Abstract
A single protein can bind and regulate many mRNAs. Multiple proteins with similar specificities often bind and control overlapping sets of mRNAs. Yet little is known about the architecture or dynamics of overlapped networks. We focused on three proteins with similar structures and related RNA-binding specificities—Puf3p, Puf4p, and Puf5p of S. cerevisiae. Using RNA Tagging, we identified a “super-network” comprised of four subnetworks: Puf3p, Puf4p, and Puf5p subnetworks, and one controlled by both Puf4p and Puf5p. The architecture of individual subnetworks, and thus the super-network, is determined by competition among particular PUF proteins to bind mRNAs, their affinities for binding elements, and the abundances of the proteins. The super-network responds dramatically: The remaining network can either expand or contract. These strikingly opposite outcomes are determined by an interplay between the relative abundance of the RNAs and proteins, and their affinities for one another. The diverse interplay between overlapping RNA–protein networks provides versatile opportunities for regulation and evolution.
Keywords: RNA control, RNA-binding proteins, PUF proteins, protein–RNA networks, protein–RNA interactions
INTRODUCTION
Proteins and RNAs form highly interconnected networks of interactions that permeate biology and cause disease (Keene 2007; Lukong et al. 2008). A single RNA-binding protein (RBP) often binds hundreds of individual RNAs, which we refer to as a “protein–RNA network.” Moreover, a single RNA molecule can be bound by multiple proteins, and its fate is dictated by the particular combination that are bound. (Muller-McNicoll and Neugebauer 2013; Singh et al. 2015). Overlapping protein–RNA networks—formed when two or more RBPs bind some of the same RNAs—are common. In particular, families of RBPs characterized by conserved structures and RNA-binding domains often overlap in binding specificity in vitro (Ray et al. 2013; Gerstberger et al. 2014). The challenge is to understand how multiple protein–RNA networks are integrated and coordinated in vivo. We sought to uncover those principles in an RNA regulatory network composed of multiple related RBPs.
PUF proteins are a versatile and exemplary family of mRNA regulators. They are conserved throughout Eukarya and play key roles in the regulation of early development, stem cells, the nervous system, and cancer (Wickens et al. 2002; Spassov and Jurecic 2003; Quenault et al. 2011). Individual PUF proteins bind hundreds of mRNAs, most commonly through specific sequences present in their 3′ untranslated regions (UTRs) (Gerber et al. 2004, 2006; Galgano et al. 2008; Hogan et al. 2008; Morris et al. 2008; Hafner et al. 2010; Kershner and Kimble 2010; Chen et al. 2012; Lapointe et al. 2015; Porter et al. 2015; Wilinski et al. 2015; Prasad et al. 2016). PUF proteins recruit other factors to the mRNA, which then control its stability, translation, and localization (Olivas and Parker 2000; Houshmandi and Olivas 2005; Goldstrohm et al. 2006, 2007; Hook et al. 2007; Lee et al. 2010).
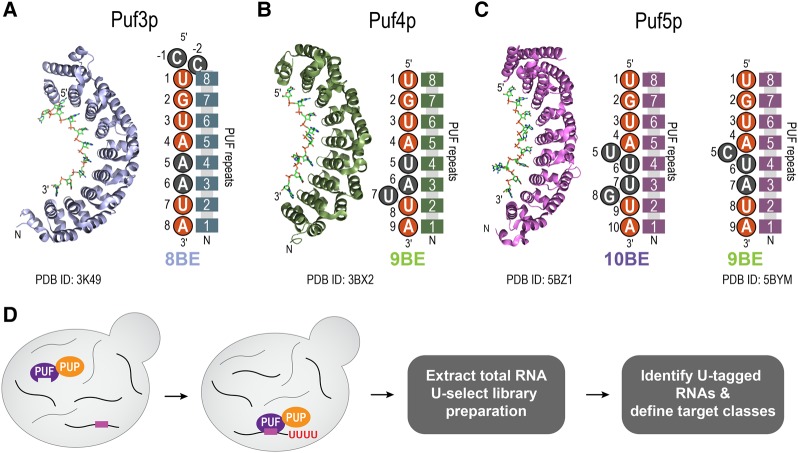
In Saccharomyces cerevisiae, three PUF proteins—Puf3p, Puf4p, and Puf5p—have similar structures and bind related but distinct RNA sequences. These “canonical” PUF proteins possess eight PUF repeats, which fold into a crescent shape (Fig. 1; Miller et al. 2008; Zhu et al. 2009; Valley et al. 2012; Wilinski et al. 2015). Each repeat possesses an RNA recognition α-helix that mediates direct binding to an RNA base. All of the proteins bind RNA elements that possess a 5′ UGUA followed by a downstream 3′ UA, termed “PUF-binding elements” (PBEs). Despite the similarity, each protein prefers PBEs of distinct length dictated by the number of nucleotides between the 5′ and 3′ features, which we refer to as “spacer nucleotides.” Puf3p preferentially binds sequence elements 8 nucleotides (nt) in length (“8BE,” two spacer nucleotides) (Fig. 1A; Gerber et al. 2004; Zhu et al. 2009; Lapointe et al. 2015). Puf4p preferentially binds sequence elements 9 nt in length (“9BE,” three spacer nucleotides) (Fig. 1B; Gerber et al. 2004; Hook et al. 2007; Miller et al. 2008; Campbell et al. 2012; Valley et al. 2012). Puf5p binds to 9BEs as well as sequence elements 10 nt in length (“10BE,” four spacer nucleotides) (Fig. 1C; Gerber et al. 2004; Campbell et al. 2012; Valley et al. 2012; Wilinski et al. 2015). This trio of PUF proteins in yeast provides a powerful model with which to determine how related networks are integrated, coordinated, and balanced in vivo.
FIGURE 1.
Puf3p, Puf4p, and Puf5p share similar structures and bind distinct yet related RNA sequences. (A) Crystal structure of Puf3p bound to RNA (PDB ID: 3K49) (Zhu et al. 2009), and a cartoon schematic of the PUF domains of Puf3p bound to an 8BE, with upstream cytosines. (B) Crystal structure of Puf4p bound to RNA (PDB ID: 3BX2) (Miller et al. 2008), and a cartoon schematic of the PUF domains of Puf4p bound to a 9BE. (C) Crystal structure of Puf5p bound to RNA (PDB ID: 5BZ1) (Wilinski et al. 2015), and a cartoon schematic of the PUF domains of Puf5p bound to a 10BE and a 9BE (based off PDB ID: 5BYM). In panels A–C, the RNA-binding domains of Puf3p, Puf4p, and Puf5p, which contain the 8 PUF repeats, are represented by the eight connected rectangles. The numbers in the rectangles indicate the PUF repeat identity, numbered from N to C terminus. The nucleotides represented by orange circles represent the common sequence typically shared by an 8BE, 9BE, and 10BE. The nucleotides in gray represent the regions that distinguish the binding elements. (D) Schematic of RNA Tagging. PUP-2 from C. elegans was fused to the C terminus of the endogenous copy of Puf3p, Puf4p, and Puf5p. Inside live yeast, the PUF–PUP fusion proteins bind to target mRNAs and deposit a covalent “U-tag” on the 3′ end of the mRNA. After extraction of total RNA under denaturing conditions, U-tagged mRNAs are preferentially enriched in the U-select library preparation and analyzed via paired-end high-throughput sequencing. U-tagged RNAs are computationally identified and analyzed to define mRNA targets and target classes.
In this report, we combine molecular, genetic, RNA Tagging, and bioinformatic approaches to define the subnetworks controlled by each protein and the “PUF super-network” into which they are integrated. Having established an overall map of the interactions, we determine how the RNA–protein interactions change upon removal of one protein. Our findings reveal that competition for RNAs is a key determinant of the super-network, but that the effects of removing one protein are not only dramatic, but vary dramatically. Removal of one protein can either expand or contract the networks that remain and reconfigure which mRNAs are controlled.
RESULTS
We used RNA Tagging to identify RNAs bound by Puf4p and Puf5p. RNA Tagging exploits a poly(U) polymerase (PUP) to covalently “U-tag” the RNAs bound by a protein of interest in vivo (Lapointe et al. 2015). The U-tagged RNAs are identified via high-throughput sequencing (Fig. 1D). We constructed “PUF4–PUP” and “PUF5–PUP” strains of S. cerevisiae, in which the open-reading frame of C. elegans PUP-2 was fused to the 3′ end of PUF4 or PUF5 at their endogenous genomic loci, respectively. We used total RNA from these strains to prepare high-throughput sequencing libraries, which were sequenced using an Illumina platform to obtain paired-end reads (Lapointe et al. 2015). Following sequencing, we identified U-tagged RNAs present in each strain, which we defined as RNAs that contained at least eight adenosines [the poly(A) tail] followed by at least one uridine (the U-tag) not encoded in the genome.
The Puf4p subnetwork
In PUF4–PUP yeast, we identified 507 mRNAs that were reproducibly U-tagged, which we hereafter refer to as “Puf4p targets” (Supplemental Fig. S1A; Supplemental Data S1). Puf4p targets were highly enriched for a 9-nt-long sequence element characterized by a 5′ UGUA and more degenerate 3′ UA (Supplemental Fig. S1B). The enriched sequence is consistent with the expected 9BE (Gerber et al. 2004; Campbell et al. 2012; Valley et al. 2012). Puf4p targets also are enriched for mRNAs encoding proteins that process rRNA and participate in ribosome biogenesis (Supplemental Fig. S1C).
To facilitate analysis of our RNA Tagging data, we separated Puf4p targets into groups, which we call “classes.” RNA Tagging provides two attributes for every Puf4p target: the number of U-tagged RNAs detected per million uniquely mapped reads (TRPM) and the number of uridines in the U-tag on each tagged RNA molecule. We leveraged these two parameters, facilitated by k-means clustering, to separate Puf4p targets into four classes based on the number of TRPM detected at increasing U-tag lengths (from at least one U to at least eight U's) (see Materials and Methods). We visualized the results via a heat map (Fig. 2A). Class I targets were detected by the most TRPMs and many U-tags of up to seven or eight U's. In contrast, class IV targets were detected by the fewest TRPMs and rarely had U-tags longer than one or two U's.
FIGURE 2.
The Puf4p subnetwork was identified via RNA Tagging. (A) Heat map displaying results of the k-means clustering analysis of all 507 Puf4p targets. Each row represents a single Puf4p target. Columns refer to the length of the U-tag detected on reads for each gene, from at least one uridine (leftmost column) to at least eight uridines (rightmost). Puf4p target classes are indicated (I, II, III, and IV). The average PBE enriched in each class of targets is also indicated with the y axis, in bits. (B) Plot showing the fraction of each Puf4p target class with PBEs enriched at least 10-fold (black) or 50-fold (gray) above background in SEQRS. (C) Plot of the enrichment of each class of Puf4p targets for 8BEs, 9BEs, and 10BEs. (D) Plot of SEQRS enrichment of Puf4p binding to 8BE, 9BE, and 10BEs in vitro. Enrichment for each PBE was calculated relative to randomers of the same length. (E) Box plot of 9BE conservation scores for all yeast mRNAs (all mRNAs) and Puf4p target classes. A score of “16” indicates the 9BE was present in all 16 species that were analyzed.
Puf4p target class correlated with enrichment for high-affinity Puf4p-binding elements. Nearly all class I targets possessed consensus 9BEs, and the 9BE progressively degenerated from class I to class IV targets (Fig. 2A). To determine whether Puf4p binding affinity was indeed correlated with target class, we analyzed data from a published Puf4p SEQRS analysis (Wilinski et al. 2015), which simultaneously determined the relative binding affinity of a single protein for millions of 10-mer sequences present in a library with 20 randomized nucleotides (Campbell et al. 2012). Class I Puf4p targets were most enriched for high-affinity 9BEs (Fig. 2B), and the enrichment progressively decreased from class I to class IV targets. We hypothesized that the degeneracy at the 3′ end of low-affinity Puf4p-binding elements was the result of a variable number of “spacer” nucleotides between the 5′ UGUA and the 3′ UA. Class I binding elements were almost entirely composed of consensus 9BEs (three spacer nucleotides), and class IV binding elements were more likely to include 8BEs (two spacer nucleotides) or 10BEs (four spacer nucleotides) (Fig. 2C). Analysis of the Puf4p SEQRS data confirmed that 9BEs were best enriched by Puf4p in vitro, with weaker enrichments for 8BEs and 10BEs (Fig. 2D).
Puf4p target class also correlated with 9BE conservation, Puf4p-dependent regulation, and biological function. To examine whether 9BEs were conserved in Puf4p targets across more than 400 million years of evolution (Taylor and Berbee 2006), we analyzed the conservation of 9BEs found in Puf4p targets with orthologs present in 16 species of budding yeast (see Materials and Methods for full details). Class I targets had the most conserved 9BEs across 16 species of budding yeast (Fig. 2E), and 9BE conservation progressively decreased from class I to the very modestly conserved class IV targets. We next examined whether Puf4p target classes were correlated with known mechanisms of Puf4p-dependent regulation by mining published data that determined the change in stability of mRNAs genome-wide with and without PUF4 (Sun et al. 2013). Class I Puf4p targets displayed the largest increase in RNA stability in the absence of PUF4 (the mRNAs have slower decay rates in a puf4Δ strain relative to a wild-type strain), and the enrichment progressively decreased from class I to class IV targets (Supplemental Fig. S1D). Class I targets were also most enriched for ribosome biogenesis-related functions, which again progressively decreased from class I to class IV targets (Supplemental Fig. S1E). Thus, Puf4p likely has a conserved role in the control of mRNAs that encode proteins important for the biogenesis of ribosomes. Taken together, these data imply that a subset of interactions we detect, hence a subset of binding events in vivo, elicit biological regulation.
The Puf5p subnetwork
To complete our map of the canonical PUF protein subnetworks, we defined the Puf5p subnetwork using RNA Tagging. PUF5–PUP reproducibly U-tagged 916 RNAs, which we refer to as “Puf5p targets” (Supplemental Fig. S2A; Supplemental Data S2). The vast majority of Puf5p targets were mRNAs (914), with two noncoding RNAs also detected (TLC1 and a tRNAAsn isoacceptor). Puf5p targets were enriched for a 5′ UGUA motif in their 3′ regulatory regions (603/916, P < 10−16). Directed searches revealed that Puf5p mRNA targets primarily were enriched for a single 9BE or 10BE (Fig. 3A,B), consistent with a previous study (Wilinski et al. 2015). Puf5p targets were enriched for a broad range of biological functions, including cytoplasmic translation, ribosome biogenesis, chromosome organization, and transcription (Supplemental Fig. S2B).
FIGURE 3.
The Puf5p subnetwork was identified via RNA Tagging. (A) Plot of the fraction of Puf5p targets and all yeast mRNAs (all mRNAs) with the indicated PBE. (B) Pie chart illustrating the number of Puf5p targets with both a 9BE and 10BE (light gray), or either a 9BE or 10BE (dark gray) in their 3′ UTR. (C) Heat map displaying results of the k-means clustering analysis of all 916 Puf5p targets. Each row represents a single Puf5p target. Columns refer to the length of the U-tag detected on reads for each gene, from at least one uridine (leftmost column) to at least eight uridines (rightmost). Puf5p target classes are indicated (I, II, III, and IV). (D) Plot showing the enrichment of each class of Puf5p targets relative to all yeast mRNAs (all mRNAs) for 9BEs and 10BEs. (E) Schematic of wild-type (WT) and a mutant PHD1 mRNA that lacks its PBEs (ΔPBE), and a plot of the number of U-tagged mRNAs detected for WT and ΔPBE PHD1 mRNAs.
Using the same strategy as with Puf4p, we separated Puf5p targets into four classes, visualized them by heat map (Fig. 3C), and analyzed their enrichment for known metrics of Puf5p function. Class I Puf5p targets were enriched for modestly conserved 10BEs (Fig. 3D; Supplemental Fig. S2C) and PUF5-dependent changes in RNA stability (Kolmogorov–Smirnov test, P < 0.00001) (Supplemental Fig. S2D). 10BE enrichment progressively decreased from class I to class IV Puf5p targets (Fig. 3D). In contrast, class II Puf5p targets were most enriched for 9BEs (Fig. 3D) and lacked enrichment for PUF5-dependent changes in RNA stability (P > 0.01).
In comparison to Puf3p (Lapointe et al. 2015) and Puf4p (this study), Puf5p target classes were less correlated with enrichments for Puf5p-binding elements, binding element conservation, and biological function. To ensure that PUF5–PUP was indeed specific for putative Puf5p mRNA targets, we tested whether U-tagging by PUF5–PUP required a Puf5p-binding element. We selected PHD1 mRNA for analysis. PHD1 is a class I target in our tagging studies, and also was strongly detected as a Puf5p target using HITS-CLIP, which pinpointed its PBEs (Wilinski et al. 2015). We first replaced the endogenous copy of PHD1 mRNA with a mutant version that lacked the PBEs (UGU to ACA substitutions) (Fig. 3E). We then analyzed the mutant PHD1 strain via RNA Tagging (Lapointe et al. 2015). Importantly, zero U-tagged PHD1 mRNAs were detected in the mutant PHD1 strain while we detected 1076 U-tagged RNAs in total across two biological replicates (51 TRPM, mean) for the wild-type allele (Fig. 3E; Supplemental Data S3). Thus, PUF5–PUP requires Puf5p-binding elements to tag mRNAs.
Our data thus define the Puf4 and Puf5p subnetworks. For Puf5p, 9BE enrichment peaked in class II rather than class I, despite Puf5p having similar in vitro binding preferences for both 9BEs and 10BEs (Valley et al. 2012; Wilinski et al. 2015). Given the correlations we observed with Puf4p and Puf3p (Lapointe et al. 2015), it was expected that both high-affinity Puf5p-binding elements would be most enriched in class I. Since Puf4p and Puf5p both bind 9BEs with high affinity (Gerber et al. 2004; Hook et al. 2007; Miller et al. 2008; Campbell et al. 2012; Valley et al. 2012; Wilinski et al. 2015), we hypothesized that Puf4p and Puf5p bind many of the same mRNAs, particularly those with 9BEs.
The PUF super-network
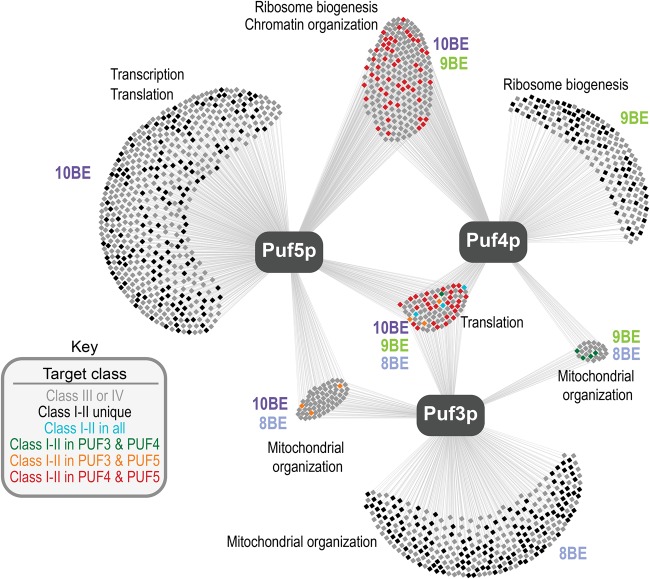
To identify mRNAs bound by multiple PUF proteins, we integrated our RNA Tagging data for each of the canonical PUF proteins: Puf3p, Puf4p, and Puf5p. We first reanalyzed our published Puf3p RNA Tagging data (Lapointe et al. 2015) using the same approaches as done here for Puf4p and Puf5p (Supplemental Data S4). Consistent with our prior analyses, Puf3p target class was highly correlated with enrichment for high-affinity and highly conserved Puf3p-binding elements, and PUF3-dependent regulation (Supplemental Fig. S3). We next constructed a map of all RNAs U-tagged by at least one canonical PUF protein (Fig. 4), which we call the “PUF super-network.” Puf3p, Puf4p, and Puf5p collectively U-tagged 1417 RNAs, thereby encompassing ∼20% of the yeast transcriptome.
FIGURE 4.
Map of the canonical PUF super-network in S. cerevisiae. Each box represents a single gene, and lines illustrate if it was U-tagged by a given PUF protein. The key indicates how genes were colored. PBEs enriched above background and broad Gene Ontology enrichments in different groups of targets are indicated.
In particular, the Puf4p and Puf5p subnetworks are highly interconnected. Three hundred and seven mRNAs were U-tagged by both Puf4p and Puf5p (Fig. 4); thus, ∼60% of the Puf4p subnetwork is included in the Puf5p subnetwork. Importantly, 82 mRNAs were class I or II targets for both Puf4p and Puf5p (red squares, Fig. 4), which represents 27% of their shared targets (Supplemental Fig. S4A). This is similar to the percentage of RNAs strongly U-tagged by individual proteins, but it is in stark contrast to the number of class I or II targets shared with Puf3p (Fig. 4; Supplemental Fig. S4A). Overlap between Puf4p and Puf5p targets was highest among class I targets and progressively decreased from class I to class IV targets (Supplemental Fig. S4B,C). To examine whether Puf4p and Puf5p overlapped in biological function, we computationally identified phenotypes most commonly associated with multiple class I or II shared targets using a public database (www.yeastgenome.org). The phenotypes included sensitivities to hydroxyurea, rapamycin, and methyl methanesulfonate (MMS). Yeast that lack both Puf4p and Puf5p (puf4Δpuf5Δ yeast strain) displayed increased sensitivity to each compound, relative to wild-type yeast or yeast that lacked either protein alone (puf4Δ and puf5Δ yeast strains) (Supplemental Fig. S4D).
We hypothesized that PUF proteins selected their RNA targets based on the presence of their preferred binding elements. Indeed, mRNAs uniquely bound by Puf3p, Puf4p, or Puf5p were most enriched for their preferred binding element (Fig. 4; Supplemental Fig. S4E). mRNAs U-tagged by both Puf4p and Puf5p were most enriched for 9BEs and weakly enriched for 10BEs (Fig. 4; Supplemental Fig. S4E), which suggested that they primarily possessed 9BEs. Indeed, the 82 mRNAs present in class I or II for both Puf4p and Puf5p primarily possessed a single PBE in their 3′ UTR (57 mRNAs) (Supplemental Fig. S4F), most of which were highly conserved (49/57) (Supplemental Fig. S4G).
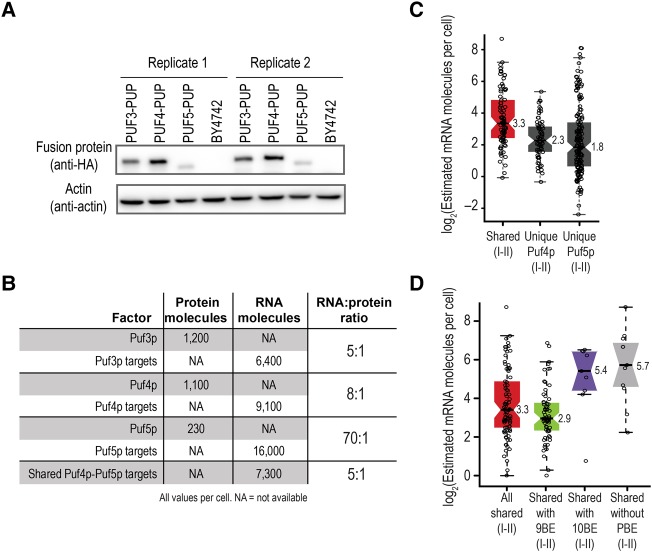
Many Puf4p targets with 9BEs are not bound by Puf5p, even though they possess a high-affinity Puf5p-binding element. To determine whether relative ratios of PUF proteins to their target mRNAs might help explain this observation, we determined the relative abundances of Puf4p, Puf5p, and the mRNAs they bind. As assessed by Western blot analyses, Puf4p was three- to ninefold more abundant than Puf5p in the RNA Tagging strains (Fig. 5A; Supplemental Fig. S5A). Our data agree with the relative abundances of the endogenous proteins and mRNAs (Hebert et al. 2014; Kulak et al. 2014; Lapointe et al. 2015). We also estimated the number of molecules present in a cell for each mRNA target of Puf4p and Puf5p. We based the estimation on published RNA-seq data (Lapointe et al. 2015) and the empirically derived value of approximately 36,000 mRNA molecules per cell (Miura et al. 2008) (see Materials and Methods). The findings, while an approximation, reveal striking differences in the molar ratios of proteins and their mRNA targets. The number of RNA target molecules exceeds that of proteins in every case, but is about nine- and 14-fold more for Puf5 than for Puf4p and Puf3p, respectively (Fig. 5B).
FIGURE 5.
Relative abundance of Puf4p, Puf5p, and their targets. (A) Western blot depicting relative protein levels of the indicated strains. PUF4–PUP and PUF5–PUP contained 3-HA epitope tags on their C termini. Actin was used as the loading control. (B) Estimated number of molecules present in a cell for Puf3p, Puf4p, Puf5p, and their mRNA targets. The estimated number of Puf3p, Puf4p, and Puf5p molecules were obtained from Kulak et al. (2014). The estimated number of mRNA molecules was calculated using our published RNA-seq data (Lapointe et al. 2015) and an estimated total number of 36,000 molecules of mRNA per cell (Miura et al. 2008). In the calculation of the RNA:protein ratio for shared Puf4p–Puf5p targets, the total number of proteins was the sum of Puf4p and Puf5p (1330). Ratios were rounded to the nearest integer. (C) Boxplot illustrating the estimated number of mRNA molecules present in each yeast cell for the indicated groups of mRNAs. The individual data points for each gene and medians are overlaid on the boxplot. “Unique Puf5p (I–II)” refers to RNAs that were uniquely U-tagged by PUF5–PUP and were present in class I or II. “Unique Puf4p (I–II)” refers to RNAs that were uniquely U-tagged by PUF4–PUP and were present in class I or II. “Shared (I–II)” refers to mRNAs U-tagged by both PUF4–PUP and PUF5–PUP and were present in class I or II of both data sets. (D) Boxplot illustrating the estimated number of mRNA molecules present in each yeast cell for the indicated groups of mRNAs. The individual data points for each gene and medians are overlaid on the boxplot. “Shared (I–II)” is defined as mRNAs U-tagged by both PUF4–PUP and PUF5–PUP and were present in class I or II of both data sets.
We hypothesized that the low abundance of Puf5p relative to Puf4p excludes Puf5p from many Puf4p targets with high-affinity binding elements. mRNAs present in class I or II (“class I–II”) of both Puf4p and Puf5p were twofold more abundant than unique class I–II Puf4p or Puf5p target mRNAs (Fisher–Pitman permutation test, P < 10−15) (Fig. 5C). The increased abundance of those mRNAs likely allows Puf5p access to them even in the presence of Puf4p. Similarly, class I–II targets of both Puf4p and Puf5p with only a 10BE, which is only weakly bound by Puf4p, or that lacked any PBE were greater than fourfold more abundant than class I–II shared targets with a 9BE (Fisher–Pitman permutation test, P < 0.001) (Fig. 5D). We therefore suggest that an interplay between mRNA abundance, protein abundance, and relative binding affinities underlies the entire PUF super-network.
Divergent effects of rewiring
Our findings suggest that Puf4p and Puf5p directly compete to bind the same pool of RNAs in vivo. We therefore examined how the Puf4p and Puf5p subnetworks were rewired in the absence of the other protein. In support of a simple competition model, we found that the Puf4p subnetwork expanded in the absence of Puf5p, and the expanded network included many additional Puf5p targets, many with relatively weak Puf4p-binding elements. However, our findings from the reciprocal experiment yielded the opposite outcome—a surprising contraction of the Puf5p subnetwork. In the absence of Puf4p, the Puf5p subnetwork lost nearly half of its targets, most of which were present in class III or IV.
To test how loss of PUF5 impacted the Puf4p subnetwork, we performed RNA Tagging in a yeast strain that expressed PUF4–PUP and lacked PUF5 (“Puf4p;puf5Δ”). We detected 1365 U-tagged mRNAs and four noncoding RNAs in Puf4p;puf5Δ yeast, which we refer to as the Puf4p;puf5Δ subnetwork (Supplemental Fig. S6A,B; Supplemental Data S5). Nearly all Puf4p targets (98%) were also Puf4p;puf5Δ targets (“retained Puf4p targets”) (Fig. 6A). They were often present in better classes in the Puf4p;puf5Δ subnetwork (Fig. 6B), suggesting they were often U-tagged better by Puf4p when Puf5p is absent.
FIGURE 6.
Rewiring of the super-network absent one component: divergent effects. (A,F) Proportional Venn diagram illustrating overlap among the indicated targets. (B,G) Plot of the fraction of the indicated target class that was present in the indicated Puf4p;puf5Δ (B) or Puf5p;puf4Δ (G) target class. For example, in panel B, all class I Puf4p targets were class I Puf4p;puf5Δ targets, and ∼45% class II Puf4p targets improved to class I Puf4p;puf5Δ targets while ∼55% class II Puf4p targets remained class II Puf4p;puf5Δ targets. (C,H) Plot of the fraction of each class of Puf4p (black) and Puf5p (gray) targets that were U-tagged in Puf4p;puf5Δ (C) or Puf5p;puf4Δ (H) yeast. (D,I) Enrichment of the indicated groups, defined in panels A and F, of genes for 8BEs, 9BEs, and 10BEs relative to all yeast mRNAs (all mRNAs). (E,J) Boxplot illustrating the estimated number of mRNA molecules present in each yeast cell for the indicated groups of mRNAs, defined in panels A and F. The individual data points for each gene and medians are overlaid on the boxplot.
As predicted by a simple competition model, the Puf4p subnetwork expanded in the absence of PUF5 to include nearly 70% of all Puf5p targets, many with relatively weak Puf4p-binding elements. The Puf4p;puf5Δ subnetwork included 322 Puf5p targets that were not bound by Puf4p in wild-type cells (“gained Puf5p targets”) (Fig. 6A). Inclusion of Puf5p targets in the Puf4p;puf5Δ subnetwork correlated with Puf5p target class (Fig. 6C). Puf5p targets absent from the Puf4p;puf5Δ subnetwork (“missed Puf5p targets”) were primarily present in class III or IV of the Puf5p subnetwork (i.e., among the weakest Puf5p targets) (Fig. 6C). Gained and missed Puf5p targets were similarly enriched for 10BEs (Fig. 6D). However, gained Puf5p targets were significantly more abundant at the mRNA level than missed Puf5p targets (Fisher–Pitman permutation test, P < 10−15) (Fig. 6E), which suggests mRNA abundance is a key factor in the determination of which Puf5p targets were gained by Puf4p. Nearly all RNAs in the Puf4p;puf5Δ subnetwork that were not Puf4p or Puf5p targets (551 RNAs) were weakly enriched for PBEs (Supplemental Fig. S6C), and they were present in class III or IV (Supplemental Fig. S6D). This suggests they were sampled by the fusion protein, perhaps due to slightly increased PUF4–PUP levels that are suggested by Western blot (Supplemental Fig. S5B). The gained mRNAs were more abundant on average, implying that their concentration influences binding events in vivo.
Our analysis of yeast that lacked Puf4p yielded dramatically different results. We performed RNA Tagging in a strain that expressed PUF5–PUP and lacked PUF4 (“Puf5p;puf4Δ”). Rather than expanding, the Puf5p subnetwork contracted in the absence of Puf4p to include only 466 U-tagged mRNAs rather than the 917 in wild-type cells (Supplemental Fig. S7A,B; Supplemental Data S6). We refer to these RNAs as the Puf5p;puf4Δ subnetwork. This subnetwork included only 50% of mRNAs (438) present in the wild-type Puf5p subnetwork (Fig. 6F), which were often detected in weaker classes (Fig. 6G). Retention of Puf5p targets in the Puf5p;puf4Δ subnetwork was highly correlated with the Puf5p target class (Fig. 6H). In the Puf5p;puf4Δ subnetwork, the Puf5p targets that were retained (438 RNAs) and lost (478 RNAs) were similarly enriched for 9BEs and 10BEs (Fig. 6I). However, the retained Puf5p targets were significantly more abundant than the lost Puf5p targets (Fisher–Pitman permutation test, P < 0.01) (Fig. 6J). The abundance of PUF5–PUP was unchanged in the presence or absence of PUF4 (Supplemental Fig. S5B).
Our RNA Tagging findings suggest that Puf5p primarily retained its “core” targets in the absence of Puf4p. To examine this further, we revisited our genetic analyses that assessed the growth of wild-type, puf4Δ, puf5Δ, and puf4Δpuf5Δ yeast strains in the presence of hydroxyurea, rapamycin, and MMS. While yeast that lack both Puf4p and Puf5p (puf4Δpuf5Δ yeast strain) displayed marked growth sensitivities in the presence of each compound, reintroduction of PUF5 on a single-copy plasmid into puf4Δpuf5Δ yeast completely restored growth on the compounds we tested (Supplemental Fig. S4D). Thus, our RNA Tagging and genetic data both suggest that Puf5p retains its core targets when Puf4p is absent. Retention of only core targets likely results from dilution of the limited quantity of Puf5p by the newly accessible Puf4p mRNA targets and binding sites. In the absence of Puf4p, very weak (and therefore undetectable) interactions with these RNAs ties up Puf5p and limits detectable binding to only the strongest and most abundant Puf5p targets (see Discussion).
DISCUSSION
Using RNA Tagging, we probed the determinants of a protein–RNA super-network to reveal principles that govern its architecture and plasticity. The U-tagging approach reveals RNAs that bind with varying efficiencies, and so distinguishes binding events that lead to biological control from those that result in transient interactions. We found that the architecture of the PUF super-network is largely governed by competition among PUF proteins for mRNAs. The outcome of the competition is dictated by their relative abundances and affinities for particular targets. These principles likely underlie other protein–RNA super-networks and provide a foundation for their analysis elsewhere in the RNA world. Indeed, families of RBPs with related binding specificities are common (Ray et al. 2013; Gerstberger et al. 2014), splicing factors compete to bind splice sites (Wang et al. 2012; Han et al. 2013; Pandit et al. 2013; Zarnack et al. 2013), and related RBPs bind many of the same mRNAs (Gerber et al. 2004; Hogan et al. 2008; Ascano et al. 2012; Porter et al. 2015; Prasad et al. 2016; Wilinski et al. 2017).
The canonical PUF super-network in yeast is composed of four major subnetworks. We demonstrated that Puf3p, Puf4p, and Puf5p each bind their own set of mRNAs, consistent with a previous study (Gerber et al. 2004). Importantly, our data also establish that Puf4p and Puf5p form a fourth subnetwork in the PUF super-network by binding many of the same mRNAs, which are often class I or II targets of both proteins. Furthermore, yeast that lack both Puf4p and Puf5p have enhanced phenotypes in comparison to yeast that lack either protein, and a recent report observed a similar effect on the destabilization of a single mRNA (Russo and Olivas 2015). The biological impetus for why some mRNAs are targeted by both Puf4p and Puf5p remains an open question, particularly since Puf4p effects only mRNA decay (Goldstrohm et al. 2006, 2007; Hook et al. 2007; Goldstrohm and Wickens 2008), while Puf5p effects both mRNA decay (Goldstrohm et al. 2006, 2007; Hook et al. 2007; Goldstrohm and Wickens 2008) and translational repression (Blewett and Goldstrohm 2012). Regardless, we suspect that Puf4p and Puf5p either bind to individual mRNA molecules sequentially or bind separate pools of mRNA molecules, since shared Puf4p–Puf5p mRNA targets most often possess a single high-affinity binding site. A dual-tagging experiment—in which one protein is fused to a PUP and the other to a different tagging enzyme (e.g., ADAR) (McMahon et al. 2016)—would provide insight into this question.
A balanced interplay between protein abundance, mRNA target abundance, and their binding affinities largely defines the architecture of the PUF super-network. Both Puf4p and Puf5p bind 9BEs with comparable, high affinity; yet the relatively high abundance of Puf4p occludes Puf5p from many mRNAs with 9BEs. To provide detectable access to Puf5p, the abundance of mRNAs with 9BEs would need to be relatively high. Indeed, mRNAs with 9BEs bound by both Puf4p and Puf5p were more abundant than those bound solely by Puf4p. High mRNA abundance also likely mediates Puf4p binding to mRNAs with weak Puf4p-binding sites (e.g., 10BEs), particularly among the mRNA targets it shares with Puf5p. In contrast, Puf5p occludes both Puf4p and Puf3p from mRNAs with 10BEs, very likely through its greater intrinsic affinity for the sequence, despite its relatively low abundance. The abundances of Puf3p and Puf4p are similar, yet both proteins have very distinct targets due to their inherent binding specificities.
Remarkably, removal of Puf5p expanded the network of Puf4p, while removal of Puf4p reduced that of Puf5p. Expansion of the Puf4p network in the absence of Puf5p supports a model in which the two compete with each other to bind mRNAs in vivo. The greater abundance of Puf4p relative to Puf5p (three- to ninefold) enables it to maintain nearly all of its targets in puf5Δ yeast while simultaneously gaining many of the best or most abundant Puf5p targets (Fig. 7). Puf4p also gained targets that were not included in the wild-type super-network, perhaps due to a slight increase in its abundance (Supplemental Fig. S5B). In parallel, we speculate that the striking contraction of Puf5p networks in the absence of Puf4p is due to its relatively low abundance. Removal of Puf4p substantially reduces the effective PUF protein concentration while simultaneously increasing the number of potential Puf5p-binding sites. This dramatically shifts the protein–RNA equilibrium of the PUF super-network. Thus, only mRNAs with high-affinity binding sites or relatively high mRNA abundances would be predicted to be bound by Puf5p at a detectable level (e.g., class I or II Puf5p targets). Indeed, our findings strongly support this model and are consistent with computational predictions (Jens and Rajewsky 2015). Alternatively, however, Puf4p could in principle stimulate Puf5p to bind its full cohort of mRNAs by direct protein–protein contacts or regulation of a regulatory factor, and vice versa.
FIGURE 7.

Proposed model for the expansion and contraction of subnetworks upon loss of a single protein. Green squares indicate Puf4p, and purple squares indicate Puf5p. Orange lines indicate a target with a high-affinity binding element, and gray lines indicate a target with a low-affinity binding element.
The architecture of the super-network provides robust opportunities for regulation and evolution. Changes in the abundance of individual proteins, their absolute affinities for a cognate site, and their relative affinities for different sites, all would rapidly switch the protein that controls a given set of functionally related RNAs. The relative cellular concentrations of the different PUF proteins in S. cerevisiae appear to vary with response to the cell cycle and metabolic state, as deduced from mRNA abundance studies (Kudlicki et al. 2007; Rowicka et al. 2007). A decrease in the RNA-binding activity of a given protein could also modulate the mRNAs bound by other proteins, including both the acquisition and loss of targets. Similarly, during evolution, the architecture of the network likely varies with the abundances of its components and their binding affinities. For example, nuclear-encoded mRNAs with functions in the mitochondria are bound by Puf3p in S. cerevisiae and by Puf5p in Neurospora crassa, due to small changes in the length of the binding sites present in orthologous RNAs (Hogan et al. 2015; Wilinski et al. 2017). It will be of interest to determine, first, how common changes in protein–RNA super-network architecture are, and second, how the drivers of those changes relate to the underlying principles we have seen in S. cerevisiae.
PUF–RNA networks are established through competition for related but divergent sites. The outcome of the competition, and thus the architecture of the PUF super-network, is determined by the balance between the levels of each of the proteins, the abundance of their targets, and the affinities of each RNA–protein interaction. These parameters provide powerful focal points for biological regulation and evolutionary change.
MATERIALS AND METHODS
Construction of yeast strains and expression plasmids
All strains were constructed in a BY4742 background (MATα his3Δ1 leu2Δ 0 lys2Δ0 ura3Δ0) as previously described (Lapointe et al. 2015). Briefly, we inserted the C. elegans pup-2 open-reading frame followed by a stop codon, the URA3 marker with its native promoter and terminator, and a 3-HA epitope tag in-frame at the 3′ end of PUF4 and PUF5. The mutant PHD1 strain that lacked Puf5p-binding elements is described in Lapointe et al. (2015). To construct the PUF4–PUP;puf5Δ (“Puf4p;puf5Δ”) and PUF5–PUP;puf4Δ (“Puf5p;puf4Δ”) RNA Tagging strains, we replaced the PUF5 and PUF4 open-reading frames, respectively, with the LEU2 marker (including its native promoter and terminator) in the appropriate RNA Tagging strain (PUF4–PUP and PUF5–PUP, respectively). To construct the puf4Δ and puf4Δ;puf5Δ yeast strain, we replaced the PUF4 open-reading frame with the LEU2 marker, including its native promoter and terminator in BY4742 and puf5Δ yeast strains, respectively. To construct puf5Δ yeast, we replaced the PUF5 open-reading frame with the HIS3 marker, including its native promoter and terminator in BY4742 yeast. URA3, LEU2, and HIS3 markers were amplified from p416tef, p415tef, and p413tef plasmids, respectively. PUF4 and PUF5 expression plasmids were constructed in a modified p416 background plasmid in which the plasmid promoter and terminators had been removed. We then inserted the PUF4 gene, including 798 upstream nucleotides (its promoter) and 457 downstream nucleotides (its 3′ UTR and terminator), and the PUF5 gene, including 1000 upstream nucleotides and 747 downstream nucleotides, into our modified p416 vector via SalI and KpnI restriction sites.
Yeast growth
Cultures were grown as previously described (Lapointe et al. 2015). Briefly, a single colony of each yeast strain was inoculated in 5 mL of yeast extract–peptone–dextrose plus adenine (YPAD) media and incubated at 30°C with 180 r.p.m. shaking for ≈24 h. Saturated cultures were used to seed 25 mL fresh YPAD at A660 ≈ 0.0002, which were grown at 30°C with 180 r.p.m. shaking until A660 ≈ 0.5–0.8. Yeast were transformed with p416-PUF4 and p416-PUF5 using standard techniques, and yeast that contained the plasmids were grown in synthetic URA3 dropout media containing dextrose (SD-URA3).
RNA Tagging library preparations
Total RNA isolations and sequencing library preparations were done as previously described (Lapointe et al. 2015).
High-throughput sequencing and raw data processing
Paired-end sequencing reads were obtained from Illumina sequencing platforms. FASTQ files were processed and aligned to the S. cerevisiae genome (version R64-1-1) as previously described (Lapointe et al. 2015).
Definition of U-tagged RNAs
As previously described (Lapointe et al. 2015), U-tagged RNAs are defined as DNA fragments that end with at least eight adenosines followed by at least one 3′ terminal thymidine (representing the U-tag) not encoded by any adapter or genomic sequence. Read 1 typically contained sequence that matched to particular genomic regions, which allowed identification of the gene. Read 2 most often identified the A–U tail sequence. The number of U-tagged RNAs per million uniquely mapped reads (TRPMs) for every gene was calculated and used to normalize data across samples.
Reproducible RNA Tagging targets
Targets of proteins were determined as previously described (Lapointe et al. 2015). Briefly, genes were called targets if they met three criteria: They were detected by at least 10-fold more TRPM in a tagging strain relative to a control nontagging strain (e.g., PUF4–PUP yeast versus BY4742); the number of TRPM detected must have been above the error rate for falsely detecting U-tagged RNAs (3%); and both of the previous criteria must have been met in all biological replicates.
Clustering analysis and class definition
TRPM values for each target were calculated for U-tags of at least 1, 2, 3, 4, 5, 6, 7, and 8 uridines in length. TRPM values were averaged (mean) across biological replicates. The order of targets was then randomized; all TRPM values were log2-transformed and separated into eight groups via k-means clustering (1000 iterations, Euclidean distance) using Gene Cluster 3.0 software. k-means groups were then sorted and ranked from longest to shortest U-tags. Heat maps were generated using MatLab (v2014a).
Classes were formed according to U-tag length. Class I targets were defined as the two groups (k-means ranked groups 1 and 2) of targets with the longest U-tags, typically including the majority of targets with U-tags up to seven or eight uridines in length. Class II targets were defined as the two groups (groups 3 and 4) with the next longest U-tags, typically including the majority of targets with U-tags up to five or six uridines. Class III was defined as groups 5 and 6, and class IV was defined as the two groups (groups 7 and 8) with the shortest U-tags, typically only one or two uridines in length.
Network map and GO analyses
The map of the PUF regulatory network was generated using Cytoscape (Shannon et al. 2003). Gene Ontology (GO) analyses were performed using YeastMine from the Saccharomyces Genome database (http://yeastmine.yeastgenome.org) using the default settings (Holm–Bonferroni correction).
Motif and directed motif analyses
Enriched sequence elements were identified using MEME as previously described (Lapointe et al. 2015). In all analyses, 3′ UTRs were defined as the longest observed isoform for a given gene (Xu et al. 2009), or 200 nt downstream from the stop codon if not previously defined. For directed PBE searches, perl regular expression searches were used to identify: 8BEs,TGTA[ATC][ATC]TA; 9BEs, TGTA[ATC][ATC][ATC]TA; 10BEs, TGTA[ATC][ATC][ATC][ATC]TA; 11BEs, TGTA[ATC][ATC][ATC][ATC][ATC]TA; and 12BEs, TGTA[ATC][ATC][ATC][ATC][ATC][ATC]TA.
Western blot analyses
Yeast strains were grown to A660 ≈ 0.5–0.7 and lysed by bead bashing in lysis buffer (Tris–HCl pH 6.8; 10% glycerol; 2% sodium dodecyl sulfate; 1.5% dithiothreitol; 0.1 mg/mL bromophenol blue). Approximately 0.3 OD of each sample was loaded onto a 4%–15% SDS–PAGE gel and transferred to a polyvinylidene difluoride (PVDF) membrane. In some instances, threefold serial dilutions were also analyzed, starting with lysate corresponding to 0.3 OD of sample. Membranes were cut in half and incubated with either mouse anti-HA.11 (clone 16B12) monoclonal antibody (Covance; MMS-101R) or mouse anti-actin (clone C4; MAB1501) monoclonal antibody overnight at 4°C using manufacturer recommended dilutions. Membranes were subsequently incubated with goat anti-mouse IgG Horseradish Peroxidase-labeled secondary antibody (KPL; 474-1806) for 1 h at room temperature. After washing with Tris-buffered saline (TBS: 20 mM Tris–HCl, pH 7.5; 150 mM NaCl) supplemented with 0.1% (v/v) tween-20, membranes were incubated with Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare) according to manufacturer's protocol. Membranes were imaged on an ImageQuant LAS 500.
RNA abundance analyses
The number of mRNA molecules present in a cell was estimated in the following way. We previously performed an RNA-seq experiment on a wild-type yeast strain (BY4742) (Lapointe et al. 2015) in which we obtained an FPKM value for every gene (FPKM, fragments per kilobase of transcript per million mapped reads). To obtain the estimated number of mRNAs present in a cell for each gene, we first summed the FPKM values for every gene (811,639 total). Next, we divided the total number of FPKM per gene by 36,000, which was an empirically determined estimate for the number of mRNA molecules present in a cell (Miura et al. 2008) and falls between two other empirically determined values (Holstege et al. 1998; Zenklusen et al. 2008). In each comparison, estimated mRNA molecules were log2-transformed, and median abundances of different groups were compared via two-tailed Student's t-tests and Fisher–Pitman permutation tests (two-sided, >10,000 repetitions). The number of Puf3p, Puf4p, and Puf5p protein molecules per cell was obtained from Kulak et al. (2014).
Mined data sets
Global changes in RNA stability for all genes in puf4Δ and puf5Δ mutants relative to wild-type yeast were obtained from Sun et al. (2013). Puf3p RNA Tagging data, and wild-type yeast RNA-seq data were recently published by our group (Lapointe et al. 2015). Puf4p SEQRS data were recently published by our group (Campbell et al. 2012). Protein subcellular localizations were obtained from Huh et al. (2003).
Yeast plate assays
Single colonies of the indicated deletion strains were grown to saturation in YPAD. Each culture was diluted to A660 ≈ 0.75, and then three 10-fold serial dilutions were made and plated on the indicated media. Plates were grown at 30°C and briefly removed to take pictures approximately every 12 h for 6 d. To select compounds to test, we systematically examined genes with mRNAs bound by both Puf4p and Puf5p for known sensitivities and focused on compounds that affected 15 or more shared Puf4p–Puf5p targets.
Conservation analysis
The 16 Saccharomycotina yeast species used to determine PBE conservation scores were chosen based on previously determined orthology (Wapinski et al. 2007). Sequences of 300 bases downstream from the translation termination codon were obtained from FungiDB (Stajich et al. 2012). Each 3′ UTR sequence was probed for putative binding elements using a custom Perl script, which is available upon request. The script determines log-likelihood scores for each k-mer (8–10 nt) based on canonical PUF-binding elements (Gerber et al. 2004; Lapointe et al. 2015; Wilinski et al. 2015). Each RNA Tagging target was assigned a “conservation score” defined as the number of orthologous genes with a PUF-binding element (positive log-likelihood value). A conservation score of 16 indicates the 9BE was present in all 16 budding yeasts, while a score of zero indicates a 9BE was absent from the 3′UTRs across all species that were analyzed.
DATA DEPOSITION
All raw sequencing data were deposited at the National Center for Biotechnology Information Sequence Read Archive with accession number PRJNA294241.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Wickens and Kimble laboratories for helpful comments and suggestions on experiments and data. We appreciate feedback on the manuscript from H. Medina, T. Hoang, B. Carrick, and C. Lopez-Anido. We thank J. Kimble (University of Wisconsin-Madison) for use of a computational server, and L. Vanderploeg of the Biochemistry Media Laboratory for help with the figures. Our research was supported by the US National Institutes of Health (GM50942) and by Wharton and Biochemistry Scholar fellowships to C.P.L.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.062687.117.
REFERENCES
- Ascano M Jr, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, et al. 2012. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 492: 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blewett NH, Goldstrohm AC. 2012. A eukaryotic translation initiation factor 4E-binding protein promotes mRNA decapping and is required for PUF repression. Mol Cell Biol 32: 4181–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell ZT, Bhimsaria D, Valley CT, Rodriguez-Martinez JA, Menichelli E, Williamson JR, Ansari AZ, Wickens M. 2012. Cooperativity in RNA–protein interactions: global analysis of RNA binding specificity. Cell Rep 1: 570–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Zheng W, Lin A, Uyhazi K, Zhao H, Lin H. 2012. Pumilio 1 suppresses multiple activators of p53 to safeguard spermatogenesis. Curr Biol 22: 420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP. 2008. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS One 3: e3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Herschlag D, Brown PO. 2004. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol 2: E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. 2006. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci 103: 4487–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. 2014. A census of human RNA-binding proteins. Nat Rev Genet 15: 829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm AC, Wickens M. 2008. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol 9: 337–344. [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Hook BA, Seay DJ, Wickens M. 2006. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol 13: 533–539. [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Seay DJ, Hook BA, Wickens M. 2007. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J Biol Chem 282: 109–114. [DOI] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Irimia M, Ross PJ, Sung HK, Alipanahi B, David L, Golipour A, Gabut M, Michael IP, Nachman EN, et al. 2013. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature 498: 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert AS, Richards AL, Bailey DJ, Ulbrich A, Coughlin EE, Westphall MS, Coon JJ. 2014. The one hour yeast proteome. Mol Cell Proteomics 13: 339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. 2008. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol 6: e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan GJ, Brown PO, Herschlag D. 2015. Evolutionary conservation and diversification of Puf RNA binding proteins and their mRNA targets. PLoS Biol 13: e1002307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728. [DOI] [PubMed] [Google Scholar]
- Hook BA, Goldstrohm AC, Seay DJ, Wickens M. 2007. Two yeast PUF proteins negatively regulate a single mRNA. J Biol Chem 282: 15430–15438. [DOI] [PubMed] [Google Scholar]
- Houshmandi SS, Olivas WM. 2005. Yeast Puf3 mutants reveal the complexity of Puf-RNA binding and identify a loop required for regulation of mRNA decay. RNA 11: 1655–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. [DOI] [PubMed] [Google Scholar]
- Jens M, Rajewsky N. 2015. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nat Rev Genet 16: 113–126. [DOI] [PubMed] [Google Scholar]
- Keene JD. 2007. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 8: 533–543. [DOI] [PubMed] [Google Scholar]
- Kershner AM, Kimble J. 2010. Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proc Natl Acad Sci 107: 3936–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlicki A, Rowicka M, Otwinowski Z. 2007. SCEPTRANS: an online tool for analyzing periodic transcription in yeast. Bioinformatics 23: 1559–1561. [DOI] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. 2014. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods 11: 319–324. [DOI] [PubMed] [Google Scholar]
- Lapointe CP, Wilinski D, Saunders HA, Wickens M. 2015. Protein-RNA networks revealed through covalent RNA marks. Nat Methods 12: 1163–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Ohn T, Chiang YC, Quigley G, Yao G, Liu Y, Denis CL. 2010. PUF3 acceleration of deadenylation in vivo can operate independently of CCR4 activity, possibly involving effects on the PAB1-mRNP structure. J Mol Biol 399: 562–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukong KE, Chang KW, Khandjian EW, Richard S. 2008. RNA-binding proteins in human genetic disease. Trends Genet 24: 416–425. [DOI] [PubMed] [Google Scholar]
- McMahon AC, Rahman R, Jin H, Shen JL, Fieldsend A, Luo W, Rosbash M. 2016. TRIBE: hijacking an RNA-editing enzyme to identify cell-specific targets of RNA-binding proteins. Cell 165: 742–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MT, Higgin JJ, Hall TM. 2008. Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p. Nat Struct Mol Biol 15: 397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura F, Kawaguchi N, Yoshida M, Uematsu C, Kito K, Sakaki Y, Ito T. 2008. Absolute quantification of the budding yeast transcriptome by means of competitive PCR between genomic and complementary DNAs. BMC Genomics 9: 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AR, Mukherjee N, Keene JD. 2008. Ribonomic analysis of human Pum1 reveals cis-trans conservation across species despite evolution of diverse mRNA target sets. Mol Cell Biol 28: 4093–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-McNicoll M, Neugebauer KM. 2013. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat Rev Genet 14: 275–287. [DOI] [PubMed] [Google Scholar]
- Olivas W, Parker R. 2000. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J 19: 6602–6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S, Zhou Y, Shiue L, Coutinho-Mansfield G, Li H, Qiu J, Huang J, Yeo GW, Ares M Jr, Fu XD. 2013. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol Cell 50: 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter DF, Koh YY, VanVeller B, Raines RT, Wickens M. 2015. Target selection by natural and redesigned PUF proteins. Proc Natl Acad Sci 112: 15868–15873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A, Porter DF, Kroll-Conner PL, Mohanty I, Ryan AR, Crittenden SL, Wickens M, Kimble J. 2016. The PUF binding landscape in metazoan germ cells. RNA 22: 1026–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenault T, Lithgow T, Traven A. 2011. PUF proteins: repression, activation and mRNA localization. Trends Cell Biol 21: 104–112. [DOI] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499: 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowicka M, Kudlicki A, Tu BP, Otwinowski Z. 2007. High-resolution timing of cell cycle-regulated gene expression. Proc Natl Acad Sci 104: 16892–16897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo J, Olivas WM. 2015. Conditional regulation of Puf1p, Puf4p, and Puf5p activity alters YHB1 mRNA stability for a rapid response to toxic nitric oxide stress in yeast. Mol Biol Cell 26: 1015–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Pratt G, Yeo GW, Moore MJ. 2015. The clothes make the mRNA: past and present trends in mRNP fashion. Annu Rev Biochem 84: 325–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassov DS, Jurecic R. 2003. The PUF family of RNA-binding proteins: does evolutionarily conserved structure equal conserved function? IUBMB Life 55: 359–366. [DOI] [PubMed] [Google Scholar]
- Stajich JE, Harris T, Brunk BP, Brestelli J, Fischer S, Harb OS, Kissinger JC, Li W, Nayak V, Pinney DF, et al. 2012. FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res 40: D675–D681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Schwalb B, Pirkl N, Maier KC, Schenk A, Failmezger H, Tresch A, Cramer P. 2013. Global analysis of eukaryotic mRNA degradation reveals Xrn1-dependent buffering of transcript levels. Mol Cell 52: 52–62. [DOI] [PubMed] [Google Scholar]
- Taylor JW, Berbee ML. 2006. Dating divergences in the Fungal Tree of Life: review and new analyses. Mycologia 98: 838–849. [DOI] [PubMed] [Google Scholar]
- Valley CT, Porter DF, Qiu C, Campbell ZT, Hall TM, Wickens M. 2012. Patterns and plasticity in RNA-protein interactions enable recruitment of multiple proteins through a single site. Proc Natl Acad Sci 109: 6054–6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Cody NA, Jog S, Biancolella M, Wang TT, Treacy DJ, Luo S, Schroth GP, Housman DE, Reddy S, et al. 2012. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell 150: 710–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A. 2007. Natural history and evolutionary principles of gene duplication in fungi. Nature 449: 54–61. [DOI] [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R. 2002. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet 18: 150–157. [DOI] [PubMed] [Google Scholar]
- Wilinski D, Qiu C, Lapointe CP, Nevil M, Campbell ZT, Tanaka Hall TM, Wickens M. 2015. RNA regulatory networks diversified through curvature of the PUF protein scaffold. Nat Commun 6: 8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilinski D, Buter N, Klocko A, Lapointe CP, Selker E, Gasch A, Wickens M. 2017. Recurrent rewiring and emergence of RNA regulatory networks. Proc Natl Acad Sci 114: E2816–E2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnack K, Konig J, Tajnik M, Martincorena I, Eustermann S, Stevant I, Reyes A, Anders S, Luscombe NM, Ule J. 2013. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell 152: 453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, Singer RH. 2008. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol 15: 1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Stumpf CR, Krahn JM, Wickens M, Hall TM. 2009. A 5′ cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs. Proc Natl Acad Sci 106: 20192–20197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.