Abstract
Recent advances in mass spectrometry-based proteomics have revealed translation of previously nonannotated microproteins from thousands of small open reading frames (smORFs) in prokaryotic and eukaryotic genomes. Facile methods to determine cellular functions of these newly discovered microproteins are now needed. Here, we couple semiquantitative comparative proteomics with whole-genome database searching to identify two nonannotated, homologous cold shock-regulated microproteins in Escherichia coli K12 substr. MG1655, as well as two additional constitutively expressed microproteins. We apply molecular genetic approaches to confirm expression of these cold shock proteins (YmcF and YnfQ) at reduced temperatures and identify the noncanonical ATT start codons that initiate their translation. These proteins are conserved in related Gram-negative bacteria and are predicted to be structured, which, in combination with their cold shock upregulation, suggests that they are likely to have biological roles in the cell. These results reveal that previously unknown factors are involved in the response of E. coli to lowered temperatures and suggest that further nonannotated, stress-regulated E. coli microproteins may remain to be found. More broadly, comparative proteomics may enable discovery of regulated, and therefore potentially functional, products of smORF translation across many different organisms and conditions.
Keywords: proteogenomics, proteomics, genomics, label-free quantitation, E. coli, cold shock, microprotein, small open reading frame, non-AUG start codon, stress response
Introduction
Small open reading frames (smORFs) of <100 amino acids are widespread in all genomes, but they remain largely nonannotated because they have been under-detected by computational genome annotation algorithms and proteomics protocols.1 In recent years, new technologies including smORF-focused computational genome analysis,1−4 liquid chromatography/tandem mass spectrometry (LC–MS/MS)-based proteomics coupled with deep sequencing,5−8 and ribosome footprinting/deep sequencing (RIBO-seq)9,10 have revealed thousands of translated smORFs in prokaryotic and eukaryotic genomes. While it has become clear that many smORF-encoded microproteins play important roles in biology,11 there remains a need to determine what fraction of newly discovered microproteins are functional, especially because many exhibit low sequence conservation with known proteins.6,11
Methods to couple discovery of nonannotated microproteins to quantitative analysis of their expression regulation may provide insights into their potential biological functions. For example, Storz and colleagues demonstrated that expression of some smORFs in bacteria is stress-inducible,2 leading to the hypothesis that smORF-encoded microproteins may function in stress responses. However, while efforts toward quantitative proteogenomics have been reported,12−17 LC–MS/MS proteogenomics has generally lagged behind RIBO-seq in differential analysis of nonannotated microprotein expression.2,9 To address this need, we have applied a label-free quantitative proteogenomic workflow to identify novel microproteins that exhibit stress-regulated expression in Escherichia coli.
We chose the cold shock response in E. coli as a model system. Cold shock is a condition under which bacteria are abruptly exposed to low temperatures (in practice, 10 °C). This causes arrest in global protein synthesis while inducing expression of a subset of proteins known as cold shock proteins. The most profoundly cold-inducible proteins are the homologues of CspA, which generally act as nucleic acid chaperones to restore transcription and protein translation at low temperatures.18 All of the nine known CspA homologues (CspA–CspI) in E. coli K12 are less than 80 amino acids in length. Therefore, we hypothesized that nonannotated small proteins could also be induced during cold shock. In this work, we compared nonannotated small protein expression in E. coli cells growing at normal and reduced temperatures. We identified four nonannotated sequences, two of which were found downstream of cspG and cspI and were upregulated by cold shock. We further characterized the noncanonical ATT start codon that initiates translation of these genes and demonstrated their conservation in closely related bacteria.
Methods
Strains and Constructs
E. coli K12 substr. MG1655 and pKD46 plasmids were a gift from Jason Crawford (Yale University). For generation of SPA tagged proteins, the tag was introduced at the C-terminal end using the method described by Uzzau et al. using bacteriophage λ recombination.19,20 Colonies on LB plates with kanamycin were screened for recombination, and the presence of the SPA tag at the C-terminus of the respective genes was verified by PCR and confirmed by sequencing. Primers for genomic tagging and integration check PCR are provided in Table S2.
For recombinant expression, the genetic region encompassing cspG–ymcF or cspI–ynfQ was PCR amplified from an E. coli K12 substr. MG1655 colony and cloned into pET 28b using restriction sites NcoI and XhoI (New England Biolabs) to yield a His6 tag at the C-terminal end of and in frame with YmcF and YnfQ proteins. All mutations were introduced by site-directed mutagenesis using inverse PCR.21
Stress Conditions for Mass Spectrometry
Stress conditions were adapted from Hemm et al.2 as follows: Approximately 500 mL of LB was inoculated with a 1:100 dilution of an overnight culture of MG1655 cells. The cells were grown at approximately 37 °C in a flask with a stir bar until they reached an OD600 between 0.4 and 0.5. The cells were split into two fractions. The control remained at 37 °C, and the cold shock sample was incubated at 10 °C for 1 h (starting from the time that the culture reached 10 °C). All cells were pelleted at 4000g for 10 min at 4 °C. The cells were resuspended in a smaller volume and transferred to a 50 mL conical tube. The cells were again pelleted at 4000g for 10 min at 4 °C. The supernatant was removed, and the pellets were flash frozen and stored at −80 °C.
Cell Lysis and Protein Size Selection
Lysis and size selection were adapted from Ma et al.5 as follows: Frozen cells from the stress conditions were resuspended in lysis buffer (50 mM HCl and 0.1% β-mercaptoethanol). The resuspension was sonicated at 35% amplitude with eighteen 10 s bursts with a 20 s rest on a Fisher Scientific model 120 sonic dismembrator. Triton X-100 was added to the sample to a final concentration of 0.05%. The sample was heated for 10 min at greater than 95 °C, allowed to cool on ice for 10 min, and then pelleted by centrifugation for 30 min at 21 100g at 4 °C. The supernatant was removed, and the pellet was discarded. The supernatant was filtered through a 5 μm filter.
A Bond Elut C8 column (Agilent) preconditioned with 1 column volume of methanol followed by 2 column volumes of triethylammonium formate (TEAF) pH 3.0 was loaded with approximately 10 mg of protein per 100 mg of bed resin and washed with 2 column volumes of TEAF pH 3.0. Size-selected proteins were eluted with two column volumes of 3:1 acetonitrile/TEAF pH 3.0 and concentrated on a Savant SPD10 SpeedVac concentrator (Thermo Scientific).
Digestion of Samples for Mass Spectrometry
The concentrated sample was redissolved in water. The resuspension was precipitated with a methanol/chloroform extraction. The precipitate was resuspended in 31 μL of a solution of 8 M urea, 0.4 M Tris-HCl, and 20 mM calcium chloride; 3 μL of 45 mM dithiothreitol (DTT) was added to the solution, and the sample was incubated at 60 °C for 10 min. The reaction was placed on ice for 30 s and then incubated at room temperature for 3 min; 3 μL of 100 mM iodoacetamide was added, and the reaction was incubated at room temperature in the dark for 30 min. The reaction was quenched with 0.67 μL of DTT; 16 μL of 1 M Tris-HCl pH 8.0 was added. Trypsin (Promega) was added at a ratio of 1:50 trypsin/protein. Water was added to bring the urea concentration to 1 M. The digest was incubated at 37 °C overnight. The following day, the reaction was brought to 1% trifluoroacetic acid (TFA). The peptides were desalted using Nest Group MicroSpin columns (C18, 300 Å) and eluted in 80% acetonitrile/0.1% TFA. The elution was concentrated on a Savant SPD1010 SpeedVac concentrator (Thermo Scientific).
Offline Fractionation of Peptides
Peptides were fractionated prior to LC–MS/MS via electrostatic repulsion–hydrophilic interaction chromatography (ERLIC).22 Desalted samples were redissolved in 50 μL of 85% acetonitrile/0.1% formic acid and loaded on a polyWAX LP column (150 × 1.0 mm; 5 μm 300 Å; PolyLC) attached to an Agilent 1100 HPLC at a 0.05 mL/min flow rate. The samples were separated over an 80 min gradient as follows (solvent A: 80% acetonitrile, 0.1% formic acid; solvent B: 30% acetonitrile, 0.1% formic acid). Isocratic flow was maintained at 100% A at a flow rate of 0.3 mL/min for 5 min, followed by a 17 min linear gradient to 8% B and a 25 min linear gradient to 45% B. Finally, a 10 min gradient to 100% B was followed by a 5 min hold at 100% B before a 10 min linear gradient back to 100% A, followed by an 8 min hold at 100% A. Fractions were collected every several minutes, resulting in 15–17 samples for further LC–MS/MS analysis. Each fraction was vacuum-dried using a Savant SPD1010 SpeedVac concentrator (Thermo Scientific).
LC–MS/MS Analysis
LC–MS/MS methods were based on a previous report.23 The fractionated samples were resuspended in approximately 7 μL of 3:8 70% formic acid/0.1% TFA. Approximately 5 μL of each sample was injected onto a 150 μm × 3 cm trap column packed in-house with ReproSil-Pur 120 Å C18 resin (Dr. Maisch). Separation was carried out on a 75 μm × 20 cm PicoFrit analytical column packed in-house using 1.9 μm ReproSil-Pur 120 Å C18 resin (Dr. Maisch). Solvents A and B (0.1% formic acid and acetonitrile/0.1% formic acid, respectively) were delivered using a Nano Acquity UPLC (Waters) in-line with an LTQ Orbitrap Velos (Thermo Scientific). Samples were trapped for 6 min at a flow rate of 2.5 μL/min at 98% A. Isocratic flow was maintained at 0.3 μL/min at 2% B for 10 min, followed by linear gradients from 2 to 10% B over 2 min, 10 to 25% B over 58 min, 25 to 40% B over 10 min, and 40 to 95% B over 2 min. Isocratic flow at 95% B was maintained for 5 min, followed by a gradient from 95 to 2% B over 10 min (MS: 30 000 resolution, 298–1750 m/z scan range; dd-MS2: top10 method, 7500 resolution, 1.0 m/z isolation window, 35 NCE).
Data Analysis
ProteoWizard MS Convert24 was used for peak picking, and files were analyzed using Mascot Version 2.5.1 (Matrix Science, Inc., London, UK).25 Carbamidomethyl (C) was set as a fixed modification. Variable modifications included carbamyl (K and N-term), oxidation (M), and phospho (STY). The peptide mass error tolerance was 20 ppm. The parameters were set to a semitryptic digest with a maximum of three missed cleavages and peptide charge states limited to +2, +3, and +4. A six-frame translation of the MG1566 genome (accession number NC_000913.3 in NCBI) and the common contaminant database were searched, and the false discovery rate was adjusted to 1% using the homology threshold. Peptides fewer than 8 amino acids in length were excluded. Identified peptides were checked for annotation against the RefSeq database for MG1655. Putative nonannotated hits were BLASTed, and those that contained only one amino acid mismatch relative to annotated proteins were discarded. Protein identifications were made on the basis of unique peptide matches that had Mascot ions scores greater than 45, with a minimum of one ion in both b and y series and at least four consecutive ions in a series or multiple unique peptides that mapped to the same ORF.
Protein Expression
To test nonannotated protein expression, 10 mL of LB was inoculated with 200 μL of the genomically SPA-tagged cultures grown overnight to saturation at 37 °C. Wild-type E. coli K12 MG1655 was used as a control. The cultures were grown at 37 °C to log phase on a shaker and split into three tubes containing 2.5 mL of the culture. Each tube was transferred to water baths at 10 °C for 1 h (cold shock), 45 °C for 20 min (heat shock), or 37 °C for 1 h. In order to assess protein expression, an aliquot from each tube corresponding to 0.2 OD600 units was taken, trichloroacetic acid (TCA) was added immediately to a final concentration of 8%, and the samples were centrifuged at 14 000g for 15 min at 4 °C. Pellets were washed with acetone, air-dried, and resuspended in SDS gel loading buffer. Samples were heated at 90 °C for 2 min, and 10 μL of each sample was loaded on a 15% SDS-PAGE gel.
To test expression of proteins from the pET vector, a single colony of E. coli BL21 (DE3) Gold cells containing the plasmid construct was inoculated into 5 mL of LB with 40 μg/mL of kanamycin and grown overnight at 37 °C. 100 μL of this culture was used to inoculate 5 mL of LB/kanamycin and grown to log phase at 37 °C on a shaker. Isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to the cultures at a final concentration of 1 mM, and growth was continued at 37 °C for 1 h, after which 0.2 OD600 units was taken and subjected to TCA precipitation followed by SDS PAGE as described above. All gels were run in duplicate so one could be stained with Coomassie and the other could be subjected to western blotting. At least three biological replicates were carried out for each experiment reported.
Western Blotting
Gels were transferred to BioTrace nitrocellulose membranes (VWR) at 30 V for 16 h or at 100 V for 1 h. Blots were blocked in 3% BSA for 1 h at room temperature on a shaker. To probe for SPA-tagged proteins, 1:1000 dilution of mouse monoclonal anti-FLAG M2 (Sigma) primary antibody was incubated with the blot for 1 h, followed by washing with Tris buffered saline containing 0.1% Tween 20 (TBS-T). Goat anti-mouse secondary antibody (Rockland) at a dilution of 1:10 000 was incubated for 1 h, followed by washing with TBS-T. Blots were developed using Clarity ECL western blotting substrate (Bio-Rad) and imaged using a ChemiDoc imaging system (Bio-Rad) and Image Lab software (BioRad). For His6-tagged proteins, His tag antibody conjugated to biotin was used. For detection, streptavidin conjugated to AlexFluor 488 was incubated with the blot for 30 min, followed by washing and analysis of the blot by a Typhoon imaging system and Image Quant software (GE Life Sciences).
Changes in protein expression of SPA-tagged proteins were assessed by quantifying the bands using Image Lab software (Bio-Rad). After background subtraction, the fold change in expression was calculated by dividing the intensity of bands at 10 °C by those at 37 °C. At least three biological replicates were carried out for each protein as well as the wild-type E. coli K12 MG1655 control.
Results
Development of a Proteomics Workflow for Discovery of Nonannotated, Cold Shock-Inducible Proteins in E. coli
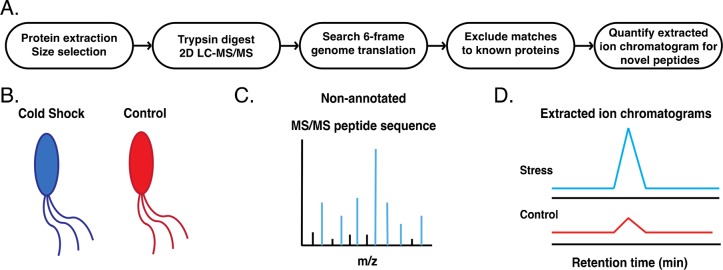
Figure 1 summarizes our comparative microprotein discovery platform. For high-sensitivity microprotein detection, we enriched the E. coli small proteome using a modification of previously reported workflows.5,6 First, we prepared stress and control samples by subjecting E. coli K12 substr. MG1655 cells growing at 37 °C in log phase to cold shock conditions (10 °C) for an hour, whereas control cells were maintained at 37 °C. Cells were lysed, and the small proteome was isolated using a C8 column that selectively retains microproteins and peptides.26 After trypsin digestion, peptides were separated by ERLIC, and each fraction was then analyzed by liquid chromatography and tandem mass spectrometry. We performed two biological replicates of the cold shock and control samples. We subsequently analyzed two additional biological replicates of the cold shock sample to assess reproducibility of protein identifications.
Figure 1.
Quantitative proteomic gene discovery in E. coli. (A) Schematic overview of the quantitative proteomics protocol. (B) Comparative analysis of nonannotated gene expression begins with parallel preparation of size-selected small proteome samples from control and experimental (cold shock) cells. (C) Nonannotated peptides are sequenced by searching their tandem mass spectrometry (MS/MS) spectra against a six-frame translation of the E. coli genome and excluding sequences matching known proteins. (D) Analysis of the peak area for the respective peptides in the extracted ion chromatograms (EICs) from MS1 spectra is used to quantify the level of upregulation relative to the control.
In order to identify all peptides in this sample, including those derived from nonannotated genomic regions, we searched these peptide fragmentation spectra against a six-frame translation of the E. coli K12 substr. MG1655 genome using MASCOT. Annotated proteins were then excluded using a string-matching algorithm6 with reference to the current E. coli K12 proteome, and, in order to conservatively exclude possible point mutants in our laboratory strain, we retained only those tryptic peptides that are at least two amino acids different from any annotated protein. Only search results yielding peptides having at least four consecutive b or y ions were considered for validation. These parameters not only greatly reduced the number of candidate peptides but also eliminated false positives. BLAST searches were performed on the candidate peptides to verify that they were unique in the E. coli genome. While single-peptide protein identifications were retained for confirmation, since many smORF-encoded microproteins yield only one detectable tryptic fragment,6 we note that two independent tryptic peptides support identification of two of our nonannotated protein hits (Table S1 and Figure S3). Tandem mass spectra for peptides that met our stringent criteria are shown in Figures 2 and S3, and peptide scores and related information are provided in Table S1.
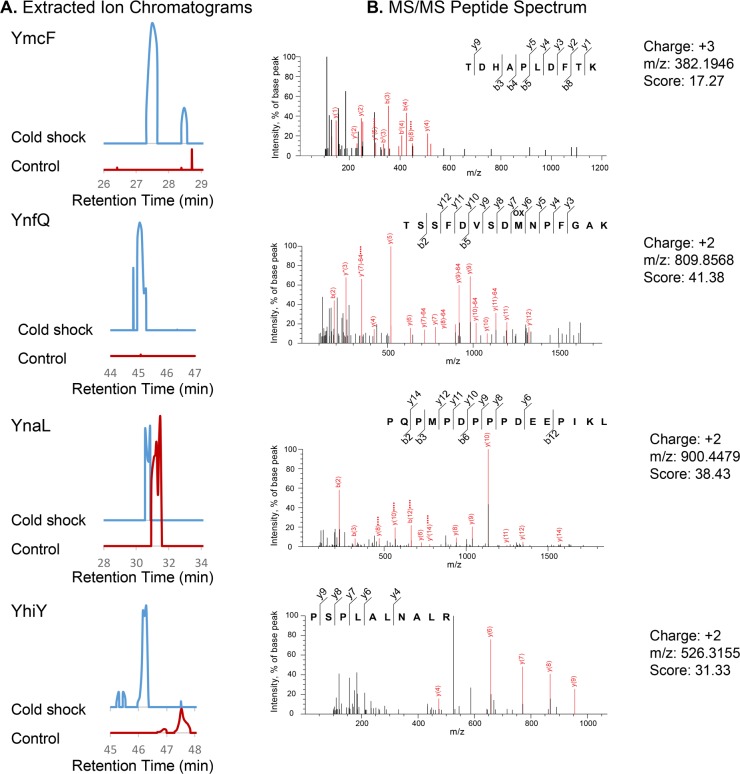
Figure 2.
Detection and semiquantitative analysis of nonannotated E. coli proteins. (A) Extracted ion chromatograms (EICs) from MS1 spectra corresponding to (B) MS/MS spectra of nonannotated tryptic peptides identified in our shotgun profiling experiments. The EIC intensity at the same retention time for a 1 Da window around the parent ion mass was compared for the control (red) vs cold shock (blue) samples. Each matched EIC pair is presented on the same y-axis scale. Because the analysis is semiquantitative, substantial intensity in both samples was taken to indicate similar expression. MS/MS spectra (right) presented correspond to the experimental EICs shown (left). Y- and b-ions are shown in red and indicated on the matched peptide scores above each spectrum. m/z, mass to charge ratio. Additional peptides corresponding to each protein as well as scores, precursor mass errors, and charge states corresponding to the MS/MS spectra in this figure can be found in Table S1.
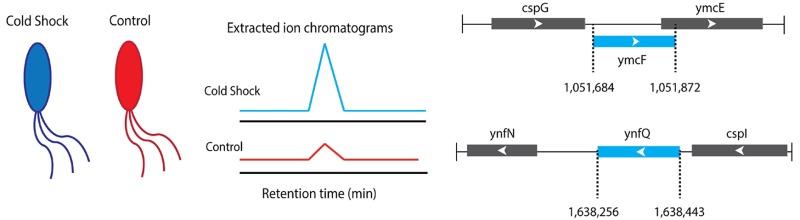
In order to identify differential expression, we utilized label-free quantitation.26−28 Briefly, we identified nonannotated proteins identified by Mascot search only in the control or stress condition. We then compared the area under the MS1 peak in the extracted ion chromatogram (EIC)29 for each of these peptides (Figure 2), providing quantitative confirmation of differential expression. As a control, we confirmed that proteins that do not change under the experimental cold shock condition exhibited constant MS1 ion intensity (Figure 2) and that upregulated MS1 intensity was observed for a peptide derived from a known cold shock protein (Figure S1). We also confirmed that, for each fraction analyzed, E. coli proteins known to be unresponsive to cold temperatures, such as ribosomal proteins, do not change in abundance in their MS1 peptide ion intensities, demonstrating that the changes we attribute to novel cold shock proteins are specific (Figure S1).
Identification of Genomic Loci Putatively Encoding Nonannotated Microproteins in E. coli
The genomic sequences corresponding to these candidate peptides were identified in order to define their full-length sequences. Our proteomics search results yielded peptides that map to four candidate nonannotated proteins in coding sequences currently annotated as intergenic. We propose to name these proteins YmcF, YnfQ, YnaL, and YhiY per convention for proteins of unknown function (Figure 3). We also identified a peptide putatively corresponding to the predicted protein YpaA (Figure S2). Comparative analysis of the EICs revealed that peptides from three of these smORFs—ynaL, yhiY, and ypaA—were present in both control and cold shocked cells (Figures 2 and S2). In contrast, peptides derived from ymcF and ynfQ were either not present in the control cells or dramatically enriched in the cold shocked cells compared to the control (Figure 2). We subsequently analyzed two cold shock sample replicates, demonstrating reproducible detection and sequencing of YmcF, YnfQ, and YnaL as well as two independent tryptic fragments supporting identification of YmcF and YnfQ, providing strong evidence for the reproducibility of their identifications (Table S1 and Figure S3). Taken together, these results suggest that comparative proteomics has the potential to identify both constitutive and regulated expression of nonannotated bacterial microproteins.
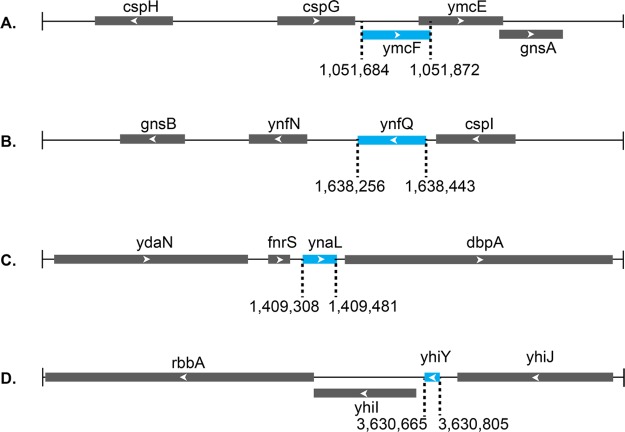
Figure 3.
Gene locus diagrams for nonannotated E. coli proteins YmcF (A), YnfQ (B), YnaL (C), and YhiY (D). Line represents chromosomal DNA, annotated protein-coding sequences are represented by gray boxes, and newly reported coding sequences are represented by blue boxes. Arrows indicate 5′–3′ directionality of the coding sequence. Sizes are proportional to length, and genomic coordinates of novel protein sequences are provided. Sizes of novel proteins were calculated either from the first in-frame ATG to stop codon (C, D) or from experimentally determined non-ATG start codons (A, B, vide infra).
Confirmation of Microprotein Expression and Cold-Shock Inducibility via Genomic Tagging
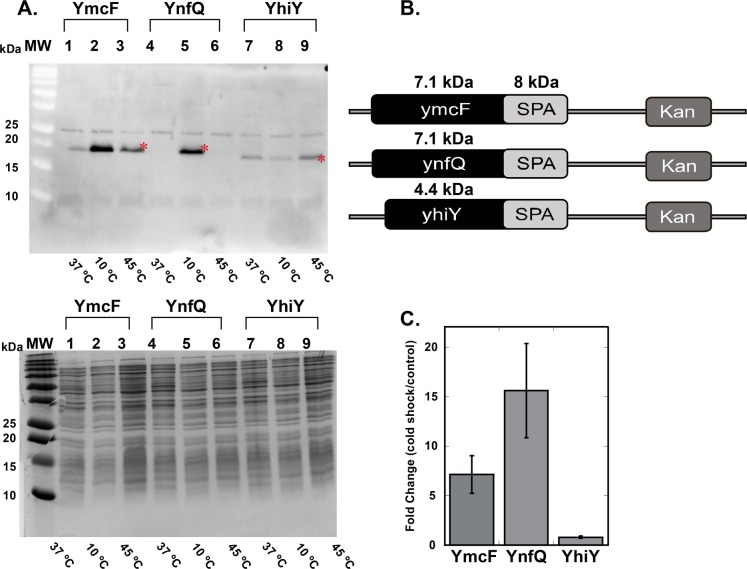
While bottom-up proteomics has proved powerful in identification of novel peptide sequences, full protein sequence coverage is rarely obtained. Therefore, this approach is insufficient to confirm assignment of observed peptides to genomic loci. Furthermore, since several of our novel protein identifications were based on single tryptic peptide-spectral matches, rigorous molecular confirmation of protein expression was required. In order to verify the smORFs encoding our putative microproteins, we generated epitope-tagged knock-in strains. The peptides identified by LC–MS/MS helped define the reading frame and stop codons for the genes that encode these proteins. For each locus, a C-terminal sequential epitope tag (SPA tag2) was added to the chromosomal copy of the candidate genes to report on expression without perturbing translation initiation (Figure S4). Protein expression under conditions of normal growth (37 °C) and cold shock (10 °C), with heat shock (42 °C) as an additional control for specificity of the cold shock response, was monitored by subjecting the respective cell lysates to SDS-PAGE followed by western blotting with an antibody against the FLAG tag that constitutes a portion of the SPA sequence.
We were able to detect robust expression of the YmcF, YnfQ, and YhiY proteins (Figure 4). Band densitometry showed that YmcF and YnfQ were significantly upregulated upon cold shock, whereas YhiY was expressed essentially equally under all conditions tested. Regarding the migration of these proteins in SDS-PAGE, the SPA tag adds 70 amino acids, or approximately 8 kDa, to the proteins of interest. Even so, YmcF, YnfQ, and YhiY migrate at slightly higher apparent molecular weights than would be expected based on their sizes, as determined by start codon mutagenesis (approximately 5–7 kDas, vide infra). This anomalous SDS-PAGE mobility has been observed for several other well-characterized microproteins6,30,31 and may be attributable to their high charge density and de-enrichment in aromatic residues.32 Despite repeated attempts, we were unable to detect expression of epitope-tagged YnaL and YpaA under any conditions. We concluded that these proteins may be post-translationally proteolyzed, so we did not consider them further. These results, combined with our proteomics analysis, confirmed that proteins YmcF, YnfQ, and YhiY are translated and that YmcF and YnfQ are upregulated during cold shock stress in E. coli.
Figure 4.
Confirmation of expression and cold shock upregulation of novel small proteins. (A) E. coli MG1655 strains with SPA epitope tags added to the C-termini of YmcF, YnfQ, and YhiY were generated. Cell lysates of strains expressing genomically tagged YmcF, YnfQ, and YhiY proteins at 37 °C, 10 °C (cold shock), and 42 °C (heat shock) were separated on a 4–20% SDS gel and stained with Coomassie blue (right). The same samples were also subjected to western blotting (left) and probed with an anti-FLAG antibody. The bands indicated by a red asterisk correspond to YmcF, YnfQ, and YhiY. (B) Bands from the blot were quantified by densitometry, and results are plotted to represent the fold change in expression for the three proteins at 10 °C (cold shock) relative to 37 °C. Error bars were calculated from three biological replicates and represent the standard error of the mean.
Identification of the Translation Initiation Sites for ymcF and ynfQ
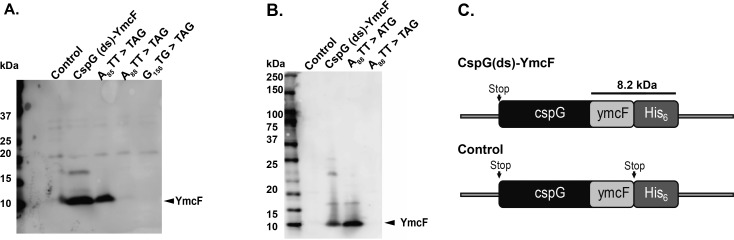
YmcF and YnfQ map to intergenic sequences downstream of the known cold shock proteins cspG and cspI, respectively. Although ymcF and ynfQ are not currently annotated in this E. coli strain, they have been predicted based on sequence conservation (Refseq accession WP_077248232.1). A closer look at the ymcF and ynfQ genes revealed that they must initiate at a noncanonical sequence due to the lack of an ATG start codon upstream of the region that produced the peptides we detected by mass spectrometry. In order to identify the translation initiation sites for ymcF and ynfQ, they were amplified along with their upstream genes and cloned into a pET expression vector to allow for the expression of a C-terminal hexa-histidine tag (His6 tag) in-frame with ymcF and ynfQ. For cspG–ymcF, the start codon for cspG and potential start codons in its vicinity were substituted with codons that would not allow for the initiation of cspG (CspG(ds)–YmcF). When expression of this construct was tested, robust translation of a small product could be observed by SDS-PAGE and subsequent blotting against the His6 tag (Figure 5A). This verified that translation of the small protein downstream of cspG occurs independently and is not a result of stop codon read-through or frame shifting during cspG translation. (Although we observed several higher molecular-weight translation products from the heterologous expression construct, these are not likely to be physiologically relevant, as they are not detectably produced from the genomically tagged strain.)
Figure 5.
Translation of YmcF initiates at an ATT start codon. (A) To verify the translation initiation site for ymcF, a cspG(ds) ymcF plasmid was cloned with a His6 tag in-frame at the C-terminus of the ymcF coding sequence. In this construct, the cspG start codon was mutated (delete start, ds) to abolish initiation of CspG. We then individually mutated candidate near-cognate ymcF start codons to stop codons. As a negative control, a stop codon was inserted before the His6 tag in the cspG(ds) ymcF plasmid. Nucleotide numbering starts immediately after the stop codon of cspG, and the sequence is provided in Figure S5. To observe expression, these constructs were introduced into BL21 cells, and IPTG induced cell lysates were subjected to SDS-PAGE followed by blotting against an antibody to the His6 tag. The YmcF protein band (carat) does not appear when the A88TT codon or the next proceeding near-cognate start codon (G156TG) is mutated to a stop codon. Nucleotide sequence and numbering for ymcF are provided in Figure S5, and additional ymcF mutagenesis experiments are presented in Figure S6. (B) Mutating the A88TT codon in ymcF to ATG results in increased expression of the same major protein product (carat).
We then sought to identify the start site for ymcF. Since there was no in-frame ATG start codon that could lead to the translation of a small YmcF protein, every near-cognate start codon downstream of cspG was mutated to a stop codon and expression of YmcF was inspected (see Figure S5 for sequence and numbering). We observed that mutations after T64TG caused a significant decrease in translation, whereas mutating A88TT to a stop codon completely abolished translation (Figures 5A and S6). Mutations of residues proceeding this also abolish translation of the major product (Figure S6), suggesting that A88TT is the translation initiation site for ymcF. Further mutation of A88TT to ATG significantly increased translation of the same product, as expected for a more efficient start codon (Figure 5B). These data are consistent with initiation of YmcF translation at A88TT.
Analysis of the genetic loci for ymcF and ynfQ revealed similar organization (Figure 3). Further, amino acid sequence alignment of YmcF and YnfQ reveals that the two proteins share 66% sequence identity (Figure 6A), suggesting that they may have arisen from a gene duplication event. On the basis of nucleotide sequence alignment of ymcF and ynfQ, we predicted the initiation site of ynfQ would be A22TT. When the preceding codon A19TT was mutated to a stop codon, YnfQ was still translated. However, when A22TT was substituted with TAG, translation of YnfQ was completely abolished, consistent with initiation of ynfQ at A22TT (Figure S7).
Figure 6.
Homology and conservation of YmcF and YnfQ. (A) Amino acid sequence alignment of YmcF and YnfQ proteins using Clustal Omega. The two proteins exhibit 66% sequence identity. (B) Nucleotide sequence alignments of ymcF (starting from position −3 relative to the first coding nucleotide) to homologous sequences from Shigella sonnei strain FORC_011 and Salmonella enterica subsp. enterica serovar Anatum str. USDA-ARS-USMARC-1677, which were identified using NCBI BLAST. The ATT start codon is underlined in red.
A BLAST search revealed the presence of ymcF homologues in some Salmonella and Shigella species, as well as conservation of the putative ATT start codon (Figure 6B). Taken together, these observations of cold-inducible synthesis and conservation in Enterobacteriaceae suggest that the YmcF and YnfQ proteins may be functional.
Discussion
While elegant genetic approaches have improved our ability to identify small proteins missed by traditional genome annotation algorithms,4 it is becoming clear that additional classes of genes have been under-annotated. For example, increasing numbers of reports have identified proteins translated by unconventional mechanisms such as initiation at noncanonical start codons,33,34 internal translation initiation sites,35,36 programmed frame-shifting,37,38 and stop codon read-through.39,40 Our comparative proteomic analysis revealed four novel E. coli proteins, all of which were previously nonannotated for at least one of the above-mentioned reasons: the encoded proteins are small, transiently expressed during stress, and/or initiate with noncanonical start codons.
The proximity of ymcF and ynfQ to known genes, in addition to their regulated expression and conservation, supports the hypothesis that they may encode functional proteins. Both are downstream of cold shock genes (cspG and cspI, respectively). The coding region of ymcF also overlaps the ymcE gene, which itself overlaps the downstream gnsA gene (Figure 3). ymcE is a suppressor of fabA6, whose gene product, FabA, catalyzes a dehydrase reaction in the synthesis of unsaturated fatty acids.41,42 Mutations in fabA6 result in a temperature-sensitive unsaturated fatty acid auxotroph phenotype which can be alleviated by overexpression of YmcE.42ynfQ is also located upstream of a homologue of gnsA named gnsB (Figure 3), which is another suppressor of the fabA6 mutant.43 The biochemical roles of YmcE, GnsA, and GnsB are yet to be determined, as these proteins remain largely uncharacterized at the molecular level. However, since our newly identified proteins are proximal in sequence space both to upstream cold shock proteins and downstream suppressors of fabA6 mutations, it is reasonable to hypothesize that these proteins may also play a role in regulating lipid synthesis during cold shock. Both YmcE and YnfQ are predicted to be structured (Figure S8A), but they have no known sequence or structural homologues. YmcF exhibits predicted structural homology to zinc-binding domains in proteins such as aspartate transcarbamoylase, largely based on five cysteine residues present in both proteins (and in YnfQ) (Figure S8B,C). Future work will focus on characterizing these proteins and testing these structural and functional hypotheses.
We utilized a molecular mutagenesis approach to identify the initiation codons for ymcF and ynfQ as A88TT and A22TT, respectively. The ATT start codon has long been known to initiate protein synthesis in bacteria, but it is thought to be rare, with only two ATT-initiating E. coli genes currently annotated: pcnB and infC.44−46 The enzyme PAP I (poly A polymerase I), which catalyzes RNA 3′ polyadenylation, is encoded by pcnB. Elevated levels of PAP I may be toxic to cells, and initiation at the noncanonical ATT start codon is proposed to be a regulatory mechanism to control PAP I production at low levels.45 Similarly, the prokaryotic translation initiation factor 3 (IF3), which is crucial for selecting the initiation codon for general protein translation, negatively regulates its own synthesis by initiating at an ATT start codon.33,46,47 It is possible that YmcF and YnfQ translation is also regulated via noncanonical start codon recognition. Our results further suggest that many more genes may remain to be identified that initiate with rare near-cognate start codons, even in eukaryotic genomes, where ATT start codons govern translation initiation of human beta-globin and frataxin.48,49
In conclusion, even though the E. coli genome has been extensively explored, our results suggest that more genes may remain to be discovered. These cryptic genes are likely to be short, may only be expressed under specific conditions, and may utilize noncanonical translation initiation mechanisms. Our quantitative proteomic workflow provides a roadmap for the discovery and characterization of these yet nonannotated genes. More broadly, we anticipate that comparative analysis of regulated smORF expression via LC/MS-based proteomics will enable the coupling of microprotein discovery to functional hypothesis generation.
Acknowledgments
We thank Jason Crawford for E. coli strain MG1655, plasmid pKD46, and advice on bacterial genetics. This work was supported by the Searle Scholars Program (S.A.S.), an American Cancer Society Institutional Research Grant Individual Award for New Investigators (IRG-58-012-57, S.A.S.), and Yale University West Campus start-up funds (to S.A.S. and J.R.). N.G.D. was supported by a Rudolph J. Anderson postdoctoral fellowship from Yale University. A.K. was in part supported by an NIH Predoctoral Training Grant (5T32GM06754 3-12). J.R. was supported by the NIH (GM117230, DK0174334). B.M.G and K.W.B. were supported by National Science Foundation GRFP grant DGE1122492.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.7b00419.
Worksheet S1: Key for proteomic analyses. Worksheet S2: Replicate 1 cold shock peptide level evidence. Worksheet S3: Replicate 1 cold shock protein level evidence. Worksheet S4: Replicate 1 control peptide level evidence. Worksheet S5: Replicate 1 control protein level evidence. Worksheet S6: Replicate 2 cold shock peptide level evidence. Worksheet S7: Replicate 2 cold shock protein level evidence. Worksheet S8: Replicate 2 control peptide level evidence. Worksheet S9: Replicate 2 control protein level evidence. Worksheet S10: Replicate 3 cold shock peptide level evidence. Worksheet S11: Replicate 3 cold shock protein level evidence. Worksheet S12: Replicate 4 cold shock peptide level evidence. Worksheet S13: Replicate 4 cold shock protein level evidence (XLSX)
Figure S1: Control MS/MS spectra. Table S1: Nonannotated peptide sequences and identification parameters. Figure S2: Mass spectrometric evidence for protein YpaA. Figure S3: Additional MS/MS spectra for nonannotated proteins. Figure S4: iPCR confirmation of knock-in strains. Table S2: Primer sequences. Figure S5: Nucleotide sequences of ymcF and ynfQ. Figure S6: Additional YmcF start codon mutagenesis experiments. Figure S7: YnfQ start codon mutagenesis. Figure S8: Cold-shock protein structural prediction (PDF)
Author Contributions
# N.G.D. and A.K. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Storz G.; Wolf Y. I.; Ramamurthi K. S. Small proteins can no longer be ignored. Annu. Rev. Biochem. 2014, 83, 753–77. 10.1146/annurev-biochem-070611-102400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemm M. R.; Paul B. J.; Miranda-Rios J.; Zhang A.; Soltanzad N.; Storz G. Small stress response proteins in Escherichia coli: proteins missed by classical proteomic studies. J. Bacteriol. 2010, 192 (1), 46–58. 10.1128/JB.00872-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemm M. R.; Paul B. J.; Schneider T. D.; Storz G.; Rudd K. E. Small membrane proteins found by comparative genomics and ribosome binding site models. Mol. Microbiol. 2008, 70 (6), 1487–501. 10.1111/j.1365-2958.2008.06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi K. S.; Storz G. The small protein floodgates are opening; now the functional analysis begins. BMC Biol. 2014, 12, 96. 10.1186/s12915-014-0096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.; Ward C. C.; Jungreis I.; Slavoff S. A.; Schwaid A. G.; Neveu J.; Budnik B. A.; Kellis M.; Saghatelian A. The Discovery of Human sORF-Encoded Polypeptides (SEPs) in Cell Lines and Tissue. J. Proteome Res. 2014, 13, 1757–1765. 10.1021/pr401280w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavoff S. A.; Mitchell A. J.; Schwaid A. G.; Cabili M. N.; Ma J.; Levin J. Z.; Karger A. D.; Budnik B. A.; Rinn J. L.; Saghatelian A. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 2012, 9 (1), 59–64. 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderperre B.; Lucier J. F.; Bissonnette C.; Motard J.; Tremblay G.; Vanderperre S.; Wisztorski M.; Salzet M.; Boisvert F. M.; Roucou X. Direct detection of alternative open reading frames translation products in human significantly expands the proteome. PLoS One 2013, 8 (8), e70698. 10.1371/journal.pone.0070698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menschaert G.; Van Criekinge W.; Notelaers T.; Koch A.; Crappe J.; Gevaert K.; Van Damme P. Deep proteome coverage based on ribosome profiling aids mass spectrometry-based protein and peptide discovery and provides evidence of alternative translation products and near-cognate translation initiation events. Mol. Cell. Proteomics 2013, 12 (7), 1780–90. 10.1074/mcp.M113.027540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N. T.; Ghaemmaghami S.; Newman J. R.; Weissman J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009, 324 (5924), 218–23. 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N. T.; Lareau L. F.; Weissman J. S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011, 147 (4), 789–802. 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvunis A. R.; Rolland T.; Wapinski I.; Calderwood M. A.; Yildirim M. A.; Simonis N.; Charloteaux B.; Hidalgo C. A.; Barbette J.; Santhanam B.; Brar G. A.; Weissman J. S.; Regev A.; Thierry-Mieg N.; Cusick M. E.; Vidal M. Proto-genes and de novo gene birth. Nature 2012, 487 (7407), 370–4. 10.1038/nature11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruana N. J.; Cooke I. R.; Faou P.; Finn J.; Hall N. E.; Norman M.; Pineda S. S.; Strugnell J. M. A combined proteomic and transcriptomic analysis of slime secreted by the southern bottletail squid, Sepiadarium austrinum (Cephalopoda). J. Proteomics 2016, 148, 170–182. 10.1016/j.jprot.2016.07.026. [DOI] [PubMed] [Google Scholar]
- Christie-Oleza J. A.; Pina-Villalonga J. M.; Bosch R.; Nogales B.; Armengaud J. Comparative proteogenomics of twelve Roseobacter exoproteomes reveals different adaptive strategies among these marine bacteria. Mol. Cell. Proteomics 2012, 11 (2), M111.013110. 10.1074/mcp.M111.013110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx H.; Hahne H.; Ulbrich S. E.; Schnieke A.; Rottmann O.; Frishman D.; Kuster B. Annotation of the Domestic Pig Genome by Quantitative Proteogenomics. J. Proteome Res. 2017, 16 (8), 2887–2898. 10.1021/acs.jproteome.7b00184. [DOI] [PubMed] [Google Scholar]
- Ogishi M.; Yotsuyanagi H.; Moriya K.; Koike K. Delineation of autoantibody repertoire through differential proteogenomics in hepatitis C virus-induced cryoglobulinemia. Sci. Rep. 2016, 6, 29532. 10.1038/srep29532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen V. K.; Steinsland H.; Wiker H. G. Improving genome annotation of enterotoxigenic Escherichia coli TW10598 by a label-free quantitative MS/MS approach. Proteomics 2015, 15 (22), 3826–34. 10.1002/pmic.201500278. [DOI] [PubMed] [Google Scholar]
- Vermillion K. L.; Jagtap P.; Johnson J. E.; Griffin T. J.; Andrews M. T. Characterizing Cardiac Molecular Mechanisms of Mammalian Hibernation via Quantitative Proteogenomics. J. Proteome Res. 2015, 14 (11), 4792–804. 10.1021/acs.jproteome.5b00575. [DOI] [PubMed] [Google Scholar]
- Phadtare S.; Alsina J.; Inouye M. Cold-shock response and cold-shock proteins. Curr. Opin. Microbiol. 1999, 2 (2), 175–80. 10.1016/S1369-5274(99)80031-9. [DOI] [PubMed] [Google Scholar]
- Uzzau S.; Figueroa-Bossi N.; Rubino S.; Bossi L. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. U. S. A. 2001, 98 (26), 15264–9. 10.1073/pnas.261348198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A.; Wanner B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 2000, 97 (12), 6640–5. 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S. N.; Hunt H. D.; Horton R. M.; Pullen J. K.; Pease L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 1989, 77 (1), 51–9. 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Hao P.; Ren Y.; Dutta B.; Sze S. K. Comparative evaluation of electrostatic repulsion-hydrophilic interaction chromatography (ERLIC) and high-pH reversed phase (Hp-RP) chromatography in profiling of rat kidney proteome. J. Proteomics 2013, 82, 254–62. 10.1016/j.jprot.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Lajoie M. J.; Rovner A. J.; Goodman D. B.; Aerni H. R.; Haimovich A. D.; Kuznetsov G.; Mercer J. A.; Wang H. H.; Carr P. A.; Mosberg J. A.; Rohland N.; Schultz P. G.; Jacobson J. M.; Rinehart J.; Church G. M.; Isaacs F. J. Genomically recoded organisms expand biological functions. Science 2013, 342 (6156), 357–60. 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers M. C.; Maclean B.; Burke R.; Amodei D.; Ruderman D. L.; Neumann S.; Gatto L.; Fischer B.; Pratt B.; Egertson J.; Hoff K.; Kessner D.; Tasman N.; Shulman N.; Frewen B.; Baker T. A.; Brusniak M. Y.; Paulse C.; Creasy D.; Flashner L.; Kani K.; Moulding C.; Seymour S. L.; Nuwaysir L. M.; Lefebvre B.; Kuhlmann F.; Roark J.; Rainer P.; Detlev S.; Hemenway T.; Huhmer A.; Langridge J.; Connolly B.; Chadick T.; Holly K.; Eckels J.; Deutsch E. W.; Moritz R. L.; Katz J. E.; Agus D. B.; MacCoss M.; Tabb D. L.; Mallick P. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30 (10), 918–20. 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D. N.; Pappin D. J.; Creasy D. M.; Cottrell J. S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20 (18), 3551–67. . [DOI] [PubMed] [Google Scholar]
- Ma J.; Diedrich J. K.; Jungreis I.; Donaldson C.; Vaughan J.; Kellis M.; Yates J. R. 3rd; Saghatelian A. Improved Identification and Analysis of Small Open Reading Frame Encoded Polypeptides. Anal. Chem. 2016, 88 (7), 3967–75. 10.1021/acs.analchem.6b00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagore D. M.; Nolte W. M.; Neveu J. M.; Rangel R.; Guzman-Rojas L.; Pasqualini R.; Arap W.; Lane W. S.; Saghatelian A. Peptidase substrates via global peptide profiling. Nat. Chem. Biol. 2009, 5 (1), 23–5. 10.1038/nchembio.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco A. D.; Tagore D. M.; Saghatelian A. Expanding the dipeptidyl peptidase 4-regulated peptidome via an optimized peptidomics platform. J. Am. Chem. Soc. 2010, 132 (11), 3819–30. 10.1021/ja909524e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantscheff M.; Schirle M.; Sweetman G.; Rick J.; Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 2007, 389 (4), 1017–31. 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- D’Lima N. G.; Ma J.; Winkler L.; Chu Q.; Loh K. H.; Corpuz E. O.; Budnik B. A.; Lykke-Andersen J.; Saghatelian A.; Slavoff S. A. A human microprotein that interacts with the mRNA decapping complex. Nat. Chem. Biol. 2016, 13 (2), 174–180. 10.1038/nchembio.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavoff S. A.; Heo J.; Budnik B. A.; Hanakahi L. A.; Saghatelian A. A human short open reading frame (sORF)-encoded polypeptide that stimulates DNA end joining. J. Biol. Chem. 2014, 289 (16), 10950–7. 10.1074/jbc.C113.533968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocca S.; Samalikova M.; Uversky V. N.; Lotti M.; Vanoni M.; Alberghina L.; Grandori R. Order propensity of an intrinsically disordered protein, the cyclin-dependent-kinase inhibitor Sic1. Proteins: Struct., Funct., Genet. 2009, 76 (3), 731–46. 10.1002/prot.22385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggerty T. J.; Lovett S. T. IF3-mediated suppression of a GUA initiation codon mutation in the recJ gene of Escherichia coli. J. Bacteriol. 1997, 179 (21), 6705–13. 10.1128/jb.179.21.6705-6713.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalut C.; Egly J. M. AUC is used as a start codon in Escherichia coli. Gene 1995, 156 (1), 43–5. 10.1016/0378-1119(95)00034-4. [DOI] [PubMed] [Google Scholar]
- Subbarayan P. R.; Sarkar M. A stop codon-dependent internal secondary translation initiation region in Escherichia coli rpoS. RNA 2004, 10 (9), 1359–1365. 10.1261/rna.7500604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarayan P. R.; Sarkar M. Escherichia coli rpoS gene has an internal secondary translation initiation region. Biochem. Biophys. Res. Commun. 2004, 313 (2), 294–9. 10.1016/j.bbrc.2003.11.132. [DOI] [PubMed] [Google Scholar]
- Atkins J. F.; Loughran G.; Bhatt P. R.; Firth A. E.; Baranov P. V. Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 2016, 44 (15), 7007–7078. 10.1093/nar/gkw530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov P. V.; Gesteland R. F.; Atkins J. F. Release factor 2 frameshifting sites in different bacteria. EMBO Rep. 2002, 3 (4), 373–377. 10.1093/embo-reports/kvf065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenthzel A. M.; Stancek M.; Isaksson L. A. Growth phase dependent stop codon readthrough and shift of translation reading frame in Escherichia coli. FEBS Lett. 1998, 421 (3), 237–42. 10.1016/S0014-5793(97)01570-6. [DOI] [PubMed] [Google Scholar]
- Williams I.; Richardson J.; Starkey A.; Stansfield I. Genome-wide prediction of stop codon readthrough during translation in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 2004, 32 (22), 6605–6616. 10.1093/nar/gkh1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y.; Zhan L.; Gao Z.; Prive G. G.; Dong Y. Crystal structure of GnsA from Escherichia coli. Biochem. Biophys. Res. Commun. 2015, 462 (1), 1–7. 10.1016/j.bbrc.2015.03.133. [DOI] [PubMed] [Google Scholar]
- Rock C. O.; Tsay J. T.; Heath R.; Jackowski S. Increased unsaturated fatty acid production associated with a suppressor of the fabA6(Ts) mutation in Escherichia coli. J. Bacteriol. 1996, 178 (18), 5382–7. 10.1128/jb.178.18.5382-5387.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugai R.; Shimizu H.; Nishiyama K.; Tokuda H. Overexpression of yccL (gnsA) and ydfY (gnsB) increases levels of unsaturated fatty acids and suppresses both the temperature-sensitive fabA6 mutation and cold-sensitive secG null mutation of Escherichia coli. J. Bacteriol. 2001, 183 (19), 5523–8. 10.1128/JB.183.19.5523-5528.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. D.; Parkinson J. S. Genetics and sequence analysis of the pcnB locus, an Escherichia coli gene involved in plasmid copy number control. J. Bacteriol. 1989, 171 (3), 1254–61. 10.1128/jb.171.3.1254-1261.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns N.; Masters M. Expression of the Escherichia coli pcnB gene is translationally limited using an inefficient start codon: a second chromosomal example of translation initiated at AUU. Mol. Microbiol. 2002, 44 (5), 1287–98. 10.1046/j.1365-2958.2002.02945.x. [DOI] [PubMed] [Google Scholar]
- Butler J. S.; Springer M.; Dondon J.; Graffe M.; Grunberg-Manago M. Escherichia coli protein synthesis initiation factor IF3 controls its own gene expression at the translational level in vivo. J. Mol. Biol. 1986, 192 (4), 767–80. 10.1016/0022-2836(86)90027-6. [DOI] [PubMed] [Google Scholar]
- Brombach M.; Pon C. L. The unusual translational initiation codon AUU limits the expression of the infC (initiation factor IF3) gene of Escherichia coli. Mol. Gen Genet 1987, 208 (1–2), 94–100. 10.1007/BF00330428. [DOI] [PubMed] [Google Scholar]
- Rahbar S.; Nozari G. A novel initiation codon mutation (ATG-->ATT) in a beta-thalassemia patient. Hemoglobin 1993, 17 (6), 557–62. 10.3109/03630269309043497. [DOI] [PubMed] [Google Scholar]
- Zuhlke C.; Laccone F.; Cossee M.; Kohlschutter A.; Koenig M.; Schwinger E. Mutation of the start codon in the FRDA1 gene: linkage analysis of three pedigrees with the ATG to ATT transversion points to a unique common ancestor. Hum. Genet. 1998, 103 (1), 102–5. 10.1007/s004390050791. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.