ABSTRACT
Vaccines still are an important way to prevent and treat acquired immunodeficiency syndrome (AIDS).1 For developing an effective T cell-based AIDS vaccine, it is critical to define the human leukocyte antigen (HLA) type and epitope that elicit the most potent responses. This study involved 29 antiretroviral therapy-naive and chronic human immunodeficiency virus (HIV)-1 subtype B-infected individuals. A polymerase chain reaction-sequence-specific primer was used to detect the HLA typing, and the enzyme-linked immunospot assay to quantify the T-cell immune function. The results showed that the HLA-DQB1*06-positive group had higher CD4 counts and lower viral load (VL) compared with the HLA-DQB1*06-negative group; A higher magnitude of HIV-1-specific T-cell response and breadth were observed in the HLA-DQB1*06-positive group; the T-cell response was proportional to VL (R2 = 0.488, P = 0.0368) in the HLA-DQB1*06-positive group. The total T-cell responses to HIV-1 Nef core region were quantified at the single-peptide level. Nine (90%) peptides were recognized in 18 (62.1%) individuals. The breath of Nef core region-specific T-cell response was correlated positively with CD4+ T cell count and inversely with VL, which improved disease outcomes. These data revealed that HLA-DQB1*06 had a protective effect on the course of HIV-1 and T-cell targeting of certain specific Nef epitopes, contributing to HIV-1 suppression. The results suggested the potential use of HLA-DQB1*06 and Nef core region in HIV-1 T-cell vaccine design.
KEYWORDS: HIV-1, HLA, NEF T-cell response, Vaccine
Introduction
It is crucial to understand accurately the influence of sequence variation among human immunodeficiency virus (HIV) subtypes and human leukocyte antigen (HLA) diversity among ethnic groups for developing a more effective vaccine candidates.2,3 The HLA-restricted T-lymphocyte immune response is one of the major factors determining the genetic diversity of HIV-1.4,5 This has been reported more frequently in the HLA-I type, but less in the HLA-II type.6 The HIV-1 Nef gene encodes 200–215 amino acids in length.1,7 Nef recognition dominates in acute HIV-1 infection, but the function of Nef T-cell responses in cohort studies of chronically infected subjects is still not clear.8 At the same time, Nef core protein is substantially more conserved.9 It has a high density of overlapping CD8+ T-cell epitopes (see Nef epitope map at www.hiv.lanl.gov). Therefore, the Nef region was chosen as the target gene for the study. HIV-1 subtype B is the most prevalent HIV-1 subtype both in China and globally. HIV viral load (VL) and CD4+ T-cell count are the key markers of HIV-1 disease progression. The emergent ascendancy of HIV-1 subtype B indicates a need for a comprehensive analysis of HIV-1 subtype B-specific immune responses, dealing with both CD4 and CD8, in the context of designing a vaccine.
Results
The Nef-core peptides of HIV-1 can cause T cell response in most HIV-1 patients
Peptide is amide combining the amino group of one amino acid with the carboxyl group of another, usually obtained by partial hydrolysis of viral protein. Activation of a T cell is initiated by the interaction of TCR with a peptide displayed by major histocompatibility complex (MHC) molecules on the surface of antigen-presenting cells (APCs). Among 29 individuals, 18 (62.1%) were found to have at least 1 peptide. The average magnitude was 973.7 SFU/106 cells (range 47.2–3783.3) (Fig. 1 Magnitude of human immunodeficiency virus (HIV)-1 subtype B Nef core specific T cell responses in 29 HIV-1 subtype B-infected individuals). Out of 10 overlapping peptides (OLPs), 9 (90%) were recognized in at least 1 individual: only 1 OLP was recognized in 1 subject, and the other 8 OLPs were recognized in more than 1 volunteer. N14 (KRQDILDLWVYHTQGYFP) was the highest ratio sum of all the responses from various individuals, with 5870 SFU/106 cells. The highest frequently recognized peptide was N12 (HFLKEKGGLEGLIYSQKR), which was recognized in 6 individuals (Fig. 2 Magnitude and Frequency of 10 HIV-1 subtype B Nef core OLPs). To visually see all peptides and patient responses, the heatmap was made out (Fig. 3 The heatmap of peptides and patient responses). These peptides can be used as a reference for the design of vaccines.
Figure 1.
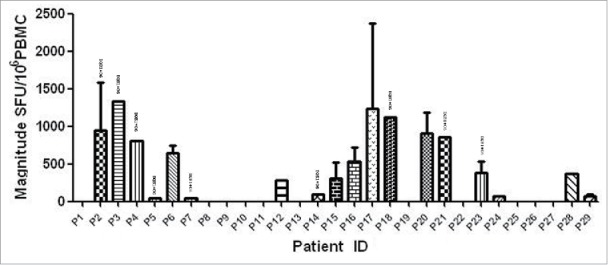
Magnitude of human immunodeficiency virus (HIV)-1 subtype B Nef core specific T cell responses in 29 HIV-1B-infected individuals. Measured as interferon (IFN)-γ enzyme-linked immunospot (ELISPOT), represented as spot-forming units (SFU)/106 peripheral blood mononuclear cells.
Figure 2.
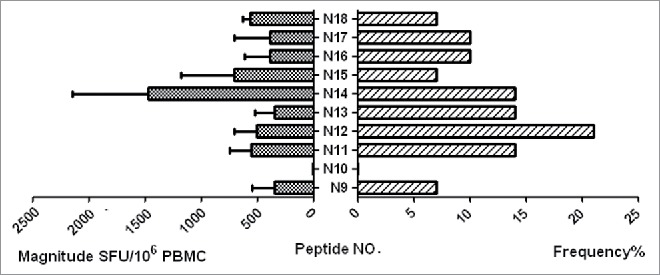
Magnitude and frequency of 10 HIV-1 subtype B Nef core OLPs. The total magnitude of T cell responses specific for each tested peptide is shown as bars in the left part of the graph. The frequency of each tested peptide is shown as bars on the right part of the graph.
Figure 3.
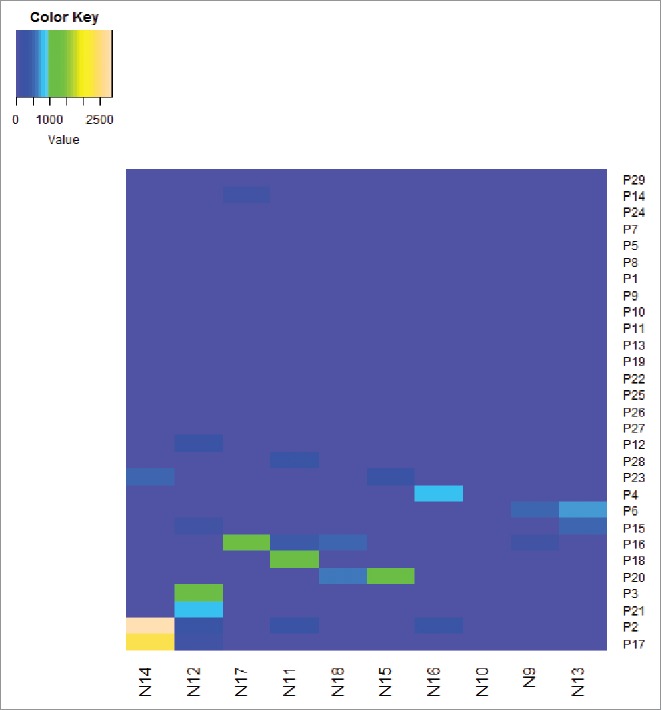
The heatmap for all patients and all peptides provide full picture with the ELISPOT responses. The right ordinate is the patient NO., The bottom abscissa is the peptide NO. The heatmap should define the cut-off as the detection limit.
ELISPOT magnitude, breadth, and clinical outcome
The correlations of ELISPOT magnitude with the CD4+ T-cell count and VL were first examined to investigate the relationship between HIV-1 Nef core region-specific T-cell function and clinical outcome. The results showed no significant correlations with the total magnitude of responses and the CD4+ T-cell count (R2 = 0.0216, P = 0.6161) (Fig. 4a ELISPOT magnitude is associated with CD4+T cell count and viral load); the same was found with the plasma VL (R2 = 0.0093, P = 0.7031) (Fig. 4b).
Figure 4.
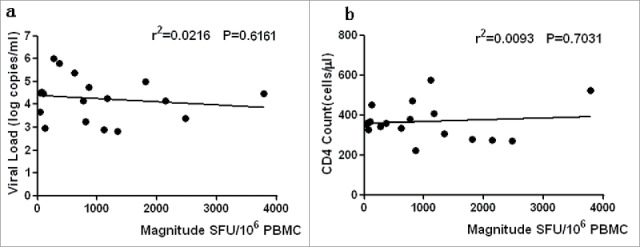
ELISPOT magnitude is associated with CD4+T cell count and viral load. The associations between ELISPOT magnitude (total spot forming units per 1 Million PBMCs and CD4+T cell count or viral load) were analyzed by Spearman's correlation. The Nef core-specific response was compared with CD4+T cell count (a) and viral load (b). P value is corrected by FDR.
Next, the relationship between the ELISPOT breadth and CD4+ T-cell count and VL was investigated. The CD4+ T-cell median count was found to be significantly higher [499 cells/mL (range 475–523)] in individuals with 4 responses compared with 329 cells/mL (range 159–543) among those with a greater range of responses (P = 0.0258)-, but not in other sites (Fig. 5a ELISPOT breadth is associated with CD4+T cell countand viral load). The VL median was significantly higher in individuals with 4 responses [2.81 log copies/mL (range 2.17–3.46)] compared with 4.55 log copies/mL (range 3.34–5.73) individuals with a greater range of responses (P = 0.0397), but not in other sites (Fig. 5b). The data showed that the more the number of reacting Nef core peptides, the higher the CD4 cell count and the lower the VL. Therefore, The Nef core region-specific immune responses could partially reduce the level of viral load.
Figure 5.
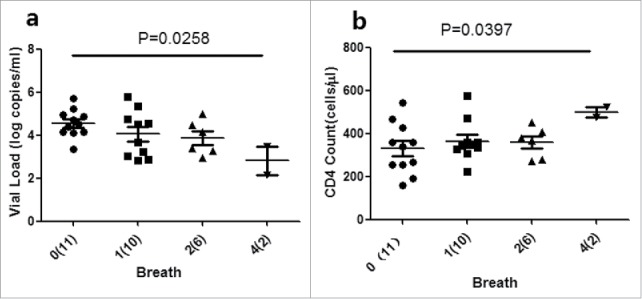
ELISPOT breadth is associated with CD4+T cell countand viral load. The associations between ELISPOT breadth [the number of reacting OLPs] and CD4+T cellcount (a) or viral load (b) were analyzed using the Kruskal-Wallis test. (n) Refers to the number of individuals. P value is corrected by FDR.
HLA-DQB1*06 and clinical outcome
The relationship between HLA-DQB1*06 and clinical outcome was examined to further clarify the role of HLA-DQB1*06 in the progression of HIV-1. The results showed that the CD4 count of the HLA-DQB1*06-positive group was higher than that of the HLA-DQB1*06-negative group, and the difference was not statistically significant (P > 0·05) (Fig. 6a Comparison of CD4 count, VL, Magnitude and Breadth between DQB1*06 positive and negative group). VL in the HLA-DQB1*06-positive group was significantly lower than that in the HLA-DQB1*06-negative group, with a statistically significant difference (P = 0·0415) (Fig. 6b). The quantitative T-cell responses were significantly higher in the HLA-DQB1*06-positive group than in the HLA-DQB1*06-negative group (P = 0.0297) (Fig. 6c). The number of reacting peptides wasn't significant difference in the HLA-DQB1*06-positive group and negative group (P > 0·05) (Fig. 6d). The correlation between HIV-1-specific T-cell responses and CD4+ T-cell count and plasma VL were analyzed. Numerical counts in the HLA-DQB1*06-positive group had a significant correlation with clinical outcome in VL (R2 = 0·4881, P = 0·0368) (Fig. 7a DQB1*06 allele-specific enzyme-linked immunospot (ELISPOT) magnitude were compared with viral load (VL) and CD4+ T cell count) and CD4+ T-cell count (R2 = 0·2131, P = 0·3121) (Fig. 7b). No statistically significant correlation was observed between either plasma VL or CD4+T-cell count and T-cell responses in the HLA-DQB1*06-negative group (Fig. 6c and d).
Figure 6.
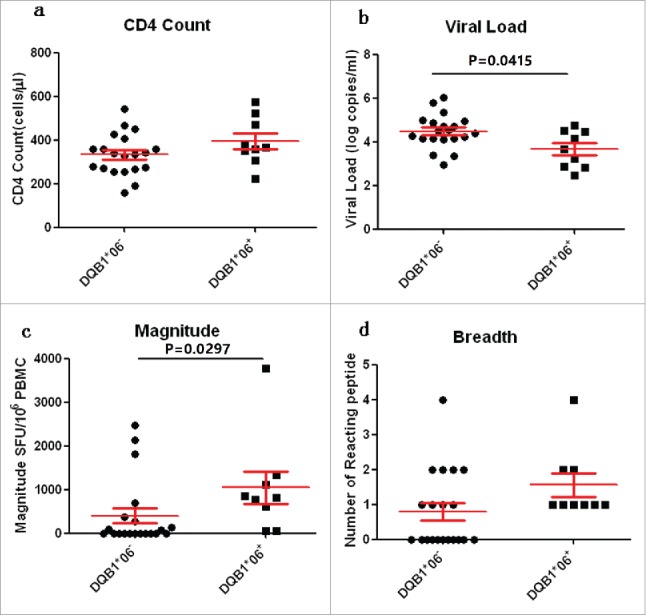
Comparison of CD4 count, VL, Magnitude and Breadth between DQB1*06 positive and negative group. P value is corrected by FDR.
Figure 7.
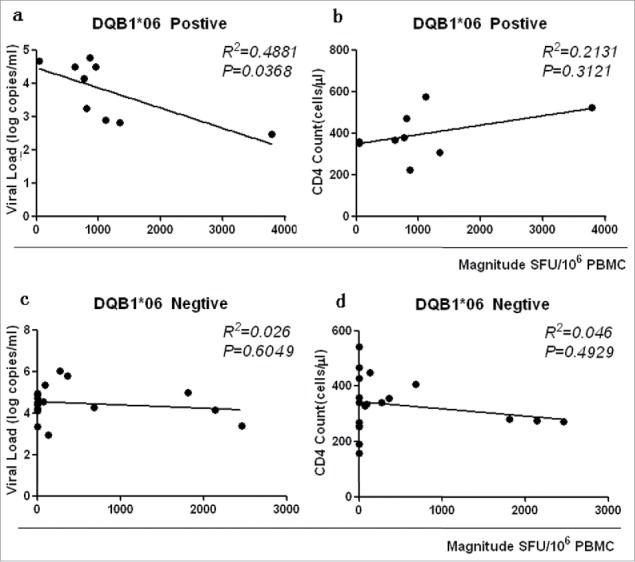
DQB1*06 allele-specific enzyme-linked immunospot (ELISPOT) magnitude were compared with viral load (VL) and CD4+ T cell count. P value is corrected by FDR.
Discussion
HIV-1 vaccine is essential in combating current HIV-1 pandemics. The HIV-1 Nef gene encodes 200–215 amino acids in length.10,11 Nef has a complex role in HIV-1 pathogenicity via several mechanisms, including downregulation of host CD4 and major histocompatibility complex (MHC) cell surface expression, modulation of T-cell function, and altering of macrophage signaling.12-14 Thus, Nef is the most versatile of all HIV-1 accessory proteins.15 This study demonstrated multiple immunodominant regions in Nef core protein. The dominant epitope peptide was screened using an HIV-1 Gag region whole-gene overlapping peptide. About 38% peptides were recognized in Gag protein.16 However, the overlap polypeptide in the Nef core region caused a higher immune response (90%) compared with the Gag region, leading to response in 62.1% individuals. These results suggested that the ability to target Nef epitopes with sufficient strength might serve as a correlate of protection. At the same time, 2 dominant epitope peptides (N12 and N14) were found in the Nef core region. Therefore, Nef core protein had extremely high immune efficiency.
This study showed statistically significant correlations between the breadth of Nef core region-specific T-cell responses and plasma VL or CD4+ T-cell count. It suggested that Nef core region-specific CD8+ T-cell responses might mediate highly effective immune control of viremia. Several further lines of evidence pointing toward the potential immunological benefit of targeting HIV-1 Nef have been reported.6,17,18 Nef has a high density of overlapping CD8+ T-cell epitopes (see Nef epitope map at www.hiv.lanl.gov), and multiple copies of Nef are produced early in the HIV-1 life cycle.10,19 Nef is the most targeted protein in acute infection,11-13 accounting for 50–90% of CD8+ T-cell responses in acutely infected subjects,8,20 as well as having the maximum CD8+ T-cell responses and the highest-magnitude IFN-gamma responses in chronic infection.21 HIV-1 vaccine-design efforts have, in large part, centered on T-cell priming immunizations, as the pivotal role of CD8+ T cells in controlling viral replication has been demonstrated in both HIV-122-24 and experimental Simian immunodeficiency virus (SIV) infections.25,26 The structure of Nef core region is relatively conservative. Therefore, the vaccine targeted at the Nef core region could improve T-cell activity against infected cells and improve immune control.
This study further strengthened the evidence regarding the involvement of DQB1*06 haplotype in the control of HIV-1 infection. Host genetic factors play an important role in mediating resistance to HIV-1 infection and may modify the course of virus infection. Many reports have shown that class I HLA alleles are closely related to disease progression. Less is known about the association of class II HLA alleles with disease progression. A few studies have cited a relationship between HLA-DRB1*13 and/or HLA-DQB1*06 and HIV-1 control27,28 The present study showed that the DQB1*06 haplotype-positive group had a higher CD4+ T-cell count and lower VL compared with the DQB1*06 haplotype-negative group. This raised the possibility of previous observations reporting a similar protective effect.27 It should be noted that most of the subjects with HLA-DQB1*06 do not have class I alleles associated with HIV-1 control: HLA-B*57, HLA-B*27, Bw4/Bw4, HLA-B*81, and so forth (Table 1 Characteristics of the study patient cohort). Therefore, the interference of other genotypes was excluded. Further exploration of mechanisms showed that the magnitude and breath of HIV-1-specific T-cell response differed among individuals with or without DQB1*06. The DQB1*06-positive group had higher magnitude and breath of HIV-1-specific T-cell response compared with the DQB1*06 haplotype-negative group. At the same time, the magnitude of HIV-1-specific T-cell response had a significant correlation with clinical outcome in the DQB1*06-positive group, suggesting that the presence of DQB1*06 haplotype influenced the T-cell immune responses to HIV-1 infection. This was consistent with data from Vyakarnam et al., who showed that HLADQB1*06 alleles had a potential protective effect on the course of HIV-1 disease.27 One of the possible explanations is that specific structural features that are shared by different HLA molecules and influence peptide-binding sites confer a protective effect. HLA-DQB1*06 is prone to combine with peptide-binding sites, mounting an efficient immune response against HIV-1. Another explanation is that patients with HLA-DQB1*06 have been reported to have strong mucosal CD4 T-cell responses.28 The last explanation is that HLA-DQB1*06 may be resistant to escape mutation if the epitope is in a region of the HIV-1 genome, the detailed structure of which is essential for viral function, and the peptide-binding groove of the allele can tolerate limited mutations in the peptide.27
Table 1.
Characteristics of the study patient cohort.
| HLA typing |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Laboratory ID | Age | Sex | Route of transmit | CD4+ T cell count | VL (Log) | A1 | A2 | B1 | B2 | Bw1 | Bw2 | DRB1-1 | DRB1-2 | DQB1-1 | DQB1-2 |
| P1 | 33 | Male | Sex | 358 | 4.7 | A*03 | A*31 | B*51 | B*51 | Bw4 | Bw4 | DRB1*14 | DRB1*04 | DQB1*05 | DQB1*03 |
| P2 | 27 | Male | Sex | 523 | 4.5 | A*01 | A*02 | B*57 | B*39 | Bw4 | Bw6 | DRB1*07 | DRB1*08 | DQB1*06 | DQB1*03 |
| P3 | 30 | Male | Sex | 306 | 2.8 | A*02 | A*11 | B*40 | B*51 | Bw4 | Bw6 | DRB1*08 | DRB1*09 | DQB1*06 | DQB1*03 |
| P4 | 39 | Female | Blood | 470 | 3.2 | A*03 | A*24 | B*07 | B*15 | Bw6 | Bw6 | DRB1*15 | DRB1*04 | DQB1*06 | DQB1*06 |
| P5 | 27 | Male | Sex | 359 | 3.7 | A*11 | A*11 | B*07 | B*52 | Bw4 | Bw6 | DRB1*01 | DRB1*15 | DQB1*05 | DQB1*06 |
| P6 | 24 | Male | Sex | 406 | 4.3 | A*02 | A*11 | B*39 | B*15 | Bw6 | Bw6 | DRB1*09 | DRB1*09 | DQB1*03 | DQB1*03 |
| P7 | 31 | Male | Sex | 357 | 6 | A*02 | A*02 | B*13 | B*15 | Bw4 | Bw6 | DRB1*15 | DRB1*12 | DQB1*06 | DQB1*03 |
| P8 | 29 | Male | Sex | 190 | 4.4 | A*33 | A*33 | B*44 | B*58 | Bw4 | Bw4 | DRB1*13 | DRB1*03 | DRB1*01 | DRB1*08 |
| P9 | 25 | Male | Sex | 255 | 4.9 | A*02 | A*11 | B*07 | B*52 | Bw4 | Bw6 | DRB1*01 | DRB1*08 | DRB1*01 | DRB1*08 |
| P10 | 28 | Male | Sex | 358 | 3.7 | A*02 | A*11 | B*46 | B*51 | Bw4 | Bw6 | DRB1*04 | DRB1*09 | DQB1*03 | DQB1*03 |
| P11 | 27 | Male | Sex | 256 | 2.7 | A*02 | A*02 | B*46 | B*15 | Bw6 | Bw6 | DRB1*09 | DRB1*12 | DQB1*03 | DQB1*03 |
| P12 | 36 | Male | Sex | 342 | 6 | A*03 | A*30 | B*13 | B*15 | Bw4 | Bw6 | DRB1*07 | DRB1*12 | DQB1*02 | DQB1*03 |
| P13 | 30 | Male | Sex | 340 | 4.9 | A*03 | A*30 | B*07 | B*52 | Bw6 | Bw6 | DRB1*09 | DRB1*12 | DQB1*03 | DQB1*03 |
| P14 | 22 | Male | Sex | 335 | 1.4 | A*02 | A*24 | B*27 | B*46 | Bw4 | Bw6 | DRB1*08 | DRB1*11 | DQB1*06 | DQB1*03 |
| P15 | 31 | Male | Sex | 569 | 2.5 | A*02 | A*24 | B*07 | B*52 | Bw4 | Bw6 | DRB1*09 | DRB1*12 | DQB1*03 | DQB1*03 |
| P16 | 25 | Male | Sex | 276 | 4.2 | A*02 | A*29 | B*40 | B*35 | Bw6 | Bw6 | DRB1*03 | DRB1*11 | DQB1*02 | DQB1*03 |
| P17 | 29 | Male | Sex | 271 | 3.4 | A*02 | A*29 | B*39 | B*15 | Bw6 | Bw6 | DRB1*09 | DRB1*12 | DQB1*03 | DQB1*03 |
| P18 | 25 | Male | Sex | 573 | 2.9 | A*02 | A*31 | B*15 | B*15 | Bw6 | Bw6 | DRB1*15 | DRB1*04 | DQB1*06 | DQB1*06 |
| P19 | 28 | Male | Sex | 429 | 4.2 | A*02 | A*33 | B*58 | B*46 | Bw4 | Bw6 | DRB1*03 | DRB1*09 | DQB1*02 | DQB1*03 |
| P20 | 35 | Male | Sex | 280 | 5 | A*02 | A*31 | B*40 | B*46 | Bw6 | Bw6 | DRB1*09 | DRB1*11 | DQB1*03 | DQB1*03 |
| P21 | 27 | Male | Sex | 250 | 4.6 | A*24 | A*24 | B*40 | B*15 | Bw6 | Bw6 | DRB1*15 | DRB1*14 | DQB1*06 | DQB1*03 |
| P22 | 39 | Male | Sex | 159 | 3.3 | A*02 | A*24 | B*40 | B*51 | Bw4 | Bw6 | DRB1*09 | DRB1*09 | DQB1*03 | DQB1*03 |
| P23 | 32 | Male | Sex | 679 | 2.1 | A*02 | A*26 | B*08 | B*40 | Bw6 | Bw4 | DRB1*07 | DRB1*08 | DQB1*06 | DQB1*03 |
| P24 | 27 | Male | Sex | 328 | 4.5 | A*02 | A*30 | B*13 | B*15 | Bw4 | Bw6 | DRB1*15 | DRB1*07 | DQB1*02 | DQB1*03 |
| P25 | 27 | Male | Sex | 224 | 4.8 | A*02 | A*11 | B*40 | B*15 | Bw6 | Bw6 | DRB1*04 | DRB1*14 | DQB1*04 | DQB1*05 |
| P26 | 50 | Male | Sex | 467 | 4.5 | A*02 | A*02 | B*48 | B*52 | Bw4 | Bw6 | DRB1*11 | DRB1*14 | DQB1*05 | DQB1*03 |
| P27 | 26 | Male | Sex | 543 | 4.2 | A*02 | A*24 | B*40 | B*15 | Bw6 | Bw6 | DRB1*16 | DRB1*04 | DQB1*05 | DQB1*05 |
| P28 | 49 | Male | Sex | 350 | 4.5 | A*02 | A*02 | B*40 | B*51 | Bw4 | Bw6 | DRB1*01 | DRB1*08 | DQB1*05 | DQB1*03 |
| P29 | 22 | Male | Sex | 449 | 3 | A*02 | A*24 | B*40 | B*38 | Bw4 | Bw6 | DRB1*01 | DRB1*08 | DQB1*05 | DQB1*03 |
As differences in correlation patterns are related to the control of viremia, differences between diverse MHC class II HLA supertypes should be taken into account in vaccine design to elicit optimal T-cell responses within HLA supertypes, as well as to design vaccine efficacy trials for participants that represent the population in which such a vaccine would be used in the future.
Materials and methods
Study subjects and clinical features
Samples were taken from participants in a previously well-characterized HIV-1-infected cohort, established in 2004, of 236 HIV-1-infected patients attending clinics in Beijing, China. The cohort consisted of chronically infected individuals with confirmed serodiagnosis of HIV-1. The HIV-1 infection was diagnosed using the solid-phase method with enzyme-linked immunosorbent assay (ELISA) kits, and infections were confirmed further by Western blot test, which was provided by the National AIDS Control Organization, located in China. Most of the subjects (95%) were homosexual men in the cohort. A total of 56 patients who remained antiretroviral treatment-naïve were identified to study the potential impact of genotype on disease progression in patients with an extended follow-up. HIV-1 subtypes were detected in 56 subjects, of which 29 patients had B subtype. Moreover, individuals who were screened for acute illnesses and infections requiring treatment were also included in the present study, although pregnant females and patients with blood dyscrasias were excluded. This study was approved specially by Beijing Youan Hospital Ethics Committee (No. 20061109V1) and conducted according to the set guidelines for research. Written informed consent was obtained from the patients after explaining the purpose and expected consequences of the study. The characteristics of the patients in the present study are detailed in Table 1.
Peripheral blood mononuclear cell isolation
Fresh blood was obtained by phlebotomy. Peripheral blood mononuclear cells(PBMCs) were then isolated using ethylenediamine tetraacetic acid, treated and heparinized. The sample blood was then placed onto a Ficoll-Hypaque density gradient (Sigma, MO, USA) and finally cryopreserved [90% fetal calf serum (FCS), 10% dimethyl sulfoxide] at −196°C liquid nitrogen until needed for testing.
Plasma VL and CD4+ T-cell counts
The HIV-1 RNA plasma VL and CD4+T-cell counts were quantified using the HIV-1 RNA 3·0 bDNA assay (Bayer, Leverkusen, Germany) and fluorescence activating cell sorter count (Becton Dickinson, CA, USA), respectively, according to the manufacturer's instructions. The threshold for RNA detection was at 50 copies/mL. All plasma VLs were presented as log10 transformed data.
Synthetic HIV-1 Nef core overlapping peptides
HIV-1 subtype B Nef core whole-gene polypeptide was obtained from the National Institute of Health, AIDS Research and Reference Reagent program (https://www.aidsreagent.org/). The peptides were synthesized by Sigma and the Medical Research Council Human Immunology Unit (WIMM, Oxford, UK); 18-mer peptides overlapped by 10 nucleotides and representing the consensus clade B proteome were pooled into the Nef core protein (Table 2 Nef Core OLPs sequence, magnitude and breath), as described previously.8,9,16
Table 2.
Nef Core OLPs sequence, magnitude and breath.
| Nef core Peptide NO | Peptide sequence | Average magnitude (SFU/106PSMC) | Number of positive patients |
|---|---|---|---|
| N9 | EVGFPVRPQVPLRPMTYK | 346.1 | 1 |
| N10 | QVPLRPMTYKGALDLSHF | 0.0 | 0 |
| N11 | YKGALDLSHFLKEKGGLE | 553.5 | 4 |
| N12 | HFLKEKGGLEGLIYSQKR | 504.4 | 6 |
| N13 | LEGLIYSQKRQDILDLWV | 346.4 | 4 |
| N14 | KRQDILDLWVYHTQGYFP | 1467.5 | 4 |
| N15 | WVYHTQGYFPDWQNYTPG | 707.8 | 2 |
| N16 | FPDWQNYTPGPGIRYPLT | 386.8 | 3 |
| N17 | PGPGIRYPLTFGWCFKLV | 387.0 | 3 |
| N18 | LTFGWCFKLVPVEPEKVE | 565.1 | 2 |
HLA typing
Genomic DNA was extracted from the patient's PBMCs using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). Genotypes, including HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1, were determined by sequence-specific primer(SSP) PCR (performed at the WeatherallInstitute of Molecular Medicine, John Radcliffe Hospital, Oxford University, UK).
Interferon-γ enzyme-linked immunospot assay
The PBMCs were thawed and cultured at 37°C for 18 h in Roswell Park Memorial Institute(RPMI)-1640 medium (Hyclone, UT, USA) with 10% FCS (Hyclone). The cultured cells (105) were then added to each well in a 96-well microtiter high-affinity plate (MAIPS4510; Millipore, MA, USA), precoated with an anti-interferon (IFN)-γantibody (10 mg/mL; Mabtech, Nacka Strand, Sweden). The single peptide was pooled, and the final concentration of each peptide was measured at 4 mg/mL. The plate was incubated for 16 h at 37°C in 5% CO2. The IFN-γ-producing cells were then made visual with a biotinylated anti-IFN-γ secondary antibody (Mabtech) and an enzyme-substrate complex. The spots with decreasing density radiating from the center were counted using the ELISPOT reader (CTL). The results were expressed as the number of spot-forming units (SFUs) per million PBMCs. More than 50 SFU/million PBMCs and 3 times more spot-forming cells compared with the negative control were considered positive.
Statistical analysis
The statistical analysis was performed using SPSS 17.0 (SPSS, IL, USA). The number of individuals with positive responses to Nef core proteins was compared using the Fisher's exact test. Then, the association between the breadth in terms of cell count and clinical outcomes (CD4+T-cell count and VL) was analyzed using the Mann-Whitney U test and the association between the magnitude of T-cell response and clinical outcomes (CD4+ T cell count and VL) Was analyzed using the Spearman's correction test. Since this study was conducted in a small subset, adjust the p-value with matlab 7.0 False Discovery Rate (FDR).
Disclosure of potential conflicts of interest
The authors declare no conflicts of interest.
Funding
This study was funded by the National Natural Science Foundation of China (81603552); Beijing health system high-level talent project (2015–3–100); and specific prediction and early-stage diagnosis of molecular markers about liver cancer screening system and mechanism research platform (2016–2).
References
- [1].Cohen KW, Frahm N. Current views on the potential for development of a HIV vaccine. Expert Opin Biol Ther 2017; 17:295-303; PMID:28095712; https://doi.org/ 10.1080/14712598.2017.1282457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Cotton LA, Kuang XT, Le AQ, Carlson JM, Chan B, Chopera DR, Brumme CJ, Markle TJ, Martin E, Shahid A, et al.. Genotypic and functional impact of HIV-1 adaptation to its host population during the North American epidemic. Plos Genet 2014; 10:e1004295; PMID:24762668; https://doi.org/ 10.1371/journal.pgen.1004295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Payne R, Muenchhoff M, Mann J, Roberts HE, Matthews P, Adland E, Hempenstall A, Huang KH, Brockman M, Brumme Z, et al.. Impact of HLA-driven HIV adaptation on virulence in populations of high HIV seroprevalence. Proc Natl Acad Sci U S A 2014; 111:E5393-400; PMID:25453107; https://doi.org/ 10.1073/pnas.1413339111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Goulder PJ, Walker BD. HIV and HLA class I: an evolving relationship. Immunity 2012; 37:426-40; PMID:22999948; https://doi.org/ 10.1016/j.immuni.2012.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Adland E, Paioni P, Thobakgale C, Laker L, Mori L, Muenchhoff M, Csala A, Clapson M, Flynn J, Novelli V, et al.. Discordant Impact of HLA on Viral Replicative Capacity and Disease Progression in Pediatric and Adult HIV Infection. Plos Pathog 2015; 11:e1004954; PMID:26076345; https://doi.org/ 10.1371/journal.ppat.1004954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Matthews PC, Listgarten J, Carlson JM, Payne R, Huang KH, Frater J, Goedhals D, Steyn D, van Vuuren C, Paioni P, et al.. Co-operative additive effects between HLA alleles in control of HIV-1. Plos One 2012; 7:e47799; PMID:23094091; https://doi.org/ 10.1371/journal.pone.0047799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Peretz Y, Tsoukas CM, Bernard NF. HIV Gag-specific immune responses predict the rate of CD4 decline. Aids 2008; 22:1222-24; PMID:18525269; https://doi.org/ 10.1097/QAD.0b013e3283021a76 [DOI] [PubMed] [Google Scholar]
- [8].Aralaguppe SG, Siddik AB, Manickam A, Ambikan AT, Kumar MM, Fernandes SJ, Amogne W, Bangaruswamy DK, Hanna LE, Sonnerborg A, et al.. Multiplexed next-generation sequencing and de novo assembly to obtain near full-length HIV-1 genome from plasma virus. J Virol Methods 2016; 236:98-104; PMID:27448822; https://doi.org/ 10.1016/j.jviromet.2016.07.010 [DOI] [PubMed] [Google Scholar]
- [9].Almeida CA, Bronke C, Roberts SG, McKinnon E, Keane NM, Chopra A, Kadie C, Carlson J, Haas DW, Riddler SA, et al.. Translation of HLA-HIV associations to the cellular level: HIV adapts to inflate CD8 T cell responses against Nef and HLA-adapted variant epitopes. J Immunol 2011; 187:2502-13; PMID:21821798; https://doi.org/ 10.4049/jimmunol.1100691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].O'Neill E, Kuo LS, Krisko JF, Tomchick DR, Garcia JV, Foster JL. Dynamic evolution of the human immunodeficiency virus type 1 pathogenic factor, Nef. J Virol 2006; 80:1311-20; PMID:16415008; https://doi.org/ 10.1128/JVI.80.3.1311-1320.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Foster JL, Garcia JV. HIV-1 Nef: at the crossroads. Retrovirology 2008; 5:84; PMID:18808677; https://doi.org/ 10.1186/1742-4690-5-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Landi A, Iannucci V, Nuffel AV, Meuwissen P, Verhasselt B. One protein to rule them all: modulation of cell surface receptors and molecules by HIV Nef. Curr Hiv Res 2011; 9:496-504; PMID:22103833; https://doi.org/ 10.2174/157016211798842116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Park SY, Mack WJ, Lee HY. Enhancement of viral escape in HIV-1 Nef by STEP vaccination. Aids 2016; 30:2449-58; PMID:27427874; https://doi.org/ 10.1097/QAD.0000000000001202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Malbec M, Sourisseau M, Guivel-Benhassine F, Porrot F, Blanchet F, Schwartz O, Casartelli N. HIV-1 Nef promotes the localization of Gag to the cell membrane and facilitates viral cell-to-cell transfer. Retrovirology 2013; 10:80; PMID:23899341; https://doi.org/ 10.1186/1742-4690-10-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Meuwissen PJ, Stolp B, Iannucci V, Vermeire J, Naessens E, Saksela K, Geyer M, Vanham G, Arien KK, et al.. Identification of a highly conserved valine-glycine-phenylalanine amino acid triplet required for HIV-1 Nef function. Retrovirology 2012; 9:34; PMID:22537596; https://doi.org/ 10.1186/1742-4690-9-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li WH, Li CY, Yang HB, Zhang HP, Zhang X, Kong LS, Xu XN, Lu SC, Yan HP. Human leucocyte antigen-Bw4 and Gag-specific T cell responses are associated with slow disease progression in HIV-1B-infected anti-retroviral therapy-naive Chinese. Clin Exp Immunol 2013; 171:298-306; PMID:23379436; https://doi.org/ 10.1111/cei.12025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Adland E, Carlson JM, Paioni P, Kløverpris H, Shapiro R, Ogwu A, Riddell L, Luzzi G, Chen F, Balachandran T, et al.. Nef-specific CD8+ T cell responses contribute to HIV-1 immune control. Plos One 2013; 8:e73117; PMID:24023819; https://doi.org/ 10.1371/journal.pone.0073117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Khalid M, Yu H, Sauter D, Usmani SM, Schmokel J, Feldman J, Gruters RA, van der Ende ME, Geyer M, Rowland-Jones S, et al.. Efficient Nef-mediated downmodulation of TCR-CD3 and CD28 is associated with high CD4+ T cell counts in viremic HIV-2 infection. J Virol 2012; 86:4906-20; PMID:22345473; https://doi.org/ 10.1128/JVI.06856-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Arguello JR, Little AM, Bohan E, Goldman JM, Marsh SG, Madrigal JA. High resolution HLA class I typing by reference strand mediated conformation analysis (RSCA). Tissue Antigens 1998; 52:57-66; PMID:9714475; https://doi.org/ 10.1111/j.1399-0039.1998.tb03024.x [DOI] [PubMed] [Google Scholar]
- [20].Goulder PJ, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol 2008; 8:619-30; PMID:18617886; https://doi.org/ 10.1038/nri2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bunce M. PCR-sequence-specific primer typing of HLA class I and class II alleles. Methods Mol Biol 2003; 210:143-71; PMID:12412454 [DOI] [PubMed] [Google Scholar]
- [22].Killian MS, Teque F, Walker RL, Meltzer PS, Killian JK. CD8(+) lymphocytes suppress human immunodeficiency virus 1 replication by secreting type I interferons. J Interferon Cytokine Res 2013; 33:632-45; PMID:23402527; https://doi.org/ 10.1089/jir.2012.0067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Demers KR, Reuter MA, Betts MR. CD8(+) T-cell effector function and transcriptional regulation during HIV pathogenesis. Immunol Rev 2013; 254:190-206; PMID:23772621; https://doi.org/ 10.1111/imr.12069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Munier CM, Kelleher AD, Kent SJ, De Rose R. The role of T cell immunity in HIV-1 infection. Curr Opin Virol 2013; 3:438-46; PMID:23747036; https://doi.org/ 10.1016/j.coviro.2013.05.009 [DOI] [PubMed] [Google Scholar]
- [25].Hansen SG, Sacha JB, Hughes CM, Ford JC, Burwitz BJ, Scholz I, Gilbride RM, Lewis MS, Gilliam AN, Ventura AB, et al.. Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science 2013; 340:1237874; PMID:23704576; https://doi.org/ 10.1126/science.1237874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hansen SG, Piatak MJ, Ventura AB, Hughes CM, Gilbride RM, Ford JC, Oswald K, Shoemaker R, Li Y, Lewis MS, et al.. Immune clearance of highly pathogenic SIV infection. Nature 2013; 502:100-04; PMID:24025770; https://doi.org/ 10.1038/nature12519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Vyakarnam A, Sidebottom D, Murad S, Underhill JA, Easterbrook PJ, Dalgleish AG, Peakman M. Possession of human leucocyte antigen DQ6 alleles and the rate of CD4 T-cell decline in human immunodeficiency virus-1 infection. Immunology 2004; 112:136-42; PMID:15096192; https://doi.org/ 10.1111/j.1365-2567.2004.01848.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ferre AL, Hunt PW, McConnell DH, Morris MM, Garcia JC, Pollard RB, Yee HF Jr, Martin JN, Deeks SG, Shacklett BL. HIV controllers with HLA-DRB1*13 and HLA-DQB1*06 alleles have strong, polyfunctional mucosal CD4+ T-cell responses. J Virol 2010; 84:11020-29; PMID:20719952; https://doi.org/ 10.1128/JVI.00980-10 [DOI] [PMC free article] [PubMed] [Google Scholar]


