SUMMARY
Pyrazinamide (PZA) is an important first-line anti-tuberculosis drug, however, there are relatively few available data on PZA resistant (PZA-R) rate in China. From June 2009 to June 2012, we selected 493 isolates from five field settings in China to investigate PZA-R by pncA gene sequencing. The result showed that PZA-R rate was 1.0% (2/196) among pan-susceptible isolates, 3.1% (4/130) among isoniazid (INH) mono-resistant isolates, 14.0% (6/43) among rifampin (RIF) mono-resistant isolates and 43.5% (54/124) among multidrug resistant (MDR) isolates. MDR tuberculosis (TB), RIF mono-resistance, and retreatment were found to be risk factors for PZA-R. Newly diagnosed PZA-R TB patients and clustered isolates with identical pncA mutations indicate that transmission of PZA-R isolates plays an important role in emergence of PZA-R TB. The results suggest that, it is necessary to conduct PZA susceptibility test among MDR isolates and modify the treatment regimens accordingly.
Keywords: Mycobacterium tuberculosis, Pyrazinamide resistance, Transmission
1. Introduction
Pyrazinamide (PZA) is a first-line drug and an important component used for the treatment of both drug-susceptible and drug-resistant tuberculosis (TB). Combined with other anti-TB drugs, PZA shows a remarkable therapeutic effect [1,2], because of its unique sterilizing activity to kill semi-dormant or persistent Mycobacterium tuberculosis [3]. Specifically, in multidrug resistant (MDR) TB therapy, it has a strong effect on the success rates of treatment [4]. So, the absence of PZA in the regimens would lead to poor treatment outcome [5–7].
At present, there are two main methods for drug susceptibility testing (DST) of M. tuberculosis: the phenotypic DST by traditional culture, and the molecular drug susceptibility test (mDST) by detecting drug resistant mutations. However, both phenotypic DST and mDST have limitations to detect PZA-R. The accuracy and reproducibility of PZA phenotypic DST are not satisfactory [8,9], because it must be conducted under acidic condition (pH 5.5–6). The growth of M. tuberculosis is inhibited at such a low pH and many factors such as inoculum size could lead to inaccurate results. For mDST, because the molecular mechanism of PZA-R is not completely understood, the PZA mDST based on detecting pncA mutations could provide only about 90% specificity and sensitivity [10]. According to studies conducted in different areas of China, the specificities of pncA sequencing to predict PZA-R were more than 90% [11–13]. Therefore, for large scale and multi-center PZA-R investigation in China, mDST is a more reliable and objective method.
China is one of the high drug resistant TB burden countries, but there are relatively few available data on PZA-R rates. Studies showed that PZA-R rates were 35.6% among drug resistant TB in Hong Kong [14] and 43.1% among MDR-TB in Zhejiang [13]. However these studies were based on samples collected from single hospital, which could not reflect the real prevalence of PZA-R in the general population. In this study, we used the pncA mutations to predict PZA-R among M. tuberculosis clinical isolates collected from a population based epidemiological study and estimated the prevalence and risk factors of PZA-R in China.
2. Materials and methods
2.1. Study population and isolates
In a previous study, we collected all M. tuberculosis isolates from five counties in different provinces of China (Songjiang in Shanghai, Wusheng in Sichuan, Pingguo in Guangxi, Wuchang in Heilongjiang, and Weishi in Henan provinces respectively) between June 2009 and June 2012 [15]. The data of patient information and isolates genotype (Beijing genotype and variable number tandem repeat (VNTR) genotype) was described in our previous study [15]. The isolates were considered to be clusters only when they shared the same VNTR genotype within each field setting. We included all drug resistant isolates (resistance to rifampin (RIF) and/or isoniazid (INH)), and randomly selected susceptible isolates (sensitivity to both RIF and INH) collected during the same period as susceptible group for comparison.
2.2. Drug susceptibility test
The isolates cultured directly from sputum were used to do phenotypic DSTand mDST. The phenotypic DST was conducted using the proportion method on Lowenstein–Jensen medium [16], and the DNA used in mDST was extracted by boiled lysis method [17]. Both phenotypic DST and mDST were performed to detect INH and RIF susceptibilityof the isolates. The mDSTof INH and RIF was performed using multiplex real-time PCR melting curve assay [18]. The isolates with consistent results of both methods were included in this study. The fluoroquinolones (FQs) and PZA susceptibility were determined by mDST using DNA sequencing. FQs mDST was performed on all MDR isolates by sequencing quinolone resistance-determining region (QRDR) of the gyrA gene [19]. PZA-R was predicted by detecting mutations among the whole pncA gene and its upstream region. We used two pairs of primers (pncA-721-F: GCTGGTCATGTTCGCGATCG, pncA-721-R: CGCTTGCGGCGAGCGCTCCA and pncA-951-F: CTGTCACCGGACGGATTTG, pncA-951-R: ATCGCGATGGAACGTGATA) to amplify and sequence 721 bp and 951 bp fragments. The pncA-951 primers were used only under the condition that the pncA-721 primers were failed to amplify the target fragment. All the mutations within the pncA gene and its upstream region were confirmed by both-direction sequencing.
According to the results of INH and RIF susceptibility, the isolates were classified into four categories: pan-susceptible (RIF and INH susceptible), INH mono-resistant, RIF mono-resistant and MDR (both RIF and INH resistant). In MDR group, the FQs resistant isolates were defined as pre-XDR. The isolates with non-synonymous mutations in pncA gene and its upstream region were considered as PZA-R.
2.3. Statistical analyses
Data were analyzed by using Stata software (version 13.1/SE, Stata Corp, College Station, Texas). We used univariate and multivariable logistic regression modeling to calculate odds ratios (OR) and adjusted odds ratio (aOR) for factors associated with PZA-R. We used a forward, stepwise approach to select the multivariable model. Factors with biological plausibility and P value < 0.2 in the univariate analysis were considered in the final model. A P value of < 0.05 was considered statistically significant.
3. Results
3.1. Characteristics of the patients and isolates
During the period from June 2009 to June 2012, 2274 culture-positive TB cases were diagnosed in the five settings [15]. With the exception of 60 isolates without DST data and 39 isolates with inconsistent results between phenotypic DST and mDST, there were 130 INH mono-resistant, 43 RIF mono-resistant and 124 MDR isolates (including 35 pre-XDR isolates). To estimate the proportion of PZA-R, we included all 297 drug-resistant isolates and randomly selected 196 isolates from 1878 pan-susceptible isolates. There were no statistically differences between selected and unselected pan-susceptible cases with regards to the characteristics of patients such as gender, age and TB treatment history. Thus, finally 493 isolates were included in this study, of which, 75.3% patients were male, the median age was 42 (interquartile range, 28–57), 78.3% were newly diagnosed cases and 77.5% isolates belonged to Beijing genotype strain.
3.2. Mutations types and prevalence of PZA resistance
To detect PZA-R related gene mutations, DNA sequencing of the pncA gene and its upstream region was performed on all isolates. In total, 66 out of the 493 isolates had non-synonymous mutations. Except of one isolate with two mutations (Y103STOP and D126G), the remaining 65 isolates carried a single mutation. Forty-eight mutation types were detected among the 66 isolates, including six insertion mutations (five frame shift mutations) and 42 point mutations (three mutations in the upstream region) (Table 1). No synonymous mutation was detected in all isolates.
Table 1.
Mutations in the pncA gene and its upstream region.
| Mutation type | Amino acid change | No. of isolates | Relationship to PZA phenotypic resistance
|
|||
|---|---|---|---|---|---|---|
| Miotto et al.* | TB-DreamDBy | MUBII-TB-DBz | Reported elsewhere [ref.]§ | |||
| A-11C | N. | 1 | Very high confidence | Reported | Reported | |
| A-11G | N. | 3 | Very high confidence | High confidence | High confidence | |
| T-7G | N. | 1 | Very high confidence | Unreported | Unreported | |
| T2C | M1T | 1 | Very high confidence | Reported | Reported | |
| T17C | I6T | 2 | Very high confidence | Unreported | Unreported | |
| G19T | V7F | 1 | Very high confidence | Reported | Reported | |
| A29C | Q10P | 3 | Very high confidence | High confidence | Reported | |
| T40G | C14G | 1 | Unreported | Unreported | Unreported | |
| G50T | G17V | 1 | Unreported | Reported | High confidence | |
| T100G | Y34D | 2 | Unreported | Unreported | Unreported | [26] |
| C102A | Y34STOP | 1 | Very high confidence | Unreported | Unreported | |
| T104C | L35P | 2 | Very high confidence | High confidence | Unreported | |
| C123A | Y41STOP | 1 | Unreported | Reported | Reported | |
| A139G | T47A | 1 | Not involved in resistance | High confidence | Reported | [12,13,27] |
| A146C | D49A | 1 | Very high confidence | High confidence | Reported | |
| A152G | H51R | 1 | Very high confidence | High confidence | High confidence | |
| 166 + G | 56Frameshift | 1 | Unreported | Unreported | Unreported | |
| C174G | F58L | 1 | Very high confidence | Reported | Reported | |
| T175C | S59P | 2 | Unreported | Reported | Reported | [26,28] |
| 186 + GGAC | 62insDY | 1 | Unreported | Unreported | Unreported | |
| TATTCCTC GTCGTG |
SSSW | |||||
| A188C | D63A | 1 | Unclear role | Unreported | Unreported | [27] |
| T192G | Y64STOP | 1 | Very high confidence | Unreported | Unreported | |
| C211T | H71Y | 1 | Very high confidence | Reported | Reported | |
| A226C | T76P | 3 | Very high confidence | High confidence | Reported | |
| T283G | Y95D | 1 | Unreported | Unreported | Unreported | |
| A286G | K96E | 1 | Very high confidence | Reported | Reported | |
| A286C | K96Q | 2 | Very high confidence | Reported | Reported | |
| T307G | Y103D | 1 | Very high confidence | Unreported | Unreported | |
| C309G | Y103STOP | 1¶ | Very high confidence | High confidence | Reported | |
| C312A | S104R | 1 | Very high confidence | Unreported | Unreported | |
| G373T | V125F | 1 | Very high confidence | Unreported | Unreported | |
| A377G | D126G | 1¶ | Unreported | Unreported | Unreported | |
| 390 + G | 130Frameshift | 1 | Very high confidence | Unreported | Unreported | |
| 390 + GG | 130Frameshift | 1 | Very high confidence | Unreported | Unreported | |
| 393 + C | 131Frameshift | 1 | Unreported | Unreported | Unreported | |
| A403C | T135P | 1 | Very high confidence | Reported | High confidence | |
| A407C | D136A | 1 | Unreported | Unreported | Unreported | |
| G413A | C138Y | 2 | Unreported | High confidence | Reported | [29,30] |
| T416G | V139G | 2 | Very high confidence | High confidence | High confidence | |
| G436A | A146T | 1 | Unreported | Reported | Reported | [11,13,28,31] |
| C442T | R148C | 1 | Unreported | Unreported | Unreported | |
| A460G | R154G | 1 | Unclear role | Reported | Reported | [22,32] |
| G463A | V155M | 1 | Very high confidence | Unreported | Unreported | |
| T467C | L156P | 7 | High confidence | Unreported | Unreported | |
| G484A | G162S | 1 | Unreported | Unreported | Unreported | [11,29] |
| T490C | S164P | 1 | Very high confidence | Unreported | Unreported | |
| 514 + C | 172Frameshift | 1 | Unreported | Unreported | Unreported | |
| A523G | M175V | 1 | Very high confidence | Reported | Reported | |
A Multicenter Study with 1950 clinical isolates [25].
TB Drug Resistance Mutation Database (TB-DreamDB) (www.tbdreamdb.com) [24].
MUBII-TB-DB (https://umr5558-bibiserv.univ-lyon1.fr/mubii/mubii-select.cgi) [23].
Previously reported in PZA-R isolates.
The two mutations (Y103STOP and D126G) coexisted in one strain.
The 66 isolates harboring non-synonymous mutations in the pncA gene and its upstream region were defined as PZA-R. Thus, the PZA-R rate was 1.0% (2/196) among pan-susceptible isolates, 3.1% (4/130) among INH mono-resistant isolates, 14.0% (6/43) among RIF mono-resistant isolates, 43.5% (54/124) among MDR and 54.3% (19/35) among pre-XDR isolates (Table 2).
Table 2.
PZA resistance among 493 M. tuberculosis isolates.
| PZA-R | Pan-susceptible | INH mono-resistant | RIF mono-resistant | MDR (including pre-XDR) | Pre-XDR | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Guangxi | 2/30 | 6.7% | 0/19 | 0% | 2/11 | 18.2% | 4/13 | 30.8% | 0/0 | N. |
| Heilongjiang | 0/21 | 0% | 1/15 | 6.7% | 0/8 | 0% | 4/14 | 28.6% | 0/2 | 0% |
| Henan | 0/42 | 0% | 1/28 | 3.6% | 1/4 | 25.0% | 21/28 | 75.0% | 8/8 | 100.0% |
| Sichuan | 0/28 | 0% | 0/20 | 0% | 3/11 | 27.3% | 10/30 | 33.3% | 2/7 | 28.6% |
| Shanghai | 0/75 | 0% | 2/48 | 4.2% | 0/9 | 0% | 15/39 | 38.5% | 9/18 | 50.0% |
| Total | 2/196 | 1.0% | 4/130 | 3.1% | 6/43 | 14.0% | 54/124 | 43.5% | 19/35 | 54.3% |
Abbreviations: PZA, pyrazinamide; INH, isoniazid; RIF, rifampin; MDR, multidrug resistant.
3.3. Risk factors of PZA resistance
In order to identify the risk factors for PZA-R, we performed univariate analysis and multivariable logistic regression analysis. Table 3 shows results of univariate analysis. Resistance of PZA significantly increased as the isolate being resistant to more anti-TB drugs (from mono-resistance to pre-XDR, P value < 0.001 for trend). In the multivariable regression analysis, retreatment (adjusted odds ratio [aOR], 2.36; 95% confidence interval [CI], 1.21–4.67, P value < 0.05), RIF mono-resistance (aOR, 7.43; 95% CI, 1.27–43.37, P value < 0.05), MDR (aOR, 45.05; 95% CI, 10.32–196.62, P value < 0.001) and pre-XDR (aOR, 86.36; 95% CI, 18.17–410.35, P value < 0.001) were independently associated with PZA resistance. These variables remained significantly associated with PZA resistance after adjustment for sex, age and Beijing genotype.
Table 3.
Univariate analysis of risk factors of PZA resistant tuberculosis.
| Characteristic | PZA-S (n = 427) No. (%) |
PZA-R (n = 66) No. (%) |
PZA-R VS PZA-S
|
|
|---|---|---|---|---|
| OR (95% CI) | P value | |||
| Sex | ||||
| Male | 319 (86.0) | 52 (14.0) | 1 | |
| Female | 108 (88.5) | 14 (11.5) | 0.80 (0.40–1.53) | 0.47 |
| Age | ||||
| < 35 | 146 (86.4) | 23 (13.6) | 1 | |
| 35–55 | 156 (84.3) | 29 (15.7) | 1.18 (0.63–2.24) | 0.58 |
| >55 | 120 (90.9) | 12 (9.1) | 0.63 (0.28–1.40) | 0.22 |
| Unknown | 5 (71.4) | 2 (28.6) | 2.54 (0.23–16.56) | 0.27 |
| TB history | ||||
| New | 354 (91.7) | 32 (8.3) | 1 | |
| Re-treated | 58 (65.9) | 30 (34.1) | 5.72 (3.09–10.50) | <0.001 |
| Unknown | 15 (78.9) | 4 (21.1) | 2.95 (0.67–9.99) | 0.06 |
| Beijing strain | ||||
| No | 103 (92.0) | 9 (8.0) | 1 | |
| Yes | 324 (85.0) | 57 (15.0) | 2.01 (0.95–4.78) | 0.06 |
| Drug resistance profile | <0.001* | |||
| Pan-susceptibility | 194 (99.0) | 2 (1.0) | 1 | |
| INH mono-resistance | 126 (96.9) | 4 (3.1) | 3.08 (0.55–17.12) | 0.18 |
| RIF mono-resistance | 37 (86.0) | 6 (14.0) | 15.73 (2.85–86.8) | <0.001 |
| MDR | 54 (60.7) | 35 (39.3) | 62.9 (11.3–350.5) | <0.001 |
| Pre-XDR | 16 (45.7) | 19 (54.3) | 115.2 (14.5–912.6) | <0.001 |
| FQs resistance in MDR (n = 124) | ||||
| No | 54 (60.7) | 35 (39.3) | 1 | |
| Yes | 16 (45.7) | 19 (54.3) | 1.83 (0.77–4.36) | 0.13 |
Abbreviations: PZA, pyrazinamide; INH, isoniazid; RIF, rifampin; MDR, multidrug resistant.
P value for trend of chi-square test.
3.4. Transmission of PZA-R isolates
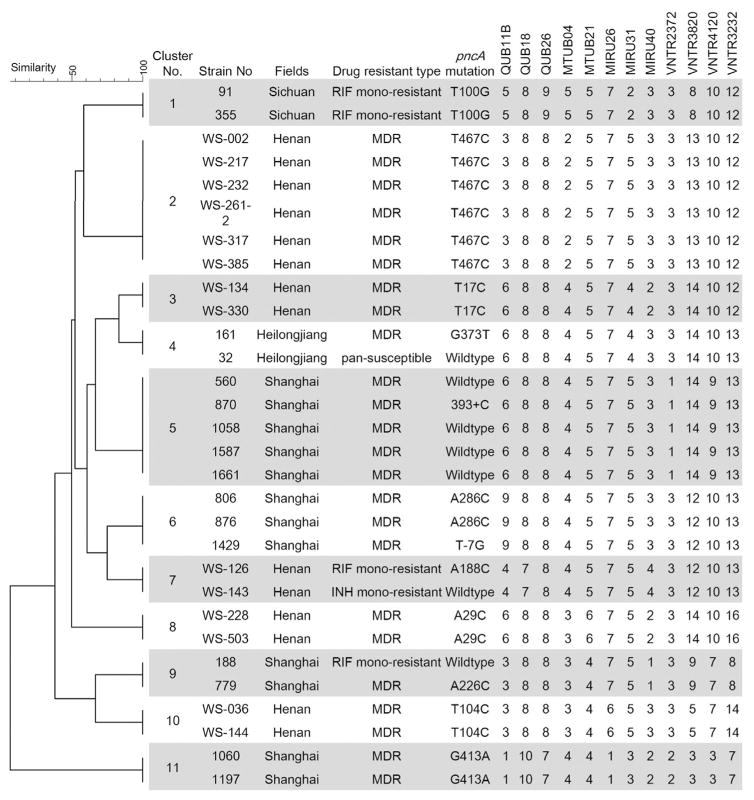
The emergence of drug resistance could be due to acquired drug resistance caused during treatment, or primary drug resistance caused by transmission of drug resistant isolates. Drug resistance in newly diagnosed TB patients is generally considered as primary drug resistance, while drug resistant TB patients with clustered isolates are suggestive of recent transmission. Among the 66 PZA-R cases, 48.5% (32/66) were newly diagnosed TB patients, indicating the occurrence of primary drug resistance. In order to identify potential recent transmission of PZA-R isolates during the study period, we analyzed the VNTR data of all 493 isolates. Results showed that PZA-R isolates were involved in 11 clusters, in which 7 clusters had at least 2 isolates with identical pncA mutations (Figure 1). Clustered isolates shared the identical pncA mutation, suggesting recent transmission of PZA-R isolates. Patients’ information showed that five PZA-R patients in cluster 2 and two patients in cluster 3 lived in the same village, thus providing probable epidemiological links for the recent transmission of PZA-R M. tuberculosis.
Figure 1.
The clustered isolates with pncA mutations. 12 loci VNTR based dendrogram and isolates profiles of 11 clusters in which pncA mutant isolates were involved. Abbreviations: INH, isoniazid; RIF, rifampin; MDR, multidrug resistant.
4. Discussion
This study covered five settings located in different provinces of China. We selected all drug resistant and randomly selected drug susceptible M. tuberculosis isolates, to predict PZA-R prevalence by mDST. The results showed that 81.8% of PZA-R isolates were MDR-TB isolates; and that MDR, pre-XDR, RIF mono-resistance and retreatment were risk factors for PZA-R. Furthermore, we observed that 48.5% of the PZA-R was primary drug resistance. Clustered isolates with identical pncA mutations were also detected, suggesting recent transmission of PZA-R isolates.
PZA is an important anti TB drug, both the WHO and the China CDC recommend PZA as a key component for treating new and re-treated patients [20]. However, PZA-R isolates can lead to serious negative effects on TB treatment [4]. Therefore, it is necessary to modify the treatment regimens of patients with PZA-R isolates. Our data showed the PZA-R rate among MDR isolates was as high as 43.5%. Hence, for MDR-TB, it is of importance to perform PZA susceptibility testing before prescribing the treatment regimens.
In this study, nearly half of the PZA-R cases were new TB patients, in the United States, the national PZA-R tuberculosis survey also found that up to 76.5% of the PZA-R MDR strains are new TB patients [21]. These PZA-R cases were regarded as primary drug resistance caused by remote or recent transmission of PZA-R strains. Our data showed that 27.3% (18/66) PZA-R isolates shared the identical pncA mutation and VNTR genotype, indicating recent transmission. For example, in Henan field site, the cluster rate of PZA-R isolates was as high as 52.2% (12/23). However, the study of MDR strains in Russia showed that identical pncA mutations existed in smaller clusters, suggesting a weak transmissibility of PZA-R isolates [22]. Whether pncA mutations induce a fitness cost that impairs transmission still need further study.
Most (81.2%, 39/48) of pncA mutation types identified in this study are associated with a high confidence of PZA resistance according to at least one TB drug resistance databases [23,24] or have been reported in PZA resistant strains [11–13,25–32] (Table 1), suggesting a strong correlation with PZA phenotypic resistance. But the mDST of PZA has an inevitable number of false positive and false negative results, there may be about 10% undetected and wrong defined PZA-R isolates by this method [10].
5. Conclusion
By detecting pncA mutation of M. tuberculosis isolates from five provinces of China, we have shown that the PZA-R rate was as high as 43.5% in MDR isolates. Our results also showed that MDR, pre-XDR, RIF mono-resistance and retreatment were risk factors for PZA-R. Finally, since transmission of PZA-R isolates play important role in emergence of PZA-R TB, it is necessary to regularly conduct PZA susceptibility testing among MDR-TB patient and modify the treatment regimens accordingly.
Acknowledgments
We thank the patients and the health-care workers of the Sichuan Wusheng CDC, Guangxi Pingguo CDC, Henan Weishi CDC, Shanghai Songjiang CDC and Heilongjaing Wuchang CDC, for their generous support and cooperation.
Funding
This study was supported by grants from Ministry of Science and Technology of China (2014DFA30340) and the National Science and Technology Major Project (2013ZX10004903-006). This study also supported by the Fogarty International Center Global Health Fellowship R25TW009343 (to C. Yang).
Footnotes
Competing interests
None declared.
Ethical approval
The Ethics Committees of the Institutes of Biomedical Sciences in Fudan University and the Shanghai Municipal Center for Disease Control and Prevention, Shanghai, approved the protocol for this study.
References
- 1.Steele MA, Des Prez RM. The role of pyrazinamide in tuberculosis chemotherapy. Chest. 1988;94:845–50. doi: 10.1378/chest.94.4.845. [DOI] [PubMed] [Google Scholar]
- 2.Tasneen R, Li SY, Peloquin CA, Taylor D, Williams KN, Andries K, et al. Sterilizing activity of novel TMC207- and PA-824-containing regimens in a murine model of tuberculosis. Antimicrob Agents Chemother. 2011;55:5485–92. doi: 10.1128/AAC.05293-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y, Mitchison D. The curious characteristics of pyrazinamide: a review. Int J Tuberc Lung Dis. 2003;7:6–21. [PubMed] [Google Scholar]
- 4.Zhang Y, Chang KC, Leung C-C, Yew WW, Gicquel B, Fallows D, et al. ‘Zs-MDR-TB’ versus ‘Zr-MDR-TB’: improving treatment of MDR-TB by identifying pyrazinamide susceptibility. Emerg Infect Dis. 2012;1:e5. doi: 10.1038/emi.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andries K, Verhasselt P, Guillemont J, Gohlmann HW, Neefs JM, Winkler H, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–7. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 6.Nuermberger E, Tyagi S, Tasneen R, Williams KN, Almeida D, Rosenthal I, et al. Powerful bactericidal and sterilizing activity of a regimen containing PA-824, moxifloxacin, and pyrazinamide in a murine model of tuberculosis. Anti-microb Agents Chemother. 2008;52:1522–4. doi: 10.1128/AAC.00074-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenthal IM, Zhang M, Williams KN, Peloquin CA, Tyagi S, Vernon AA, et al. Daily dosing of rifapentine cures tuberculosis in three months or less in the murine model. PLoS Med. 2007;4:e344. doi: 10.1371/journal.pmed.0040344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chedore P, Bertucci L, Wolfe J, Sharma M, Jamieson F. Potential for erroneous results indicating resistance when using the Bactec MGIT 960 system for testing susceptibility of Mycobacterium tuberculosis to pyrazinamide. J Clin Microbiol. 2010;48:300–1. doi: 10.1128/JCM.01775-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Permar S, Sun Z. Conditions that may affect the results of susceptibility testing of Mycobacterium tuberculosis to pyrazinamide. J Med Microbiol. 2002;51:42–9. doi: 10.1099/0022-1317-51-1-42. [DOI] [PubMed] [Google Scholar]
- 10.Chang KC, Yew WW, Zhang Y. Pyrazinamide susceptibility testing in Myco-bacterium tuberculosis: a systematic review with meta-analyses. Antimicrob Agents Chemother. 2011;55:4499–505. doi: 10.1128/AAC.00630-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui Z, Wang J, Lu J, Huang X, Zheng R, Hu Z. Evaluation of methods for testing the susceptibility of clinical Mycobacterium tuberculosis isolates to pyrazinamide. J Clin Microbiol. 2013;51:1374–80. doi: 10.1128/JCM.03197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan Y, Hu Z, Zhang T, Cai X, Kuang H, Liu Y, et al. Role of pncA and rpsA gene sequencing in detection of pyrazinamide resistance in Mycobacterium tuberculosis isolates from southern China. J Clin Microbiol. 2014;52:291–7. doi: 10.1128/JCM.01903-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia Q, Zhao LL, Li F, Fan YM, Chen YY, Wu BB, et al. Phenotypic and genotypic characterization of pyrazinamide resistance among multidrug-resistant Mycobacterium tuberculosis isolates in Zhejiang, China. Antimicrob Agents Chemother. 2015;59:1690–5. doi: 10.1128/AAC.04541-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan RC, Hui M, Chan EW, Au TK, Chin ML, Yip CK, et al. Genetic and phenotypic characterization of drug-resistant Mycobacterium tuberculosis isolates in Hong Kong. J Antimicrob Chemother. 2007;59:866–73. doi: 10.1093/jac/dkm054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang C, Shen X, Peng Y, Lan R, Zhao Y, Long B, et al. Transmission of Mycobacterium tuberculosis in China: a population-based molecular epidemiologic study. Clin Infect Dis. 2015;61(2):219–27. doi: 10.1093/cid/civ255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang C, Luo T, Sun G, Qiao K, Sun G, DeRiemer K, et al. Mycobacterium tuberculosis Beijing strains favor transmission but not drug resistance in China. Clin Infect Dis. 2012;55:1179–87. doi: 10.1093/cid/cis670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lan R, Yang C, Lan L, Ou J, Qiao K, Liu F, et al. Mycobacterium tuberculosis and non-tuberculous mycobacteria isolates from HIV-infected patients in Guangxi, China. Int J Tuberc Lung Dis. 2011;15:1669–75. doi: 10.5588/ijtld.11.0036. [DOI] [PubMed] [Google Scholar]
- 18.Luo T, Jiang L, Sun W, Fu G, Mei J, Gao Q. Multiplex real-time PCR melting curve assay to detect drug-resistant mutations of Mycobacterium tuberculosis. J Clin Microbiol. 2011;49:3132–8. doi: 10.1128/JCM.02046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ginsburg AS, Grosset JH, Bishai WR. Fluoroquinolones, tuberculosis, and resistance. Lancet Infect Dis. 2003;3:432–42. doi: 10.1016/s1473-3099(03)00671-6. [DOI] [PubMed] [Google Scholar]
- 20.WHO. Treatment of tuberculosis: guidelines. World Health Organization; 2010. [PubMed] [Google Scholar]
- 21.Kurbatova EV, Cavanaugh JS, Dalton T, CES, Cegielski JP. Epidemiology of pyrazinamide-resistant tuberculosis in the United States, 1999–2009. Clin Infect Dis. 2013;57:1081–93. doi: 10.1093/cid/cit452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casali N, Nikolayevskyy V, Balabanova Y, Harris SR, Ignatyeva O, Kontsevaya I, et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet. 2014;46:279–86. doi: 10.1038/ng.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flandrois JP, Lina G, Dumitrescu O. MUBII-TB-DB: a database of mutations associated with antibiotic resistance in Mycobacterium tuberculosis. BMC Bioinform. 2014;15:107. doi: 10.1186/1471-2105-15-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, Murray MB. Tuberculosis drug resistance mutation database. PLoS Med. 2009;6:e2. doi: 10.1371/journal.pmed.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miotto P, Cabibbe AM, Feuerriegel S, Casali N, Drobniewski F, Rodionova Y, et al. Mycobacterium tuberculosis pyrazinamide resistance determinants: a multicenter study. MBio. 2014:5. doi: 10.1128/mBio.01819-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ando H, Mitarai S, Kondo Y, Suetake T, Sekiguchi JI, Kato S, et al. Pyrazinamide resistance in multidrug-resistant Mycobacterium tuberculosis isolates in Japan. Clin Microbiol Infect. 2010;16:1164–8. doi: 10.1111/j.1469-0691.2009.03078.x. [DOI] [PubMed] [Google Scholar]
- 27.Cuevas-Cordoba B, Xochihua-Gonzalez SO, Cuellar A, Fuentes-Dominguez J, Zenteno-Cuevas R. Characterization of pncA gene mutations in pyrazinamide-resistant Mycobacterium tuberculosis isolates from Mexico. Infect Genet Evol. 2013;19:330–4. doi: 10.1016/j.meegid.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 28.Jnawali HN, Hwang SC, Park YK, Kim H, Lee YS, Chung GT, et al. Characterization of mutations in multi- and extensive drug resistance among strains of Mycobacterium tuberculosis clinical isolates in Republic of Korea. Diagn Microbiol Infect Dis. 2013;76:187–96. doi: 10.1016/j.diagmicrobio.2013.02.035. [DOI] [PubMed] [Google Scholar]
- 29.Somoskovi A, Dormandy J, Parsons LM, Kaswa M, Goh KS, Rastogi N, et al. Sequencing of the pncA gene in members of the Mycobacterium tuberculosis complex has important diagnostic applications: identification of a species-specific pncA mutation in “Mycobacterium canettii” and the reliable and rapid predictor of pyrazinamide resistance. J Clin Microbiol. 2007;45:595–9. doi: 10.1128/JCM.01454-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou L, Osei-Hyiaman D, Zhang Z, Wang B, Yang A, Kano K. Molecular characterization of pncA gene mutations in Mycobacterium tuberculosis clinical isolates from China. Epidemiol Infect. 2000;124:227–32. doi: 10.1017/s0950268899003635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jonmalung J, Prammananan T, Leechawengwongs M, Chaiprasert A. Surveillance of pyrazinamide susceptibility among multidrug-resistant Mycobacterium tuberculosis isolates from Siriraj Hospital, Thailand. BMC Microbiol. 2010;10:223. doi: 10.1186/1471-2180-10-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell PJ, Morlock GP, Sikes RD, Dalton TL, Metchock B, Starks AM, et al. Molecular detection of mutations associated with first- and second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2011;55:2032–41. doi: 10.1128/AAC.01550-10. [DOI] [PMC free article] [PubMed] [Google Scholar]