Abstract
Primate herpesviruses express more noncoding RNAs (ncRNAs) than any other class of mammalian viruses during either latency or the lytic phase of the viral life cycle. T cells transformed by the monkey virus Herpesvirus saimiri (HVS) express seven viral U-rich ncRNAs called HSURs. Conserved sequences in HSURs1 and 2 exhibit complementarity to three host-cell microRNAs (miRNAs). The predicted interactions of HSURs1 and 2 with these miRNAs were confirmed by coimmuno-precipitation experiments performed on extracts of marmoset T cells transformed by a wild-type or a mutant HVS lacking these two HSURs. Mutational analyses demonstrated that the binding of miR-27 to HSUR1 and that of miR-16 to HSUR2 involves base pairing. One of these miRNAs, miR-27, is dramatically lowered in abundance in HVS-transformed cells, with consequent effects on the expression of miR-27 target genes. Transient knockdown and ectopic expression of HSUR1 demonstrated that degradation of mature miR-27 occurs in a sequence-specific and binding-dependent manner but does not occur by AU-rich element (ARE)-mediated decay, which controls the intracellular level of HSUR1 itself. This viral strategy exemplifies the use of an ncRNA to control host-cell gene expression via the miRNA pathway and has potential applications both experimentally and therapeutically.
Herpesvirus saimiri (HVS) is an oncogenic γ-herpesvirus that innocuously infects the T cells of its natural host, the squirrel monkey. But in New World primates, T-cell transformation leads to aggressive leukemias, lymphomas, and lymphosarcomas (Ensser and Fleckenstein 2005). HVS encodes seven Sm-class small nuclear RNAs (snRNAs) called HSURs, for Herpesvirus saimiri U-rich RNAs, which are the most abundantly expressed gene products in HVS-transformed T cells (Murthy et al. 1986; Lee et al. 1988). In addition to 5′ trimethyl caps, Sm protein-binding sites and 3′-terminal stem loops common to snRNAs assembled into Sm ribonucleoproteins (Sm snRNPs), HSURs1, 2, and 5 contain AREs near their 5′ ends (Myer et al. 1992; Fan et al. 1997; Albrecht 2000; Cook et al. 2004). Among different HVS subgroups, HSURs1 and 2 are the most highly conserved and the only snRNAs expressed by the closely related Herpesvirus ateles (Albrecht 2000).
Previously, we showed that the expression of HSURs1 and 2 in transformed marmoset T cells does not alter the abundance of the ~8% of host mRNAs that contain AREs in their 3′-untranslated regions (3′UTRs) (Cook et al. 2004). AREs are known to induce rapid decay of the messenger RNAs (mRNAs) in which they reside (Gingerich et al. 2004). Because ARE-mRNAs often encode protooncoproteins or growth factors, manipulation of the levels of their protein products in latently infected T cells would make sense for the virus. Instead, we found that the presence of HSURs1 and 2 correlates with posttranscriptional up-regulation of host genes that are hallmarks of T-cell activation, but the molecular mechanism of this regulation in HVS-transformed T cells remained elusive (Cook et al. 2005). Here, we review results that indicate that the HSUR1 snRNP binds a host miRNA, miR-27, and targets it for destruction (Cazalla et al. 2010) This strategy represents an example of ncRNA warfare, where the virus produces an ncRNA that manipulates the miRNA pathway of the host cell apparently to the advantage of the virus.
EVIDENCE THAT HSUR1 DIRECTS THE DEGRADATION OF MIR-27 IN HVS-TRANSFORMED T CELLS
Bioinformatic searches revealed potential RNA–RNA interactions between conserved sequences in HSURs1 and 2 and the critical seed regions of three miRNAs expressed in T cells (Fig. 1). Immunoprecipitation experiments using anti-AGO2 antibodies to select miRNAs packaged into miRNPs revealed an association with HSURs1 and 2, whereas HSURs3–7 were not immunoprecipitated above background levels. Conversely, use of anti-Sm antibodies confirmed that the three predicted miRNAs (miR-27, miR-16, and miR-142-3p) can be detected interacting with Sm snRNPs in extracts of wild-type but not mutant HVS-transformed marmoset T cells.
Figure 1.
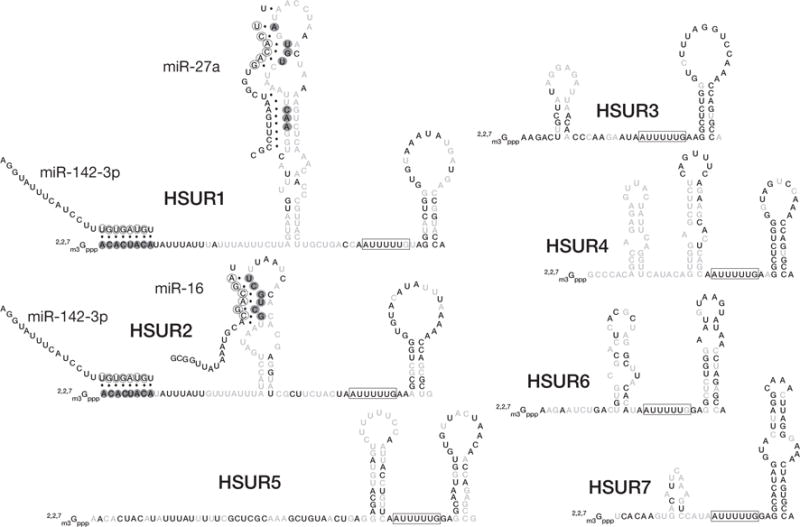
Sequences of HSURs of subgroup A HVS are shown with their Sm-binding sites boxed. Bold nucleotides are absolutely conserved in all available genome sequences from various isolates of HVS A, B, and C and in H. ateles. Also pictured are human miRNAs miR-142-3p, miR-16, and miR-27a. Complementarity between miRNAs and HSURs is represented by dots. Perfectly conserved HSUR residues involved in miRNA binding are highlighted in gray, and seed regions are circled within the miRNAs.
Mutational analyses then highlighted the importance of predicted Watson–Crick base pairing in the interactions of HSUR1 with miR-27 and of HSUR2 with miR-16. These experiments were conducted using human Jurkat T cells stably transformed with a plasmid containing the HSUR-encoding region of HVS, enabling the introduction of mutations expected to disrupt HSUR–miRNA interactions. Changing the miR-binding sites in either HSUR1 or 2 to sequences predicted to base pair with a control non-binding miRNA, miR-20, satisfyingly revealed that a novel interaction with this miRNA could be successfully engineered, confirming the base-pairing nature of miR binding to HSURs.
While conducting the above experiments, we noticed marked increases in the overall levels of miR-27 in T cells transformed by the mutant HVS lacking HSURs1 and 2 versus the wild-type virus. A fourfold difference was confirmed by quantitative real-time polymerase chain reaction (PCR) measurements, with both of the genetic loci—for miR-27a and miR-27b—contributing to the up-regulation. In contrast, neither of the other two miRNAs bound by HSURs (miR-16 and miR-142-3p) were increased in abundance in the mutant-transformed cells nor were two other miRNAs that are included in the same pri-miRNA transcript as miR-27a (miR-23a and miR-24). Moreover, levels of both the pri- and pre-miRs for the two miR-27 isoforms were not altered between the wild-type and mutant-transformed cells. These observations suggested that mature miR-27 was being selectively targeted for degradation.
We tested degradation by a pulse-chase type of assay (Hwang et al. 2007) in which synthetic miRNAs labeled at their 5′ ends were transfected into marmoset T cells transformed by either wild-type HVS or the mutant lacking HSURs1 and 2; levels of remaining miRNAs were then assessed at various times. In the wild-type transformed cells, miR-27a exhibited a considerably shorter half-life than either miR-16 (bound by HSUR2 but not diminished in levels) or miR-20 (a control non-HSUR-binding miRNA). To confirm that this decreased half-life is caused by the presence of HSUR1 rather than other accumulated mutations in the wild-type transformed T-cell line, we knocked down HSUR1 by transfecting a chimeric RNaseH-targeting oligonucleotide and observed increases in miR-27 levels correlating with decreased HSUR1.
Unfortunately, only a handful of mRNA targets of miR-27 have been validated (Ben-Ami et al. 2009; Crist et al. 2009; Guttilla and White 2009; Wang et al. 2009; Jennewein et al. 2010). We tested the levels of these proteins in marmoset T cells transformed by the wild-type versus the mutant HVS lacking HSURs1 and 2. Indeed, reproducible up-regulation of the level of one of these proteins, FOXO1, a transcription factor implicated in oncogenesis and cell cycle control, was observed in the wild-type versus the mutant-transformed marmoset T cells.
Finally, we explored whether the ARE-mediated decay pathway might be involved in miR-27 decay. This was an attractive idea because HSUR1 itself undergoes ARE-mediated degradation (Fan et al. 1997) and could conceivably carry a bound miRNA along into the same destructive milieu. However, experiments in transiently transformed Jurkat T cells indicated that mutation of HSUR1’s ARE, which produces higher levels of HSUR1, increased (rather than alleviated) the down-regulation of miR-27a. Thus, the HSUR-bound miRNA does not appear to undergo degradation via the ARE-mediated decay pathway.
CONCLUSIONS
We demonstrated that HSURs1 and 2 snRNPs directly bind certain host miRNAs in virally transformed T cells, resulting in the degradation of one of these miRNAs (Cazalla et al. 2010). This constitutes the first example of a functional consequence of the interaction between an miRNA and another naturally occurring ncRNA, although such associations have been previously reported. It is also the first example of a virus using this strategy to regulate host-cell gene expression.
Newly synthesized Sm snRNAs transit to the cytoplasm for initial assembly with the heteroheptamer of Sm proteins, assisted by the SMN (survival of motor neurons) complex (Patel and Bellini 2008). They then return to and concentrate in the nucleus for function; a nuclear location has accordingly been verified for HSURs (Golembe et al. 2005). Moreover, evidence for the nuclear presence of miRNAs and the core miRNP protein AGO2 is accumulating (Castanotto et al. 2009; Weinmann et al. 2009). Thus, the binding of miRNAs to HSUR snRNPs could occur in either the nucleus or the cytoplasm.
Likewise, neither the location nor the mechanism of HSUR1-induced miR-27 degradation is yet known. It has been recently reported that extensive pairing between a target RNA and an miRNA triggers miRNA tailing and 3′-to-5′ trimming, affecting the stability of the miRNA. Ameres et al. (2010) propose that extensive pairing of the 3′ half of the miRNA with its target RNA leads to release of the 3′ end from the Argonaute PAZ domain and subsequent exposure to hitherto unknown nucleotidyl transferases and 3′-to-5′ exonucleases. Thus, because HSUR1 can form extensive base pairs with the 3′ end of miR-27, its destabilization could result from 3′-end trimming and tailing, as described by Ameres et al. (2010). Alternatively, subcellular redistribution of miR-27 to the nucleus following binding of HSUR1 could effect destabilization of the miRNA (Hwang et al. 2007). Further experimentation will be required to test these hypotheses.
The interactions of HSURs1 and 2 with miR-142-3p and miR-16 do not lower the levels of these miRNAs in HVS-transformed T cells. The fact that HSURs bind the seed regions of these miRNAs (as well as those of miR-27) suggests that HSURs could compete with mRNA targets by acting as miRNA “sponges.” Down-regulating the activity of miR-142-3p and miR-16 might be advantageous for the virus, particularly in the case of miR-16, which is reported to target cell cycle and apoptosis regulators such as Bcl-2 and cyclins D1 and E1 (Cimmino et al. 2005; Liu et al. 2008). Indeed, the only known phenotype of marmoset T cells transformed with HVS lacking HSURs1 and 2 is that they grow more slowly than those transformed by wild-type virus (Murthy et al. 1989; Cook et al. 2005). However, we did not observe differences in the abundance of miR-16 target proteins in the presence or absence of HSURs1 and 2.
It is not yet clear why HVS down-regulates the expression of miR-27 and how down-regulation of this miRNA is beneficial for the virus. It was recently reported that miR-27 levels are also dramatically reduced following infection of mouse cell lines with murine cytomegalovirus (MCMV), a β-herpesvirus (Buck et al. 2010). As we have documented for HVS, down-regulation of miR-27 by MCMV occurs posttranscriptionally, most likely via rapid decay of the mature miRNA, suggesting a conserved mechanism between these two herpesviruses. Only a few targets of miRNA-27, including the transcription factors FOXO1, RUNX1, and PAX3, have been validated (Ben-Ami et al. 2009; Crist et al. 2009; Guttilla and White 2009). In particular, none of the mRNAs for the eight T-cell activation proteins that are up-regulated by HSURs1 and 2 in HVS-transformed T cells (Cook et al. 2005) possess obvious sites for miR-27 in their 3′UTRs. Thus, the important effects of the virus on the T-cell state may be indirect, and identification of additional targets of miR-27 in T cells transformed with HVS is much needed.
Targeting a bound miRNA for decay is a novel function for an Sm snRNP The most abundant Sm snRNPs in eukaryotic cells have pivotal roles in pre-mRNA splicing and—in metazoans—also in the 3′-end processing of histone pre-mRNAs (Schumperli and Pillai 2004; Wachtel and Manley 2009). Evolutionarily, HVS clearly acquired Sm snRNAs from its host cell. Interestingly, there are no other viruses expressing Sm snRNAs described so far. Perhaps, elucidation of the remaining functions of HSURs will reveal yet other novel roles. Whether the mammalian hosts of HVS encode low-abundance Sm snRNPs that function comparably to control miRNA abundance likewise remains an open question.
HSUR1 expression potentially provides a tool for targeted destruction of any particular miRNA because its miR–27–binding site can be mutated to select another miRNA that then undergoes down-regulation. This strategy is effective in T and B cells but has not been observed in trials with HeLa or HEK cells (D Cazalla, unpubl.), perhaps implicating lymphoid-specific factors in the decay. Our findings increase the potential for experimental manipulation of specific miRNA function. Similarly, stable expression of a mutated HSUR1 could provide enduring knockdown of specific miRNAs in T or B cells as a therapeutic strategy.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grant CA16038. The content is solely the responsibility of the investigators and does not necessarily represent the official views of the NIH. J.A.S is an investigator of the Howard Hughes Medical Institute.
References
- Albrecht JC. Primary structure of the Herpesvirus ateles genome. J Virol. 2000;74:1033–1037. doi: 10.1128/jvi.74.2.1033-1037.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ami O, Pencovich N, Lotem J, Levanon D, Groner Y. A regulatory interplay between miR-27a and Runx1 during megakaryopoiesis. Proc Natl Acad Sci. 2009;106:238–243. doi: 10.1073/pnas.0811466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck AH, Perot J, Chisholm MA, Kumar DS, Tuddenham L, Cognat V, Marcinowski L, Dolken L, Pfeffer S. Post-transcriptional regulation of miR-27 in murine cytomegalovirus infection. RNA. 2010;16:307–315. doi: 10.1261/rna.1819210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D, Lingeman R, Riggs AD, Rossi JJ. CRM1 mediates nuclear-cytoplasmic shuttling of mature microRNAs. Proc Natl Acad Sci. 2009;106:21655–21659. doi: 10.1073/pnas.0912384106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Yario T, Steitz J. Down-regulation of a host micro-RNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook HL, Mischo HE, Steitz JA. The Herpesvirus saimiri small nuclear RNAs recruit AU-rich element-binding proteins but do not alter host AU-rich element-containing mRNA levels in virally transformed T cells. Mol Cell Biol. 2004;24:4522–4533. doi: 10.1128/MCB.24.10.4522-4533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook HL, Lytle JR, Mischo HE, Li MJ, Rossi JJ, Silva DP, Desrosiers RC, Steitz JA. Small nuclear RNAs encoded by Herpesvirus saimiri upregulate the expression of genes linked to T cell activation in virally transformed T cells. Curr Biol. 2005;15:974–979. doi: 10.1016/j.cub.2005.04.034. [DOI] [PubMed] [Google Scholar]
- Crist CG, Montarras D, Pallafacchina G, Rocancourt D, Cumano A, Conway SJ, Buckingham M. Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc Natl Acad Sci. 2009;106:13383–13387. doi: 10.1073/pnas.0900210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensser A, Fleckenstein B. T-cell transformation and oncogenesis by γ2-herpesviruses. Adv Cancer Res. 2005;93:91–128. doi: 10.1016/S0065-230X(05)93003-0. [DOI] [PubMed] [Google Scholar]
- Fan XC, Myer VE, Steitz JA. AU-rich elements target small nuclear RNAs as well as mRNAs for rapid degradation. Genes Dev. 1997;11:2557–2568. doi: 10.1101/gad.11.19.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingerich TJ, Feige JJ, LaMarre J. AU-rich elements and the control of gene expression through regulated mRNA stability. Anim Health Res Rev. 2004;5:49–63. doi: 10.1079/ahr200460. [DOI] [PubMed] [Google Scholar]
- Golembe TJ, Yong J, Battle DJ, Feng W, Wan L, Dreyfuss G. Lymphotropic Herpesvirus saimiri uses the SMN complex to assemble Sm cores on its small RNAs. Mol Cell Biol. 2005;25:602–611. doi: 10.1128/MCB.25.2.602-611.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 2009;284:23204–23216. doi: 10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- Jennewein C, von Knethen A, Schmid T, Brune B. MicroRNA-27b contributes to lipopolysaccharide-mediated peroxisome proliferator-activated receptor γ (PPARγ) mRNA destabilization. J Biol Chem. 2010;285:11846–11853. doi: 10.1074/jbc.M109.066399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SI, Murthy SC, Trimble JJ, Desrosiers RC, Steitz JA. Four novel U RNAs are encoded by a herpesvirus. Cell. 1988;54:599–607. doi: 10.1016/s0092-8674(88)80004-7. [DOI] [PubMed] [Google Scholar]
- Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36:5391–5404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy S, Kamine J, Desrosiers RC. Viral-encoded small RNAs in herpes virus saimiri induced tumors. EMBO J. 1986;5:1625–1632. doi: 10.1002/j.1460-2075.1986.tb04405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy SC, Trimble JJ, Desrosiers RC. Deletion mutants of herpesvirus saimiri define an open reading frame necessary for transformation. J Virol. 1989;63:3307–3314. doi: 10.1128/jvi.63.8.3307-3314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myer VE, Lee SI, Steitz JA. Viral small nuclear ribonucle-oproteins bind a protein implicated in messenger RNA destabilization. Proc Natl Acad Sci. 1992;89:1296–1300. doi: 10.1073/pnas.89.4.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SB, Bellini M. The assembly of a spliceosomal small nuclear ribonucleoprotein particle. Nucleic Acids Res. 2008;36:6482–6493. doi: 10.1093/nar/gkn658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumperli D, Pillai RS. The special Sm core structure of the U7 snRNP: Far-reaching significance of a small nuclear ribonucleoprotein. Cell Mol Life Sci. 2004;61:2560–2570. doi: 10.1007/s00018-004-4190-0. [DOI] [PubMed] [Google Scholar]
- Wachtel C, Manley JL. Splicing of mRNA precursors: The role of RNAs and proteins in catalysis. Mol Biosyst. 2009;5:311–316. doi: 10.1039/b820828j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Rathinam R, Walch A, Alahari SK. ST14 (suppression of tumorigenicity 14) gene is a target for miR-27b, and the inhibitory effect of ST14 on cell growth is independent of miR-27b regulation. J Biol Chem. 2009;284:23094–23106. doi: 10.1074/jbc.M109.012617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann L, Hock J, Ivacevic T, Ohrt T, Mutze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G. Importin 8 is a gene silencing factor that targets Argonaute proteins to distinct mRNAs. Cell. 2009;136:496–507. doi: 10.1016/j.cell.2008.12.023. [DOI] [PubMed] [Google Scholar]


