ABSTRACT
The gene regulatory network comprised of LEAFY (LFY), APETALA1 (AP1), the AP1 paralog CAULIFLOWER (CAL), and TERMINAL FLOWER1 (TFL1) is a major determinant of the flowering process in Arabidopsis thaliana. TFL1 activity in the shoot apical meristem provides inflorescence identity while the transcription factors LFY and AP1/CAL confer floral identity to emerging floral primordia. It has been thought that LFY and AP1/CAL control the onset of flowering in part by repressing TFL1 expression in flowers. However, in the June issue of Plant Physiology, we reported that LFY and AP1 act antagonistically in the regulation of several key flowering regulators, including TFL1. Specifically, TFL1 transcription was suppressed by AP1 but promoted by LFY. Here, we present additional evidence for the role of LFY as an activator of TFL1 and propose that this regulatory activity is pivotal for the indeterminate growth of the SAM during the reproductive phase of development.
KEYWORDS: APETALA1, Arabidopsis, CAULIFLOWER, flowering, inflorescence development, LEAFY, meristem identity, TERMINAL FLOWER1
Angiosperms integrate a multitude of endogenous and environmental signals to determine a time for flowering that ensures reproductive success. Research conducted over the past 25 years, especially in Arabidopsis thaliana (Arabidopsis), has revealed many of the genes that orchestrate the flowering process.1-3 These include inflorescence meristem identity genes, such as TERMINAL FLOWER1 (TFL1), and floral meristem identity genes, including the transcription factor-coding genes LEAFY (LFY), APETALA1 (AP1) and the AP1-paralog CAULIFLOWER (CAL). Because these genes together determine when and where flowers will be formed, it has been proposed that differences in their expression patterns or in the functions of the corresponding proteins can explain much of the diversity of inflorescence architectures observed among the angiosperms.4
The inflorescence of Arabidopsis is characterized by a main axis (shoot) with an indeterminate shoot apical meristem (SAM) that laterally produces flowers. A number of elegant studies showed that TFL1 and LFY/AP1 are essential for building the Arabidopsis inflorescence. TFL1 is expressed in the central region of the SAM, providing inflorescence identity and allowing the indeterminate growth of the shoot apex, while LFY and AP1/CAL are expressed in the flanks of the SAM, providing floral identity to emerging primordia.5-7 In tfl1 mutants, LFY/AP1 expression expands into the SAM that, consequently, acquires floral identity and abruptly terminates with the formation of a flower-like structure.7-9 Conversely, in lfy and ap1 mutants, flowers are substituted by shoot-like structures.7,10-11 An even more dramatic conversion of flowers into inflorescence-like meristems is observed in ap1 cal double-mutant plants.12 In recent years, several studies have provided a molecular basis for the antagonism between TFL1 and LFY/AP1: both LFY and AP1 proteins were shown to bind to essential cis-regulatory elements in the TFL1 promoter.13-15
LFY and AP1 regulate floral development in a partially redundant manner and share many target genes.13-17 However, it is not known whether LFY and AP1 act together or provide independent inputs to these targets. We addressed this question by analyzing the transcriptional activity of LFY in the absence of AP1/CAL function.18 To this end, we used a p35S:LFY-GR line introgressed into an ap1 cal double-mutant background to determine the gene expression changes caused by LFY activation in the inflorescence. We found that LFY can regulate some of its known target genes independently of AP1/CAL activity. In contrast, other LFY targets, including the floral homeotic genes APETALA3 and AGAMOUS, appear to require functional AP1/CAL. In agreement with the results of a previous meta-analysis of published data sets,18,19 we further found that LFY and AP1/CAL regulate certain targets antagonistically. These included regulators of floral initiation such as FLOWERING LOCUS D, TEMPRANILLO1, APETALA2 and, notably, TFL1. TFL1 was upregulated in response to LFY-GR activation but downregulated by AP1-GR in ap1 cal inflorescences. In agreement with the transcriptional response of TFL1, activation of LFY-GR in ap1 cal plants led to a significant inhibition of flower formation while activation of AP1-GR caused an immediate and synchronized onset of flowering, as previously reported.17
These results, as well as a set of previous observations, led us to reconsider the nature of the relationship between LFY and TFL1. Firstly, the expression domains of TFL1 and LFY overlap in the inflorescence-like meristems of ap1 cal plants.20-21 Moreover, weak LFY expression has been detected in the stem of wild-type inflorescences,22 where TFL1 is also expressed.23 The reanalysis of published transcriptomics data sets further showed that activation of LFY-GR in seedlings leads to upregulation of TFL1 expression.3 Taken together, these results imply that TFL1 and LFY are not necessarily antagonists and that LFY may be able to activate TFL1, at least, in the absence of AP1/CAL activity. In line with this idea, LFY was shown to bind to a region approximately 2.8 kilobases (kb) downstream of TFL1, which is essential for the maintenance of TFL1 expression in the inflorescence meristem and, consequently, for SAM indeterminacy.14,23 Although LFY itself does not appear to be expressed in the SAM, it has been demonstrated that LFY protein is mobile and can travel to the inflorescence meristem.24 Thus, in addition to its role in flower development, LFY might be needed for indeterminate growth of the SAM. This would not be the first described function of LFY in a shoot meristem, as it has been shown previously that LFY stimulates axillary meristem growth.25
To test the putative role of LFY in SAM identity via activation of TFL1, we made use of a previously generated set of pTFL1:GUS reporter lines23 and monitored TFL1 expression in genetic backgrounds with modified LFY activity (Fig. 1). First, we tested whether the LFY binding sites located in the 3′ region of the TFL1 promoter14 were essential for the transcriptional response of TFL1 to LFY. To this end, we analyzed the activity of two pTFL1:GUS reporters – one containing the LFY binding sites, the other one lacking them – in plants that express a fusion protein between LFY and the VP16 transcriptional activator under the control of a heat-shock inducible promoter (pHS:LFY-VP16).26,27 As described in our recent publication,18 the activity of the pTFL1:GUS reporter containing the full length promoter of TFL1 was broadly activated in wild-type seedlings after the heat-shock treatment but restricted to the shoot apex in plants grown under normal conditions (Fig. 1A-C). In contrast, plants carrying a truncated version of pTFL1:GUS without the LFY binding sites (ΔLFY in Fig. 1) did not exhibit ectopic GUS activity after induction of LFY-VP16 expression (Fig. 1D). Therefore, the LFY binding sites in the 3′ region of the TFL1 promoter appear to be necessary for the response of pTFL1:GUS to LFY-VP16.
Figure 1.
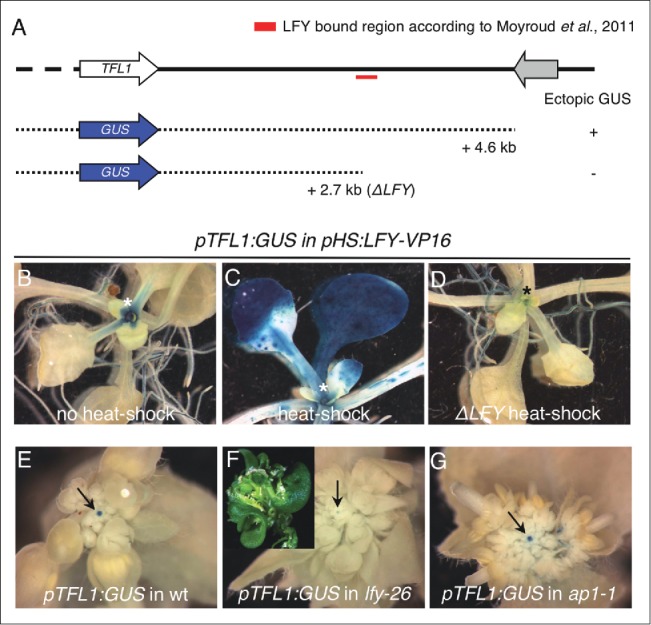
Analysis of pTFL1:GUS reporter lines in genetic backgrounds with modified LFY activity. (A) Summary of the experiments performed to test the functional relevance of the LFY binding sites in the 3′ region of the TFL1 promoter. The activity of 2 pTFL1:GUS constructs was assayed in pHS:LFY-VP16 seedlings: one with the full length TFL1 promoter (2.2 kb of the 5′ region plus 4.6 kb of the 3′) and one with a truncated version of the promoter lacking the LFY binding region (2.2 kb of the 5′ region plus 2.7 kb of the 3′, ΔLFY). ‘Ectopic GUS (+)’ denotes staining in roots, cotyledons and developing leaves after a heat-shock treatment. Growth conditions, heat-shock treatments (incubation for 3 h at 37°C during 3 consecutive days) and GUS staining were conducted as described previously.18 (B-D) Representative images of pHS:LFY-VP16 seedlings containing a pTFL1:GUS reporter and stained for GUS: (B) reporter activity of the full length TFL1 promoter in a pHS:LFY-VP16 seedling grown under control conditions without heat-shock, (C) reporter activity of the full length TFL1 promoter in a pHS:LFY-VP16 seedling after heat-shock, (D) reporter activity of the truncated version of the TFL1 promoter (ΔLFY) in a pHS:LFY-VP16 seedling after heat-shock. Asterisks point to the position of the SAM. (E-F) Reporter activity of the full length TFL1 promoter in representative inflorescence apices of wild type (accession Landsberg erecta) (E), lfy-26 (F) and ap1–1 (G) plants. The inset in (F) shows the terminal carpelloid structure in lfy-26 shoots. Arrows in the main panels point to the position of the SAM. GUS staining was conducted as described previously.23
Next, we asked whether a loss of LFY function affects TFL1 transcription in the SAM. To test this, we monitored the activity of pTFL1:GUS in the strong lfy-26 allele.27 Compared to the wild type, the intensity of the GUS signal significantly decreased in the center of lfy-26 inflorescences (Fig. 1E-F). This result is in agreement with the absence of TFL1 expression in the SAM of lfy-7 mutant plants.20 In contrast, pTFL1:GUS activity was not apparently affected in the inflorescence apex of ap1–1 mutants (Fig. 1E, G), which also exhibit impaired floral meristem identity.10 As described previously,7 we observed that the inflorescences of lfy-26 mutants terminated in carpelloid structures (Fig. 1F inset). This determinacy phenotype of lfy-26 plants may be caused by the low levels of TFL1 expression in the inflorescence apex we detected with the pTFL1:GUS reporter. Taken together, these results suggest that LFY promotes TFL1 expression in the SAM to ensure indeterminate growth.
LFY may also activate TFL1 in flowers, at least under conditions where AP1/CAL are non-functional. In fact, the results presented in our Plant Physiology paper suggest that LFY, AP1/CAL and TFL1 may be part of an incoherent feedback loop28 during early establishment flower development, where LFY activates both TFL1 and the repressors of TFL1, AP1/CAL.18 This regulatory loop might ensure that flower formation commences only when AP1/CAL levels are sufficiently high to repress TFL1 expression and to trigger the genetic program required for flower development. A characterization of the protein complexes that regulate inflorescence and floral development may be required to explain the antagonistic activities of LFY and AP1 in the control of TFL1 and other flowering regulators.
Acknowledgments
A.S.-M. is funded by the European Union's Horizon 2020 Research and Innovation program under Marie Skłodowska-Curie grant agreement No. 746396. F.W. and E.G. are supported by grants from Science Foundation Ireland. F.M. is supported by MINECO and FEDER, grant No. BIO2015–64307-R. B.Z. is supported by the Irish Research Council.
References
- 1.Blázquez MA, Ferrándiz F, Madueño F, Parcy F. How floral meristems are built. Plant Mol Biol. 2006;60:855–70. doi: 10.1007/s11103-006-0013-z. [DOI] [PubMed] [Google Scholar]
- 2.Fornara F, de Montaigu A, Coupland G. SnapShot: Control of flowering in Arabidopsis. Cell. 2010;141:550. doi: 10.1016/j.cell.2010.04.024. [DOI] [PubMed] [Google Scholar]
- 3.Denay G, Chahtane H, Tichtinsky G, Parcy F. A flower is born: an update on Arabidopsis floral meristem formation. Curr Opin Plant Biol. 2017;35:15–22. doi: 10.1016/j.pbi.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Benlloch R, Berbel A, Serrano-Mislata A, Madueño F. Floral initiation and inflorescence architecture: a comparative view. Ann Bot. 2007;100:659–76. doi: 10.1093/aob/mcm146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E. Inflorescence commitment and architecture in Arabidopsis. Science 1997;275:80–3. doi: 10.1126/science.275.5296.80. [DOI] [PubMed] [Google Scholar]
- 6.Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 1992;360:273–7. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- 7.Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell 1992;69:843–59. doi: 10.1016/0092-8674(92)90295-N. [DOI] [PubMed] [Google Scholar]
- 8.Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 1993;119:721–43 [Google Scholar]
- 9.Gustafson-Brown C, Savidge B, Yanofsky MF. (1994). Regulation of the Arabidopsis floral homeotic gene APETALA1. Cell. 1994;76:131–43. doi: 10.1016/0092-8674(94)90178-3. [DOI] [PubMed] [Google Scholar]
- 10.Irish VF, Sussex IM. Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 1990;2:741–53. doi: 10.1105/tpc.2.8.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huala E, Sussex IM. (1992). LEAFY interacts with floral homeotic genes to regulate Arabidopsis floral development. Plant Cell 1992;4:901–13. doi: 10.1105/tpc.4.8.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kempin SA, Savidge B, Yanofsky MF. Molecular basis of the cauliflower phenotype in Arabidopsis. Science. 1995;267:522–5. doi: 10.1126/science.7824951. [DOI] [PubMed] [Google Scholar]
- 13.Kaufmann K, Wellmer F, Muiño JM, Ferrier T, Wuest SE, Kumar V, Serrano-Mislata A, Madueño F, Krajewski P, Meyerowitz EM et al.. Orchestration of floral initiation by APETALA1. Science. 2010;328:85–9. doi: 10.1126/science.1185244. [DOI] [PubMed] [Google Scholar]
- 14.Moyroud E, Minguet EG, Ott F, Yant L, Posé D, Monniaux M, Blanchet S, Bastien O, Thévenon E, Weigel D et al.. Prediction of regulatory interactions from genome sequences using a biophysical model for the Arabidopsis LEAFY transcription factor. Plant Cell. 2011;23:1293–306. doi: 10.1105/tpc.111.083329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winter CM, Austin RS, Blanvillain-Baufume S, Reback MA, Monniaux M, Wu MF, Sang Y, Yamaguchi A, Yamaguchi N, Parker JE et al.. LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev Cell. 2011;20:430–43. doi: 10.1016/j.devcel.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 16.William DA, Su Y, Smith MR, Lu M, Baldwin DA, Wagner D. Genomic identification of direct target genes of LEAFY. Proc Natl Acad Sci USA. 2004;101:1775–80. doi: 10.1073/pnas.0307842100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wellmer F, Alves-Ferreira M, Dubois A, Riechmann JL, Meyerowitz EM. Genome-wide analysis of gene expression during early Arabidopsis flower development. PLoS Genet. 2006;2:e117. doi: 10.1371/journal.pgen.0020117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goslin K, Zheng B, Serrano-Mislata A, Rae L, Ryan PT, Kwaśniewska K, Thomson B, O'Maoileidigh D, Madueño F, Wellmer F, Graciet E. Transcription factor Interplay between LEAFY and APETALA1/ CAULIFLOWER during floral initiation. Plant Physiol. 2017;174:1097–109. doi: 10.1104/pp.17.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winter CM, Yamaguchi N, Wu MF, Wagner D. (2015). Transcriptional programs regulated by both LEAFY and APETALA1 at the time of flower formation. Physiol Plant. 2015;155:55–73. doi: 10.1111/ppl.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ratcliffe OJ, Bradley DJ, Coen ES. Separation of shoot and floral identity in Arabidopsis. Development. 1999;126:1109–20. [DOI] [PubMed] [Google Scholar]
- 21.Ferrandiz C, Gu Q, Martienssen R, Yanofsky MF. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development. 2000;127:725–34. [DOI] [PubMed] [Google Scholar]
- 22.Blázquez MA, Soowal LN, Lee I, Weigel D. LEAFY expression and flower initiation in Arabidopsis. Development. 1997;124:3835–44. [DOI] [PubMed] [Google Scholar]
- 23.Serrano-Mislata A, Fernández-Nohales P, Doménech MJ, Hanzawa Y, Bradley D, Madueño F. Separate elements of the TFL1 cis-regulatory region integrate pathways to control flowering time and shoot meristem identity. Development. 2016;143:3315–27. doi: 10.1242/dev.135269. [DOI] [PubMed] [Google Scholar]
- 24.Sessions A, Yanofsky MF, Weigel D. Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science. 2000;289:779–82. doi: 10.1126/science.289.5480.779. [DOI] [PubMed] [Google Scholar]
- 25.Chahtane H, Vachon G, Le Masson M Thévenon E, Périgon S, Mihajlovic N, Kalinina A, Michard R, Moyroud E, Monniaux M et al.. A variant of LEAFY reveals its capacity to stimulate meristem development by inducing RAX1. Plant J. 2013;74:678–89. doi: 10.1111/tpj.12156. [DOI] [PubMed] [Google Scholar]
- 26.Benlloch R, Kim MC, Sayou C, Thevenon E, Parcy F, Nilsson O. Integrating long-day flowering signals: a LEAFY binding site is essential for proper photoperiodic activation of APETALA1. Plant J. 2011;67:1094–102. doi: 10.1111/j.1365-313X.2011.04660.x. [DOI] [PubMed] [Google Scholar]
- 27.Lee I, Wolfe DS, Nilsson O, Weigel D. A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr Biol. 1997;7:95–104. doi: 10.1016/S0960-9822(06)00053-4. [DOI] [PubMed] [Google Scholar]
- 28.Kim D, Kwon YK, Cho KH. The biphasic behavior of incoherent feed-forward loops in biomolecular regulatory networks. BioEssays. 2008;30:1204–11. doi: 10.1002/bies.20839. [DOI] [PubMed] [Google Scholar]


