Abstract
We demonstrated a general protection-deprotection strategy for the design of fluorescent protein biosensors through the construction of a turn-on Hg2+ sensor. A combination of fluorescent protein engineering and unnatural amino acid mutagenesis was used. Unlike previously reported fluorescent protein-based Hg2+ sensors that relied on the binding of Hg2+ to the sulfhydryl group of cysteine residues, a well-established chemical reaction, oxymercuration, was transformed into biological format and incorporated into our sensor design. This novel Hg2+ sensor displayed good sensitivity and selectivity both in vitro and in live bacterial cells. Over 60 folds change in fluorescence signal output was observed in the presence of 10 μM Hg2+, while such change was undetectable when nine other metal ions were tested. This new design strategy could expand the repertoire of fluorescent protein-based biosensors for the detection of small-molecule analytes.
Keywords: fluorescent protein biosensor, unnatural amino acid, mercury sensor, fluorescent probe, turn-on probe
Graphical Abstract
In the past two decades, a large number of fluorescent protein (FP)-based biosensors have been developed for various biological applications in live cells.1–4 These FP biosensors provide real-time information of cellular activities with high spatial resolution. In comparison to small-molecule-based sensors, FP biosensors can be introduced into live cells in the form of DNA and become an intrinsic part of the cellular system. They are suitable for applications in sub-cellular compartments, cells, tissues, or even whole animals. Both protein directed evolution and rational design have been applied to the development of FP biosensors.1–3 While the traditional strategies are based on the standard genetic code, recent efforts have been made towards the incorporation of unnatural amino acids (unAAs) into the design of FP biosensors. This is mainly achieved by replacing the chromophore-forming tyrosine residue with an unnatural analog.3–10 As the structure of the chromophore plays the key role in defining the fluorescence signature of a FP, this approach strives to produce the maximum signal output by directly modifying chemical compositions of the chromophore. Prior efforts have been made to design sensors for Cu(II),5 H2O2,6 H2S,7 peroxinitrite,8, 9 and Zn(II).10
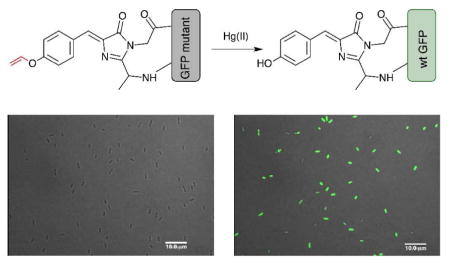
While the unAA mutagenesis enabled a straightforward adoption of several well-established aqueous phase chemical reactions onto the design board, the construction of unAA-based FP biosensor is still in its infancy and faces multiple challenges.3, 4 In seek of a general principle to design unAA-based FP biosensors, we were inspired by the widely employed functional group protection-deprotection strategy in the development of small-molecule fluorescent probes.11 We envisaged that a blockage of the phenolate formation using a protecting group could dramatically affect photophysical properties of the FP chromophore (Figure 1). Upon deprotection, the wild-type chromophore can be recovered, which would lead to a strong fluorescence change as the signal output. In this work, we exploited this protection-deprotection strategy and constructed a FP-based turn-on probe for the detection of Hg2+.
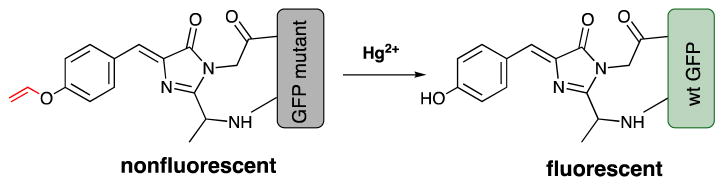
Figure 1.
The construction of a green fluorescent protein (GFP)-based Hg2+ sensor using a general protection-deprotection design strategy.
As one of the widespread heavy metal pollutants in the environment, mercury poses lethal threats to the ecosystem and human health. Sensitive and selective detection of mercury in both environmental and biological settings are highly desirable. Among a variety of methods, fluorescent chemosensors have attracted considerable attentions due to their high sensitivity, good selectivity, and simplicity.12–15 In general, a Hg2+-specific fluorescent probe consists of a fluorescent reporter and a mercury responsive (e.g., chelating) unit, which leads to changes in photophysical properties of the reporter upon its interaction with Hg2+. Several of these probes can be used to detect Hg2+ in live cells.12–15 Besides small-molecule probes,12–17 protein-based,18, 19 in particular fluorescent protein (FP)-based,20–22 detection methods have been developed in recent years. The FP biosensors have the advantages in its genetic encodability and the flexibility of facile targeting into different sub-cellular compartments.1, 2 All previously reported designs rely on the specific and high-affinity binding of Hg2+ to the sulfhydryl group of cysteine residues that are in close proximity to the FP chromophore. Upon binding to Hg2+, the fluorescence of FP could be either quenched20, 21 or moderately enhanced.22 In the present work, we devised a reaction-based Hg2+ FP sensor, which was highly selective and displayed a large fluorescence enhancement in the presence of Hg2+.
RESULTS AND DISCUSSION
General design strategy
In order to apply the protection-deprotection strategy to the design of a FP-based Hg2+ sensor, we examined the highly specific and Hg2+-promoted hydrolysis of a vinyl ether (Figure 1).23–25 In this design, the hydroxyl group of the chromophore-forming Tyr66 is protected as a vinyl ether (Figure 1). The protecting group can be indirectly installed by the genetic incorporation of p-vinyloxy-L-phenylalanine (ViP) at position Tyr66 of the FP. We hypothesized that the resulting unnatural chromophore should display a different fluorescence property from that of the wild-type FP. After the deprotection of the phenolic hydroxyl group through oxymercuration reaction in the presence of Hg2+, the wild-type FP chromophore can be restored (Figure 1).
Genetic incorporation of ViP
To implement the above design, we have synthesized ViP (Figure 2A) and observed a fast deprotection rate in the presence of Hg (II) at room temperature. After deprotection, the formation of tyrosine from ViP was confirmed by NMR (Figure S1). In order to achieve selective incorporation of ViP into proteins using the amber suppression strategy,26 a library of previously reported aminoacyl-tRNA synthetase (aaRS) mutants were screened. In such screening, the amber suppression efficiency was directly linked to the expression level of a superfolder GFP mutant (sfGFP-Asn149TAG) that has an amber nonsense codon at position Asn149.27, 28 Among all the aaRS variants examined, mutant PrFRS (Tyr32Ala, Glu107Pro, Asp158Ala, and Leu162Ala)29 supported an efficient synthesis of full-length sfGFP (Figure 2B). This synthetase also displayed the best fidelity towards ViP and was chosen for the following sensor construction.
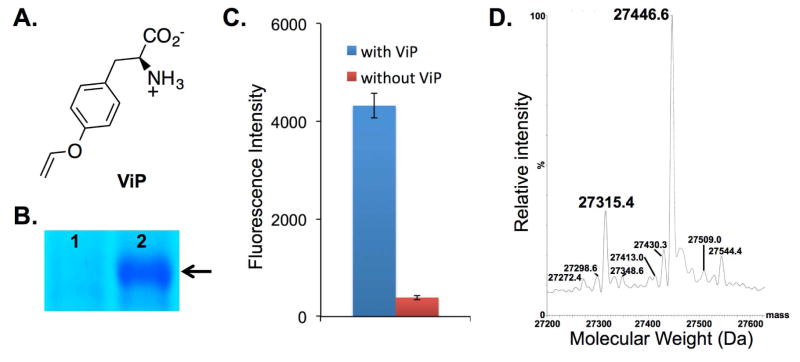
Figure 2.
Genetic incorporation of p-vinyloxy-L-phenylalanine (ViP). (A) The structure of ViP; (B) SDS-PAGE analysis of sfGFP-Asn149TAG mutant (arrow) expressed either in the absence (lane 1) or in the presence (lane 2) of 1 mM ViP; (C) Fluorescence readings of cells expressing PrFRS and sfGFP-Asn149TAG mutant. The expressions were conducted either in the presence or in the absence of 1 mM ViP. Fluorescence intensity was normalized to cell growth; (D) Deconvoluted ESI-MS spectra of the sfGFP-Tyr66ViP mutant. Expected masses: 27315.5 Da (without the N-terminal Met) and 27446.7 (with the N-terminal Met); observed masses: 27315.4 Da (without the N-terminal Met) and 27446.6 Da (with the N-terminal Met). The other signals do not correspond to sfGFP mutant that contains tyrosine (the major background incorporation; 27290.7 or 27421.7) or any other proteinogenic amino acids at position Tyr66.
We next examined the efficiency and fidelity for the genetic incorporation of ViP into the chromophore of a sfGFP mutant (sfGFP-Tyr66TAG)10 that contains an amber (TAG) nonsense mutation at position Tyr66. Protein expression was conducted in E. coli in the presence of 1 mM ViP. A yield of 15.5 mg/L of protein was obtained after a purification using affinity chromatography. The purity of the protein sample was estimated to be higher than 90% (Figure S13; The impurities were mainly E. coli native proteins that bind to Ni-NTA resin.). Mass spectrometry analyses (Figure 2D) of the purified protein (sfGFP-Tyr66ViP) confirmed that ViP was site specifically incorporated at position 66 of sfGFP with high fidelity. The mass spectrometry data also indicated that the chromophore is matured or partially matured (expected mass for a fully matured sfGFP-Tyr66ViP: 27446.7 Da; observed mass: 27446.6). This observation is consistent with prior reports of FPs containing unnatural chromophores.6, 9, 10
Detection of Hg2+ using purified sensor protein
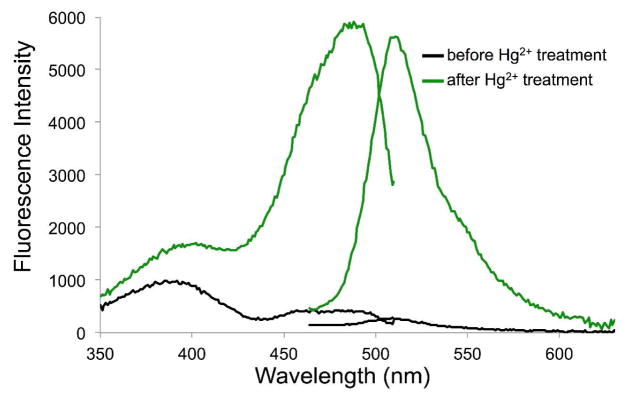
We demonstrated that sfGFP-Tyr66ViP could change its fluorescence emission profile in response to Hg2+. As shown in Figure 3, before the addition of Hg2+, a very weak fluorescence signal was detected. The fluorescence quantum yield (ϕ) of sfGFP-Tyr66ViP was measured as 0.07 (Table S1). After the addition of 10 μM Hg2+ and 90 min of incubation, a strong fluorescence signal (ϕ = 0.58) that was identical to that of the wild-type sfGFP was observed, which indicated that the vinyl ether was hydrolyzed and the wild-type sfGFP chromophore was restored. This notion was further confirmed by the mass spectrometry analysis (Figure S12). As a control, no increase of fluorescence intensity was observed when wild-type sfGFP was incubated with a range of different concentrations of Hg2+ under the same conditions (Figure S2). Next, we investigated the Hg2+-induced fluorescence changes of sfGFP-Tyr66ViP at different concentrations of Hg2+. As shown in Figure S3B, the magnitudes of the fluorescence changes were proportional to the concentration of Hg2+. Significant fluorescence signal increase could be detected when Hg2+ concentration was as low as 5 μM. Little time-dependent fluorescence increase of sfGFP-Tyr66ViP was observed when Hg2+ concentration was 2.5 μM or lower. This likely resulted from the shielded chromophore that could not be easily accessed by Hg2+.
Figure 3.
Fluorescence excitation and emission spectra of sfGFP-Tyr66ViP before (black lines) and after (green lines) the addition of Hg2+.
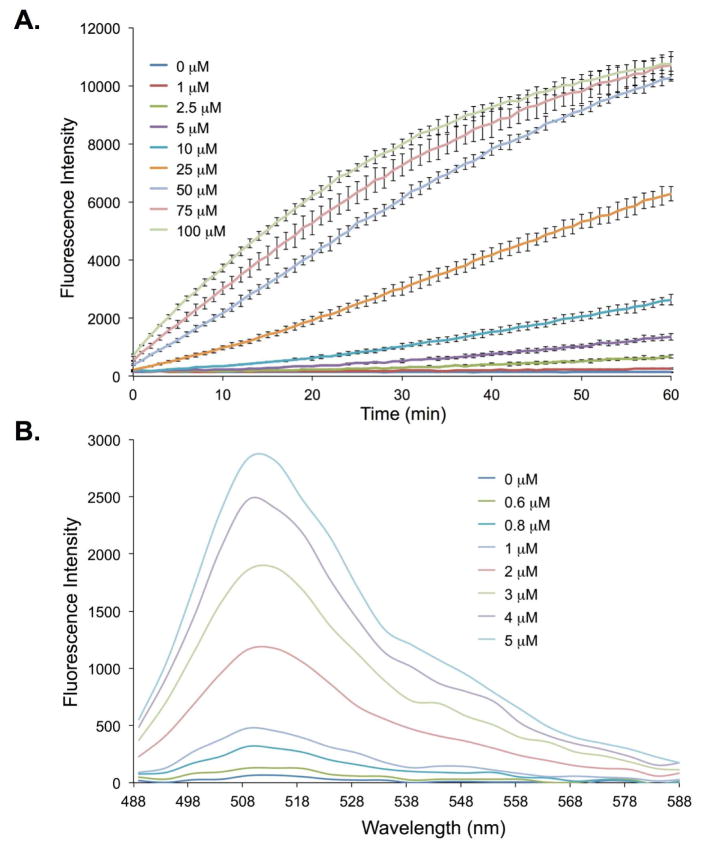
To further improve the Hg2+ sensing kinetics as well as the detection limit, we explored a circularly permuted sfGFP (cpsfGFP-Tyr66ViP; (ϕ = 0.02) variant in which the original N- and C-termini of the sfGFP were connected through a flexible peptide linker and the new N- and C-termini were formed by introducing a terminal break between residues 144 and 145 of the parent protein.10 The new N- and C-termini were spatially close to the chromophore of cpsfGFP-Tyr66ViP to enable a potentially better access by Hg2+. In comparison to sfGFP-Tyr66ViP (Figures S3B), a much improved sensing kinetics of cpsfGFP-Tyr66ViP was observed (Figures S3C). By using 5 μM Hg2+, a 4.5-fold increase in fluorescence intensity was observed at 60 min for cpsfGFP-Tyr66ViP (Figure 4A). As a comparison, the sfGFP-Tyr66ViP displayed a 1.7-fold increase in fluorescence intensity under the same conditions (Figure S3A). The cpsfGFP-Tyr66ViP also displayed a lower detection limit. A 5-fold increase in fluorescence intensity could be detected when 0.8 μM of Hg2+ was incubated with cpsfGFP-Tyr66ViP (Figure 4B). To ensure that the observed fluorescence increases of the sensors are not derived from a UV-associated deprotection of vinyl ether, we conducted photostability studies on the two sensors. As shown in Figure S11, no obvious fluorescence increase was observed under UV irradiation, indicating that both sfGFP-Tyr66ViP and cpsfGFP-Tyr66ViP have good photostability.
Figure 4.
Fluorescence changes of cpsfGFP-Tyr66ViP in the presence of varied concentrations of Hg2+. (A) Time courses monitored at λex = 485 nm and λem = 515 nm; (B) Emission spectrum after 5 h of incubations. The protein concentration was 0.5 μM.
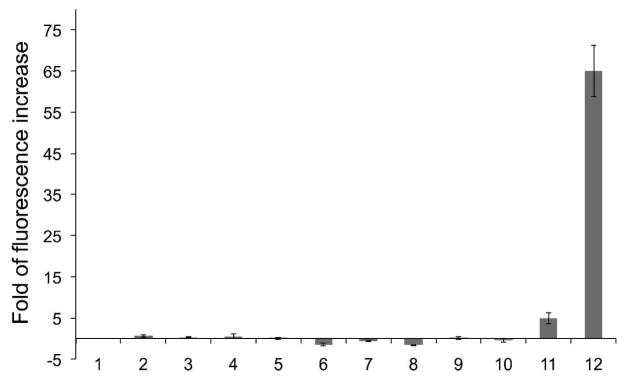
Next, we examined the selectivity of sfGFP-Tyr66ViP (Figure S4) and cpsfGFP-Tyr66ViP (Figure 5) towards Hg2+ over other common metal ions. No significant fluorescence enhancement of the two sensors was observed in the presence of 100 μM of Zn2+, Mg2+, Fe2+, Mn2+, Cu2+, Co2+, Pd2+, Cr3+, or Pb2+ (Figures 5, S4). On the other hand, over 20-fold enhancement in fluorescence intensity was observed (Figure S4) when sfGFP-Tyr66ViP was incubated with 100 μM of Hg2+. Over 60-fold enhancement was observed for cpsfGFP-Tyr66ViP with 10 μM of Hg2+ (Figure 5). In comparison, a previously reported GFP-based Hg2+ sensor showed a maximal fluorescence enhancement of ~1.6 fold in vitro.22 The significantly higher fold of fluorescence enhancement of our sensors benefited from the novel design principle, which targets direct changes in the chemical composition of the chromophore upon its reaction with the analyte, Hg2+.
Figure 5.
Fluorescence changes of cpsfGFP-Tyr66ViP in response to different metal ions after 90 minutes of incubation. 1, blank (no metal ion added); 2, Zn2+; 3, Mg2+; 4, Fe2+; 5, Mn2+; 6, Cu2+; 7, Co2+; 8, Pd2+; 9, Cr3+; 10, Pb2+ (2–10, 100 μM); 11, 1 μM HgCl2; 12, 10 μM HgCl2.
Detection of Hg2+ in live cells
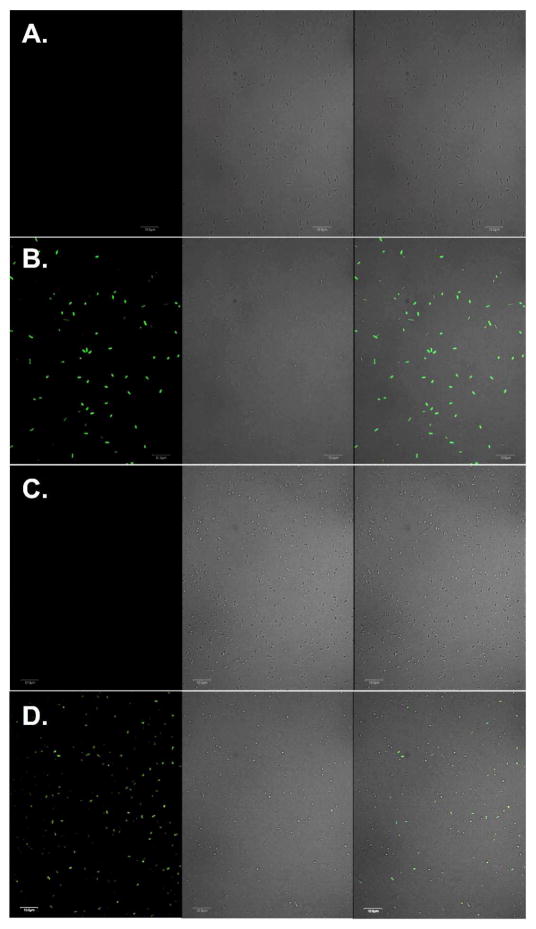
Finally, we demonstrated that both sfGFP-Tyr66ViP and cpsfGFP-Tyr66ViP could be used to detect Hg2+ in live cells. As shown in Figure 6, a drastic increase in fluorescence intensity was clearly observed when E. coli cells expressing either sfGFP-Tyr66ViP (Figures 6B) or cpsfGFP-Tyr66ViP (Figure 6D) were incubated in PBS buffer containing 100 μM Hg2+. Fluorescence of sfGFP was undetectable when Hg2+ was not included in the solution (Figures 6A and 6C). In addition, no fluorescence of sfGFP was detected when E. coli cells cultured in the absence of either ViP (Figure S5A) or PrFRS (Figure S5B) were incubated with Hg2+. Again, faster kinetics and higher sensitivity was observed in cells expressing the circular permutated sensor, cpsfGFP-Tyr66ViP (Figure S6). However, a relatively lower expression level did lead to smaller change in absolute fluorescence in cpsfGFP-Tyr66ViP-expressing cells than that of the sfGFP-Tyr66ViP-expressing cells (Figure S7). We also measured fluorescence intensity of live cells in the presence of different concentrations of Hg2+ in the buffer. A clear concentration dependency was observed (Figure S10). It should be noted that the intracellular concentrations of Hg2+ could be much lower than those calculated values in the imaging buffer. As Hg2+ transport into and out of bacteria has not been fully understood, this FP-based sensor could be applied to the study of transport and/or detoxification mechanisms in live cells.
Figure 6.
Fluorescence imaging of live E. coli cells expressing sfGFP variants. (A) E. coli cells expressing sfGFP-Tyr66ViP before the addition of Hg2+; (B) E. coli cells expressing sfGFP-Tyr66ViP after the addition of Hg2+; (C) E. coli cells expressing cpsfGFP-Tyr66ViP before the addition of Hg2+; (D) E. coli cells expressing cpsfGFP-Tyr66ViP after the addition of Hg2+. Left-panels are fluorescent images of E. coli cells in FITC channel (488 nm excitation and 525±25 nm emission), middle panels are bright-field images of the same E. coli cells, right panels are composite images of bright-field and fluorescent images. Scale bars, 10 μm.
Conclusion
In conclusion, we demonstrated a new strategy for the design of unAA-based FP sensor by implementing the widely used protection-deprotection approach in small-molecule sensors. We rationally designed and successfully constructed a Hg2+ sensor, in which the hydroxyl group of the sfGFP chromophore was protected as a vinyl ether. The sensing element was installed using a genetically encoded unnatural amino acid (ViP). In the presence of Hg2+, the vinyl ether side chain was hydrolyzed and led to a drastic increase of fluorescence signal. This new class of FP-based Hg2+ sensor possesses excellent selectivity and high sensitivity. It is also anticipated that the protection-deprotection strategy, with a genetically encoded unnatural amino acid as the sensing element, could be applied to the creation of FP-based chemosensors for other analytes.
METHODS
Materials and general methods
Unless otherwise noted, starting materials, solvents and reagents for chemical synthesis were obtained from commercial suppliers (Acros, Alfa Aesar, Sigma-Aldrich, Chem-impex) and used without further purification. Dry solvents were either freshly distilled by following standard methods or directly purchased from Acros. Deuterated solvents were obtained from Sigma-Aldrich. Flash chromatography (FC) was carried out using SiliaFlash P60 (0.04–0.063 mm, 230–400 mesh) from Silicycle or Amberlite XAD4 from Sigma-Aldrich. Thin layer chromatography (TLC) was performed on glass-backed, precoated silica gel plates (Analtech). NMR spectra were recorded at 25 °C using a Bruker Advance III-HD 400 MHz NMR. Chemical shifts were reported in ppm with deuterated solvents as internal standards (CDCl3, H 7.26, C 77.16; D2O, 4.79). Multiplicity was reported as follows: s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, b = broad. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on Bio-Rad mini-PROTEAN electrophoresis system using 15% or 18% homemade SDS-PAGE gels. Bio-Rad Prestained Protein Ladder was applied to at least one lane of each gel for the estimation of apparent molecular weights. Protein gels were stained by Coomassie Brilliant Blue staining solution and visualized under Bio-rad Molecular Imager ChemiDoc XRS+ System. Anti-his6 tag antibody was used in Western blot. Fluorescence characterization was performed on BioTek Synergy H1 Hybrid Multimode Monochromater Fluorescence Microplate reader. Live cells were imaged on Olympus FV500 inverted (Olympus IX-81) confocal microscope.
Screening of aminoacyl-tRNA synthetase variants for the genetic incorporation of ViP
A plasmid encoding an aminoacyl-tRNA synthetase (PylRS or TyrRS) mutant of interest was co-transformed with either plasmid pGFPuv-N149TAG28 or plasmid psfGFP-N149TAG28 into E. coli GeneHogs. The resulting strains were inoculated into 5 mL of GMML media containing kanamycin (Kan, 50 mg/L) and chloramphenicol (Cm, 34 mg/L). The cultures were incubated at 37 °C with shaking until OD600 reached 0.6, and then protein expression was induced by the addition of 0.5 mM IPTG and 1 mM ViP (control cultures were only induced with 0.5 mM IPTG). After being incubated at 37 °C with shaking for 54 hours, cells (1 mL) from of each culture were then pelleted down and washed with PBS buffer (1x). The cell pellet was re-suspended with 1 mL PBS buffer for fluorescence and OD600 measurements using a Synergy H1 Hybrid plate reader. The fluorescence of GFPuv was monitored with λEx = 390 nm and λEm = 510 nm. The fluorescence of sfGFP was monitored with λEx = 485 nm and λEm = 515 nm. The cell density was estimated by measuring the sample absorbance at 600 nm. Fluorescence intensities were normalized to cell growth. An aaRS variant that could lead to the synthesis of full-length GFPuv or sfGFP only in the presence of ViP was considered as a hit.
Protein expression and purification
The pEVOL plasmid harboring PrFRS gene was co-transformed into E. coli BL21(DE3) cells with plasmid psfGFP-Tyr66TAG (or pcpsfGFP-Tyr66TAG). Cells were grown on a LB agar plate containing 100 mg/L ampicillin and 34 mg/L chloramphenicol at 37 °C overnight. A single colony from co-transformation was inoculated into 5 mL LB media supplemented with 100 mg/L ampicillin and 34 mg/L chloramphenicol. After incubation with shaking at 37 °C overnight, cells were pelleted down by centrifugation at 5000 g and 4 °C for 15 min and washed once with PBS buffer. Subsequently, the washed cells were transferred into 50 mL GMML containing 100 mg/L ampicillin and 34 mg/L chloramphenicol, and grown with shaking at 37 °C. The protein expression was induced at OD600 of 0.8 by the additions of IPTG (0.5 mM), arabinose (0.02%), and ViP (1 mM). After being cultivated at 37 °C (30 °C for the expression of cpsfGFP) for an additional 16 h, cells were collected by centrifugation at 5000 g and 4 °C for 15 min. Harvested cells were re-suspended in the loading buffer for affinity chromatography and were lysed by sonication. After centrifugation (21000 g, 30 min, 4 °C) to remove cellular debris, the cell-free supernatant was applied to Ni Sepharose 6 Fast Flow resin (GE Healthcare) and the protein was purified by following the manufacture’s instruction. The purified protein was buffer exchanged into 10 mM PBS buffer prior to the assays and MS analysis. Protein concentrations were determined by Bradford assay (Bio-Rad).
Protein mass spectrometry
The sample was analyzed by a Q-Exactive HF mass spectrometer under the positive mode.
Characterization of spectroscopic properties
The fluorescence excitation spectra were scanned from 350 nm to 510 nm with the emission wavelength set at 540 nm. The emission spectra were scanned from 470 nm to 630 nm at the fixed excitation wavelength of 440 nm. For any single-point measurement of fluorescence intensity, excitation at 485 nm and emission at 515 nm were used. The quantum yields of sfGFP-Tyr66ViP and cpsfGFP-Tyr66ViP were calculated by comparison of the integrated emission intensity of these two proteins with the known fluorescence quantum yield of fluorescein (0.93).30
In vitro and In vivo kinetic study of the mercury biosensors
All of the in vitro kinetic studies were performed on the Synergy H1 plate reader with slow shaking at 30 °C. Fluorescence intensity was recorded as the single point measurement every minute. A 150 μL of protein solution (500 nM) was immediately mixed with 1.5 μL of different concentration of Hg2+ stock solutions to obtain the indicated concentration before the fluorescence readings were recorded.
A similar method was applied to the in vivo kinetic studies. After induction of protein expression, E. coli BL21(DE3) cells expressing sfGFP-Tyr66ViP (or cpsfGFP-Tyr66ViP) were cultivated for 16 h and harvested by centrifugation at 4 °C. In order to remove free ViP, cells were washed three time with PBS buffer, re-suspended in PBS buffer, and incubated at 37 °C for 30 minutes. Cells were then collected and re-suspended in PBS buffer. A 150 μL of cell suspension was mixed with 1.5 μL of different concentration of Hg2+ stock solutions to obtain the indicated concentration before the fluorescence readings were recorded.
Selectivity study of the Hg2+ biosensors
Purified protein stock solution was diluted with PBS buffer to obtain a final concentration of 500 nM. Metal ion stock solutions (10 mM) was prepared from their corresponding chloride salts; except for K2PdCl4, FeSO4, and Pb(NO3)2. Metal ion stock solutions were added to the diluted protein solution at the ratio of 1 to 100 to afford the final concentration of 100 μM. For the blank control, the same volume of PBS buffer was added instead of metal ion stock solutions. The metal ion and protein mixtures were then incubated at 30 °C or 37 °C for indicated time. Fluorescence intensity was measured on the Synergy H1 plate reader.
Fluorescent imaging of live E COLI cells
The E. coli cells expressing sfGFP-Tyr66ViP (or cpsfGFP-Tyr66ViP) were collected by centrifugation for 5 min at 21000 g and 4 °C. In order to remove free ViP, cells were washed three time with PBS buffer, re-suspended in PBS buffer, incubated at 37 °C for 30 minutes. The collected cells were resuspended in PBS buffer. Stock solution of Hg2+ was added to reach the final concentrations of 100 μM. In the blank control, the same volume of PBS buffer was added. After incubation for 90 minutes at 37 °C, cells was immediately imaged under Olympus FV500 inverted (Olympus IX-81) confocal microscope. FITC channel was excited at 488 nm and imaged at 525±25 nm.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Health (grant 1R01AI111862 to J.G. and W.N.) and National Science Foundation (grant 1553041 to J.G.). The authors thank Dr. Y. Zhou for help in fluorescence microscope analysis.
Footnotes
Notes
The authors declare no competing financial interest.
Supporting Information. Synthetic procedures, primer list, additional figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Newman RH, Fosbrink MD, Zhang J. Genetically encodable fluorescent biosensors for tracking signaling dynamics in living cells. Chem Rev. 2011;111:3614–3666. doi: 10.1021/cr100002u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frommer WB, Davidson MW, Campbell RE. Genetically encoded biosensors based on engineered fluorescent proteins. Chem Soc Rev. 2009;38:2833–2841. doi: 10.1039/b907749a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niu W, Guo J. Expanding the chemistry of fluorescent protein biosensors through genetic incorporation of unnatural amino acids. Mol BioSyst. 2013;9:2961–2970. doi: 10.1039/c3mb70204a. [DOI] [PubMed] [Google Scholar]
- 4.Tamura T, Hamachi I. Recent progress in design of protein-based fluorescent biosensors and their cellular applications. ACS Chem Biol. 2014;9:2708–2717. doi: 10.1021/cb500661v. [DOI] [PubMed] [Google Scholar]
- 5.Ayyadurai N, Prabhu NS, Deepankumar K, Lee SG, Jeong HH, Lee CS, Yun H. Development of a selective, sensitive, and reversible biosensor by the genetic incorporation of a metal-binding site into green fluorescent protein. Angew Chem Int Ed Engl. 2011;50:6534–6537. doi: 10.1002/anie.201008289. [DOI] [PubMed] [Google Scholar]
- 6.Wang F, Niu W, Guo J, Schultz PG. Unnatural amino acid mutagenesis of fluorescent proteins. Angew Chem Int Ed Engl. 2012;51:10132–10135. doi: 10.1002/anie.201204668. [DOI] [PubMed] [Google Scholar]
- 7.Chen S, Chen Z-j, Ren W, Ai H-w. Reaction-based genetically encoded fluorescent hydrogen sulfide sensors. J Am Chem Soc. 2012;134:9589–9592. doi: 10.1021/ja303261d. [DOI] [PubMed] [Google Scholar]
- 8.Chen Z-j, Ren W, Wright QE, Ai H-w. Genetically encoded fluorescent probe for the selective detection of peroxynitrite. J Am Chem Soc. 2013;135:14940–14943. doi: 10.1021/ja408011q. [DOI] [PubMed] [Google Scholar]
- 9.Chen ZJ, Tian ZQ, Kallio K, Oleson AL, Ji A, Borchardt D, Jiang DE, Remington SJ, Ai HW. The N-B interaction through a water bridge: Understanding the chemoselectivity of a fluorescent protein based probe for peroxynitrite. J Am Chem Soc. 2016;138:4900–4907. doi: 10.1021/jacs.6b01285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, Li J, Hu C, Zhou Q, Zhang W, Hu M, Zhou J, Wang J. Significant expansion of the fluorescent protein chromophore through the genetic incorporation of a metal-chelating unnatural amino acid. Angew Chem Int Ed Engl. 2013;52:4805–4809. doi: 10.1002/anie.201301307. [DOI] [PubMed] [Google Scholar]
- 11.Tang Y, Lee D, Wang J, Li G, Yu J, Lin W, Yoon J. Development of fluorescent probes based on protection-deprotection of the key functional groups for biological imaging. Chem Soc Rev. 2015;44:5003–5015. doi: 10.1039/c5cs00103j. [DOI] [PubMed] [Google Scholar]
- 12.Nolan EM, Lippard SJ. Tools and tactics for the optical detection of mercuric ion. Chem Rev. 2008;108:3443–3480. doi: 10.1021/cr068000q. [DOI] [PubMed] [Google Scholar]
- 13.Chen G, Guo Z, Zeng G, Tang L. Fluorescent and colorimetric sensors for environmental mercury detection. Analyst. 2015;140:5400–5443. doi: 10.1039/c5an00389j. [DOI] [PubMed] [Google Scholar]
- 14.Domaille DW, Que EL, Chang CJ. Synthetic fluorescent sensors for studying the cell biology of metals. Nat Chem Biol. 2008;4:168–175. doi: 10.1038/nchembio.69. [DOI] [PubMed] [Google Scholar]
- 15.Kim HN, Ren WX, Kim JS, Yoon J. Fluorescent and colorimetric sensors for detection of lead, cadmium, and mercury ions. Chem Soc Rev. 2012;41:3210–3244. doi: 10.1039/c1cs15245a. [DOI] [PubMed] [Google Scholar]
- 16.Lin W, Cao X, Ding Y, Yuan L, Long L. A highly selective and sensitive fluorescent probe for Hg2+ imaging in live cells based on a rhodamine-thioamide-alkyne scaffold. Chem Commun. 2010;46:3529–3531. doi: 10.1039/b927373e. [DOI] [PubMed] [Google Scholar]
- 17.Lin W, Cao X, Ding Y, Yuan L, Yu Q. A reversible fluorescent Hg2+ chemosensor based on a receptor composed of a thiol atom and an alkene moiety for living cell fluorescence imaging. Organic & Biomolecular Chemistry. 2010;8:3618–3620. doi: 10.1039/c0ob00081g. [DOI] [PubMed] [Google Scholar]
- 18.Suresh M, Mishra SK, Mishra S, Das A. The detection of Hg2+ by cyanobacteria in aqueous media. Chem Commun. 2009:2496–2498. doi: 10.1039/b821687h. [DOI] [PubMed] [Google Scholar]
- 19.Wegner SV, Okesli A, Chen P, He C. Design of an emission ratiometric biosensor from MerR family proteins: A sensitive and selective sensor for Hg2+ J Am Chem Soc. 2007;129:3474–3475. doi: 10.1021/ja068342d. [DOI] [PubMed] [Google Scholar]
- 20.Chapleau RR, Blomberg R, Ford PC, Sagermann M. Design of a highly specific and noninvasive biosensor suitable for real-time in vivo imaging of mercury(II) uptake. Protein Sci. 2008;17:614–622. doi: 10.1110/ps.073358908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu Z, Zhao M, Sheng Y, Bentolila LA, Tang Y. Detection of mercury ion by infrared fluorescent protein and its hydrogel-based paper assay. Anal Chem. 2011;83:2324–2329. doi: 10.1021/ac103236g. [DOI] [PubMed] [Google Scholar]
- 22.Jiang T, Guo DP, Wang Q, Wu X, Li Z, Zheng ZH, Yin BY, Xia L, Tang JX, Luo WX, Xia NS, Jiang YB. Developing a genetically encoded green fluorescent protein mutant for sensitive light-up fluorescent sensing and cellular imaging of Hg(II) Anal Chim Acta. 2015;876:77–82. doi: 10.1016/j.aca.2015.03.026. [DOI] [PubMed] [Google Scholar]
- 23.Santra M, Ryu D, Chatterjee A, Ko SK, Shin I, Ahn KH. A chemodosimeter approach to fluorescent sensing and imaging of inorganic and methylmercury species (pg 2115: 2009) Chem Commun. 2009:7599–7599. doi: 10.1039/b900380k. [DOI] [PubMed] [Google Scholar]
- 24.Santra M, Roy B, Ahn KH. A “reactive” ratiometric fluorescent probe for mercury species. Organic letters. 2011;13:3422–3425. doi: 10.1021/ol2011693. [DOI] [PubMed] [Google Scholar]
- 25.Kim I, Kim D, Sambasivan S, Ahn KH. Synthesis of pi-extended coumarins and evaluation of their precursors as reactive fluorescent probes for mercury ions. Asian J Org Chem. 2012;1:60–64. [Google Scholar]
- 26.Wu X, Schultz PG. Synthesis at the interface of chemistry and biology. J Am Chem Soc. 2009;131:12497–12515. doi: 10.1021/ja9026067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song X, Shang X, Ju T, Cerny R, Niu W, Guo J. A photoactivatable Src homology 2 (SH2) domain. RSC Adv. 2016;6:51120–51124. [Google Scholar]
- 28.Shang X, Song X, Faller C, Lai R, Li H, Cerny R, Niu W, Guo J. Fluorogenic protein labeling using a genetically encoded unstrained alkene. Chem Sci. 2017;8:1141–1145. doi: 10.1039/c6sc03635j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deiters A, Schultz PG. In vivo incorporation of an alkyne into proteins in Escherichia coli. Bioorg Med Chem Lett. 2005;15:1521–1524. doi: 10.1016/j.bmcl.2004.12.065. [DOI] [PubMed] [Google Scholar]
- 30.Brouwer AM. Standards for photoluminescence quantum yield measurements in solution (IUPAC technical report) Pure Appl Chem. 2011;83:2213–2228. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.