Prog. Brain Res. 2012;195:237–254. doi: 10.1016/B978-0-444-53860-4.00011-8. [DOI] [PubMed] [Google Scholar]
- Shi P, Bakewell MA, Zhang J. Did brain-specific genes evolve faster in humans than in chimpanzees? Trends Genet. 2006;22:608–613. doi: 10.1016/j.tig.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Shibata M, Gulden FO, Sestan N. From trans to cis: transcriptional regulatory networks in neocortical development. Trends Genet. 2015;31:77–87. doi: 10.1016/j.tig.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S, Kwan KY, Li M, Lefebvre V, Sestan N. Cis-regulatory control of corticospinal system development and evolution. Nature. 2012;486:74–79. doi: 10.1038/nature11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinawi M, Schaaf CP, Bhatt SS, Xia Z, Patel A, Cheung SW, Lanpher B, Nagl S, Herding HS, Nevinny-Stickel C, et al. A small recurrent deletion within 15q13.3 is associated with a range of neurodevelopmental phenotypes. Nat. Genet. 2009;41:1269–1271. doi: 10.1038/ng.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- Shulha HP, Crisci JL, Reshetov D, Tushir JS, Cheung I, Bharadwaj R, Chou H-J, Houston IB, Peter CJ, Mitchell AC, et al. Human-specific histone methylation signatures at transcription start sites in prefrontal neurons. PLoS Biol. 2012;10:e1001427. doi: 10.1371/journal.pbio.1001427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigaard RK, Kjær M, Pakkenberg B. Development of the cell population in the brain white matter of young children. Cereb. Cortex. 2016;26:89–95. doi: 10.1093/cercor/bhu178. [DOI] [PubMed] [Google Scholar]
- Silbereis JC, Pochareddy S, Zhu Y, Li M, Sestan N. The cellular and molecular landscapes of the developing human central nervous system. Neuron. 2016;89:248–268. doi: 10.1016/j.neuron.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DL. Genomic divergence and brain evolution: How regulatory DNA influences development of the cerebral cortex. BioEssays. 2016;38:162–171. doi: 10.1002/bies.201500108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart IH, Dehay C, Giroud P, Berland M, Kennedy H. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb. Cortex. 2002;12:37–53. doi: 10.1093/cercor/12.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Franz H, Yan Z, Lorenc A, Guo S, Giger T, Kelso J, Nickel B, Dannemann M, Bahn S, et al. Transcriptional neoteny in the human brain. Proc. Natl. Acad. Sci. USA. 2009;106:5743–5748. doi: 10.1073/pnas.0900544106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Liu X, Tang L, Yan Z, Hu H, Guo S, Jiang X, Zhang X, Xu G, Xie G, et al. MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol. 2011;9:e1001214. doi: 10.1371/journal.pbio.1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Liu X, Khaitovich P. Human brain evolution: transcripts, metabolites and their regulators. Nat. Rev. Neurosci. 2013;14:112–127. doi: 10.1038/nrn3372. [DOI] [PubMed] [Google Scholar]
- Sousa AMM, Meyer KA, Sestan N. Molecular and cellular mechanisms of human brain development and evolution. In: Gazzaniga MS, Mangun GR, editors. The Cognitive Neurosciences. Fifth. The MIT Press; 2015. pp. 67–74. [Google Scholar]
- Spiers H, Hannon E, Schalkwyk LC, Smith R, Wong CCY, O’Donovan MC, Bray NJ, Mill J. Methylomic trajectories across human fetal brain development. Genome Res. 2015;25:338–352. doi: 10.1101/gr.180273.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spocter MA, Hopkins WD, Barks SK, Bianchi S, Hehmeyer AE, Anderson SM, Stimpson CD, Fobbs AJ, Hof PR, Sherwood CC. Neuropil distribution in the cerebral cortex differs between humans and chimpanzees. J. Comp. Neurol. 2012;520:2917–2929. doi: 10.1002/cne.23074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squair J. Craniopagus: Overview and the implications of sharing a brain. University of British Columbia’s Undergraduate Journal of Psychology 2012;1 [Google Scholar]
- Stephan H, Frahm H, Baron G. New and revised data on volumes of brain structures in insectivores and primates. Folia Primatol. (Basel) 1981;35:1–29. doi: 10.1159/000155963. [DOI] [PubMed] [Google Scholar]
- Stevens CF. How cortical interconnectedness varies with network size. Neural Comput. 1989;1:473–479. [Google Scholar]
- Stone JL, Goodrich JT. The craniopagus malformation: classification and implications for surgical separation. Brain. 2006;129:1084–1095. doi: 10.1093/brain/awl065. [DOI] [PubMed] [Google Scholar]
- Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J, Eichler EE 1000 Genomes Project. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Huddleston J, Catacchio CR, Malig M, Hillier LW, Baker C, Mohajeri K, Kondova I, Bontrop RE, Persengiev S, et al. Great Ape Genome Project. Evolution and diversity of copy number variation in the great ape lineage. Genome Res. 2013;23:1373–1382. doi: 10.1101/gr.158543.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Nyengaard JR, De Groot DM, Gundersen HJ. Total regional and global number of synapses in the human brain neocortex. Synapse. 2001;41:258–273. doi: 10.1002/syn.1083. [DOI] [PubMed] [Google Scholar]
- Taverna E, Götz M, Huttner WB. The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu. Rev. Cell Dev. Biol. 2014;30:465–502. doi: 10.1146/annurev-cellbio-101011-155801. [DOI] [PubMed] [Google Scholar]
- Todorov AB, Cohen KL, Spilotro V, Landau E. Craniopagus twins. J. Neurol. Neurosurg. Psychiatry. 1974;37:1291–1298. doi: 10.1136/jnnp.37.12.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Wildman DE, Liu G, Xu W, Johnson RM, Hof PR, Kapatos G, Grossman LI, Goodman M. Sister grouping of chimpanzees and humans as revealed by genome-wide phylogenetic analysis of brain gene expression profiles. Proc. Natl. Acad. Sci. USA. 2004;101:2957–2962. doi: 10.1073/pnas.0308725100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Goodman M, Erez O, Romero R, Liu G, Islam M, Opazo JC, Sherwood CC, Grossman LI, Wildman DE. Distinct genomic signatures of adaptation in pre- and postnatal environments during human evolution. Proc. Natl. Acad. Sci. USA. 2008;105:3215–3220. doi: 10.1073/pnas.0712400105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Bullmore ET, Sporns O. Comparative Connectomics. Trends Cogn. Sci. 2016;20:345–361. doi: 10.1016/j.tics.2016.03.001. [DOI] [PubMed] [Google Scholar]
- Van Essen DC. A tension-based theory of morphogenesis and compact wiring in the central nervous system. Nature. 1997;385:313–318. doi: 10.1038/385313a0. [DOI] [PubMed] [Google Scholar]
- Van Schaik CP. The Primate Origins of Human Nature. Hoboken, New Jersey, USA: John Wiley & Sons; 2016. [Google Scholar]
- Vermunt MW, Tan SC, Castelijns B, Geeven G, Reinink P, de Bruijn E, Kondova I, Persengiev S, Bontrop R, Cuppen E, et al. Netherlands Brain Bank. Epigenomic annotation of gene regulatory alterations during evolution of the primate brain. Nat. Neurosci. 2016;19:494–503. doi: 10.1038/nn.4229. [DOI] [PubMed] [Google Scholar]
- Vernot B, Tucci S, Kelso J, Schraiber JG, Wolf AB, Gittelman RM, Dannemann M, Grote S, McCoy RC, Norton H, et al. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science. 2016;352:235–239. doi: 10.1126/science.aad9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Lynch VJ. The gene regulatory logic of transcription factor evolution. Trends Ecol. Evol. 2008;23:377–385. doi: 10.1016/j.tree.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Walløe S, Eriksen N, Dabelsteen T, Pakkenberg B. Aneurological comparative study of the harp seal (Pagophilus groenlandicus) and harbor porpoise (Phocoena phocoena) brain. Anat. Rec. (Hoboken) 2010;293:2129–2135. doi: 10.1002/ar.21295. [DOI] [PubMed] [Google Scholar]
- Wang H-Y, Chien H-C, Osada N, Hashimoto K, Sugano S, Gojobori T, Chou C-K, Tsai S-F, Wu C-I, Shen C-KJ. Rate of evolution in brain-expressed genes in humans and other primates. PLoS Biol. 2007;5:e13. doi: 10.1371/journal.pbio.0050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Watson SK, Reamer LA, Mareno MC, Vale G, Harrison RA, Lambeth SP, Schapiro SJ, Whiten A. Socially transmitted diffusion of a novel behavior from subordinate chimpanzees. Am. J. Primatol. 2017;79 doi: 10.1002/ajp.22642. http://dx.doi.org/10.1002/ajp.22642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiten A, Goodall J, McGrew WC, Nishida T, Reynolds V, Sugiyama Y, Tutin CE, Wrangham RW, Boesch C. Cultures in chimpanzees. Nature. 1999;399:682–685. doi: 10.1038/21415. [DOI] [PubMed] [Google Scholar]
- Williams RW, Herrup K. The control of neuron number. Annu. Rev. Neurosci. 1988;11:423–453. doi: 10.1146/annurev.ne.11.030188.002231. [DOI] [PubMed] [Google Scholar]
- Wood B, Baker J. Evolution in the genus Homo. Annu. Rev. Ecol. Evol. Syst. 2011;42:47–69. [Google Scholar]
- Wrangham RW, Jones JH, Laden G, Pilbeam D, Conklin-Brittain N. The raw and the stolen. Cooking and the ecology of human origins. Curr. Anthropol. 1999;40:567–594. [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Wu D-D, Irwin DM, Zhang Y-P. De novo origin of human protein-coding genes. PLoS Genet. 2011;7:e1002379. doi: 10.1371/journal.pgen.1002379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich S, Kircher M, Vieth B, Haase A, Merkert S, Beier J, Göhring G, Glage S, Schambach A, Curnow EC, et al. Primate iPS cells as tools for evolutionary analyses. Stem Cell Res(Amst.) 2014;12:622–629. doi: 10.1016/j.scr.2014.02.001. [DOI] [PubMed] [Google Scholar]
- Xie C, Zhang YE, Chen J-Y, Liu C-J, Zhou W-Z, Li Y, Zhang M, Zhang R, Wei L, Li C-Y. Hominoid-specific de novo protein-coding genes originating from long non-coding RNAs. PLoS Genet. 2012;8:e1002942. doi: 10.1371/journal.pgen.1002942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Jensen-Seaman MI, Chemnick L, Kidd JR, Deinard AS, Ryder O, Kidd KK, Li WH. Low nucleotide diversity in chimpanzees and bonobos. Genetics. 2003;164:1511–1518. doi: 10.1093/genetics/164.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X-J, Zheng H-K, Wang J, Wang W, Su B. Detecting lineage-specific adaptive evolution of brain-expressed genes in human using rhesus macaque as outgroup. Genomics. 2006;88:745–751. doi: 10.1016/j.ygeno.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Yu F, Jiang QJ, Sun XY, Zhang RW. A new case of complete primary cerebellar agenesis: clinical and imaging findings in a living patient. Brain. 2015;138:e353. doi: 10.1093/brain/awu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Konopka G, Hunt BG, Preuss TM, Geschwind D, Yi SV. Divergent whole-genome methylation maps of human and chimpanzee brains reveal epigenetic basis of human regulatory evolution. Am. J. Hum. Genet. 2012;91:455–465. doi: 10.1016/j.ajhg.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Barres BA. A smarter mouse with human astrocytes. BioEssays. 2013;35:876–880. doi: 10.1002/bies.201300070. [DOI] [PubMed] [Google Scholar]
- Zhang K, Sejnowski TJ. A universal scaling law between gray matter and white matter of cerebral cortex. Proc. Natl. Acad. Sci. USA. 2000;97:5621–5626. doi: 10.1073/pnas.090504197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Webb DM, Podlaha O. Accelerated protein evolution and origins of human-specific features: Foxp2 as an example. Genetics. 2002;162:1825–1835. doi: 10.1093/genetics/162.4.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Li M, Sousa AMM, Sestan N. XSAnno: a framework for building ortholog models in cross-species transcriptome comparisons. BMC Genomics. 2014;15:343. doi: 10.1186/1471-2164-15-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollikofer CP, De León MS. Pandora’s growing box: Inferring the evolution and development of hominin brains from endocasts. Evol. Anthropol. 2013;22:20–33. doi: 10.1002/evan.21333. [DOI] [PubMed] [Google Scholar]