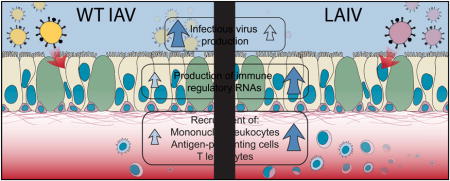
Abstract
The host innate immune response to influenza virus is a key determinant of pathogenic outcomes and long-term protective immune responses against subsequent exposures. Here, we present a direct contrast of the host responses in primary differentiated human nasal epithelial cell (hNEC) cultures following infection with either a seasonal H3N2 influenza virus (WT) or the antigenically-matched live-attenuated vaccine (LAIV) strain. Comparison of the transcriptional profiles obtained 24 and 36 hours post-infection showed that the magnitude of gene expression was greater in LAIV infected relative to that observed in WT infected hNEC cultures. Functional enrichment analysis revealed that the antiviral and inflammatory responses were largely driven by type III IFN induction in both WT and LAIV infected cells. However, the enrichment of biological pathways involved in the recruitment of mononuclear leukocytes, antigen-presenting cells, and T lymphocytes was uniquely observed in LAIV infected cells. These observations were reflective of the host innate immune responses observed in individuals acutely infected with influenza viruses. These findings indicate that cell-intrinsic type III IFN-mediated innate immune responses in the nasal epithelium are not only crucial for viral clearance and attenuation, but may also play an important role in the induction of protective immune responses with live-attenuated vaccines.
Keywords: influenza virus, host response, live-attenuated vaccine, inflammation, interferon, epithelial cells
Graphical Abstract
INTRODUCTION
Influenza A viruses (IAV) remain one of the most relevant human pathogens with annual epidemics resulting in 250,000 to 500,000 deaths worldwide (Girard et al., 2005). Annual vaccination against influenza viruses is required due to frequent antigenic drift and inefficient cross-protective immunity from prior infections or vaccination. Although vaccination remains the best approach against infection and disease, the identification of new circulating strains and the time for production and distribution present constraints on vaccine development. Additionally, other host-driven factors can contribute to the efficacy of influenza vaccines (Del Giudice and Rappuoli, 2015). Defining host molecular responses—and the viral and host factors interactions that trigger and regulate them—is therefore essential to developing effective vaccines.
Live-attenuated influenza vaccine (LAIV) strains are based on the cold-adapted master donor virus A/Ann Arbor/6/1960 (H2N2). The 6:2 reassortant vaccine viruses contain the RNA segments encoding the hemagglutinin (HA) and neuraminidase (NA) surface antigens derived from circulating wild type influenza virus (WT) and the six internal RNA segments derived from the master donor virus. LAIV has been described to have three phenotypic changes that relate to its reduced ability to cause disease: i) it causes reduced disease in animal models compared to seasonal strains of influenza (attenuation, or at) ii) has restricted replication at temperatures at or above 39°C (temperature sensitivity or ts) and iii) is able to efficiently replicate at temperatures as low as 25°C (cold-adaptation or ca). Although several pre-clinical (Chen et al., 2010; Cheng et al., 2013; Lanthier et al., 2011; Lau et al., 2012) and clinical studies (He et al., 2006; Hoft et al., 2011; Nakaya et al., 2011) have demonstrated that LAIVs are safe, efficient, and an effective means of vaccination, the mediators and correlates of protection still remain poorly understood.
In this study, we use a whole-genome transcriptional approach to elucidate the early response to wild-type (WT) A/Victoria/361/2011 (H3N2) influenza A virus (A/Victoria/361/2011) and an antigenically matched LAIV strain in primary differentiated human nasal epithelial cell (hNEC) cultures. We demonstrate that LAIV elicits robust antiviral responses, which may also restrict viral replication. LAIV also induces enhanced chemokine secretion and that downstream transcriptional profiles following infection with vaccine strains are predictive of increased leukocyte and lymphocyte recruitment to sites of virus replication which may contribute to the generation of a strong adaptive immune responses. These responses are reflective of those observed in the nasal epithelium during acute infection of humans with IAV. Thus, primary differentiated hNEC cultures can model the innate immune responses present in influenza-infected individuals and provide insight into the molecular mechanisms of LAIV efficacy.
MATERIALS AND METHODS
Viruses
Seasonal influenza A virus A/Victoria/361/2011 (H3N2) and an antigenically-matched live attenuated influenza vaccine virus A/Victoria/361/2011 LAIV (LAIV; HA and NA RNA segments from A/Victoria/361/2011, and other RNA segments from A/Ann Arbor/6/1960) were utilized in this study. Virus seed stocks were obtained from Medimmune and virus sequences verified using MiSeq to obtain sequences for all virus segments. The viruses are referred to as H3N2 WT or LAIV in the manuscript. Working stocks were generated by infecting confluent MDCK cells at an MOI of 0.01 for 48 to 72 hours in DMEM supplemented with 0.3% bovine serum albumin, 2mM GlutaMax, 100 U/ml Penicillin, 100 μg/ml Streptomycin, and 4 μg/ml N-acetyl Trypsin (NAT). The supernatants were then harvested, clarified by low speed centrifugation, and aliquoted for storage at −70°C.
Cell culture conditions
Madin-Darby canine kidney (MDCK) cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS), 2mM GlutaMax, 100 U/ml Penicillin and 100 μg/ml Streptomycin and maintained in a humidified environment at 37°C with 5% CO2.
Human nasal epithelial cell (hNECs) cultures (male and female donors) were obtained from non-diseased tissue during endoscopic sinus surgery for non-infection related conditions and grown in culture at the air-liquid interface (ALI) as previously described (Lane et al., 2004; Ramanathan and Lane, 2007; Ramanathan et al., 2007). Female and male donors were used and the ages were between 18–49 years old. Tissue processing, differentiation medium, and culture conditions have been previously described in detail (Fischer et al., 2015).
Infection of primary differentiated hNECs
Differentiated hNEC cultures were infected at a multiplicity of infection (MOI) of one (high MOI) or 0.03 (low MOI) 50% tissue culture infectious dose (TCID50) per cell. All incubations were performed at 32°C in a humidified environment with 5% CO2. Prior to infection, the apical surface of the cultures was washed with 200 μl DMEM (supplemented with 0.3% BSA, 2mM Glutamax, 100 U/ml Penicillin, and 100 μg/ml Streptomycin) and the basolateral media was replaced with 500–1,000 μl of fresh LHC Basal Medium:DMEM-H. The virus inoculum was added to the apical compartment in a volume of 100 μl and incubated for 1 hour, after which the inoculum was aspirated and the apical surface was washed three times with 200 μl phosphate buffered saline containing calcium and magnesium (PBS+). The plates were then returned to the incubator. Both apical washes and basolateral medium were collected and stored at −70°C. Virus production in apical washes was quantified by determining the 50% tissue culture infectious dose (TCID50) using MDCK cells (Lalime and Pekosz, 2014) and the Reed-Muench algorithm (Reed, 1938).
Microarray experiments and data processing
RNA was isolated from Trizol homogenates of mock, WT or LAIV infected cells harvested at 24hpi and 36hpi Fluorescent-labeled probes were generated from each sample using Agilent one-color LowInput Quick Amp Labeling Kit (Agilent Technologies). All infected samples were confirmed to express viral M2 mRNA by quantitative RT-PCR (Forero et al., 2015; McCown and Pekosz, 2005). Individual cRNA samples were then hybridized to oligonucleotide microarrays for gene expression profiling using SurePrint G3 Human Gene Expression v2 Microarray Kit (G4851A; Agilent Technologies).
The primary transcriptomic data was extracted and quantile normalized using the ‘normalize Between Arrays’ method available in the ‘limma’ package of the R statistical computing software suite and adjusted for batch effects using the ComBat software (Johnson et al., 2007). Differential expression (DE) of H3N2 WT and LAIV was determined by comparing the average ratio of virus-infected replicates to time-matched and donor-matched mock-infected samples based on a linear model fit using the ‘limma’ package. Criteria for differential expression were an absolute fold-change of 1.5 and an adjusted p-value of <0.05, calculated by Benjamini-Hochberg correction (Benjamini and Hochberg, 1995).
Functional enrichment analysis
Functional analysis of DE genes was done using Ingenuity Pathway Analysis (IPA, Ingenuity Systems). IPA canonical pathway enrichment was calculated using a right-tailed Fisher’s exact test with a threshold of significance set at p-values 0.05. Enrichment of Diseases and Biological Functions and upstream regulators analysis were based on activation |Z-scores| >2 and p-values < 0.05. Virus-infected cells were compared to time- and donor-matched mock-infected samples.
Gene expression profile correlations
Correlation analysis of influenza signatures to cytokine response gene expression profiles derived from hNECs was performed using data available from the NCBI-GEO database under the following accession number: GSE19182. Spearman correlation coefficients (r) and confidence intervals were calculated using the ‘psych’ package in R statistical computing software suite. Virus-infected cells were compared to time- and donor-matched mock-infected samples.
Chemokine and cytokine measurements
Secreted chemokines and proinflammatory cytokines were quantified from the basolateral samples of both low and high MOI infections using multiplex ELISAs (MesoScale Diagnostics): V-PLEX Human Chemokine Panel 1 (Eotaxin, Eotaxin-3, IP-10 (CXCL10), MCP-1, MCP-4, MDC, MIP-1α, MIP-1β, TARC, IL-8) and Human ProInflammatory Tissue Culture 9-Plex (GM-CSF, IFN-γ, IL-1β, IL-10, IL-12p70, IL-2, IL-6, IL-8, TNF-α), respectively, following manufacturer instructions. Samples were diluted 1:2 for the proinflammatory cytokine panel and 1:4 for the chemokine panel in the provided reagent diluents and run in duplicate. Plates were washed as described above. All incubations were performed sealed at room temperature with shaking (3000 rpm). Before reading, 2x read buffer was added and the plates were incubated for 5 minutes (ProInflammatory 9-Plex) or read immediately (Chemokine Panel 1 V-Plex) on a Meso Scale Discovery SECTOR Imager 2400. Protein concentrations were calculated using the MesoScale Diagnostics Discovery Workbench (version 4.0.12).
Interferon ELISAs
IFN-λ and IFN-β secretion were quantified from both the apical and basolateral compartments by singleplex enzyme-linked immunosorbant assay (ELISA). The IFN-β ELISA (VeriKine Human Interferon Beta ELISA Kit; Pestka Biomedical Laboratories, Inc) was performed according to manufacturer’s instructions. IFN-λ ELISAs (DIY Human IFN Lambda 3/1/2 (IL-28B/29/28A, Pestka Biomedical Laboratories, Inc.) were performed following manufacturer’s instructions with the following modifications: Costar 96-well Half-Area Plates (Corning) were coated with 50 μl of IFN-λ capture antibody overnight at room temperature, washed, and then blocked for at least an hour in 100 μl PBS with 1% BSA. Due to the reduced volume of the plates, all volumes from the protocol were halved.
For both assays, samples were diluted 1:4 in PBS with 1% BSA and analyzed in duplicate. The sealed plates were incubated at room temperature (with the exception of the tetramethyl-benzidine, TMB, development). Plates were washed three times with 150 ul of wash buffer (PBS and 0.05% Tween-20) on an AquaMax 4000 plate washer and then blotted dry between sample, detection antibody, and streptavidin-horseradish peroxidase incubations. Results were read on a FilterMax F5 microplate reader at 450 nm (with a 540 nm reference for IFN-λ) and protein concentrations were determined using SoftMax Pro 6.4.
RNA sequencing
Total RNA was extracted from nasal swabs collected from three individuals (2 males and 1 female) confirmed to be infected with H3N2 viruses by PCR in the 2012–13 influenza season. The individuals showed symptoms of acute respiratory infection and either fever or cough/sore throat at time of enrollment and symptom onset occurred no longer than 4 days before enrollment. Samples from the time of enrollment (day 0) or three days after enrollment (day 3) were used to assess the host responses during acute infection. Samples from day 7 post enrollment were used as baseline values because the patients were symptom free at that time. Libraries for RNA sequencing were constructed using TruSeq RNA Access kit (Illumina). The libraries were clonally amplified and sequenced on a NextSeq 500 sequencing system (Illumina). General quality control of the raw reads was performed using FastQC. Ribosomal RNA reads were removed via mapping by Bowtie (v2.1.0) (Langmead and Salzberg, 2012) using an index of human, mouse, and rat rRNA sequences. The number of read pairs following ribosomal RNA removal in each sample varied from 20 million to 100 million read pairs. Each sample was randomly sampled down to 20 million reads to adjust for these differences. For each sample, about 90% of remaining reads were mapped against the human reference genome (hg19, build GRCh37, from the UCSC genome browser using STAR (v2.4.0h1) (Dobin et al., 2013). Quantitative gene counts were produced utilizing the human annotation associated with the genome using the python package HT-Seq (Anders et al., 2015). Differential gene expression analysis was done using edgeR (Robinson et al., 2010). Statistical significance cutoff was set at a fold change > |2| and an adjusted p-value of 0.05.
Statistical analysis
Statistical analyses for viral RNA expression and high MOI virus titers were performed using t-tests. Data from virus replication kinetics and protein secretion were analyzed using a Multiple Analysis of Variance (MANOVA), with time and virus as variables in GraphPad Prism version 5.0 (GraphPad software Inc. La Jolla, CA); p-values of < 0.05 were considered significant. * indicates a significant differences between WT and LAIV samples while # indicates a difference from mock-infected cultures.
Data dissemination
The hNEC microarray expression data and the nasal swab RNA-seq expression data generated in this study are available via the following accession identifiers on the NCBI-GEO database: (GSE83285, GSE83215 and GSE81951).
RESULTS
Influenza A virus replication in primary differentiated human nasal epithelial cells
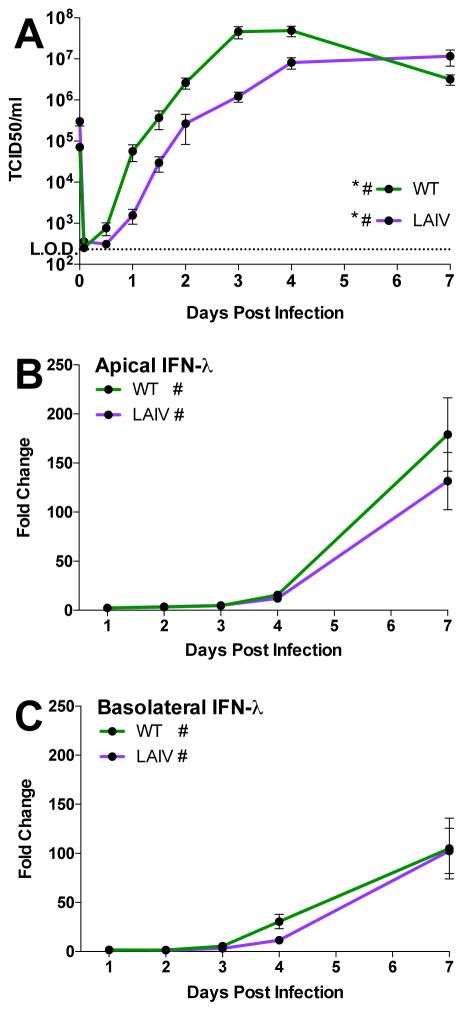
To better understand the response to WT and LAIV infection in differentiated hNEC cultures, cells were infected with a seasonal H3N2 isolate (A/Victoria/361/2011; WT) and the antigenically-matched LAIV strain at a high multiplicity of infection (MOI= 1). Infectious virus production was determined at at 24 and 36 hours post-infection (hpi). There was a ~10-fold decrease in infectious virus production at 24 hpi (Fig. 1A, left) for LAIV compared to WT virus. At 36 hpi, there were no observable differences in infectious virus production (Fig 1B, right). No differences in viral mRNA transcription were detected in infected hNEC cultures at either 24 hpi (Fig. 1A, left) or 36 hpi (Fig. 1B, right). These data confirm that LAIV infection in hNEC cultures leads to reduced infectious virus particle production but equivalent viral gene expression when compared to WT infection which has been previously demonstrated with older LAIV strains (Fischer et al., 2014; Fischer et al., 2015).
Figure 1. Gene expression and replication of WT H3N2 and antigenically-matched live-attenuated viruses in differentiated hNEC cultures.
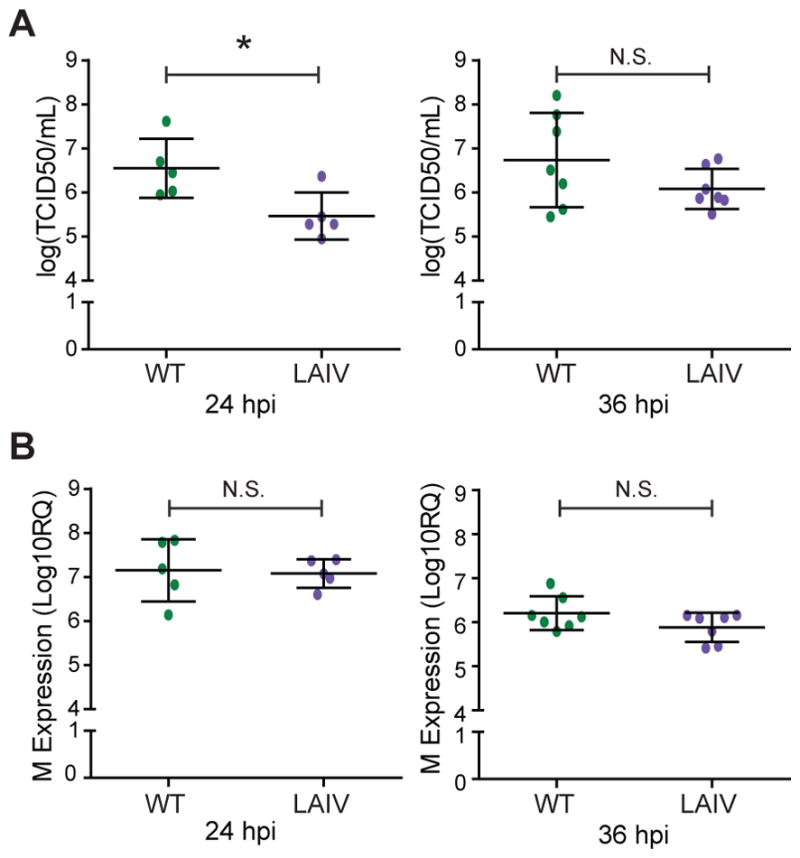
Differentiated human nasal epithelial cell cultures were infected at a multiplicity of infection (MOI) of 1 and incubated at 32°C. (A) Measurement of infectious virus particle production in apical supernatant of infected hNEC cells. (B) Expression of cell-associated viral M2 mRNA expression was determined by quantitative RT-PCR. * indicates p>0.05. Three to four infected wells were averaged for the data points shown and each data point represents one donor with 5–7 donors were used for each time point.
The host transcriptional response to LAIV infection is more robust than that observed with WT virus infection
Having confirmed that replication of the H3N2 LAIV strain is attenuated in hNECs, the transcriptional responses to WT and LAIV strains were evaluated to better understand host-driven factors that could contribute to viral restriction. Significant changes in gene expression (fold change >|1.5|) in virus- compared to mock-infected cells were detected at both 24 hpi (Fig. 2A) and 36 hpi (Fig. 2B). Multidimensional scaling was used to visualize the expression of the 432 differentially-expressed genes (DE) across all infections. The molecular signatures that correspond to both WT and LAIV infection are robust, as they diverge from the transcriptional profiles of mock-infected samples (Fig. 2C–D). Importantly, gene expression across donors showed a high degree of reproducibility across replicates as well as concordance amongst distinct donors. Between the virus strains, consistent responses were identified at both 24 hpi (Fig. 2C) and 36hpi (Fig. 2D), largely due to the observation that a majority of DE genes were shared amongst strains (Fig. 2A–B). However, LAIV infection elicited a greater number of significant DE genes, which was more pronounced by 36hpi (Fig. 2B).
Figure 2. Global analysis of the transcriptional responses to WT and antigenically-matched LAIV in differentiated hNEC cultures.
(A and B) Venn Diagrams of overlap in the response to infection with WT and LAIV in hNEC cultures. Number of differentially expressed (DE) genes following virus infection relative to time-matched mocks. Differential gene expression cutoff was set to fold change >1.5 and a q-value < 0.05 calculated using a moderated t-test with subsequent Benjamini-Hochberg correction. Multidimensional scaling (MDS) representation of the similarities in the differential transcriptional profiles elicited by viral infection at (C) 24 hpi (D) and 36 hpi. Each donor is represented by a distinct shape. Biological replicates are represented as single points and unique color distinguishes infectious conditions. The quality of the representation is provided by the Kruskal Stress value, with the low percentage of Kruskal stress suggesting a faithful 2D representation of global transcriptional differences between viral strains. (E) Average ratio of gene expression between LAIV and WT infected cells. Heatmap represents the log2 fold expression changes of 253 genes displaying a 1.2 fold-change of expression in LAIV- over WT-infected cells relative to donor-matched, mock-infected cells.
Host responses to viral infection are aimed at preventing viral replication and promoting pathogen clearance. To better understand the differences in the host response to infection with distinct viral strains, the subset of genes within the previously defined DE subset that showed greater than |1.2| fold-change differences between LAIV and WT viruses was examined further. Almost 60 percent of the DE genes (253 total) were found to be more impacted by LAIV infection (Fig. 2E). Together, these data suggest that while the host response to both circulating H3N2 viruses and antigenically-matched LAIV strains is likely regulated by similar molecular pathways across distinct donors, the magnitude of these responses is enhanced in LAIV-infected cells.
LAIV infection results in enhanced type III interferon-mediated responses relative to WT virus infection
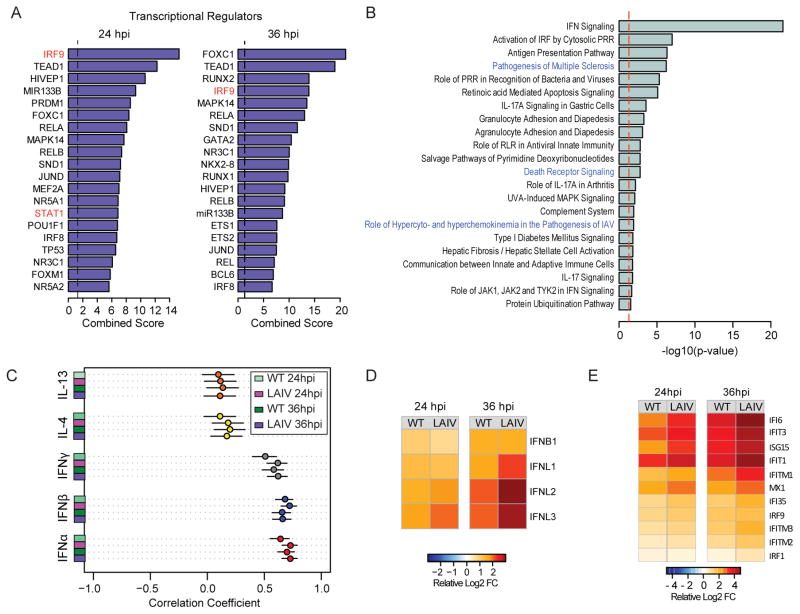
To define the host regulatory pathways involved in attenuation of LAIV strains, the transcription factors mediating the expression of the 253 genes found to be more differentially expressed in LAIV infection at both 24hpi (Fig. 3A) and 36hpi (Fig. 3B) were evaluated. Overall, the transcriptional factors predicted to regulate the LAIV signature (IRF9, RELA, RELB, STAT1, JUN) have been previously associated with the regulation of innate antiviral responses (reviewed by (Hoffmann et al., 2015)). Canonical pathway enrichment analysis revealed that the most significantly enriched (Enrichment score >1.3) pathways are involved in pathogen recognition and interferon (IFN) synthesis, response to IFN, and antigen presentation (Fig. 3B).
Figure 3. The host response to influenza is regulated by type I and III interferon responses in hNEC cultures.
(A) Prediction of upstream regulatory transcription factors that control the enhanced LAIV response following infection in hNEC cultures. ChEA was utilized to predict transcription factors previously found to be associated with the regulation of gene expression of the 253 genes showing enhanced differential expression following LAIV infection. Bar graphs represent combined enrichment scores for each factor. (B) Gene set enrichment of canonical pathways of the 253 genes showing enhanced differential expression following LAIV infection derived using IPA. Bar graph represents pathways with significant enrichment scores (enrichment score > 1.3) Bar length represents the enrichment scores, which are based on the −log10 p-value as determined by Fisher’s exact test. (C) Meta-analysis of hNEC transcriptional responses following cytokine stimulation. Responses to WT and LAIV infection were correlated to those observed following treatment with type I IFN (IFNα and IFNβ; red and blue circles, respectively), type II IFN (IFNγ; grey) and Th2-polarizing cytokines, IL-4 and IL-13 (yellow and orange circles, respectively). Each bubble represents the Spearman correlation coefficient, and the confidence interval is represented by the black bar. (D) Relative RNA expression of type I and type III IFN genes. Heatmaps represent the average ratio of differential expression (log2 fold change) relative to donor-matched, mock-infected samples. (E) Relative RNA expression of interferon responsive genes. Heatmap represent the average ratio of differential expression (log2 fold change) relative to donor-matched, mock-infected samples.
IFN secretion exerts pleiotropic antiviral effects by autocrine and paracrine activation of cell membrane-associated receptors, coordinating the late phase of viral recognition and further amplifying antiviral effector functions. To identify key antiviral cytokines that coordinate the response to IAV and LAIV in hNEC cultures, the transcriptional signature captured in our study was compared with those observed following stimulation of primary differentiated respiratory epithelial cells by type I IFN, type II IFN or Th-2 polarizing cytokines (Giovannini-Chami et al., 2012) (Fig. 3C). Genes responsive to both WT and LAIV viruses were highly, positively correlated to genes responsive to type I IFN (IFNα and IFNβ). On the other hand, a moderate positive correlation with type II IFN responses, and no correlation with the transcriptional responses to Th2 polarizing cytokines, IL-4 and IL-13 was observed.
The type I IFN response profiles are similar to those elicited by type III IFN genes and are coordinated by overlapping signal transduction molecules (Crotta et al., 2013; Zhou et al., 2007), IRF9 and STAT1, which we have predicted to drive the more robust LAIV-specific response in hNECs. While the secretion of type I and type III IFNs occurs in multiple cell types, type III IFN receptor mediated responses are restricted to dendritic and epithelial cells which express the type III IFN receptor (Ank et al., 2008). To elucidate the role of type I and III IFN in the transcriptional response to infection in hNECs, the relative expression of IFN genes following infection (Fig. 3D) was evaluated. Amongst all the DE genes following infection, significant increases were detected in IFNβ and the IFNλ genes by 24 hpi. While there was no difference in the magnitude of IFNB1 gene expression following WT or LAIV infection, induction of IFNλ genes was greater in LAIV-infected compared to WT-infected cultures. The contrast in the IFNλ genes was most evident by 36 hpi. Concomitant with the increased expression of these genes, was the enhanced induction of downstream IFN-stimulated genes in LAIV-infected cells (Fig. 3E). IFNλ receptor expression was not altered in response to infection. Together, these findings suggest that the attenuation of LAIV infection in hNEC cultures is correlated with enhanced induction of type III IFN-dependent innate antiviral responses.
LAIV induces greater pro-inflammatory and chemotactic responses in infected hNEC cultures
Studies into the correlates of protection of LAIV have demonstrated that unlike the inactivated vaccines, the protective nature of LAIV is independent of serum antibodies (Nakaya et al., 2011; Sasaki et al., 2007). Moreover, it has been demonstrated that the immunoprotection by LAIV vaccines is associated with the induction of T cell-mediated immunity (He et al., 2006; Hoft et al., 2011; Subbramanian et al., 2010). To assess whether pro-inflammatory and chemotactic responses induced by WT virus infection differ from those induced by LAIV infection, the enrichment of genes known to affect biological functions associated with the crosstalk between innate and adaptive immune responses was examined. A unique, significant enrichment of genes involved in the chemotaxis, activation, and maturation of immune cells was observed, which dominated by 36hpi in LAIV-infected cultures (Fig. 4A).
Figure 4. LAIV infection is associated with enhanced cytokine and chemokine induction in hNEC cultures.
(A) Gene set enrichment of biological functions pertaining to immune cell recruitment and regulation derived from IPA. Bubble plot representation of significant enrichment scores (activation z-score >2) in at least one timepoint in cells infected with LAIV. Significant enrichment is denoted by red color. Activation z-scores < 2 are denoted in white and grey. Crosses signify a lack of enrichment. Bubble diameter represents the −log10 p-value as determined by Fisher’s exact test. (B) Measurement of CXCL10 production in the basolateral medium in infected hNEC cells at 36 hpi. Graphs represent the log2 fold change in cytokine expression relative to donor-matched, mock-infected cells (left). Ratio of CXCL10 expression in LAIV-infected cells relative to WT- infected cells (right). Values greater than zero denote increased cytokine production in LAIV-infected cells relative to WT-infected cells. (C) Differential expression of cytokine and chemokine genes. Heatmap represent the average ratio of differential expression (log2 fold change) relative to donor-matched, mock-infected samples.
Transcription factors that regulate cytokine and chemokine expression (Fig. 3A) and pro-inflammatory canonical pathways (Fig. 3B, blue) were amongst the subset of genes that were more highly expressed in LAIV-infected cultures. At 36 hpi, where the induction of chemotactic responses was most robust, there was an increase in the secretion of the CXCR3 ligand CXCL10 in both WT and IAV-infected hNEC cultures with a trend toward more production in LAIV-infected cultures (Fig. 4B). This was consistent with a significant increase in mRNA expression (Fig. 4C), of CXCR3 targeting genes CXCL9 and CXCL10. Together, these data demonstrate that LAIV induces epithelial cell responses that may drive the increased recruitment of lymphocytes and leukocytes to the infected nasal epithelium.
Differentiated hNECs recapitulate the response H3N2 observed during acute infection
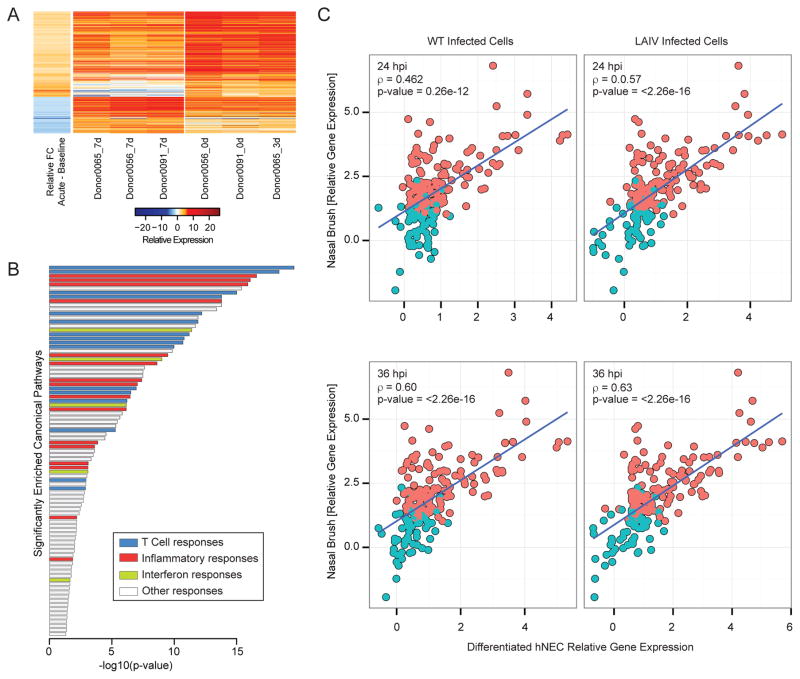
Given that hNEC cultures can capture distinct transcriptional changes following virus infection, we evaluated whether these responses were reflective of the transcriptional changes observed in the infected nasal epithelium of IAV-infected individuals. Genome-wide transcriptional profiles were determined from nasal swabs of individuals acutely infected with H3N2 IAV (Fig. 5A). Acute gene responses (day 0 or 3 after influenza diagnosis) were compared to responses at day 7 post infection (considered baseline responses). Genes that underwent significant changes in expression (fold change >|2|, p-value < 0.05) were identified in pathways involved in T cell-specific, inflammatory, and antiviral functions, amongst other immune and metabolic functions (Fig. 5B). The concordance of the in vivo gene expression changes with those identified in virus-infected hNEC cultures was then determined (Fig. 5C). Overall, the expression changes captured in hNEC cultures following infection with either WT or LAIV, showed moderate correlation with the gene expression patterns observed in the nasal epithelium during acute influenza infection. These results demonstrate that primary differentiated hNEC cultures are a relevant model for evaluating the host transcriptional response to WT and LAIV infection.
Figure 5. Host transcriptional profiling in nasal swabs of patients infected with H3N2 influenza virus.
Gene expression profile in nasal washes derived from H3N2-infected donors. (A) Heatmap represents the log2-transformed gene expression at baseline (7 days post-visit) or during acute infection (time of visit or 3 days post initial visit, as indicated) across three donors (2 male and 1 female). The average ratio of differential expression (donor-matched acute infection samples vs. baseline samples) is presented in the first column. Columns 2–7 represent the log2-transformed gene expression at baseline (columns 2–4) or at time of initial clinical visit (columns 5–7). (B) Gene set enrichment of canonical pathways of the 577 genes differentially expressed early during acute infection relative to baseline using IPA. Bar graph represents pathways with significant enrichment scores (enrichment score > 1.3). Bar length represents the enrichment scores, which are based on the −log10 p-value as determined by Fisher’s exact test. Bar color determines the distinct biological functions that the enriched canonical pathways are predicted to affect. (C) Correlation of hNEC and nasal epithelium-derived transcriptional responses following IAV infection. Responses to WT and LAIV infection in hNEC cultures (x-axis) were correlated to those observed during acute infection with H3 IAV (y-axis). Each bubble in the graph represents a DE gene in hNEC cultures. Pink color represents genes that were determined to be DE in the nasal brush, while blue color represents genes that did not pass statistical cut-off for differential expression in the nasal brush. The Spearman correlation coefficient, r, is depicted for each contrast.
LAIV replicates less efficiently, but induces a proportionally greater innate immune response than WT at late times post infection in a differentiated hNEC model of a natural infection
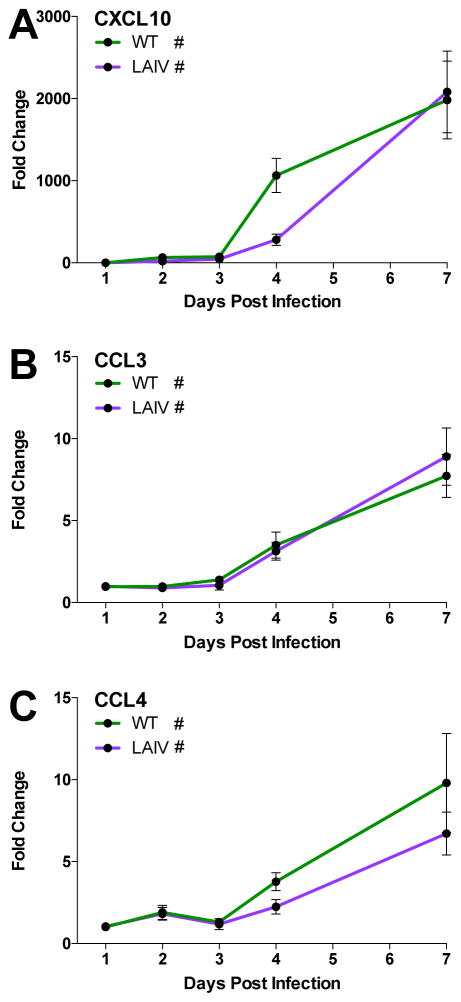
To determine if transcriptional responses observed in high MOI hNEC infections and from in vivo influenza infected human samples were reflected in the production of IFN, chemokines and cytokines, hNEC cultures were infected with WT and LAIV virus at a low MOI and virus replication as well as interferon and chemokine protein production was assessed at various times post infection. As in high MOI infections, LAIV infectious virus production is reduced in hNEC cultures relative to WT (Fig. 6A), both in kinetics and peak titer. Given the reduced virus production in LAIV-infected cells, we then assessed the induction of various innate immune factors after WT or LAIV infection. IFNλ production is significantly elevated following infection with both viruses, in both the apical and basolateral media relative to mock-infected cultures (Figure 6B and 6C), mirroring the predicted type III IFN production. There was no statistically significant difference in IFNλ production between WT and LAIV-infected cultures despite reduced infectious virus production in LAIV infections. Basolateral secretion of the chemokines CXCL10, CCL3, and CCL4 increases after 3days post infection in WT- and LAIV-infected hNEC cultures compared to mock infected cultures, but there is no statistical difference in production between WT and LAIV-infected cells (Fig. 7A–C) – again, despite reduced LAIV virus replication compared to WT virus. CXCL10 was detected during high MOI infections (Fig. 4B) and showed induction in both WT and LAIV infections (Fig. 7A). Secretion of the macrophage inflammatory proteins CCL3 and CCL4 (Fig. 7B and C) confirm the transcriptional responses involving activation of leukocyte recruitment pathways (Fig. 4A). Therefore, while the overall induction of innate immune factors is similar in LAIV and WT infected hNEC cultures at the protein level, the response is proportionally greater during LAIV infection given the reduced infectious virus production seen in LAIV-infected hNEC cultures.
Figure 6. LAIV- and WT-infected hNEC cultures produce similar amounts of type III IFNs despite different levels of infectious virus production.
Apical and basolateral media from hNEC cultures infected with a low MOI of WT or LAIV were harvested and assessed for viral replication (A) or interferon lambda production (B, C). During exponential replication (until 4 days post infection), LAIV replication is significantly reduced relative to WT (p<0.0001, MANOVA). (B) Apical and (C) basolateral type III IFN secretion is significantly elevated relative to donor-matched mock-infected cells but is similar between the two viral infections. Cultures from 6 donors (3 males and 3 females) were used with n=3–4 wells per timepoint per culture. Data represented as fold change over mock. (* = p≤0.05 for WT to LAIV comparison, # = p<0.05 compared to mock-infected samples, MANOVA)
Figure 7. Chemokine secretion into the basolateral media of WT- and LAIV-infected hNEC cultures.
(A) CXCL10, (B) CCL3, and (C) CCL4 secretion is significantly greater than in donor-matched mock infected cells for both WT and LAIV infections, however there is no significant difference in the fold change of production induced between the two viruses. Basolateral supernatants from hNEC cultures infected with the indicated viruses were harvested at the indicated times and assessed for chemokine production using the MSD chemokine analysis platform. Cultures from 6 donors (3 males and 3 females) were used with n=3–4 wells per timepoint per culture. Data represented as fold change over mock. (* = p≤0.05 for WT to LAIV comparison, # = p<0.05 compared to mock-infected samples, MANOVA)
DISCUSSION
In this study, the response of differentiated human nasal epithelial cell cultures to WT and LAIV infection in vitro was examined at the transcriptional and protein level to identify the factors associated with LAIV-mediated immune protection. The hNEC cultures faithfully recapitulated the transcriptional responses present in nasal swabs of humans infected with influenza, particularly in modeling T cell-specific, inflammatory, and antiviral functions, amongst other immune and metabolic pathways. The isolation and differentiation of nasal epithelial cells provides a model to study the host epithelial cell responses at the initial site of replication, allowing both transcript analysis and confirmation of protein synthesis and secretion. This provides a targeted focus for investigating host factors involved in early immune recognition and response to WT and LAIV.
While both WT and LAIV replicate in the upper airways, only WT is able to reach the lower lungs. Despite this, many in vitro studies have used cells derived from the lower airways, such as the A549 alveolar epithelial cell carcinoma line or primary cell cultures derived from the trachea, bronchioles and alveoli (Chan et al., 2008; Ilyushina et al., 2012; Jin et al., 2003; Pyo and Zhou, 2014; Snyder et al., 1988; Snyder et al., 1985). While these studies are useful for examining pathogenesis following lung infection, they fail to faithfully recreate the environment initially encountered by WT and LAIV. There are significant differences between the cells of the upper and lower airways, including factors such as sialic acid distribution (reviewed in (Kumlin et al., 2008)) and innate immune response (Comer et al., 2012).
Importantly, there is also a temperature difference between the two environments: the lower airways are maintained at 37°C but the nasal epithelium is cooler, with an average temperature of 32°C. Influenza infections are often done at 37°C and this may impact the induction of innate immune responses (Foxman et al., 2015). The experiments in this study were conducted at 32°C to mimic the temperature normally experienced by nasal epithelial cells.
As previously demonstrated for older LAIV strains (Fischer et al., 2014; Fischer et al., 2015), the replication of recent H3N2 LAIV strains was impaired relative to that of WT viruses even at a permissive temperature, suggestive of additional, epithelial-specific attenuations mediating LAIV replication efficiency. Infection with WT and LAIV resulted in a significant induction of genes primarily involved in pathogen-recognition, IFN synthesis, response to IFN stimulation, and inflammation. The stronger induction of innate immune responses in LAIV- compared to WT-infected hNECs may be one factor contributing to LAIV attenuation in hNEC cultures.
In accordance with previous studies comparing seasonal H1N1 IAV and antigenically-matched LAIV (Fischer et al., 2014), we demonstrated that LAIV can elicit enhanced innate immune responses as compared to a seasonal H3N2 IAV (WT) infection. Our study expands upon the previous finding by demonstrating that heightened innate immune responses are conserved in LAIV strains in an HA and NA antigen-independent manner. Furthermore, the current study provides a comprehensive view on the transcriptional landscape in response to infection with WT and LAIV strains. Specifically, we reveal that in hNEC cultures, the innate immune response to influenza infection is largely driven by type I and type III IFN. However, while viral recognition results in the induction of both IFNB1 and IFNL1-3 genes, LAIV infection results in a unique enhancement of the IFNL genes. While expression and secretion of type I and type III IFN following viral recognition is ubiquitous amongst cell types, the specific response to type III IFN is restricted to dendritic and epithelial cells (Ank et al., 2008).
We demonstrate a differential induction of CXCR3 ligands, CXCL10, CXCL9, and CXCL11, which have been shown to play an important role in the recruitment of monocytes and lymphocytes to the infected tissue (Wareing et al., 2004). Importantly, CXCL10 also plays an important role in coordinating the generation of effector and memory CD8+T cells. Prior studies aimed at uncovering the correlates of protection for LAIV have demonstrated that unlike TIV vaccination, where serum HAI antibodies correlate well with protection from infection, LAIV vaccination does not elicit robust neutralizing antibody responses (Nakaya et al., 2011). Rather, LAIV is thought to induce robust cellular responses and protective mucosal immune responses (He et al., 2006; Sasaki et al., 2007; Subbramanian et al., 2010). This agrees with our observation of increased secretion of the macrophage inflammatory proteins CCL3 and CCL4 following viral infection.
Our data demonstrate that viral titer may not be solely responsible for the degree of innate immune activation. The reduced infectious virus production in LAIV-infected hNEC cultures was still able to induce as strong of a response as WT infection and suggests a proportionally greater immune response to LAIV. This stronger activation of epithelial cell innate immune responses may play a role in the induction of LAIV-induced immune responses and compensate for the reduced replication of LAIV.
Acknowledgments
The research in this manuscript was supported by the NIAID network of Centers of Excellence in Influenza Research and Surveillance (CEIRS), under contracts HHSN272201400005C and HHSN272201400007C, NIH Office of the Director P51OD010425, T32AI007417 and R01AI097417. We acknowledge MedImmune Inc. for providing LAIV for use in these experiments. The findings and conclusions in this report are those of the authors and do not necessarily reflect the views of the funding agency.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ank N, Iversen MB, Bartholdy C, Staeheli P, Hartmann R, Jensen UB, Dagnaes-Hansen F, Thomsen AR, Chen Z, Haugen H, Klucher K, Paludan SR. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. J Immunol. 2008;180:2474–2485. doi: 10.4049/jimmunol.180.4.2474. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- Chan W, Zhou H, Kemble G, Jin H. The cold adapted and temperature sensitive influenza A/Ann Arbor/6/60 virus, the master donor virus for live attenuated influenza vaccines, has multiple defects in replication at the restrictive temperature. Virology. 2008;380:304–311. doi: 10.1016/j.virol.2008.07.027. [DOI] [PubMed] [Google Scholar]
- Chen GL, Lamirande EW, Jin H, Kemble G, Subbarao K. Safety, immunogencity, and efficacy of a cold-adapted A/Ann Arbor/6/60 (H2N2) vaccine in mice and ferrets. Virology. 2010;398:109–114. doi: 10.1016/j.virol.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Zengel JR, Suguitan AL, Jr, Xu Q, Wang W, Lin J, Jin H. Evaluation of the humoral and cellular immune responses elicited by the live attenuated and inactivated influenza vaccines and their roles in heterologous protection in ferrets. J Infect Dis. 2013;208:594–602. doi: 10.1093/infdis/jit207. [DOI] [PubMed] [Google Scholar]
- Comer DM, Elborn JS, Ennis M. Comparison of Nasal and Bronchial Epithelial Cells Obtained from Patients with COPD. PLoS ONE. 2012;7:e32924. doi: 10.1371/journal.pone.0032924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotta S, Davidson S, Mahlakoiv T, Desmet CJ, Buckwalter MR, Albert ML, Staeheli P, Wack A. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog. 2013;9:e1003773. doi: 10.1371/journal.ppat.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Giudice G, Rappuoli R. Inactivated and adjuvanted influenza vaccines. Curr Top Microbiol Immunol. 2015;386:151–180. doi: 10.1007/82_2014_406. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer WA, 2nd, Chason KD, Brighton M, Jaspers I. Live attenuated influenza vaccine strains elicit a greater innate immune response than antigenically-matched seasonal influenza viruses during infection of human nasal epithelial cell cultures. Vaccine. 2014;32:1761–1767. doi: 10.1016/j.vaccine.2013.12.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer WA, 2nd, King LS, Lane AP, Pekosz A. Restricted replication of the live attenuated influenza A virus vaccine during infection of primary differentiated human nasal epithelial cells. Vaccine. 2015;33:4495–4504. doi: 10.1016/j.vaccine.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forero A, Tisoncik-Go J, Watanabe T, Zhong G, Hatta M, Tchitchek N, Selinger C, Chang J, Barker K, Morrison J, Berndt JD, Moon RT, Josset L, Kawaoka Y, Katze MG. The 1918 Influenza Virus PB2 Protein Enhances Virulence through the Disruption of Inflammatory and Wnt-Mediated Signaling in Mice. J Virol. 2015;90:2240–2253. doi: 10.1128/JVI.02974-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxman EF, Storer JA, Fitzgerald ME, Wasik BR, Hou L, Zhao H, Turner PE, Pyle AM, Iwasaki A. Temperature-dependent innate defense against the common cold virus limits viral replication at warm temperature in mouse airway cells. Proc Natl Acad Sci U S A. 2015;112:827–832. doi: 10.1073/pnas.1411030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannini-Chami L, Marcet B, Moreilhon C, Chevalier B, Illie MI, Lebrigand K, Robbe-Sermesant K, Bourrier T, Michiels JF, Mari B, Crenesse D, Hofman P, de Blic J, Castillo L, Albertini M, Barbry P. Distinct epithelial gene expression phenotypes in childhood respiratory allergy. Eur Respir J. 2012;39:1197–1205. doi: 10.1183/09031936.00070511. [DOI] [PubMed] [Google Scholar]
- Girard MP, Cherian T, Pervikov Y, Kieny MP. A review of vaccine research and development: human acute respiratory infections. Vaccine. 2005;23:5708–5724. doi: 10.1016/j.vaccine.2005.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XS, Holmes TH, Zhang C, Mahmood K, Kemble GW, Lewis DB, Dekker CL, Greenberg HB, Arvin AM. Cellular immune responses in children and adults receiving inactivated or live attenuated influenza vaccines. J Virol. 2006;80:11756–11766. doi: 10.1128/JVI.01460-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HH, Schneider WM, Rice CM. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 2015;36:124–138. doi: 10.1016/j.it.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoft DF, Babusis E, Worku S, Spencer CT, Lottenbach K, Truscott SM, Abate G, Sakala IG, Edwards KM, Creech CB, Gerber MA, Bernstein DI, Newman F, Graham I, Anderson EL, Belshe RB. Live and inactivated influenza vaccines induce similar humoral responses, but only live vaccines induce diverse T-cell responses in young children. J Infect Dis. 2011;204:845–853. doi: 10.1093/infdis/jir436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyushina NA, Ikizler MR, Kawaoka Y, Rudenko LG, Treanor JJ, Subbarao K, Wright PF. Comparative study of influenza virus replication in MDCK cells and in primary cells derived from adenoids and airway epithelium. J Virol. 2012;86:11725–11734. doi: 10.1128/JVI.01477-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Lu B, Zhou H, Ma C, Zhao J, Yang CF, Kemble G, Greenberg H. Multiple amino acid residues confer temperature sensitivity to human influenza virus vaccine strains (FluMist) derived from cold-adapted A/Ann Arbor/6/60. Virology. 2003;306:18–24. doi: 10.1016/s0042-6822(02)00035-1. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Kumlin U, Olofsson S, Dimock K, Arnberg N. Sialic acid tissue distribution and influenza virus tropism. Influenza Other Respir Viruses. 2008;2:147–154. doi: 10.1111/j.1750-2659.2008.00051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalime EN, Pekosz A. The R35 residue of the influenza A virus NS1 protein has minimal effects on nuclear localization but alters virus replication through disrupting protein dimerization. Virology. 2014;458–459:33–42. doi: 10.1016/j.virol.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane AP, Saatian B, Yu XY, Spannhake EW. mRNA for genes associated with antigen presentation are expressed by human middle meatal epithelial cells in culture. Laryngoscope. 2004;114:1827–1832. doi: 10.1097/00005537-200410000-00028. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanthier PA, Huston GE, Moquin A, Eaton SM, Szaba FM, Kummer LW, Tighe MP, Kohlmeier JE, Blair PJ, Broderick M, Smiley ST, Haynes L. Live attenuated influenza vaccine (LAIV) impacts innate and adaptive immune responses. Vaccine. 2011;29:7849–7856. doi: 10.1016/j.vaccine.2011.07.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau YF, Wright AR, Subbarao K. The contribution of systemic and pulmonary immune effectors to vaccine-induced protection from H5N1 influenza virus infection. J Virol. 2012;86:5089–5098. doi: 10.1128/JVI.07205-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCown MF, Pekosz A. The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging. J Virol. 2005;79:3595–3605. doi: 10.1128/JVI.79.6.3595-3605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR, Kasturi SP, Khan N, Li GM, McCausland M, Kanchan V, Kokko KE, Li S, Elbein R, Mehta AK, Aderem A, Subbarao K, Ahmed R, Pulendran B. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12:786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo HM, Zhou Y. Protective efficacy of intranasally administered bivalent live influenza vaccine and immunological mechanisms underlying the protection. Vaccine. 2014;32:3835–3842. doi: 10.1016/j.vaccine.2014.04.065. [DOI] [PubMed] [Google Scholar]
- Ramanathan M, Jr, Lane AP. Innate immunity of the sinonasal cavity and its role in chronic rhinosinusitis. Otolaryngol Head Neck Surg. 2007;136:348–356. doi: 10.1016/j.otohns.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Ramanathan M, Jr, Lee WK, Dubin MG, Lin S, Spannhake EW, Lane AP. Sinonasal epithelial cell expression of toll-like receptor 9 is decreased in chronic rhinosinusitis with polyps. Am J Rhinol. 2007;21:110–116. doi: 10.2500/ajr.2007.21.2997. [DOI] [PubMed] [Google Scholar]
- Reed LJMH. A simple method of estimating fifty percent endpoints. The American Journal of Hygiene. 1938;27:493–497. [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, Jaimes MC, Holmes TH, Dekker CL, Mahmood K, Kemble GW, Arvin AM, Greenberg HB. Comparison of the influenza virus-specific effector and memory B-cell responses to immunization of children and adults with live attenuated or inactivated influenza virus vaccines. J Virol. 2007;81:215–228. doi: 10.1128/JVI.01957-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder MH, Betts RF, DeBorde D, Tierney EL, Clements ML, Herrington D, Sears SD, Dolin R, Maassab HF, Murphy BR. Four viral genes independently contribute to attenuation of live influenza A/Ann Arbor/6/60 (H2N2) cold-adapted reassortant virus vaccines. J Virol. 1988;62:488–495. doi: 10.1128/jvi.62.2.488-495.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder MH, Clements ML, De Borde D, Maassab HF, Murphy BR. Attenuation of wild-type human influenza A virus by acquisition of the PA polymerase and matrix protein genes of influenza A/Ann Arbor/6/60 cold-adapted donor virus. J Clin Microbiol. 1985;22:719–725. doi: 10.1128/jcm.22.5.719-725.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbramanian RA, Basha S, Shata MT, Brady RC, Bernstein DI. Pandemic and seasonal H1N1 influenza hemagglutinin-specific T cell responses elicited by seasonal influenza vaccination. Vaccine. 2010;28:8258–8267. doi: 10.1016/j.vaccine.2010.10.077. [DOI] [PubMed] [Google Scholar]
- Wareing MD, Lyon AB, Lu B, Gerard C, Sarawar SR. Chemokine expression during the development and resolution of a pulmonary leukocyte response to influenza A virus infection in mice. J Leukoc Biol. 2004;76:886–895. doi: 10.1189/jlb.1203644. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Hamming OJ, Ank N, Paludan SR, Nielsen AL, Hartmann R. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. J Virol. 2007;81:7749–7758. doi: 10.1128/JVI.02438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]