Maezawa et al. show that Polycomb-repressive complex 1 (PRC1) directs timely activation of germline genes during spermatogenesis.
Keywords: germline, spermatogenesis, spermatogonia, Polycomb, gene activation
Abstract
During spermatogenesis, a large number of germline genes essential for male fertility are coordinately activated. However, it remains unknown how timely activation of this group of germline genes is accomplished. Here we show that Polycomb-repressive complex 1 (PRC1) directs timely activation of germline genes during spermatogenesis. Inactivation of PRC1 in male germ cells results in the gradual loss of a stem cell population and severe differentiation defects, leading to male infertility. In the stem cell population, RNF2, the dominant catalytic subunit of PRC1, activates transcription of Sall4, which codes for a transcription factor essential for subsequent spermatogenic differentiation. Furthermore, RNF2 and SALL4 together occupy transcription start sites of germline genes in the stem cell population. Once differentiation commences, these germline genes are activated to enable the progression of spermatogenesis. Our study identifies a novel mechanism by which Polycomb directs the developmental process by activating a group of lineage-specific genes.
Timely activation of spermatogenesis-specific genes is essential for male fertility (Kimmins and Sassone-Corsi 2005). During spermatogenesis, spermatogonial stem cells, which are capable of self-renewal, develop into differentiating spermatogonia, enter meiosis, and undergo further differentiation into haploid sperm (Fig. 1A; Griswold 2016). The gene expression program of male germ cells is largely distinct from those of somatic lineages, and a large number of germline genes are activated specifically during spermatogenesis (Hasegawa et al. 2015; Sin et al. 2015). Although regulation of gene expression is fundamental for spermatogenesis, the underlying mechanism that directs the coordinated activation of germline genes is largely unexplored.
Figure 1.

PRC1 is required for spermatogonial maintenance and spermatogenic differentiation. (A) Schematic of spermatogenesis and stage markers. (B) Immunostaining of PLZF in testicular sections at postnatal day 7 (P7) and 6 wk. PLZF-positive cells are shown with arrowheads in the right panels. Quantitative data are shown at the far right. Data are represented as mean ± SD. Four independent mice were examined for each time point. (C) Immunostaining of γH2AX in testicular sections at 6 wk. Bars, 20 µm. A testicular tubule devoid of germ cells is shown with an asterisk. The regions with yellow squares are magnified in the insets (bottom left corners). Quantitative data are shown at the far right. Data are represented as mean ± SEM. Three independent mice were examined. Bars, 20 µm. (**) P < 0.001; (***) P < 0.0001, unpaired t-test.
Polycomb proteins regulate heritable gene silencing and are responsible for stem cell renewal and development (Aloia et al. 2013; Simon and Kingston 2013). Polycomb-based heritable silencing safeguards cell type-specific gene expression by suppressing non-lineage-specific gene expression, thereby defining cellular identity (Geisler and Paro 2015). One of the Polycomb complexes in mammals, Polycomb-repressive complex 1 (PRC1), is a major modifier of histone H2A through monoubiquitination of Lys119 (H2AK119ub), a modification usually—but not always—associated with gene repression (Illingworth et al. 2015; Pengelly et al. 2015). During germ cell development, PRC1 coordinates timing of sexual differentiation in female primordial germ cells (Yokobayashi et al. 2013) and establishes developmental competence of oocytes for the following generation by silencing genes that induce somatic differentiation (Posfai et al. 2012). However, the function of PRC1 in spermatogenesis remains unknown.
To test the function of PRC1 in spermatogenesis, in this study, we generated a loss-of-function mouse model of RNF2 (also known as RING1B), a component in the heterodimeric E3 ubiquitin ligases of PRC1 (Simon and Kingston 2013). In the germline stem (GS) cell population, we demonstrate that RNF2 activates the transcription of Sall4, which codes for a transcription factor essential for subsequent spermatogenic differentiation (Hobbs et al. 2012). Furthermore, RNF2 and SALL4 together occupy the transcription start sites (TSSs) of a variety of germline genes in the stem cell population. Once differentiation commences, the occupied germline genes are activated. Our study identifies a novel epigenetic mechanism by which PRC1 directs the timely activation of germline genes during spermatogenesis.
Results
PRC1 is required for the maintenance of undifferentiated spermatogonia and for spermatogenic differentiation
In postnatal male germ cells, RNF2 is abundantly expressed in PLZF+ undifferentiated spermatogonia, which includes a subpopulation of self-renewing spermatogonial stem cells, and in c-Kit+ differentiating spermatogonia, which undergo committed differentiation and mitotic proliferation (Supplemental Fig. S1; Hasegawa et al. 2015). Although RNF2 appears to be the most active component in the heterodimeric E3 ligases of PRC1, the RNF2 paralog RING1 (also known as RING1A) can partially compensate for the loss of RNF2 (Endoh et al. 2012). Ring1 knockout mice are viable and do not have fertility defects (del Mar Lorente et al. 2000), but Rnf2 knockout mice are embryonic-lethal (Voncken et al. 2003). Thus, to determine the global functions of PRC1 in spermatogenesis, we pursued a genetic strategy to eliminate PRC1's catalytic activity for histone H2AK119 ubiquitination: We generated a conditional deletion of Rnf2 (Rnf2 conditional knockout [cKO]) with Ddx4-Cre, which is expressed specifically in germ cells after embryonic day 14 (Gallardo et al. 2007), on a Ring1 knockout background (Ddx4-Cre; Rnf2cKO; Ring1 knockout). We refer to this mouse line as the PRC1cKO. The phenotype was compared with littermate controls that harbored floxed alleles for Rnf2 on a Ring1 knockout background without Ddx4-Cre (termed PRC1 control [PRC1ctrl]). This mouse model enabled us to define the function of RNF2 without compensation from RING1 while also representing a “complete” loss of function of PRC1 as shown in other biological contexts (Endoh et al. 2012; Posfai et al. 2012; Yokobayashi et al. 2013).
PRC1cKO males were infertile and had small testes (Supplemental Fig. S2A–C). We confirmed a near-complete rate of Ddx4-Cre-mediated recombination by observing the depletion of H2AK119ub, a readout of PRC1 activity, in GCNA (a germ cell marker)+ germ cells of PRC1cKO testes at postnatal day 7 (P7) and at 6 wk; confirming the germline-specific nature of the conditional deletion, H2AK119ub deposition was preserved in somatic cells of PRC1cKO testes (Supplemental Fig. S2D–F). Furthermore, PRC1cKO males showed smaller testicular tubules at 6 wk, suggesting germ cell depletion in the PRC1cKO (Supplemental Fig. S2G,H).
To determine the function of PRC1 in spermatogenesis, we sought to identify and investigate the stages of spermatogenesis affected by the loss of PRC1. In the PRC1cKO, the proportion of PLZF+ undifferentiated spermatogonia was comparable with that of the PRC1ctrl at P2 but slowly decreased over a period of 6 wk (Fig. 1B), suggesting that PRC1 is required for the maintenance of undifferentiated spermatogonia. Furthermore, γH2AX+ meiotic cells, which represent early stages of meiotic prophase, such as the leptotene and zygotene stages, were greatly diminished in the PRC1cKO at 6 wks of age (Fig. 1C), suggesting that a large population of germ cells undergoes progressive depletion prior to entry into meiotic prophase. Therefore, we conclude that PRC1 loss of function leads to germ cell depletion prior to meiosis. γH2AX+ meiotic cells of the PRC1cKO did not exhibit XY bodies (Fig. 1C, inset), which represent transcriptionally silent sex chromosomes during meiotic prophase; additionally, histone variant H1T, a marker of mid-pachytene spermatocytes, was not detected (Supplemental Fig. S2I). These results indicate that PRC1cKO spermatocytes cannot progress into the mid-pachytene stage. Therefore, PRC1 is required not only for the maintenance of undifferentiated spermatogonia but also for spermatogenic differentiation.
To determine the cause of germ cell depletion in the PRC1cKO, we analyzed germ cells for the occurrence of apoptosis. We found that the frequency of tubules containing apoptotic cells in the PRC1cKO, as judged by immunostaining of cleaved Caspase-3, was significantly higher than that of the PRC1ctrl at P7 (Supplemental Fig. S3A). The population of PLZF+ undifferentiated spermatogonia was higher among cleaved Caspase-3+ cells of the PRC1cKO versus the PRC1ctrl (Supplemental Fig. S3B), suggesting that the loss of PRC1 causes apoptosis in undifferentiated spermatogonia. We next examined whether the cell cycle is altered in spermatogonia of the PRC1cKO. By immunostaining against Ki67, a marker of cell proliferation, and Histone H3 phosphorylated at Ser10 (phospho-Histone H3), a marker of the M phase of the cell cycle, we observed that the populations of Ki67+ and phospho-Histone H3+ germ cells in the PRC1cKO testes were normal (Supplemental Fig. S3C,D). These data exclude the possibility that germ cell depletion in the PRC1cKO is due to cell cycle arrest at a particular cell cycle stage, supporting our assertion that increased apoptosis of spermatogonia is a cause of germ cell depletion in the PRC1cKO.
PRC1 is required for the timely activation of germline genes in spermatogenesis
To investigate gene expression changes underlying the spermatogenic defects caused by PRC1 inactivation, we performed RNA sequencing (RNA-seq) using Thy1+ undifferentiated spermatogonia and c-Kit+ differentiating spermatogonia purified from P7 testes by magnetic-activated cell sorting (MACS). Our RNA-seq data demonstrate that 69 genes were up-regulated in Thy1+ spermatogonia of the PRC1cKO as compared with littermate controls, while we observed a more extensive up-regulation of 269 genes after differentiation into c-Kit+ spermatogonia in the PRC1cKO (Fig. 2A). Gene ontology (GO) analysis revealed that up-regulated genes were enriched for those that function in cell adhesion, the morphogenesis of epithelial and endothelial tubes, and programmed cell death (Supplemental Fig. 4A,B). These broad functions are not directly related to spermatogenesis, since they are suppressed in normal spermatogenesis. Thus, these results suggest that PRC1 suppresses the expression of non-lineage-specific genes.
Figure 2.
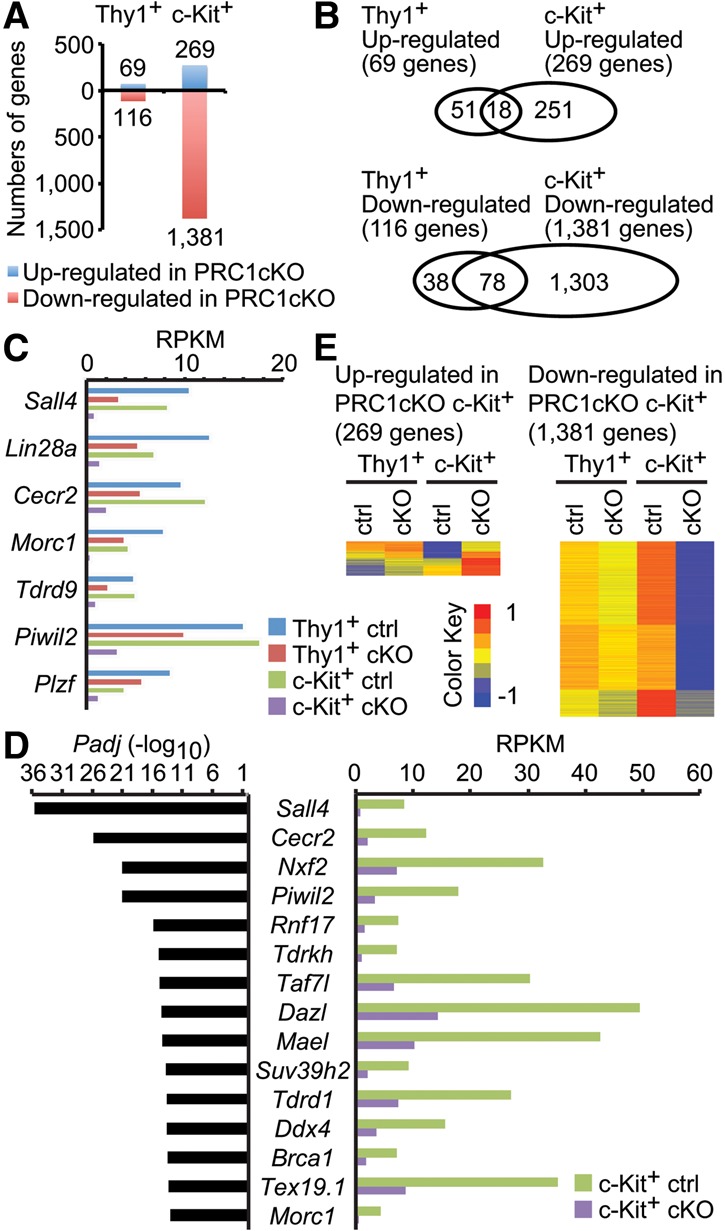
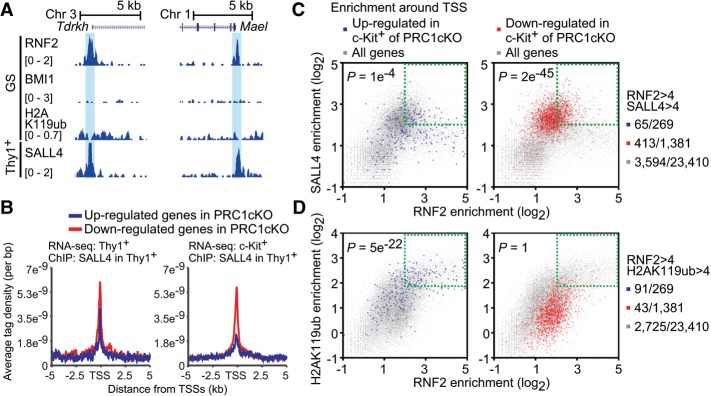
PRC1 is required for timely activation of germline genes in spermatogonia. (A) The number of differentially expressed genes detected by RNA-seq (at least a 1.5-fold change) in Thy1+ and c-Kit+ spermatogonia (two biological replicates) between the PRC1ctrl and the PRC1cKO. (B) Venn diagrams showing the numbers of regulated genes between Thy1+ and c-Kit+ spermatogonia. (C) Representative down-regulated germline genes in Thy1+ spermatogonia of the PRC1cKO and their average reads per kilobase million (RPKM) values. (D) Fifteen representative germline genes among the top 50 down-regulated genes in c-Kit+ spermatogonia from the lowest P-values. (Left) P-values. (Right) Average RPKM values. P-values represent false discovery rate (FDR)-adjusted P-values by DESeq2. (E) Heat maps showing gene expression patterns of both up-regulated and down-regulated genes in c-Kit+ spermatogonia. Relative expression normalized to a range of −1 to 1 is shown.
Notably, we observed that 116 genes were down-regulated in Thy1+ spermatogonia, and the down-regulation became more extensive in c-Kit+ spermatogonia of the PRC1cKO, with decreased expression of 1381 genes (Fig. 2A). Many of the genes down-regulated in Thy1+ cells were also consistently down-regulated in c-Kit+ cells (78 out of 116 genes) (Fig. 2B). Interestingly, the lowest P-value for down-regulation in both gene sets was Sall4, a gene that encodes a transcription factor essential for spermatogenic differentiation (Fig. 2C,D; Supplemental Table 1; Hobbs et al. 2012). In addition to Sall4, several genes that are required for spermatogenesis were down-regulated in both Thy1+ and c-Kit+ spermatogonia of the PRC1cKO (Fig. 2C). For example, we observed the shared down-regulation of Plzf, which functions in the maintenance of undifferentiated spermatogonia, and Lin28a, which functions in the expansion of undifferentiated spermatogonia (Buaas et al. 2004; Costoya et al. 2004; Chakraborty et al. 2014). We also observed the shared down-regulation of Morc1, Tdrd9, and Piwil2 (also known as Mili), all of which are required for repression of transposable elements in spermatogenesis (Kuramochi-Miyagawa et al. 2004; Shoji et al. 2009; Pastor et al. 2014). Furthermore, the down-regulated genes in c-Kit+ spermatogonia of the PRC1cKO include genes required for several key processes of spermatogenesis (Fig. 2D; Supplemental Fig. S4E); in normal spermatogenesis, these genes are highly activated during the differentiation from Thy1+ to c-Kit+ spermatogonia (Fig. 2E). The down-regulated genes in c-Kit+ spermatogonia of the PRC1cKO include those that function in the differentiation/maintenance of spermatogonia (Dmrt1, Nxf2, Sohlh1, and Sohlh2) (Pan et al. 2009; Matson et al. 2010; Suzuki et al. 2012), meiotic initiation (Stra8 and Dazl) (Anderson et al. 2008; Lin et al. 2008), and transposon repression and/or regulation of piRNA (Tdrkh, Mael, Tdrd1, Ddx4, and Tex19.1) (Chuma et al. 2006; Ollinger et al. 2008; Soper et al. 2008; Kuramochi-Miyagawa et al. 2010; Saxe et al. 2013; Pastor et al. 2014). This group also includes Scml2, which encodes a germline-specific Polycomb protein that is required for the suppression of somatic genes in spermatogenesis (Hasegawa et al. 2015); the tumor suppressor gene Rb, which encodes an essential factor in cell cycle regulation that is also required for stem cell maintenance (Hu et al. 2013); and several DNA damage response, repair, and recombination genes that are required for spermatogonia and/or meiosis, including Brca1 (Turner et al. 2004; Broering et al. 2014), Tex15 (Yang et al. 2008a), Fancd2 (Alavattam et al. 2016), Atr (Royo et al. 2013), Brca2 (Sharan et al. 2004), Tex11 (Yang et al. 2008b), Atm (Takubo et al. 2008), Rad51 (Dai et al. 2017), and Blm (Holloway et al. 2010). Therefore, we conclude that PRC1 has an essential role in the timely activation of germline genes during spermatogonial differentiation.
Since many of the genes involved in the suppression of retrotransposons were down-regulated in the PRC1cKO, we examined the expression of retrotransposons by analyzing RNA-seq data. However, we found that expression of retrotransposons did not change between PRC1cKO and PRC1ctrl spermatogonia (Supplemental Fig. S5A). On the other hand, we found a modest up-regulation of GSAT, a representative of major satellite DNA, in both Thy1+ and c-Kit+ cells and a modest up-regulation of SATMIN, a representative of minor satellite DNA, in Thy1+ cells. To further evaluate changes in retrotransposon expression, we performed immunostaining against LINE1 ORF1 protein (ORF1p). We found that LINE1 ORF1p was not derepressed in PRC1cKO testes but was expressed at a similar level between PRC1cKO and PRC1ctrl testes (Supplemental Fig. S5B); this expression level was similar to previous observations of expression in wild-type testes (Soper et al. 2008). These results suggest that PRC1 is required for the suppression of major and minor satellite DNA but not for the suppression of retrotransposons in spermatogenesis, although PRC1 activates genes involved in the suppression of retrotransposons.
RNF2 directly binds to TSSs of activated genes in spermatogonia
Although Polycomb complexes are known as gene suppressors, recent studies have revealed an additional function of Polycomb in direct gene activation (Creppe et al. 2014; Gao et al. 2014). To determine whether PRC1 directly activates germline genes during spermatogenesis, we analyzed the genomic distribution of Polycomb proteins by investigating ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) data of GS cells (Hasegawa et al. 2015), which are a cultured spermatogonial stem cell population that retains stem cell potential. In GS cells, RNF2 was present near TSSs of genes down-regulated in PRC1cKO spermatogonia, such as Sall4 (Fig. 3A). Average tag density analysis revealed that in GS cells, RNF2 was highly associated with TSSs of genes down-regulated in Thy1+ spermatogonia of the PRC1cKO and was also associated with TSSs and gene bodies of genes up-regulated in the PRC1cKO (Fig. 3B), consistent with a canonical role of PRC1 in gene repression. These results suggest that, in addition to its canonical function in gene repression, RNF2 directly binds a group of transcriptionally active germline genes. The specificity of these peaks was confirmed with the analysis of input chromatin using GS cells (Supplemental Fig. S6A). Importantly, in GS cells, RNF2 was preloaded to genes that are either down-regulated or up-regulated in subsequent c-Kit+ differentiating spermatogonia (Fig. 3C). This result suggests that RNF2 binding in the stem cell phase predicts gene regulation in later stages, which is consistent with the notion that unidirectional differentiation of spermatogenesis is preset by epigenetic mechanisms in the stem cell stage (Hasegawa et al. 2015; Sin et al. 2015).
Figure 3.
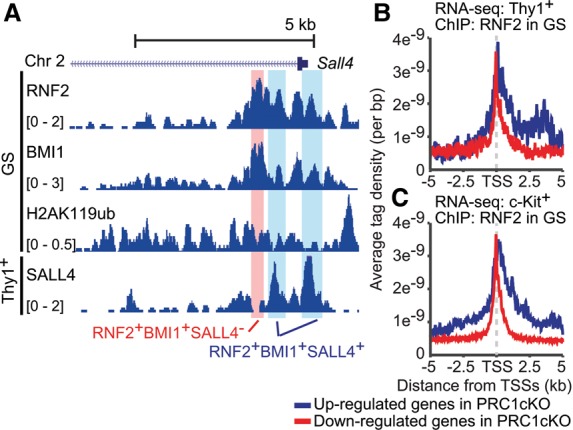
RNF2 directly binds to TSSs of activated genes in spermatogonia. (A) Binding peaks of RNF2, BMI1, H2AK119ub, and SALL4 across the Sall4 gene locus in GS cells and cultured Thy1+ spermatogonia. A double-positive peak (RNF2 and BMI1) lacking SALL4 is highlighted in red. Triple-positive peaks (RNF2, BMI1, and SALL4) are highlighted in blue. (B,C) Occupancy of RNF2 around TSSs in wild-type GS cells. ChIP-seq data of GS cells are shown for the indicated genes based on RNA-seq analyses.
In contrast to RNF2, H2AK119ub and another PRC1 subunit, BMI1, were not highly associated with the TSSs of down-regulated genes in the PRC1cKO, although they were highly associated with up-regulated genes (Supplemental Fig. S6B–D), suggesting that the function of RNF2 in gene activation is at least in part independent of H2AK119ub and BMI1. Between up-regulated and down-regulated genes in the PRC1cKO, the distinct binding profiles of RNF2, BMI1, and H2AK119ub suggest that the mechanisms underlying RNF2-dependent gene activation and suppression are likely to be distinct.
RNF2 and SALL4 together occupy TSSs of germline genes for activation
Because our data suggest that RNF2 functions in gene activation in a mechanism distinct from its role in gene suppression, we next searched for a cofactor working with RNF2 in gene activation. Since Sall4 is the gene with the lowest P-value for down-regulation in both the Thy1+ and c-Kit+ spermatogonia of the PRC1cKO, we examined the genomic localization of the SALL4 transcription factor using published ChIP-seq data of in vitro cultures of Thy1+ spermatogonia (Lovelace et al. 2016). SALL4 occupies its own regulatory element in the vicinity of its TSS at the Sall4 gene locus, and SALL4 peaks largely overlapped with those of RNF2, although we observed some RNF2 peaks independent of SALL4 (Fig. 3A). We also found that, together, RNF2 and SALL4 occupy TSSs of RNF2-dependent germline genes, such as Tdrkh and Mael (Fig. 4A). Average tag density analysis revealed that SALL4 is highly associated with down-regulated genes in Thy1+ spermatogonia of the PRC1cKO as compared with up-regulated genes, and this tendency is more evident for genes down-regulated in c-Kit+ spermatogonia of the PRC1cKO (Fig. 4B). In addition, enrichment analysis around TSSs revealed that down-regulated genes in Thy1+ and c-Kit+ spermatogonia of the PRC1cKO were enriched with both SALL4 and RNF2, while up-regulated genes were not (Fig. 4C). Additional enrichment analysis confirmed that the coenrichment of H2AK119ub and RNF2 occurred only on TSSs of up-regulated genes in c-Kit+ spermatogonia of the PRC1cKO and not on TSSs of down-regulated genes (Fig. 4D).
Figure 4.
RNF2 and SALL4 together occupy TSSs of germline genes for activation. (A) Binding peaks of RNF2, BMI1, H2AK119ub, and SALL4 across the Tdrkh and Mael gene loci in GS cells and cultured Thy1+ spermatogonia. Double-positive peaks (RNF2 and SALL4) are highlighted in blue. (B) Occupancy of SALL4 around TSSs in wild-type GS cells. ChIP-seq data of cultured Thy1+ spermatogonia are shown for the indicated genes based on RNA-seq analysis. (C,D) Scatter plots showing enrichment of ChIP-seq intensity (±2 kb around TSSs) on genes regulated in c-Kit+ spermatogonia (up-regulated genes are in the left panel, and down-regulated genes are in the right panel). Distribution of all genes is shown with gray dots. P-values are based on hypergeometric probability tests for genes with enrichment greater than four for both RNF2 and SALL4 (C) and for both RNF2 and H2AK119ub (D). The numbers of genes are shown at the right.
Consistent with the enrichment of SALL4 in ChIP-seq data of Thy1+ spermatogonia, SALL4 localized in the nuclei of PLZF+ undifferentiated spermatogonia in PRC1ctrl testes (Fig. 5A). RNF2 localized in the nuclei of prospermatogonia through to meiotic pachytene spermatocytes; SALL4, on the other hand, gradually accumulated in the nuclei of spermatogonia (Hobbs et al. 2012; Gassei and Orwig 2013), with its greatest enrichment in differentiating spermatogonia, after which it disappeared from nuclei during the leptotene/zygotene stages of meiotic prophase (Supplemental Fig. S7). However, SALL4 localization was absent in PLZF+ undifferentiated spermatogonia of the PRC1cKO (Fig. 5A), in accord with the RNF2-dependent transcriptional activation of Sall4 (Fig. 2). These results suggest that RNF2 promotes transcriptional activation and subsequent nuclear accumulation of SALL4, and then RNF2 and SALL4 together occupy a cohort of germline genes for activation during spermatogenesis. Consistent with the depletion of SALL4 in the PRC1cKO, the spermatogenic defects reported for Sall4cKO mice (Hobbs et al. 2012) are similar to the phenotypes of PRC1cKO mice with regard to the gradual depletion of the stem cell compartment and the decreased numbers of differentiating spermatogonia. Thus, it is conceivable that the apoptotic cell death observed in the PRC1cKO is due to defective RNF2-dependent activation of germline genes such as Sall4.
Figure 5.
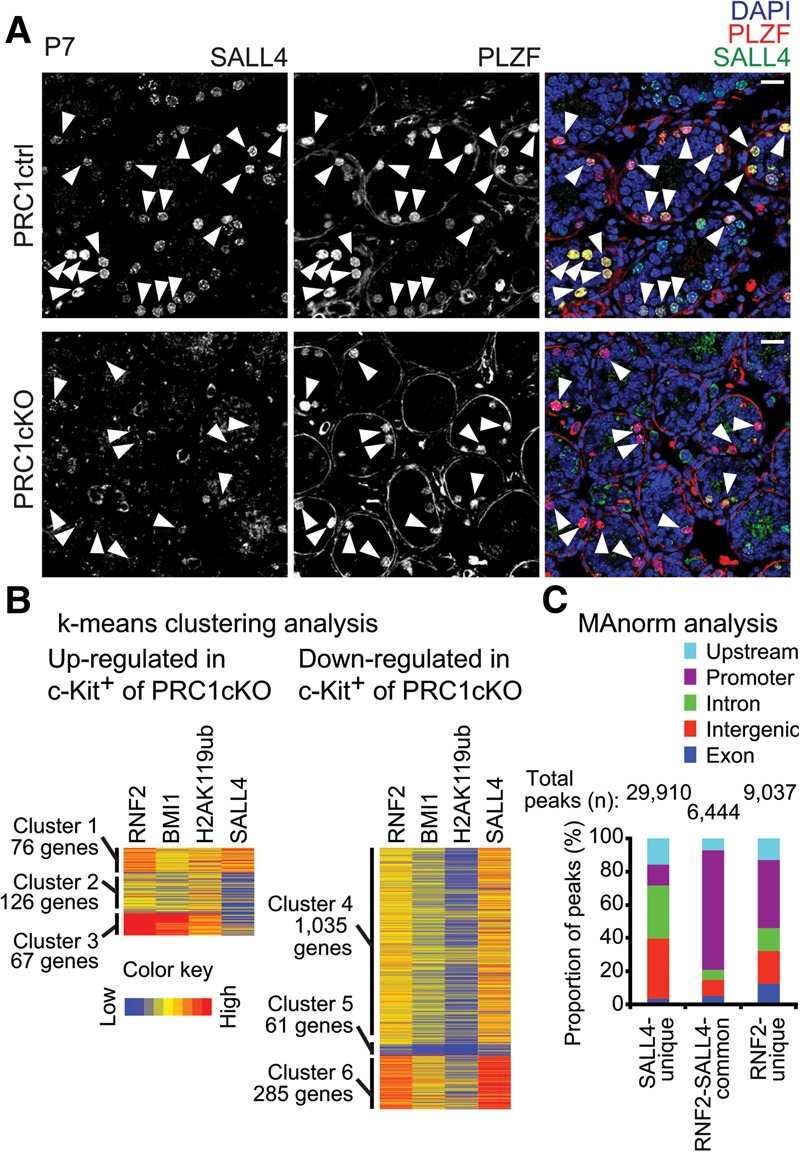
RNF2-dependent nuclear localization of SALL4 and their genomic distribution. (A) Immunostaining of SALL4 and PLZF in a testicular section at P7. Slides were counterstained with DAPI. PLZF-positive cells are shown with arrowheads. Bar, 20 µm. (B) k-means clustering analysis of the indicated genes based on ChIP-seq analysis. (C) MAnorm analysis of ChIP-seq data of RNF2 and SALL4.
To further define the mode of gene regulation by RNF2 and SALL4, we performed k-means clustering analysis of genes up-regulated and down-regulated in the PRC1cKO (Fig. 5B; Supplemental Fig. S8A; Supplemental Table 2). Up-regulated genes in c-Kit+ spermatogonia of the PRC1cKO comprised three clusters: Cluster 3 (67 genes) was highly enriched with genes bound by PRC1, including BMI1 and H2AK119ub, but not with SALL4, thus representing genes targeted for suppression by PRC1. Cluster 1 (76 genes) was enriched with genes bound by RNF2 and SALL4, suggesting that RNF2 and SALL4 together occupy a small subset of suppressed genes. Genes in cluster 2 (126 genes) were not highly enriched with PRC1 and SALL4 and were thus likely to be indirect targets. Contrastingly, in down-regulated genes in c-Kit+ spermatogonia of the PRC1cKO, RNF2 and SALL4 enrichment was found extensively, as seen in clusters 4 and 6 (respectively, 1035 and 285 genes). Cluster 5 (61 genes) consisted of genes indirectly activated by RNF2. To determine the gene expression profiles of PRC1-regulated genes in stages of spermatogenesis beyond that of differentiating spermatogonia, we analyzed RNA-seq data of wild-type meiotic pachytene spermatocytes and post-meiotic spermatocytes (Supplemental Fig. S8B). Our analyses revealed that genes in clusters 4 and 6 tend to be expressed in pachytene spermatocytes and round spermatids, although expression levels vary for each gene.
Finally, we independently confirmed the co-occupancy of RNF2 and SALL4 using peaks detected through MAnorm analysis (Shao et al. 2012). While RNF2 and SALL4 peaks were distributed in different places throughout the genome, RNF2 and SALL4 common peaks were enriched in promoter regions (Fig. 5C), suggesting that large sets of RNF2 and SALL4 peaks occupy TSSs together in undifferentiated spermatogonia.
RNF2 interacts with SALL4 to work together for gene activation
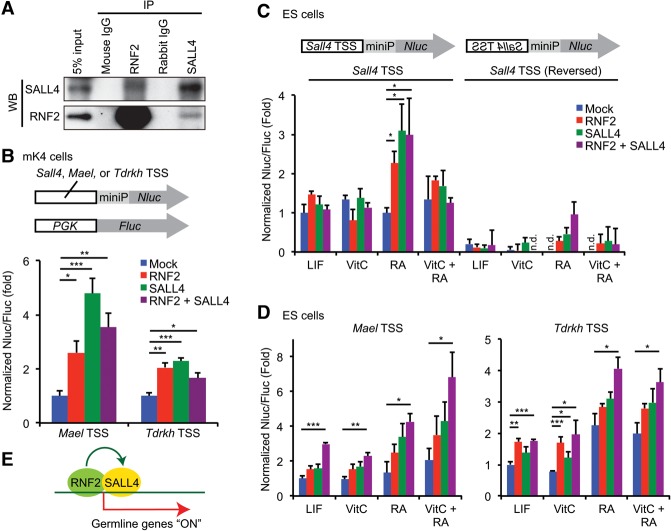
The above results suggested the existence of an RNF2–SALL4 complex for gene activation. To test for an in vivo interaction between RNF2 and SALL4, we performed coimmunoprecipitation of RNF2 and SALL4 using wild-type testicular extracts. Our results confirmed an in vivo interaction between RNF2 and SALL4 (Fig. 6A). To validate the direct function of RNF2 and SALL4 in gene activation, we performed luciferase reporter assays that measure the activity of regulatory elements under conditions where RNF2 and SALL4 are expressed. We performed a dual-luciferase reporter assay, in which the activity of regulatory elements was indicated by the expression of NanoLuc luciferase (Nluc) driven by a minimal promoter and normalized to the expression of control firefly luciferase (Fluc) driven by a PGK promoter (Fig. 6B). First, we performed the dual-luciferase reporter assay using kidney-derived mK4 cells. While Sall4 regulatory regions (the regions near TSSs, shown as the Sall4 TSSs) did not respond with RNF2 and/or SALL4 in mK4 cells (data not shown), RNF2 and SALL4 act on the regulatory regions of Mael and Tdrkh (Mael TSS and Tdrkh TSS) for gene activation (Fig. 6B).
Figure 6.
RNF2 interacts with SALL4, and RNF2 and SALL4 work together for gene activation. (A) Coimmunoprecipitation of RNF2 and SALL4 using wild-type testes at 14–19 d of age. (B) Dual-luciferase reporter assays using mK4 cells. Regulatory regions near TSSs of the Sall4, Mael, and Tdrkh genes were examined. (miniP) Minimal promoter. The relative luciferase activities (Nluc/Fluc) were normalized to Nluc/Fluc ratios from mock experiments in mK4 cells. (C,D) Dual-luciferase reporter assays using embryonic stem (ES) cells. Leukemia inhibitory factor (LIF) was used to maintain undifferentiated ES cells, and differentiation was induced through the withdrawal of LIF and the addition of vitamin C (VitC) and/or retinoic acid (RA). Relative luciferase activities (Nluc/Fluc) were normalized to Nluc/Fluc ratios from mock experiments in undifferentiated ES cells. (*) P < 0.05; (**) P < 0.005; (***) P < 0.001, unpaired t-test. Data are represented as mean ± SEM. Three to five independent experiments were performed. (E) Model of action of RNF2 and SALL4 for activation of germline genes, in which RNF2 activates transcription of Sall4, and RNF2 and SALL4 together occupy target genes for activation.
Next, we sought to perform dual-luciferase reporter assays with GS cells to test the gene activation function of an RNF2–SALL4 complex in a context that resembles the in vivo activation of germline genes. However, we found that the transient transfection of GS cells was very inefficient (data not shown). Instead, we tested embryonic stem (ES) cells treated with vitamin C or retinoic acid (RA), an active metabolite of vitamin A, both of which induce differentiation and the expression of germline genes (Blaschke et al. 2013; Suzuki et al. 2016). In ES cells, the Sall4 TSS was activated by RNF2 or SALL4 only after RA treatment, while vitamin C treatment did not change the responsiveness of the Sall4 TSS (Fig. 6C). Reverse orientation of the Sall4 TSS did not work in the assay (Fig. 6C), confirming the specific regulatory action of the Sall4 TSS in cis. Intriguingly, the Mael TSS and Tdrkh TSS were highly activated when both RNF2 and SALL4 were expressed, and responsiveness increased when ES cells were treated with RA (Fig. 6D). For the Mael TSS, responsiveness further increased when ES cells were treated with both RA and vitamin C. These data indicate that RNF2 and SALL4 act on regulatory elements of germline genes for activation during differentiation, supporting the notion that an RNF2–SALL4-activating complex promotes activation of germline genes.
Together, our study identifies a mechanism by which a cohort of germline genes is directly activated in spermatogonia, in which RNF2 activates transcription of Sall4, and RNF2 and SALL4 then act together on target genes for activation (Fig. 6E).
Discussion
Our study establishes a framework for the regulation of germline genes during spermatogenesis. In particular, we demonstrate that RNF2 is required for the activation of many critical regulators of spermatogenesis. Importantly, the role of RNF2 in the activation of Stra8 in spermatogonia is opposite to its role in female primordial germ cells, in which RNF2 suppresses the expression of Stra8 to prevent precocious entry into meiosis (Yokobayashi et al. 2013). Our luciferase assays demonstrated that regulatory elements of germline genes become responsive when ES cells are treated with RA, the major chemical determinant of spermatogenic differentiation (Griswold 2016). Since Stra8 is induced by RA, the distinct functions of RNF2 to regulate Stra8 likely depend on the presence of RA.
The function of RNF2 in the activation of germline genes may likely be independent of PRC2, the other major set of Polycomb complexes, as dysregulation of germline genes was not observed in spermatogenesis lacking components of PRC2 (Mu et al. 2014). A notable common feature between PRC1 and PRC2 mutant mice is that Cdkn2a, which encodes an essential cell cycle suppressor, is a major Polycomb target in somatic cells yet remains repressed in germ cells deficient for PRC1 (data not shown) and PRC2 (Mu et al. 2014). These results suggest that the cell cycle undergoes distinct regulation in spermatogonia versus somatic cells. Because multiple Polycomb subunits are exchanged to acquire specific functions for different biological contexts (Gao et al. 2012; Tavares et al. 2012), it is possible that distinct Polycomb complexes containing RNF2 serve as an activator of germline genes.
One intriguing possibility is that SALL4 dissociates the PRC1 complex, transforming it from a repressive complex into an “activating” complex. Since RNF2 stably associates with nuclei in male germ cells, it is possible that RNF2 is preloaded to target genes, and, later, SALL4 is recruited to these target loci. In support of this, SALL4 localization is dynamic during spermatogenesis. Furthermore, literature supports the possibility that RNF2 is preloaded to target genes. A recent report demonstrated that the same class of germline genes identified in the present study (and which includes Mael and Tdrkh) is suppressed by PCGF6-containing PRC1 (PCGF6–PRC1), a noncanonical PRC1, in ES cells (Endoh et al. 2017). In primordial germ cells, PRC1 suppresses germline genes (Yokobayashi et al. 2013). Therefore, it is conceivable that PCGF6–PRC1 targets germline genes in early development and continues to suppress them in early germ cell stages until such time that the germline genes are required for later stages in germ cell development. This possible function of PCGF6 may explain why germline genes are not fully associated with BMI1 (also known as PCGF4): The diversity of PRC1 complexes could depend, in part, on the presence of distinct PCGF factors (Gao et al. 2012).
The PCGF6–PRC1 complex forms a stable complex with the core PRC1 subunit RNF2, the epigenetic regulator L3MBTL2, and the transcription factor MAX (Gao et al. 2012). Both L3MBTL2 and MAX are known to suppress germline genes in ES cells (Qin et al. 2012; Maeda et al. 2013), and, furthermore, the loss of MAX promotes spermatogenic differentiation and meiotic entry in ES cells and GS cells (Suzuki et al. 2016). Thus, we speculate that PCGF6–PRC1 is transformed into an RNF2–SALL4-activating complex through the progressive removal of PCGF6, L3MBTL2, and MAX during spermatogenic differentiation. Interestingly, the suppression of germline genes in ES cells depends on the deposition of H2AK119ub (Endoh et al. 2017), and H2AK119ub is depleted from germline genes in undifferentiated spermatogonia when they are activated. Together, these results raise the compelling possibility that PRC1 has context-dependent functions through changing subunit compositions and H2AK119ub deposition. In future studies, it would be intriguing to identify Polycomb proteins working with RNF2 for gene activation.
Materials and methods
Animals
Generation of mutant Ring1 and Rnf2 floxed alleles was reported previously (del Mar Lorente et al. 2000; Cales et al. 2008). Ddx4-Cre transgenic mice were purchased from the Jackson Laboratory (Gallardo et al. 2007). For each experiment, a minimum of three independent mice was analyzed. The Institutional Animal Care and Use Committee approved this work (protocol no. IACUC2015-0032).
Antibodies
A list of antibodies used in this study is in the Supplemental Material.
Germ cell fractionation
Spermatogonia, isolated as described previously (Hasegawa et al. 2015), were collected from C57Bl/6N mice at 6–8 d of age. Testes were collected in a 24-well plate in Dulbecco's modified Eagle's medium (DMEM) supplemented with GlutaMax (Thermo Fisher Scientific), nonessential amino acids (NEAA) (Thermo Fisher Scientific), penicillin, and streptomycin (Thermo Fisher Scientific). After removing the tunica albuginea, testes were digested with 1 mg/mL collagenase for 20 min at 34°C to remove interstitial cells and then centrifuged at 188g for 5 min. Tubules were washed with the medium and then digested with 2.5 mg/mL trypsin for 20 min at 34°C to obtain a single-cell suspension. Cells were filtered with a 40-µm strainer to remove Sertoli cells, and the cell suspension was plated in a 24-well plate for 1 h in medium supplemented with 10% fetal bovine serum (FBS), which promoted the adhesion of the remaining somatic cells. Germ cells were washed with MACS buffer (PBS supplemented with 0.5% BSA, 5 mM EDTA) and incubated with CD117 (c-Kit) MicroBeads (Miltenyi Biotec) for 20 min on ice. Cells were washed and resuspended with MACS buffer and filtered with a 40-µm strainer. Cells were separated by autoMACS Pro Separator (Miltenyi Biotec) with the program “possel.” Cells in the flowthrough fraction were washed with MACS buffer and incubated with CD90.2 (Thy1) MicroBeads (Miltenyi Biotec) for 20 min on ice. Cells were washed and resuspended with MACS buffer, filtered with a 40-µm strainer, and then separated by autoMACS Pro Separator (Miltenyi Biotec) with the program “posseld.” Purity was confirmed by RNA-seq analysis and immunostaining.
Histology and immunohistochemistry
For preparation of testicular paraffin blocks, testes were fixed with 4% paraformaldehyde (PFA) overnight at 4°C. Testes were dehydrated and embedded in paraffin. For histological analysis, 4-µm-thick paraffin sections were deparaffinized and stained with hematoxylin and eosin. For immunohistochemistry, 7-µm-thick paraffin sections were deparaffinized and autoclaved in target retrieval solution (DAKO) for 10 min at 121°C. Sections were blocked with Blocking One Histo (Nacalai) for 1 h at room temperature and then incubated with primary antibodies overnight at 4°C. The resulting signals were detected by incubation with secondary antibodies conjugated to fluorophores (Thermo Fisher Scientific, Biotium, or Jackson ImmunoResearch). Sections were counterstained with DAPI. Images were obtained by confocal laser scanning microscope (A1R, Nikon) and processed with NIS-Elements (Nikon) and ImageJ (National Institutes of Health).
Code availability: analysis of ChIP-seq results
ChIP-seq data for RNF2, input, BMI1, and H2AK119ub in GS cells were downloaded from the Gene Expression Omnibus (accession no. GSE55060) (Hasegawa et al. 2015). ChIP-seq data for SALL4 in Thy1+ spermatogonia were downloaded from the Gene Expression Omnibus (accession no. GSE73390) (Lovelace et al. 2016). Data analysis was performed in the Wardrobe experiment management system (https://code.google.com/p/genome-tools) (Kartashov and Barski 2015). Briefly, reads were aligned to the mouse genome (mm10) with Bowtie version 1.0.0 (Langmead et al. 2009) before being displayed as coverage on a local mirror of the University of California at Santa Cruz Genome Browser. Peaks of RNF2, input, BMI1, H2AK119ub, and SALL4 enrichment were identified using MACS2 version 2.1.0.20140616 (Zhang et al. 2008). MAnorm, software designed for quantitative comparison of ChIP-seq data sets (Shao et al. 2012), was applied to compare genome-wide SALL4 and RNF2 profiles. Unique peaks were defined using the following criteria: (1) defined as “unique” by the MAnorm algorithm, (2) P-value of <0.01, and (3) raw read counts of unique peaks >20. Common peaks between the SALL4 and RNF2 data sets were defined using the following criteria: (1) defined as “common” by MAnorm algorithm and (2) raw read counts of both stages >20. Average tag density profiles were calculated around RefSeq TSSs. The resulting graphs were smoothed in 200-base-pair windows. Enrichment levels for ChIP-seq experiments were calculated for 4-kb windows at promoter regions of the genes (±2 kb surrounding TSSs). Tag values were normalized as follows: Read counts were multiplied by 1,000,000 and then divided by the total number of reads in each nucleotide position. Total amounts of tag values in promoter regions were calculated as enrichment.
RNA-seq
Total RNA was purified from Thy1+ or c-Kit+ spermatogonia using an RNeasy microkit (Qiagen) according to the manual provided. RNA quality and quantity were checked using Bioanalyzer (Agilent) and Qubit (Life Technologies), respectively. The initial amplification step was performed with the NuGEN Ovation RNA-seq system version 2. The assay was used to amplify RNA samples and create double-stranded cDNA. Libraries were then created with the Nextera XT DNA sample preparation kit (Illumina) and sequenced with an Illumina HiSeq 2500. RNA-seq data analysis was performed in the Wardrobe experiment management system (https://code.google.com/p/genome-tools) (Kartashov and Barski 2015). Briefly, reads were mapped to the mm10 genome using TopHat version 2.0.9 (Trapnell et al. 2012) and assigned to the RefSeq genes (or isoforms) using the Wardrobe algorithm. Differential gene expression analysis was performed using DESeq2 (Love et al. 2014) in the Wardrobe environment. For GO analysis, the Database for Annotation, Visualization, and Integrated Discovery (DAVID) was used to identify the GO terms (Huang et al. 2009).
RNA-seq analysis of repetitive elements
All mouse repetitive element sequences (313 loci) were downloaded from Repbase (http://www.girinst.org/repbase) (Bao et al. 2015). A Bowtie2 index was created using the function “bowtie2-build.” Raw RNA-seq reads were aligned to repetitive sequences using the default parameters of Bowtie2. A table of aligned read counts per repetitive element was extracted from sorted .bam files using the SAMtools function “idxtats.” A read count output file was input to the DESeq2 package (Love et al. 2014); comparison of expression levels of each repetitive element between the PRC1ctrl and the PRC1cKO was performed using the functions “DESeqDataSetFromMatrix” and “DESeq.” Differentially expressed repetitive elements between the PRC1ctrl and the PRC1cKO were identified by two criteria: more than twofold change and Wald test (P < 0.05).
Cell culture
The mouse ES cell line J1 was maintained in DMEM (Thermo Fisher Scientific) supplemented with 20% FBS (HyClone), 25 mM HEPES (pH 7.2–7.5) (Thermo Fisher Scientific), 1% MEM NEAA (Thermo Fisher Scientific), 1% GlutaMAX-I (Thermo Fisher Scientific), 100 U/mL penicillin–streptomycin (Thermo Fisher Scientific), 0.1 mM β-mercaptoethanol (Sigma), and 500 U/mL leukemia inhibitory factor (LIF). The cells were cultured in feeder-free conditions on 0.1% gelatin-coated tissue culture plates. For differentiation experiments, ES cells were treated for 72 h with 100 µg/mL vitamin C and/or 1 µM all-trans RA without LIF (Blaschke et al. 2013; Suzuki et al. 2016). The mK4 cells (Valerius et al. 2002) were cultured in DMEM with GlutaMAX (Thermo Fisher Scientific), 10% FBS, and 100 U/mL penicillin–streptomycin (Thermo Fisher Scientific).
Plasmid constructions
For construction of expression vectors for RNF2 and SALL4 with an N-terminal 3xFlag tag, the full-length RNF2 or SALL4 coding sequence was amplified by PCR using Q5 High-Fidelity 2X master mix (New England Biolabs) with primers containing the 3xFlag tag sequence. The PCR product was inserted into the pGL3.54 vector (Promega) at the luc2 gene locus. To generate reporter vectors, DNA fragments around TSSs of Sall4, Mael, or Tdrkh were designed based on RNF2- and SALL4-binding sites detected by MAnorm analysis and synthesized by IDT as gBlocks gene fragments. We chose the minus strand of chromosome 2 (168,766,670–168,767,574) for the Sall4 TSS, the minus strand of chromosome 1 (166,238,666–166,239,118) for the Mael TSS, and the plus strand of chromosome 3 (94,412,744–94,413,466) for the Tdrkh TSS. The gBlocks gene fragments were inserted between the XhoI and BglII sites of the pNL3.2 vector (Promega). For a positive control reporter vector, a DNA fragment of the PGK promoter was inserted into the pNL3.2 vector.
Dual-luciferase reporter assays
The Nano-Glo dual-luciferase reporter assay system (Promega) was used to measure promoter activities. Briefly, 5 × 104 ES cells or 3 × 104 mK4 cells were seeded in TC-treated 96-well solid white polystyrene microplates (Corning) 1 d before transfection. Transient transfections were performed in OptiMEM (Thermo Fisher Scientific) with Lipofectamine 3000 reagent (Thermo Fisher Scientific) according to the manufacturer's instructions. The cells were transfected with combinations of pGL3.54, a control reporter; pNL3.2 inserted with or without the Sall4 TSS, Mael TSS, Tdrkh TSS, or PGK promoter as an experimental reporter; and expression vectors for RNF2 and SALL4. Three to five replicates were used for each condition. For ES cells, the medium was replaced with vitamin C- and/or RA-containing medium or LIF-containing medium after 5 h of transfection. The medium was replaced every 24 h. After 72 h of treatment, dual-luciferase assays were performed according to the manufacturer's instructions (Promega). For mK4 cells, dual-luciferase assays were performed 48 h after transfection. Light signals were acquired with the Synergy H1 hybrid multimode microplate reader (BioTek), and the relative response ratio (RRR) was calculated according to the manufacturer's instructions (Promega). Normalized RRR is shown as normalized Nluc/Fluc luminescence.
Coimmunoprecipitation
Single-cell suspension from juvenile whole testes (P12–P19; five to eight mice) was prepared by treatment with collagenase and trypsin as described above. Cells were suspended in 1 mL of buffer A (10 mM HEPES-KOH at pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, protease inhibitors [Roche]) and disrupted with 35 strokes in a Dounce homogenizer with a tight-fitting pestle. Lysates were centrifuged at 1700g for 10 min at 4°C. Nuclear pellets were washed with buffer A and suspended with 500 µL of buffer B (20 mM HEPES-KOH at pH 7.9, 50 mM KCl, 5 mM CaCl2, 1 mM MgCl2, 10% glycerol, 0.5 mM DTT, protease inhibitors, 1 µL MNase [New England Biolabs], 100 U of benzonase [Sigma]). Nuclei were incubated for 10 min at 37°C and then further disrupted by sonication. EGTA (10 mM final), 300 mM NaCl (final), and 0.2% NP-40 (final) were added to the lysates. The lysates were incubated with rotation at 20 rpm for 1 h at 4°C and centrifuged at 8000g for 10 min at 4°C. Supernatant was transferred to a new tube; mixed with an equal volume of buffer C (20 mM HEPES-KOH at pH 7.9, 10% glycerol, 0.5 mM DTT, protease inhibitors); incubated with anti-RNF2 (MBL), anti-SALL4 (Abcam) antibody, or control IgG overnight at 4°C; and mixed with Dynabeads Protein G (Thermo Fisher Scientific). The beads were washed four times with buffer D (20 mM HEPES-KOH at pH 7.9, 150 mM NaCl, 10% glycerol, 0.05% NP-40, 0.5 mM DTT). The bound proteins were eluted with Laemmli buffer for 5 min at 95°C and analyzed by Western blotting.
Data availability
RNA-seq data reported in this study were deposited to the Gene Expression Omnibus (accession no. GSE102783).
Supplementary Material
Acknowledgments
We thank Ho-Su Sin and Andrey V. Kartashov for technical assistance; Tony DeFalco, Yueh-Chiang Hu, and members of the Namekawa laboratory for discussion and helpful comments; Alex Bortvin and Sandy Martin for the LINE1-ORF1p antibody; and Mary Ann Handel for the H1T antibody. This work was supported by a research grant from the March of Dimes Foundation (FY13-510 to S.H.N.) and National Institutes of Health grants (DP2GM119134 to A.B., and R01GM 098605 to S.H.N.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.302000.117.
References
- Alavattam KG, Kato Y, Sin HS, Maezawa S, Kowalski IJ, Zhang F, Pang Q, Andreassen PR, Namekawa SH. 2016. Elucidation of the Fanconi anemia protein network in meiosis and its function in the regulation of histone modifications. Cell Rep 17: 1141–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloia L, Di Stefano B, Di Croce L. 2013. Polycomb complexes in stem cells and embryonic development. Development 140: 2525–2534. [DOI] [PubMed] [Google Scholar]
- Anderson EL, Baltus AE, Roepers-Gajadien HL, Hassold TJ, de Rooij DG, van Pelt AM, Page DC. 2008. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc Natl Acad Sci 105: 14976–14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O. 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke K, Ebata KT, Karimi MM, Zepeda-Martinez JA, Goyal P, Mahapatra S, Tam A, Laird DJ, Hirst M, Rao A, et al. 2013. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 500: 222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broering TJ, Alavattam KG, Sadreyev RI, Ichijima Y, Kato Y, Hasegawa K, Camerini-Otero RD, Lee JT, Andreassen PR, Namekawa SH. 2014. BRCA1 establishes DNA damage signaling and pericentric heterochromatin of the X chromosome in male meiosis. J Cell Biol 205: 663–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, de Rooij DG, Braun RE. 2004. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet 36: 647–652. [DOI] [PubMed] [Google Scholar]
- Cales C, Roman-Trufero M, Pavon L, Serrano I, Melgar T, Endoh M, Perez C, Koseki H, Vidal M. 2008. Inactivation of the polycomb group protein Ring1B unveils an antiproliferative role in hematopoietic cell expansion and cooperation with tumorigenesis associated with Ink4a deletion. Mol Cell Biol 28: 1018–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty P, Buaas FW, Sharma M, Snyder E, de Rooij DG, Braun RE. 2014. LIN28A marks the spermatogonial progenitor population and regulates its cyclic expansion. Stem Cells 32: 860–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Kitamura K, Kasai S, Fujioka M, Hiyoshi M, Takamune K, Noce T, Nakatsuji N. 2006. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc Natl Acad Sci 103: 15894–15899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP. 2004. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet 36: 653–659. [DOI] [PubMed] [Google Scholar]
- Creppe C, Palau A, Malinverni R, Valero V, Buschbeck M. 2014. A Cbx8-containing polycomb complex facilitates the transition to gene activation during ES cell differentiation. PLoS Genet 10: e1004851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Voloshin O, Potapova S, Camerini-Otero RD. 2017. Meiotic knockdown and complementation reveals essential role of RAD51 in mouse spermatogenesis. Cell Rep 18: 1383–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Mar Lorente M, Marcos-Gutierrez C, Perez C, Schoorlemmer J, Ramirez A, Magin T, Vidal M. 2000. Loss- and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development 127: 5093–5100. [DOI] [PubMed] [Google Scholar]
- Endoh M, Endo TA, Endoh T, Isono K, Sharif J, Ohara O, Toyoda T, Ito T, Eskeland R, Bickmore WA, et al. 2012. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet 8: e1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh M, Endo TA, Shinga J, Hayashi K, Farcas A, Ma KW, Ito S, Sharif J, Endoh T, Onaga N, et al. 2017. PCGF6–PRC1 suppresses premature differentiation of mouse embryonic stem cells by regulating germ cell-related genes. Elife 6: e21064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo T, Shirley L, John GB, Castrillon DH. 2007. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 45: 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D. 2012. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell 45: 344–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Lee P, Stafford JM, von Schimmelmann M, Schaefer A, Reinberg D. 2014. An AUTS2–Polycomb complex activates gene expression in the CNS. Nature 516: 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassei K, Orwig KE. 2013. SALL4 expression in gonocytes and spermatogonial clones of postnatal mouse testes. PLoS One 8: e53976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler SJ, Paro R. 2015. Trithorax and Polycomb group-dependent regulation: a tale of opposing activities. Development 142: 2876–2887. [DOI] [PubMed] [Google Scholar]
- Griswold MD. 2016. Spermatogenesis: the commitment to meiosis. Physiol Rev 96: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K, Sin HS, Maezawa S, Broering TJ, Kartashov AV, Alavattam KG, Ichijima Y, Zhang F, Bacon WC, Greis KD, et al. 2015. SCML2 establishes the male germline epigenome through regulation of histone H2A ubiquitination. Dev Cell 32: 574–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs RM, Fagoonee S, Papa A, Webster K, Altruda F, Nishinakamura R, Chai L, Pandolfi PP. 2012. Functional antagonism between Sall4 and Plzf defines germline progenitors. Cell Stem Cell 10: 284–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway JK, Morelli MA, Borst PL, Cohen PE. 2010. Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination. J Cell Biol 188: 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YC, de Rooij DG, Page DC. 2013. Tumor suppressor gene Rb is required for self-renewal of spermatogonial stem cells in mice. Proc Natl Acad Sci 110: 12685–12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Illingworth RS, Moffat M, Mann AR, Read D, Hunter CJ, Pradeepa MM, Adams IR, Bickmore WA. 2015. The E3 ubiquitin ligase activity of RING1B is not essential for early mouse development. Genes Dev 29: 1897–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartashov AV, Barski A. 2015. BioWardrobe: an integrated platform for analysis of epigenomics and transcriptomics data. Genome Biol 16: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmins S, Sassone-Corsi P. 2005. Chromatin remodelling and epigenetic features of germ cells. Nature 434: 583–589. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. 2004. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131: 839–849. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, Shiromoto Y, Asada N, Toyoda A, Fujiyama A, et al. 2010. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev 24: 887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Gill ME, Koubova J, Page DC. 2008. Germ cell-intrinsic and -extrinsic factors govern meiotic initiation in mouse embryos. Science 322: 1685–1687. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovelace DL, Gao Z, Mutoji K, Song YC, Ruan J, Hermann BP. 2016. The regulatory repertoire of PLZF and SALL4 in undifferentiated spermatogonia. Development 143: 1893–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda I, Okamura D, Tokitake Y, Ikeda M, Kawaguchi H, Mise N, Abe K, Noce T, Okuda A, Matsui Y. 2013. Max is a repressor of germ cell-related gene expression in mouse embryonic stem cells. Nat Commun 4: 1754. [DOI] [PubMed] [Google Scholar]
- Matson CK, Murphy MW, Griswold MD, Yoshida S, Bardwell VJ, Zarkower D. 2010. The mammalian doublesex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Dev Cell 19: 612–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu W, Starmer J, Fedoriw AM, Yee D, Magnuson T. 2014. Repression of the soma-specific transcriptome by Polycomb-repressive complex 2 promotes male germ cell development. Genes Dev 28: 2056–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollinger R, Childs AJ, Burgess HM, Speed RM, Lundegaard PR, Reynolds N, Gray NK, Cooke HJ, Adams IR. 2008. Deletion of the pluripotency-associated Tex19.1 gene causes activation of endogenous retroviruses and defective spermatogenesis in mice. PLoS Genet 4: e1000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Eckardt S, Leu NA, Buffone MG, Zhou J, Gerton GL, McLaughlin KJ, Wang PJ. 2009. Inactivation of Nxf2 causes defects in male meiosis and age-dependent depletion of spermatogonia. Dev Biol 330: 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Stroud H, Nee K, Liu W, Pezic D, Manakov S, Lee SA, Moissiard G, Zamudio N, Bourc'his D, et al. 2014. MORC1 represses transposable elements in the mouse male germline. Nat Commun 5: 5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pengelly AR, Kalb R, Finkl K, Muller J. 2015. Transcriptional repression by PRC1 in the absence of H2A monoubiquitylation. Genes Dev 29: 1487–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posfai E, Kunzmann R, Brochard V, Salvaing J, Cabuy E, Roloff TC, Liu Z, Tardat M, van Lohuizen M, Vidal M, et al. 2012. Polycomb function during oogenesis is required for mouse embryonic development. Genes Dev 26: 920–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Whyte WA, Anderssen E, Apostolou E, Chen HH, Akbarian S, Bronson RT, Hochedlinger K, Ramaswamy S, Young RA, et al. 2012. The polycomb group protein L3mbtl2 assembles an atypical PRC1-family complex that is essential in pluripotent stem cells and early development. Cell Stem Cell 11: 319–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo H, Prosser H, Ruzankina Y, Mahadevaiah SK, Cloutier JM, Baumann M, Fukuda T, Hoog C, Toth A, de Rooij DG, et al. 2013. ATR acts stage specifically to regulate multiple aspects of mammalian meiotic silencing. Genes Dev 27: 1484–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxe JP, Chen M, Zhao H, Lin H. 2013. Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J 32: 1869–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Zhang Y, Yuan GC, Orkin SH, Waxman DJ. 2012. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol 13: R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan SK, Pyle A, Coppola V, Babus J, Swaminathan S, Benedict J, Swing D, Martin BK, Tessarollo L, Evans JP, et al. 2004. BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development 131: 131–142. [DOI] [PubMed] [Google Scholar]
- Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, Kondoh G, Okawa K, Chujo T, Suzuki T, et al. 2009. The TDRD9–MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell 17: 775–787. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. 2013. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell 49: 808–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin HS, Kartashov AV, Hasegawa K, Barski A, Namekawa SH. 2015. Poised chromatin and bivalent domains facilitate the mitosis-to-meiosis transition in the male germline. BMC Biol 13: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper SF, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, Bortvin A. 2008. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell 15: 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Ahn HW, Chu T, Bowden W, Gassei K, Orwig K, Rajkovic A. 2012. SOHLH1 and SOHLH2 coordinate spermatogonial differentiation. Dev Biol 361: 301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Hirasaki M, Hishida T, Wu J, Okamura D, Ueda A, Nishimoto M, Nakachi Y, Mizuno Y, Okazaki Y, et al. 2016. Loss of MAX results in meiotic entry in mouse embryonic and germline stem cells. Nat Commun 7: 11056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takubo K, Ohmura M, Azuma M, Nagamatsu G, Yamada W, Arai F, Hirao A, Suda T. 2008. Stem cell defects in ATM-deficient undifferentiated spermatogonia through DNA damage-induced cell-cycle arrest. Cell Stem Cell 2: 170–182. [DOI] [PubMed] [Google Scholar]
- Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, et al. 2012. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148: 664–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JM, Aprelikova O, Xu X, Wang R, Kim S, Chandramouli GV, Barrett JC, Burgoyne PS, Deng CX. 2004. BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr Biol 14: 2135–2142. [DOI] [PubMed] [Google Scholar]
- Valerius MT, Patterson LT, Witte DP, Potter SS. 2002. Microarray analysis of novel cell lines representing two stages of metanephric mesenchyme differentiation. Mech Dev 112: 219–232. [DOI] [PubMed] [Google Scholar]
- Voncken JW, Roelen BA, Roefs M, de Vries S, Verhoeven E, Marino S, Deschamps J, van Lohuizen M. 2003. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci 100: 2468–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Eckardt S, Leu NA, McLaughlin KJ, Wang PJ. 2008a. Mouse TEX15 is essential for DNA double-strand break repair and chromosomal synapsis during male meiosis. J Cell Biol 180: 673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Gell K, van der Heijden GW, Eckardt S, Leu NA, Page DC, Benavente R, Her C, Hoog C, McLaughlin KJ, et al. 2008b. Meiotic failure in male mice lacking an X-linked factor. Genes Dev 22: 682–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobayashi S, Liang CY, Kohler H, Nestorov P, Liu Z, Vidal M, van Lohuizen M, Roloff TC, Peters AH. 2013. PRC1 coordinates timing of sexual differentiation of female primordial germ cells. Nature 495: 236–240. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data reported in this study were deposited to the Gene Expression Omnibus (accession no. GSE102783).