Abstract
Clostridium species (particularly Clostridium difficile, Clostridium botulinum, Clostridium tetani and Clostridium perfringens) are associated with a range of human and animal diseases. Several other species including Clostridium tertium, Clostridium cadaveris, and Clostridium paraputrificum have also been linked with sporadic human infections, however there is very limited, or in some cases, no genomic information publicly available. Thus, we isolated one C. tertium strain, one C. cadaveris strain and three C. paraputrificum strains from preterm infants residing within neonatal intensive care units and performed Whole Genome Sequencing (WGS) using Illumina HiSeq. In this report, we announce the open availability of the draft genomes: C. tertium LH009, C. cadaveris LH052, C. paraputrificum LH025, C. paraputrificum LH058, and C. paraputrificum LH141. These genomes were checked for contamination in silico to ensure purity, and we confirmed species identity and phylogeny using both 16S rRNA gene sequences (from PCR and in silico) and WGS-based approaches. Average Nucleotide Identity (ANI) was used to differentiate genomes from their closest relatives to further confirm speciation boundaries. We also analysed the genomes for virulence-related factors and antimicrobial resistance genes, and detected presence of tetracycline and methicillin resistance, and potentially harmful enzymes, including multiple phospholipases and toxins. The availability of genomic data in open databases, in tandem with our initial insights into the genomic content and virulence traits of these pathogenic Clostridium species, should enable the scientific community to further investigate the disease-causing mechanisms of these bacteria with a view to enhancing clinical diagnosis and treatment.
Keywords: Clostridium, functional annotation, whole genome sequencing, virulence
Medical Relevance
Clostridium, which means “a small spindle” in Greek (due to its rod-shaped morphology), is classified as a genus under the phylum Firmicutes and class Clostridia, and comprises 221 species to date (September 2017) (Parte 2014). Clostridium spp. are Gram-positive spore-forming anaerobes found ubiquitously in the environment (soil and water) and the intestinal tract of humans and animals (Yamagishi etal. 1964; Miwa 1975; de Vos etal. 1982). There are several significant human and animal disease causing Clostridium species including Clostridium difficile (pseudomembranous colitis), Clostridium botulinum (infant botulism), Clostridium tetani (tetanus), and Clostridium perfringens (acute watery diarrhea/necrotising enterocolitis [NEC]), with associated pathology ascribed to the toxins they produce (Bruggemann and Gottschalk 2004; Awad etal. 2014; Carter and Peck 2015; Sim etal. 2015). There are also several less well-studied species including Clostridium tertium, Clostridium paraputrificum, and Clostridium cadaveris, which have been sporadically reported in the literature to be associated with human infection.
C. cadaveris (formerly Clostridium capitovale), is thought to be a key tissue-decomposing bacterium in dead carcasses, and is generally not considered pathogenic in living individuals (Poduval etal. 1999). However, this bacterium has infrequently been associated with human systemic diseases, including intraperitoneal infection (Leung etal. 2009) and bacteremia (Poduval etal. 1999; Schade etal. 2006).
C. tertium, is an aerotolerant and nontoxin-producing species. During The First World War, it was the third most frequently isolated bacteria from war wounds, after C. perfringens and C. sporogenes (Henry 1917). This organism was officially recognized as a pathogen in 1963, when the first C. tertium-associated septicemia case was reported (King etal. 1963). C. tertium has also been associated with infections including peritonitis (Butler and Pitt 1982) and pneumonia (Johnson and Tenover 1988). Importantly, C. tertium is also linked with cattle enteritis (Silvera etal. 2003), preterm NEC (Cheah etal. 2001) and adult enterocolitis (Coleman etal. 1993).
C. paraputrificum has previously been isolated from formula-fed infants within their first weeks of life (Stark and Lee 1982). This pathogen has been associated with paediatric infection (sepsis) (Brook 1995), adult necrotizing enterocolitis (Shandera etal. 1988), bacteremia (Shinha and Hadi 2015), and preterm NEC (Smith etal. 2011). Interestingly, this organism was shown to produce NEC-like lesions, including gas cysts, in an animal model and thus supports their disease-causing link (Waligora-Dupriet etal. 2005).
Whole genome sequencing (WGS) has contributed significantly to biomedical and veterinary research through our increased understanding of pathogens at a genomic level. Despite the medical importance of these three pathogenic Clostridium species, there is currently no sequenced genomes of C. tertium or C. cadaveris available to the research community (apart from 16S rRNA gene sequences) and only four genomes of C. paraputrificum accessible on NCBI databases as of September 2017 (Geer etal. 2010). In this study, we sequenced one C. cadaveris isolate, one C. tertium isolate and three C. paraputrificum isolates from preterm infant faecal samples obtained from two neonatal intensive care units (NICUs) units in England. We identified these using their 16S rRNA gene sequences (both full-length PCR and in silico) and WGS-based k-mer phylogenetic assignment, thus contributing new genomic data on these pathogenic bacteria. We also verified their phylogenetic positions using WGS data, measured genetic distances via Average Nucleotide Identity (ANI), and performed genome-wide functional annotation (COG classification). These genomic data and analyses increases our understanding of the virulence potentials and functionalities of these pathogenic bacteria, with a future view to unraveling disease-causing mechanisms.
Genome Description
Here, we report the release of draft genomes sequenced on Illumina HiSeq 2500 platform as stated in table 1. C. paraputrificum isolates have a genome size between 3.6 and 3.7 million bases and a stable GC content from 29.6 to 29.9%, which is in line with the four public genomes (Geer etal. 2010). C. tertium has a larger genome (3.9 million bases) and relatively lower GC content of 27.8%, whilst C. cadaveris has a smaller genome (3.4 million bases) compared with C. paraputrificum, and a significantly higher GC content of 31.2%. All draft genomes were assembled using Prokka de novo assembler and 80% (four out of five) of the genomes analyzed were <50 contigs, except for C. paraputrificum LH058 with 84 contigs.
Table 1.
Genome Description, Assembly Statistics, and Clinical Information of Isolates Used in This Study
| C. tertiumLH009 | C. cadaverisLH052 | C. paraputrificum LH025 | C. paraputrificum LH058 | C. paraputrificum LH141 | |
|---|---|---|---|---|---|
| Genome size (bp) | 3,970,462 | 3,460,249 | 3,797,748 | 3,776,795 | 3,630,606 |
| No. of contigs | 49 | 46 | 40 | 84 | 29 |
| Genes | 3,910 | 3,395 | 3,896 | 3,823 | 3,655 |
| CDS | 3,821 | 3,310 | 3,813 | 3,745 | 3,565 |
| N50 (bp) | 258,765 | 118,391 | 479,233 | 101,241 | 390,404 |
| tRNAs | 89 | 84 | 83 | 77 | 90 |
| GC content (%) | 27.8 | 31.2 | 29.6 | 29.9 | 29.7 |
| Origin of isolates | 29-week preterm infant | 32-week preterm infant | 29-week preterm infant | 32-week preterm infant | 27-week preterm infant |
| Hospital | RH | NNUH | RH | NNUH | RH |
RH: Rosie Hospital, Cambridge, UK; NNUH: Norfolk and Norwich Hospital, Norwich, UK.
These five strains were isolated from preterm infants residing at two different NICUs (table 1), which is in line with previous findings that report frequent detection of C. paraputrificum (16–22%) and C. tertium (4–9%) in infant cohorts (Tonooka etal. 2005; Ferraris etal. 2012). However, to date there are no reports of C. cadaveris isolation from infants.
Phylogenetic Positions
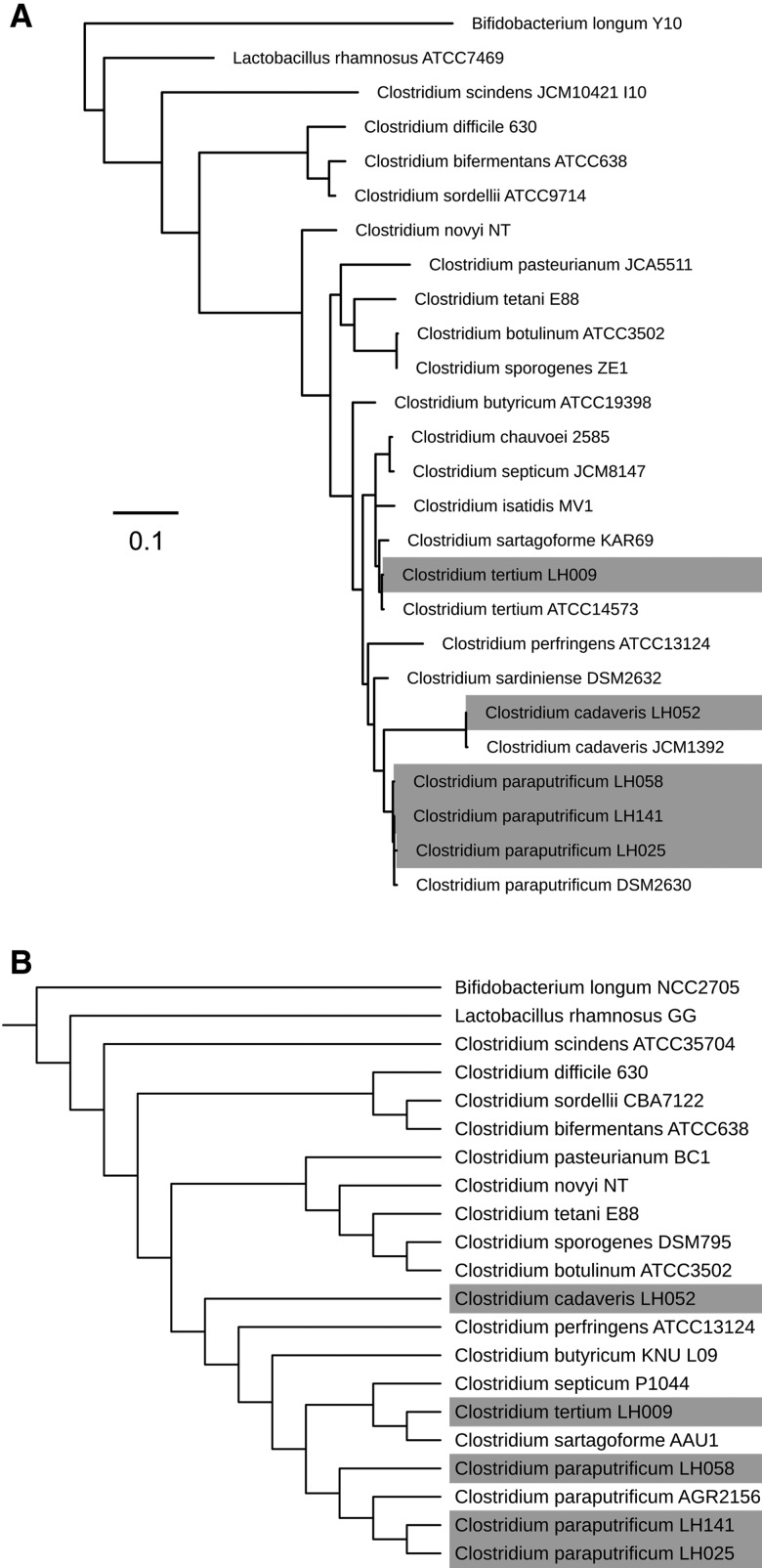
To assign phylogenetic position, and identify these isolates, we computationally extracted 16S rRNA sequences from genomes to construct a Clostridium 16S rRNA phylogeny (based on 19 isolates in the NCBI nucleotide database) as in figure 1A. Here, we coupled three genomic approaches to confirm taxonomic position of these newly released genomes. We firstly, performed a PCR targeting almost the full length of the 16S rRNA gene, and predicted the whole 16S rRNA gene sequence in silico. Secondly, we employed Average Nucleotide Identity (ANI) to confirm species boundaries; ANI cut-offs for species discrimination is known to be approximately 95%, and this value has been reported to mirror the traditional taxonomic gold standard method DNA–DNA hybridization (DDH) to define species (Richter and Rossello 2009). Lastly, we performed CVTree—an alignment-free whole genome-based phylogenetic construction method, which is known for speed and accuracy for taxonomic assignment (Xu and Hao 2009).
Fig. 1.
—(A) 16S rRNA maximum-likelihood (ML) phylogenetic tree of 19 species of Clostridium. (B) WGS-based alignment-free cladogram of representative Clostridium species. Lactobacillus rhanmosus and Bifidobacterium longum have been used as outgroups. Grey labels indicate newly sequenced isolates in this study.
At a 16S rRNA level, LH058, LH141, and LH025 fall in the same lineage as C. paraputrificum DSM2630, indicating species-level relatedness (fig. 1A), with LH052 clustering with C. cadaveris JCM1392, and LH009 within the same lineage as C. tertium ATCC14573 and Clostridium sartagoforme KAR69. CVTree phylogenetic analysis, providing greater resolution based on sequence comparison, showed similar relationships. (fig. 1B); all C. paraputrificum isolates grouped within the same lineage as C. paraputrificum DSM2630, when compared with other Clostridium species, indicating correct species assignment for isolates LH058, LH141, and L025. C. cadaveris LH052 is most closely related to C. perfringens ATCC13124, and LH009 (C. tertium as assigned according to 16S data) is closely related to C. sartagoforme AAU1 (fig. 1B).
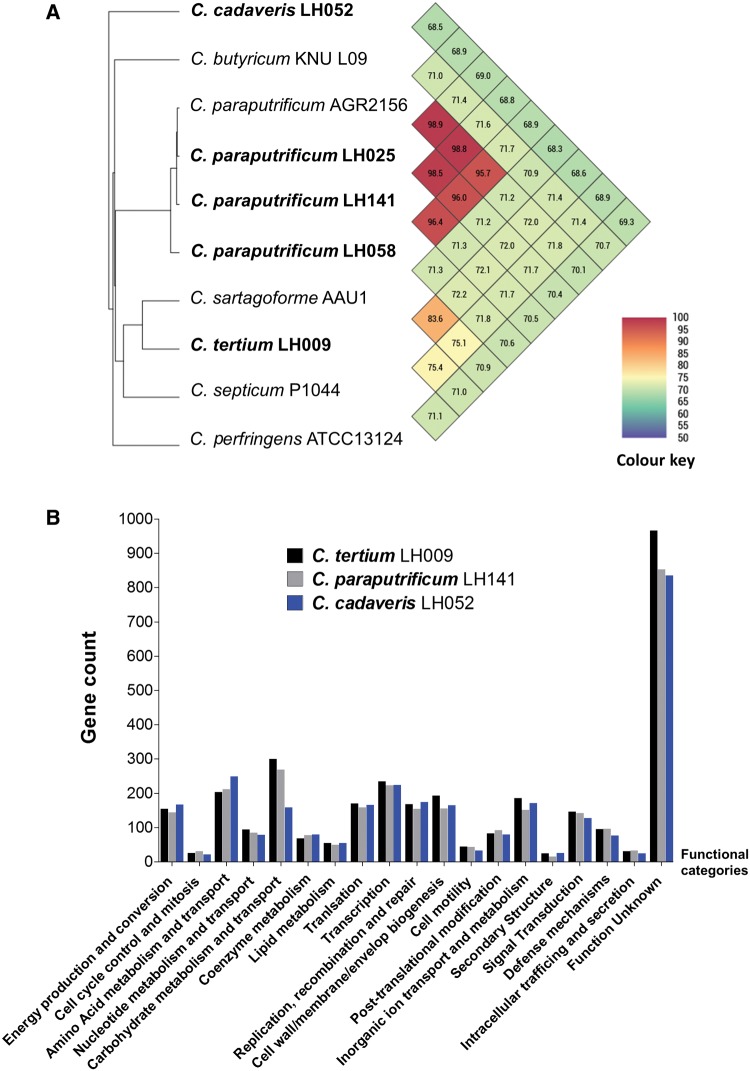
We next used ANI analysis to provide higher phylogenetic resolution (fig. 2A). C. paraputrificum AGR2156 are identical to LH025, LH141, and LH058 in terms of nucleotide sequences, sharing ANI of >95.7%, thus determined to be the same species. Although LH009 is closely related to C. sartagoforme AAU1, the ANI calculation does not allocate these two within the same species (ANI = 83.6%, < 95% as species cut-off), which indicates LH009 is distinct from its closest relatives, and may be identified as the species C. tertium. LH052 is also evolutionarily distant (based on ANI calculation, 68.5%) from other Clostridium, indicating this isolate is a separate species, C. cadaveris.
Fig. 2.
—(A) Average Nucleotide Identity (ANI) values in representative Clostridium genomes. (B) Comparison of functional annotations based on COG classifications on three representative genomes.
Virulence Traits and Genome-Wide Functional Analyses
Using genome annotations, we performed a thorough search on virulence-related terms including “phospholipase,” “hemolysin,” “resistance,” “lactamase,” “drug,” and “toxin” to provide initial insights into the potential virulence-linked genes encoded within these genomes (table 2).
Table 2.
Virulence-Related Genes Detected in Selected Clostridium Genomes
| Isolate | Gene Names | Gene Description and Functions |
|---|---|---|
| C. tertium LH009 | ytpA | Phospholipase |
| vanW | Vancomycin B-type resistance | |
| stp | Multidrug resistance | |
| mdtK | Multidrug resistance | |
| tetM | Tetracycline resistance | |
| marA | Multiple antibiotic resistance | |
| mecR1 | Methicillin resistance | |
| norM | Multidrug resistance | |
| mepA | Multidrug export protein | |
| hcpC | Beta-lactamase precursor | |
| sme-1 | Carbapenem-hydrolyzing beta-lactamase precursor | |
| C. cadaveris LH052 | ytpA | Phospholipase |
| (n/a) | Patatin-like phospholipase | |
| (n/a) | Phospholipase C precursor | |
| toxA | Toxin A | |
| marA | Multiple antibiotic resistance | |
| vanW | Vancomycin B-type resistance | |
| norm | Multidrug resistance protein | |
| tetM | Tetracycline resistance | |
| marR | Multiple antibiotic resistance | |
| mdtA | Multidrug resistance | |
| mepA | Multidrug export protein | |
| (n/a) | Beta-lactamase precursor | |
| C. paraputrificum LH141 | ytpA | Phospholipase |
| toxA | Toxin A | |
| marR | Multiple antibiotic resistance | |
| tetM | Tetracycline resistance | |
| norM | Multidrug resistance | |
| vanW | Vancomycin B-type resistance | |
| mdtK | Multidrug resistance | |
| mepA | Multidrug export protein | |
| (n/a) | Beta-lactamase precursor | |
| sme-1 | Carbapenem-hydrolyzing beta-lactamase precursor |
C. tertium LH009, C. cadaveris LH052, and C. paraputrificum LH141 harbour phospholipase genes (ytpA) that are homologous to phospholipases encoded in other pathogen genomes including Bacillus subtilis, Pseudomonas aeruginosa, and Streptococcus pneumoniae. Phospholipases are known to possess hydrolytic activity against eukaryotic cell membranes, and are thus considered key virulence factors. C. perfringens produces homologous phospholipase C (also known as alpha toxin) that has previously been reported to damage epithelial cells (Verherstraeten etal. 2013), and which shares >58% protein sequence identity with the phospholipase encoded by gene ytpA. Importantly, LH052 and LH141 also possess toxA gene, which encodes C. difficile-associated Toxin A, known to be one of the main virulence factors during infection having cytotoxic and proinflammatory activities (Awad etal. 2014).
Notably, antimicrobial resistance genes are encoded in all three genomes, including vancomycin (vanW) and tetracycline resistance (tetM) (Evers and Courvalin 1996; Donhofer etal. 2012). Other resistance traits include multidrug efflux pumps; that is, those encoded by mdtK and norM (fluoroquinolones) (Horiyama etal. 2011; Golparian etal. 2014), mdtA (aminocoumarin) (Guerrero etal. 2012), and efflux pump transcriptional regulators marA and marR (Maira-Litran etal. 2000). In addition, methicillin resistance gene mecR1 was detected in LH009 (Shore etal. 2011), whilst beta-lactamase (penicillins and carbapenems) precursor (inactive protein sequence that could potentially be activated via posttranslational modification) was encoded in all genomes (Marciano etal. 2007). The prevalence of multiple antimicrobial resistance genes in these clinical strains may correspond to the environment in which they were isolated; preterm infants residing in NICUs where antimicrobial usage is extensive (Albrich etal. 2004).
From the COG-based genome-wide annotation, most genes (>40% in each genome) did not map with any known functional orthologs, which highlights the limitation of genomic tools and current databases, for understanding these bacteria at a functional level. Gene counts in most categories of these three genomes did not differ significantly from one another (fig. 2B). However, the number of genes involved in carbohydrate metabolism and transport is lower in C. cadaveris LH052 (n = 159), than encoded in C. tertium LH009 (n = 300) and C. paraputrificum LH141 (n = 269), whereas LH052 possesses more genes (n = 249) involved in amino acid metabolism and transport as compared with LH009 (n = 203) and LH141 (n = 212). These functional differences may correspond to divergent modes of metabolism and nutritional substrates for C. cadaveris, which is distinct from C. tertium and C. paraputrificum (correlates to WGS phylogeny positions), and may link to previous isolations of this species from additional environmental niches, i.e. dead carcasses. Therefore, we conclude that these three Clostridium species are similar in terms of genomic functionalities, however due to the high number of function-unknown genes, this somewhat reduces in-depth comparison between genomes and will require further experimental work.
Materials and Methods
Faecal Sample Collection
Fecal sample collection was performed under an on-going preterm infant study (BAMBI) which is approved by University of East Anglia (UEA) Faculty of Medical and Health Sciences (FMH) Ethics Committee. Sample collection was done in accordance with the procedures outlined by National Research Ethics Service (NRES) approved UEA Biorepository (Licence no.: 11208). Participating infants were given written consent by their parents for fecal sample collection at Norfolk and Norwich University Hospital (Norwich, UK) and Rosie Hospital (Cambridge, UK). Fecal samples were routinely collected from infant nappies in the NICUs into sterile stool containers and stored at 4 °C.
Bacterial Isolates and Preliminary 16S rRNA PCR Identification
A total of five Clostridium isolates (including C. tertium, C. cadaveris, and C. paraputrificum) were analyzed in this study. Isolates were preliminarily identified using 16S rRNA full-length PCR (Weisburg etal. 1991). Primers used as in table 3. Near 1kbp PCR products were subsequently sequenced (Eurofins, Luxembourg) and compared with 16S rRNA bacteria sequence database on NCBI using BLASTn (optimized for megablast) search algorithm (Camacho etal. 2009).
Table 3.
Sequence of Primers Used for PCR Amplification of 16S rRNA Gene
| Primers | Sequence |
|---|---|
| fD1 | 5′-AGA GTT TGA TCC TGG CTC AG-3′ |
| fD2 | 5′-AGA GTT TGA TCA TGG CTC AG-3′ |
| rP1 | 5′-ACG GTT ACC TTG TTA CGA CTT-3′ |
Genomic DNA Extraction
Overnight 10 ml pure cultures in BHI were harvested for phenol-chloroform DNA extraction. Briefly, bacterial pellets were resuspended in 2 ml 25% sucrose in 10 mM Tris and 1 mM EDTA at pH 8.0. Cells were lysed using 50 μl 100 mg/ml lysozyme (Roche). 100 μl 20 mg/ml Proteinase K (Roche), 30 μl 10 mg/ml RNase A (Roche), 400 μl 0.5 M EDTA (pH 8.0) and 250 μl 10% Sarkosyl NL30 (Fisher) were added subsequently into the lysed bacterial suspension. This follows by 1-h ice incubation and 50 °C overnight water bath.
Second-day protocol comprises three rounds of phenol-chloroform-isoamyl alcohol (Sigma) extraction using 15 ml gel-lock tubes (Qiagen). Chloroform-Isoamyl alcohol (Sigma) extraction was performed to remove residual phenol, followed by ethanol precipitation and 70% ethanol wash. DNA pellets were finally resuspended in 200–300 μl of 10 mM Tris (pH 8.0). DNA concentration was quantified using Qubit dsDNA BR assay kit (Invitrogen) and DNA quality assessed by Nanodrop spectrophotometer.
Whole Genome Sequencing, Genome Assembly and Annotation
Isolated DNA of pure cultures was subjected to multiplex standard Illumina library preparation protocol followed by sequencing via Illumina HiSeq 2500 platform with read length 2 × 125 bp (paired-end reads) and an average sequencing coverage of 60×. Draft genome assemblies were generated using an assembly and annotation pipeline as described previously (Page etal. 2016). All genomes were annotated using Prokka v1.11 (Seemann 2014).
Contamination Estimation
Webtool ContEst16S was used to check for potential contamination of the draft genomes based on Genbank database (Lee etal. 2017).
16S rRNA Phylogeny
Publicly available16S rRNA genes were retrieved from NCBI nucleotide database (Geer etal. 2010). 16S rRNA genes from our isolates were predicted using Barrnap v0.7 (https://github.com/Victorian-Bioinformatics-Consortium/barrnap, last accessed September 20, 2017) and extracted using BEDTools getfasta utility (Quinlan and Hall 2010). All 16S rRNA sequences were subsequently concatenated as a multisequence fasta, and sequences were aligned with MUSCLE (Edgar 2004). Neighbor-joining (NJ) tree was generated in 1000 bootstrap replicates using Juke-Cantor distance (Gouy etal. 2010). Maximum-likelihood (ML) tree was produced by PhyML GTR model with 1000 bootstrap replicates (Guindon etal. 2010). Trees were edited using iTOL (Letunic and Bork 2016).
Alignment-Free WGS Phylogeny
Selected Clostridium genome sequences were retrieved from NCBI genome database. Annotated multiple protein sequences were used as input for CVTree v5.0 to generate alignment-free WGS-based phylogeny using the optimized six as the k-tuple length (Xu and Hao 2009). Tree was edited using iTOL as described in previous section.
Average Nucleotide Identity (ANI)
OrthoANI Tool v.093 (OAT) was employed to calculate the ANI (both directions) between genomes (Lee etal. 2016). Identity >95% was used as cut-off for species delineation.
Genome-Wide Functional Assignment (COG)
Functional assignments were implemented using eggNOG-mapper v0.99.3 (Huerta-Cepas etal. 2017), based on eggNOG orthology data (Huerta-Cepas etal. 2016). Sequence searches were performed using HMMER3 (Eddy 2011). Data were extracted using Shell scripts (https://github.com/raymondkiu/eggnog-mapper_COGextraction, last accessed September 20, 2017) and visualized in GraphPad PRISM v5.04.
Ethics Approval and Consent for Participation
This study was approved by the University of East Anglia (UEA) Faculty of Medical and Health Sciences (FMH) Ethics Committee. Sample collection follows the protocols outlined by NRES approved UEA Biorepository (Licence no.: 11208). Written consent was given by the parents for their infants for participation in this study.
Acknowledgments
This work was supported by a Wellcome Trust Investigator Award to LJH (grant number 100974/C/13/Z). RK is a PhD student at the Norwich Medical School of University of East Anglia (UEA), partially funded by UEA international bursary.
Literature Cited
- Albrich WC, Monnet DL, Harbarth S.. 2004. Antibiotic selection pressure and resistance in Streptococcus pneumoniae and Streptococcus pyogenes. Emerg Infect Dis. 10(3): 514–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad MM, Johanesen PA, Carter GP, Rose E, Lyras D.. 2014. Clostridium difficile virulence factors: insights into an anaerobic spore-forming pathogen. Gut Microbes 5(5): 579–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook I. 1995. Clostridial infection in children. J Med Microbiol. 42(2): 78–82. [DOI] [PubMed] [Google Scholar]
- Bruggemann H, Gottschalk G.. 2004. Insights in metabolism and toxin production from the complete genome sequence of Clostridium tetani. Anaerobe 10(2): 53–68. [DOI] [PubMed] [Google Scholar]
- Butler T, Pitt S.. 1982. Spontaneous bacterial peritonitis due to Clostridium tertium. Gastroenterology 82(1): 133–134. [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AT, Peck MW.. 2015. Genomes, neurotoxins and biology of Clostridium botulinum Group I and Group II. Res Microbiol. 166(4): 303–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah FC, Lim KE, Boo NY.. 2001. Clostridium tertium in cerebrospinal fluid of a premature neonate with necrotizing enterocolitis: contamination or real? Acta Paediatr. 90(6): 704–705. [PubMed] [Google Scholar]
- Coleman N, et al. 1993. Neutropenic enterocolitis associated with Clostridium tertium. J Clin Pathol. 46(2): 180–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos N, Mevissen-Verhage E, van Amerongen WH, Marcelis J.. 1982. A new selective medium for the culture of Clostridia from human faeces. Eur J Clin Microbiol. 1(5): 267–271. [DOI] [PubMed] [Google Scholar]
- Donhofer A, et al. 2012. Structural basis for TetM-mediated tetracycline resistance. Proc Natl Acad Sci U S A. 109(42): 16900–16905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol. 7(10): e1002195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5): 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers S, Courvalin P.. 1996. Regulation of VanB-type vancomycin resistance gene expression by the VanS(B)-VanR (B) two-component regulatory system in Enterococcus faecalis V583. J Bacteriol. 178(5): 1302–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraris L, et al. 2012. Clostridia in premature neonates’ gut: incidence, antibiotic susceptibility, and perinatal determinants influencing colonization. PLoS One 7(1): e30594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geer LY, et al. 2010. The NCBI BioSystems database. Nucleic Acids Res. 38(Database issue): D492–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golparian D, Shafer WM, Ohnishi M, Unemo M.. 2014. Importance of multidrug efflux pumps in the antimicrobial resistance property of clinical multidrug-resistant isolates of Neisseria gonorrhoeae. Antimicrob Agents Chemother. 58(6): 3556–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O.. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 27(2): 221–224. [DOI] [PubMed] [Google Scholar]
- Guerrero P, et al. 2012. Characterization of the BaeSR two-component system from Salmonella Typhimurium and its role in ciprofloxacin-induced mdtA expression. Arch Microbiol. 194(6): 453–460. [DOI] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3): 307–321. [DOI] [PubMed] [Google Scholar]
- Henry H. 1917. An investigation of the cultural reactions of certain anaerobes found in wounds. J Pathol Bacteriol. 21: 344–385. [Google Scholar]
- Horiyama T, Nikaido E, Yamaguchi A, Nishino K.. 2011. Roles of Salmonella multidrug efflux pumps in tigecycline resistance. J Antimicrob Chemother. 66(1): 105–110. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, et al. 2017. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, et al. 2016. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44(D1): D286–D293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JR, Tenover FC.. 1988. Clostridium tertium bacteremia in a patient with aspiration pneumonia: an elusive diagnosis. J Infect Dis. 157(4): 854–855. [DOI] [PubMed] [Google Scholar]
- King BM, Ranck BA, Daugherty FD, Rau CA.. 1963. Clostridium tertium septicemia. N Engl J Med. 269: 467–469. [DOI] [PubMed] [Google Scholar]
- Lee I, et al. 2017. ContEst16S: an algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int J Syst Evol Microbiol. 67(6): 2053–2057. [DOI] [PubMed] [Google Scholar]
- Lee I, Kim YO, Park SC, Chun J.. 2016. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 66(2): 1100–1103. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1): W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Sasson M, Patel SR, Viveiros K.. 2009. Clostrium cadaveris intra-peritoneal abscess. Am J Gastroenterol. 104(10): 2635–2636. [DOI] [PubMed] [Google Scholar]
- Maira-Litran T, Allison DG, Gilbert P.. 2000. An evaluation of the potential of the multiple antibiotic resistance operon (mar) and the multidrug efflux pump acrAB to moderate resistance towards ciprofloxacin in Escherichia coli biofilms. J Antimicrob Chemother. 45(6): 789–795. [DOI] [PubMed] [Google Scholar]
- Marciano DC, Karkouti OY, Palzkill T.. 2007. A fitness cost associated with the antibiotic resistance enzyme SME-1 beta-lactamase. Genetics 176(4): 2381–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa T. 1975. Clostridia in soil of the Antarctica. Jpn J Med Sci Biol. 28(4): 201–213. [DOI] [PubMed] [Google Scholar]
- Page AJ, et al. 2016. Robust high-throughput prokaryote de novo assembly and improvement pipeline for Illumina data. Microb Genom. 2(8): e000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parte AC. 2014. LPSN—list of prokaryotic names with standing in nomenclature. Nucleic Acids Res. 42(Database issue): D613–D616. doi: 10.1093/nar/gkt1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduval RD, Mohandas R, Unnikrishnan D, Corpuz M.. 1999. Clostridium cadaveris bacteremia in an immunocompetent host. Clin Infect Dis. 29(5): 1354–1355. doi: 10.1086/313491 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6): 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Rossello MR.. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 106(45): 19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schade RP, et al. 2006. Clostridium cadaveris bacteraemia: two cases and review. Scand J Infect Dis. 38(1): 59–62. [DOI] [PubMed] [Google Scholar]
- Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30(14): 2068–2069. [DOI] [PubMed] [Google Scholar]
- Shandera WX, Humphrey RL, Stratton LB.. 1988. Necrotizing enterocolitis associated with Clostridium paraputrificum septicemia. South Med J. 81(2): 283–284. [DOI] [PubMed] [Google Scholar]
- Shinha T, Hadi C.. 2015. Clostridium paraputrificum bacteremia associated with colonic necrosis in a patient with AIDS. Case Rep Infect Dis. 2015: 312919. doi: 10.1155/2015/312919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore AC, et al. 2011. Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 55(8): 3765–3773. doi: 10.1128/AAC.00187-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvera M, Finn B, Reynolds KM, Taylor DJ.. 2003. Clostridium tertium as a cause of enteritis in cattle. Vet Rec. 153(2): 60.. [DOI] [PubMed] [Google Scholar]
- Sim K, et al. 2015. Dysbiosis anticipating necrotizing enterocolitis in very premature infants. Clin Infect Dis. 60(3): 389–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, et al. 2011. Community analysis of bacteria colonizing intestinal tissue of neonates with necrotizing enterocolitis. BMC Microbiol. 11: 73. doi: 10.1186/1471-2180-11-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark PL, Lee A.. 1982. Clostridia isolated from the feces of infants during the first year of life. J Pediatr. 100(3): 362–365. [DOI] [PubMed] [Google Scholar]
- Tonooka T, et al. 2005. Detection and quantification of four species of the genus Clostridium in infant feces. Microbiol Immunol. 49(11): 987–992. [DOI] [PubMed] [Google Scholar]
- Verherstraeten S, et al. 2013. The synergistics necrohemorrhagic action of Clostridium perfringens perfringolysin and alpha toxin in the bovine intestine and against bovine endothelial cells. Vet Res. 44(1): 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waligora-Dupriet AJ, Dugay A, Auzeil N, Huerre M, Butel MJ.. 2005. Evidence of clostidial implication in necrotizing enterocolitis through bacterial fermentation in a gnotobiotic quail model. Pediatr Res. 58(4): 629–635. [DOI] [PubMed] [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ.. 1991. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 173(2): 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Hao B.. 2009. CVTree update: a newly designed phylogenetic study platform using composition vectors and whole genomes. Nucleic Acids Res. 37(Web Server issue): W174–W178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi T, Ishida S, Nishida S.. 1964. Isolation of toxigenic strains of Clostridium perfringens from soil. J Bacteriol. 88: 646–652. [DOI] [PMC free article] [PubMed] [Google Scholar]