Abstract
The moose (Alces alces) is a ruminant that harvests energy from fiber-rich lignocellulose material through carbohydrate-active enzymes (CAZymes) produced by its rumen microbes. We applied shotgun metagenomics to rumen contents from six moose to obtain insights into this microbiome. Following binning, 99 metagenome-assembled genomes (MAGs) belonging to 11 prokaryotic phyla were reconstructed and characterized based on phylogeny and CAZyme profile. The taxonomy of these MAGs reflected the overall composition of the metagenome, with dominance of the phyla Bacteroidetes and Firmicutes. Unlike in other ruminants, Spirochaetes constituted a significant proportion of the community and our analyses indicate that the corresponding strains are primarily pectin digesters. Pectin-degrading genes were also common in MAGs of Ruminococcus, Fibrobacteres and Bacteroidetes and were overall overrepresented in the moose microbiome compared with other ruminants. Phylogenomic analyses revealed several clades within the Bacteriodetes without previously characterized genomes. Several of these MAGs encoded a large numbers of dockerins, a module usually associated with cellulosomes. The Bacteroidetes dockerins were often linked to CAZymes and sometimes encoded inside polysaccharide utilization loci, which has never been reported before. The almost 100 CAZyme-annotated genomes reconstructed in this study provide an in-depth view of an efficient lignocellulose-degrading microbiome and prospects for developing enzyme technology for biorefineries.
Introduction
Improved processing of lignocellulose from plant biomass is considered a promising avenue for production of alternative biofuels, green chemicals and biomaterials. Lignocellulose is a highly abundant renewable resource that can be obtained from agricultural and forestry residues and energy crops, as well as from industrial and household wastes. In forest-rich countries, biofuel from forest residues could contribute substantially to fuel supplies. A key process open to improvement is the enzymatic hydrolysis of lignocellulose into fermentable sugars. By enhancing enzyme activities and tailoring enzyme cocktails for complete breakdown of lignocelluloses, the cost of this rate-limiting step could be lowered substantially (Mohanram et al., 2013; Goldbeck et al., 2014).
The Carbohydrate-Active enZyme database (CAZy) organizes enzymes required for lignocellulose polysaccharide degradation into sequence-based families covering large classes, in particular glycoside hydrolases (GHs), polysaccharide lyases (PLs) and carbohydrate esterases (CEs) (Lombard et al., 2014). The action of these enzymes is further enhanced in nature by anchoring a carbohydrate-binding module (CBM) to the catalytic module. Many microbes living in plant biomass-degrading environments such as soil, composts and herbivore digestive tracts express large pools of carbohydrate-active enzymes (CAZymes) for lignocellulose conversion.
Ruminants (for example, cattle, sheep, goats, cervids) have a vast enlargement of their gastrointestinal tract, known as the forestomachs, which are comprised of the rumen, reticulum and omasum. The rumen contains large amounts of anaerobic microorganisms (bacteria, protists and fungi) and functions as a highly efficient bioreactor for lignocellulose conversion. The microbial community composition varies with diet and host, but a core rumen microbiome is found that include Prevotella, Butyrivibrio, Ruminococcus, Bacteriodales and Clostridiales (Henderson et al., 2015). The rumen microbes ferment feed to form volatile fatty acids that are the major energy source for all ruminants.
Different bacterial taxa utilize different cellulose-degradative strategies in the ruminal ecosystem (Flint, 2008). The firmicute Ruminococcus flavefaciens uses the cellulosome, an extracellular multienzyme complex. The typical cellulosome architecture is composed of dockerin-bearing enzymes that bind the multiple cohesin domains found on a scaffoldin subunit (Bayer et al., 2004). The latter is attached to the cell-surface, exposing the catalytic domains to the substrate. The efficient cellulolytic machinery of the Gram-negative bacterium Fibrobacter succinogenes represents a less understood system for cellulose depolymerization, involving attachment to the substrate by a yet unknown protein followed by cleavage of the polymer by a distinct set of cellulases (Wilson, 2009; Suen et al., 2011b; Ransom-Jones et al., 2014). Recent enzyme characterization efforts have also shed light on cellulolytic functions in species from the Bacteroidetes phylum (Naas et al., 2014; Mackenzie et al., 2015). Within the Bacteroidetes, including the rumen-based Prevotella ruminicola, CAZymes are organized in physically linked gene clusters that encode the entire functional chain involving binding, degrading and importing defined glycan structures. These gene clusters are referred to as polysaccharide utilization loci (PULs) (Bjursell et al., 2006; Terrapon and Henrissat, 2014). A PUL can be dedicated to breakdown of a specific polysaccharide, such as mannan or xylan (Cuskin et al., 2015; Rogowski et al., 2015).
Genome sequencing of the isolated rumen species F. succinogenes and R. albus has successfully revealed sequence-encoding enzymes for the breakdown of plant material (Brumm et al., 2011; Suen et al., 2011a, b). However, a vast majority of rumen microbes has not been successfully isolated in pure cultures and therefore not yet genetically characterized. Shotgun metagenomics circumvents the limitation of culturing and has been used to reveal a large number of genes coding for CAZymes in the guts of animals and insects (Warnecke et al., 2007; Duan and Feng, 2010; Pope et al., 2010, 2012; Hess et al., 2011; Muegge et al., 2011; Patel et al., 2014). Furthermore, shotgun metagenomics in combination with binning methods enable reconstruction of nearly complete single genomes that could be explored for new lignocellulose-degrading CAZymes (Herlemann et al., 2013; Kishi et al., 2015).
The moose (Alces alces) is the largest browsing wild ruminant of Northern Europe (referred to as the ‘King of the Forest’ in Nordic countries) and according to physiological and anatomical characteristics of the rumen, classified as a ‘concentrate selector’ (Hofmann, 1989). The moose diet varies with season but is in winter dominated by woody plants where Scots pine (Pinus sylvestris) ranks as the quantitatively most important during the coldest season (Cederlund et al., 1980; Bergström and Hjeljord, 1987). Pine residues found in the rumen include needles, twigs and bark. During the rest of the year, the diet is complex and is composed mainly of leaves from deciduous trees such as rowan (Sorbus aucuparia), aspen (Populus tremula), willows (Salix spp.) and birches (Betula spp.). Also shrubs and herbaceous plants, for example, heather (Calluna vulgaris), European blueberry (Vaccinium myrtillus) and fireweed (Chamaenerion angustifolium), are important components of the diet. Moreover, small quantities of grasses, ferns, mosses and fungi are consumed over the year. Consequently, microbial communities of the moose rumen are expected to be able to process cellulose, pectins, arabinogalactan and different types of hemicelluloses found in diverse plant families such as galactomannan, glucomannan, galactoglucomannan, arabinoglucuronoxylan, glucuronoxylan, xyloglucan and mixed-linkage glucan. Although it has been shown that the moose rumen harbors bacterial families that include fibrolytic species (Ishaq and Wright, 2012, 2014), the actual CAZyme content of the moose rumen remains to be explored. The fact that the moose manages to utilize energy from fiber-rich twigs and branches of pine and birch is critical for its survival in harsh winter conditions. It also motivates the detailed exploration of its rumen microbiome, as presented in this study.
Materials and methods
Sampling, DNA extraction and sequencing
Rumen content was collected during October 2011 from six individual moose. The moose were shot by licensed hunters during Swedish hunting season. No animals were killed for the purpose of this study. Rumen content was sampled postmortem, for which no ethical permission is required under Swedish law. Immediately after sampling, the material was put in a coolbox and frozen within 7 h at −80 °C. Subsamples were taken for sequencing after thawing and then shipped (<24 h transport) on dry ice and then stored again at −80 °C. Three moose (103, 104 and 105) were from a densely forested area near Växjö in southern Sweden and three (101B, 102 and 106) from the island Öland dominated by farmland where forests are scarce. The average temperatures in October 2011 were for Växjö and Öland 7.7 and 8.1 °C, respectively, according to data from the Swedish Meteorological and Hydrological Institute. All moose were of adult age and all except 104 and 105 were male. Microbial DNA was extracted from the rumen samples based on a protocol adapted from Roume et al. (2013). Briefly, for each extraction, 200 mg±10% frozen moose rumen samples were subsampled and homogenized by cryomilling in an Oscillating Mill MM 400 (Retsch, Haan, Germany) for 2 min at 30 Hz using two 7 mm stainless steel milling balls. For cell lysis, 400 μl of 1:2 (v/v) methanol:chloroform solution was added to the frozen homogenized samples and subsequently cryomilled (Oscillating Mill MM 400) for 2 min at 20 Hz using five additional 1 mm stainless steel milling balls. The cryolysis procedure ensures indiscriminate lysis of cells in the sample (Roume et al., 2013). The lysed sample was centrifuged for 5 min at 10 000 g at 4 °C. Following centrifugation, polar and non-polar phases were removed and the interphase pellet (along with the steel milling balls) was kept on ice for the sequential extraction of total RNA (enriched in large RNA), genomic DNA and protein sequential isolations using the Qiagen AllPrep DNA/RNA/Protein Mini kit-based method (QA, Qiagen, Venlo, The Netherlands). Purified, high molecular weight DNA extracts were obtained and sequenced on Illumina’s (San Diego, CA, USA) HiSeq system (2 × 100 bp reads). For sample 102, an additional sequencing run was made owing to an initial low yield.
Taxonomic profiling using rRNA reads
Reads from small subunit ribosomal RNA genes were extracted from subsets of each sample. These subsets contained the same number of reads and were produced by randomly picking 25 680 424 sequencing read pairs from each sample. These were subsequently classified with Metaxa2 using the default cutoff score (Bengtsson-Palme et al., 2015).
Assembly, binning and genome annotation
After quality filtering using Sickle (Joshi and Fass, 2011), each sample was de novo assembled separately with Ray (Boisvert et al., 2010) using different values for k (=41, 51, 61, 71, 81). To form one assembly per sample, Newbler (Margulies et al., 2005) was used in the same manner as described in Hugerth et al. (2015). The sequencing reads and genome assemblies have been deposited at the European Nucleotide Archive (www.ebi.ac.uk) under accession number PRJEB12797. Binning was performed using CONCOCT (Alneberg et al., 2014) on each sample individually but using contig coverage information obtained by mapping reads from each sample, and putative genomes, where at least 31 out of the 36 single copy genes were present and maximum 2 genes were present in >2 copies, were considered as metagenome-assembled genomes (MAGs) (Hugerth et al., 2015). Annotation was made using PROKKA (Seemann, 2014). Completeness was further assessed using CheckM (Parks et al., 2015). The abundances of MAGs were estimated by calculating the average coverage depth over the assembled genome and normalizing with the number of million read pairs in each sample, respectively. The resulting number then corresponds to coverage depth per million read pairs in the sample.
Phylogenomic analysis
Taxonomy of MAGs was assigned in a combination of 16S rRNA analysis and by whole-genome characterization. First, phylogeny for MAGs was set from the taxonomic ranking table and by viewing the phylogenomic tree both produced by Phylophlan (Segata et al., 2013). The tree was visualized using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). For MAGs that did not cluster with a reference genome, 16S rRNA sequences were extracted using RNAmmer (Lagesen et al., 2007) and blast searched (Altschul et al., 1990) and annotated with SINA/SILVA (Preuse et al., 2012). Related genomes (from isolates, single cells and metagenomes) were then added and the Phylophlan tree reconstruction was repeated. Phylogenomic clusters were produced by amino-acid comparisons (http://enve-omics.ce.gatech.edu/aai/), and clades were set based on >60% amino-acid identity in >50 genes. Members within the same clusters were considered to be of same genus (Rodriguez-R and Konstantinos, 2014). All MAGs that belonged to Spirochaetes were considered as Treponema on the basis on their phylogenomic positions even though they did not fulfill the >60% amino-acid criterion. Taxonomy was also assessed for all contigs using DIAMOND (Buchfink et al., 2015) to search protein sequences against NCBI’s non-redundant protein database and evaluated with MEGAN5 (Huson et al., 2011). The taxonomic annotation for each contig was given by the lowest common ancestor shared by at least 90% of the assigned genes on that contig.
CAZyme annotation and PUL prediction
CAZy module assignments were produced by first comparing queries to the full-length sequences of the CAZy database using BLAST (Altschul et al., 1990). Query sequences that did not obtain an e-value<10−6 were discarded. Retained sequences that aligned over their entire length with a protein in the database with >50% identity were directly assigned to the same family as the subject sequence. The remaining retained sequences were subjected in parallel to (i) a BLAST search against a library built with partial sequences corresponding to individual GH, PL, CE and CBM modules and (ii) a HMMER2 search using hidden Markov models built for each CAZy module family, allowing both family assignment and a view of CAZyme modularity. The CAZymes of each MAG were separately validated by human curation. Modules on open reading frames truncated by contig ends were included in the analysis. PULs were predicted in the 99 MAGs and to cope with possibly fragmented loci, a procedure less stringent than that used for PULDB (Terrapon et al., 2015) was used by starting from each susD gene and without the requirement of the presence of susC. The bovine (Hess et al., 2011) and the reindeer (Pope et al., 2012) rumen metagenomes were downloaded and analyzed in the same way as the moose data. Contigs (from the original article) were analyzed for the bovine metagenome, while shotgun 454 reads were analyzed for the reindeer metagenome, as the low coverage of this data set precluded significant assembly (Pope et al., 2012). As a consequence, no PUL analysis was conducted on the reindeer data. Fisher’s exact test was employed to test for overrepresentation/under-representation of a CAZy in one microbiome vs another. P-values were false-discovery rate adjusted to account for multiple testing.
Results and Discussion
Bacteroidetes, Firmicutes and Spirochaetes are abundant in the moose rumen microbiome
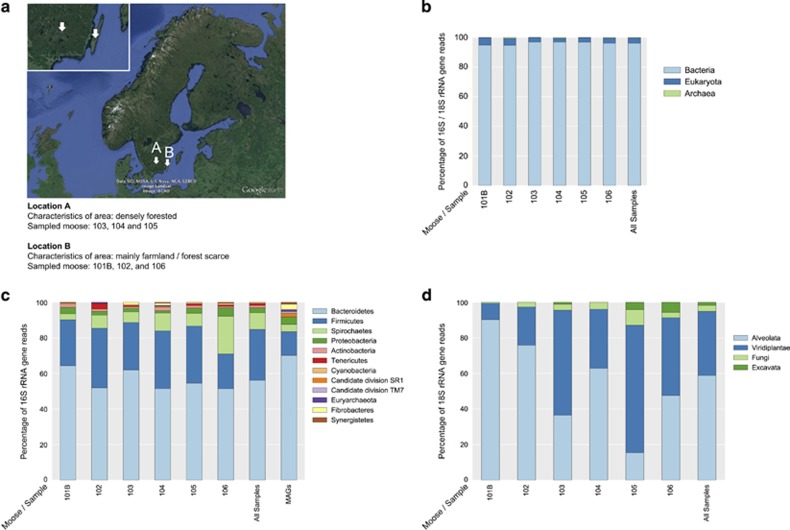
The rumen contents from six moose killed during the annual moose hunt in Sweden were subjected to DNA extraction and shotgun metagenomic sequencing. Samples 103, 104 and 105 corresponds to moose from forest-rich habitats, whereas 101B, 102 and 106 were from locations consisting of mainly farmland (Figure 1a). The yield of read-pairs was on average 83 million per sample (range: 45–139). Overall taxonomic composition was assessed by characterizing rRNA gene-encoding reads extracted from the data set (Figures 1b–d). Of the classified reads, on average 0.35% were of archaeal, 3.62% of eukaryotic and 96.03% of bacterial origin. Fungi constituted a minor fraction (3.41%) of the eukaryotic sequences, plants an intermediate (36.14%) and protists the dominating part (60.44%) (Figure 1d). The vast majority of protists in the present study were ciliates of superphylum Alveolata, with Entodinium being the dominating genus among reads classified to this level. This is in agreement with a recent investigation of eukaryotic diversity in moose, where Entodinium dominated the protists not only in Norwegian moose but also in an individual moose from Alaska (Ishaq et al., 2015). In general, the rumen is dominated by bacteria in terms of cell numbers, as observed in this study; however, protists may reach 50% of the biomass and be important for plant matter degradation (Weimer, 2013; Wright, 2015). One gene in the metagenome, predicted on a contig that was manually checked for assembly errors, encoded a protein of protist origin with an unexpected high number of eight GH5_4 modules. Based on BLAST searches, there was no protein matching the entire open reading frame; instead different regions aligned to different peptides of the ciliate Polyplastron multivesiculatum with approximate amino-acid identities of 55%.
Figure 1.
Locations where samples were collected and area characteristics (a). Overall taxonomic composition based on 16S and 18S rRNA reads (b). Prokaryotic taxonomic composition based on 16S rRNA reads and MAG phylogeny (c). Eukaryotic taxonomic composition based on 18S rRNA reads (d).
The prokaryotic communities were dominated by the phyla Bacteroidetes (in total 56.04% of the 16S rRNA reads), Firmicutes (28.71%) and Spirochaetes (9.41%) (Figure 1c). At considerably lower abundance, taxa belonging to Proteobacteria (2.83%) and Actinobacteria (1.32%) were observed, followed by Tenericutes (1.06%) and Cyanobacteria (0.62%). We did not find any correlation between the habitats of the moose (forest rich vs forest scarce/farmland) and the taxonomic profiles; however, variation between samples was apparent (Figures 1c and d). The dominance of Bacteroidetes and Firmicutes agrees with an earlier study of North American moose rumen prokaryotic diversity (Ishaq and Wright, 2014) and is a general characteristic of mammalian gut microbiomes (Ley et al., 2008). Interestingly, Spirochaetes was the third most common phylum in the present investigation, even outnumbering the Firmicutes in moose 106 whose habitat has typical farmland characteristics. This contrasts to other rumen studies that generally determine spirochetes at levels around or below 1% (Pope et al., 2010, 2012; Wallace et al., 2015). The majority (78%) of the spirochete reads were classified as genus Treponema. Treponemes dominate the biomass-degrading environment of the hindgut of termites (Warnecke et al., 2007). The observed low abundance of the phyla Proteobacteria, Actinobacteria and Synergistetes agrees well with results from a previous moose rumen study investigating samples from North America and Norway (Ishaq and Wright, 2014).
The moose rumen microbiome feature a wide range of CAZymes
In total, 55 800 GHs, 3409 PLs, 5320 CEs and 5793 CBMs were identified in the moose rumen metagenome contigs (see Supplementary Table S1 for assembly statistics and Supplementary Table S2 for complete list of CAZymes). The 10 most frequently occurring GH families (Table 1) included many families involved in plant carbohydrate deconstruction, such as those containing starch-degrading enzymes (GH13, GH77 and GH97), pectin-depolymerizing hydrolases (GH28), pectin- and arabinoxylan-degradation enzymes (GH43 and GH51), and family GH5, which contains mainly cellulases and mannanases. Enzymes in the abundant families GH2 and GH3 act as accessory enzymes not only to complete plant polysaccharide digestion but may also be involved in the degradation of carbohydrates originating from microbes in the rumen. GH92 was the most abundant GH family (Table 1 and Supplementary Table S2) not involved in the breakdown of plant material. It consists of α-mannosidases suggesting a role in host glycan utilization or degradation of fungal carbohydrates. A number of families that have relatively low representation in the CAZy database were present, such as GH97, GH98, GH106, GH123, GH124 and GH128 (Table 1).
Table 1. Numbers of selected GH modules predicted in the total metagenome and in the MAGs, as well as their representation in the CAZy database at the time of submission.
|
Number of GH modules |
|||
|---|---|---|---|
| Total metagenome | MAGs | CAZy | |
| Most common GH families | |||
| GH13 | 5992 | 508 | 32 607 |
| GH43 | 4965 | 641 | 5425 |
| GH2 | 3776 | 488 | 6657 |
| GH3 | 3378 | 353 | 10 457 |
| GH97 | 2310 | 236 | 582 |
| GH5 | 2147 | 216 | 7129 |
| GH28 | 1903 | 236 | 3781 |
| GH92 | 1731 | 171 | 1801 |
| GH51 | 1573 | 202 | 1549 |
| GH77 | 1400 | 109 | 3459 |
| GH families involved in cellulose degradation | |||
| GH5 | 2147 | 216 | 7129 |
| GH9 | 459 | 70 | 2026 |
| GH44 | 30 | 3 | 109 |
| GH45 | 49 | 9 | 357 |
| GH48 | 7 | 2 | 765 |
| GH families with low representation in the CAZy database | |||
| GH97 | 2310 | 236 | 582 |
| GH98 | 203 | 19 | 83 |
| GH106 | 1171 | 107 | 188 |
| GH123 | 47 | 7 | 83 |
| GH124 | 12 | 1 | 4 |
| GH128 | 190 | 46 | 182 |
| Total GHs | |||
| 55 800 | 6283 | 361 691 | |
| GH5 subfamilies | |||
| GH5_4 | 1097 | 88 | 160 |
| GH5_7 | 239 | 33 | 133 |
| GH5_2 | 223 | 33 | 245 |
| GH5_46 | 86 | 12 | 24 |
| GH5_44 | 81 | 1 | 15 |
| GH5_38 | 70 | 4 | 10 |
| GH5_21 | 69 | 15 | 10 |
| GH5_37 | 51 | 1 | 19 |
| GH5_1 | 33 | 4 | 133 |
| GH5_13 | 29 | 6 | 59 |
| GH5_27 | 22 | 0 | 5 |
| GH5_8 | 16 | 0 | 71 |
| GH5_36 | 12 | 0 | 23 |
| GH5_22 | 7 | 0 | 12 |
| GH5_52 | 6 | 2 | 6 |
| GH5_10 | 5 | 3 | 19 |
| GH5_28 | 1 | 0 | 9 |
| GH5_25 | 1 | 0 | 16 |
| GH5_45 | 1 | 0 | 5 |
| GH5_42 | 1 | 0 | 42 |
| GH5_12 | 1 | 0 | 8 |
Abbreviations: CAZy, Carbohydrate-Active enZyme; GH, glycoside hydrolase; MAG, metagenome-assembled genome.
Various non-GH CAZymes involved in the degradation of polysaccharides were also found. For the PL class, the most prevalent families were PL1, PL10 and PL11 (Supplementary Table S2). This suggests the presence of many microbial strains capable of pectin degradation. Members of 13 out of the 16 defined CE families were found (Supplementary Table S2). CE1, CE12 and CE8 all contain deacetylases targeting plant carbohydrates. CBM50 was the most populated CBM family, followed by CBM48, CBM6, CBM32 and CBM20 (Supplementary Table S2). Modular enzymes with CBM50 connected to catalytic modules of either GH23 or GH73 were frequently observed confirming their involvement in peptidoglycan degradation. No auxiliary redox enzymes (AAs) were present in the metagenome, as expected in an anaerobic environment such as the moose rumen.
For comparison, we reanalyzed the metagenome data of two other well-sampled rumen microbiomes: the bovine (Hess et al., 2011) and the reindeer (Pope et al., 2012) microbiomes. These showed overall high correlations in CAZyme content to the moose (Supplementary Figure S1). Among the CAZyme families displaying highest differences in representation, primarily genes encoding pectin-degrading enzymes were observed (Supplementary Figure S1; Supplementary Table S2). Families PL1, PL11, CE8 and CE12 were highly enriched (Fisher’s exact test, false-discovery rate adjusted P<10−40) in the moose rumen compared with the bovine rumen, and these were also more abundant in the moose rumen compared with the reindeer rumen, although not significantly so due to the much lower sequencing depth of the reindeer microbiome. Family GH98, recently shown to include endoxylanases involved in degradation of complex xylans (Rogowski et al., 2015), was also enriched in the moose rumen compared with the other data sets. Interestingly, another xylanase-containing family, GH8, was more frequent in the bovine and reindeer rumens, and GH11 was more frequent in the bovine rumen. These observations suggest that the moose rumen microbiome is more specialized in degrading pectin-rich diets and tailored toward a different xylan composition compared with the bovine and reindeer microbiomes.
Ninety-nine genomes from 10 prokaryotic phyla were reconstructed
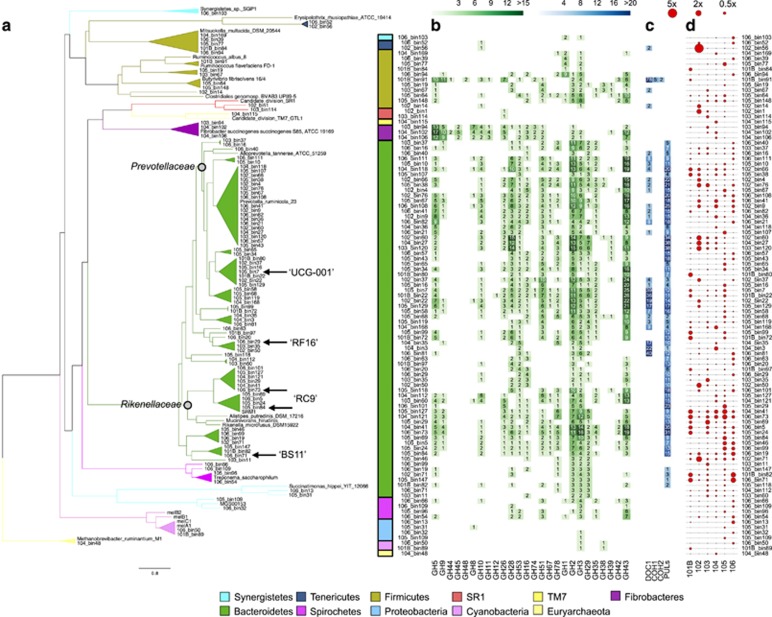
In order to reconstruct genomes of individual strains from the shotgun metagenomic data, contigs for each sample were binned using CONCOCT (Alneberg et al., 2014) as previously described (Hugerth et al., 2015). Based on the presence of single-copy genes, 99 genome bins were approved as MAGs. The average estimated completeness of these MAGs was 90% and average contamination was estimated to be 1.7% (Supplementary Table S3 gives detailed information on each MAG). According to the assessment framework proposed by Parks et al. (2015), 64 of the MAGs were near complete (⩾90% completeness), 31 were substantially complete (⩾70%) and the remaining 4 were moderately complete (⩾50%). Of the four moderately complete, three belong to the candidate divisions TM7 and SR1 with few sequenced genomes, and completeness in these MAGs may therefore have been underestimated; average N50 length values were actually higher in this group (43 131 bp) as compared with the average of all MAGs (15 206 bp). Ninety-three out of the 99 MAGs displayed low contamination levels according to the criteria (⩽5%) by Parks et al. (2015). Phylogenetic reconstruction of the 99 MAGs affiliated 68 to the Bacteroidetes, 12 to the Firmicutes, 4 to the Spirochaetes, 4 to the Proteobacteria, 3 to the Fibrobacteres, 2 to the non-photosynthetic Cyanobacteria proposed as Melainabacteria, 2 to the Tenericutes and 2 to the candidate phylum ‘SR1’. Synergistetes, candidate phylum ‘TM7’ and the archaeal phylum Euryarchaeota were represented by singletons (Figure 2a). As this study was focused on prokaryotic genome analyses, the evaluation criteria of the bins were optimized for prokaryotes, which may explain the lack of identified reconstructed eukaryotic genomes. The overall MAG diversity resembled that obtained by 16S rRNA profiling (Figure 1c) which suggests a rather unbiased binning procedure and that the reconstructed genomes are representatives for the moose rumen prokaryotic community. However, no high-quality genomes belonging to the Actinobacteria were assembled, while multiple contigs from this phylum were present in the assembly (data not shown). Based on the average amino-acid identity of the encoded proteins, the genomes were grouped into clades corresponding to approximately the genus level (Rodriguez-R and Konstantinos, 2014) (Figure 2). Of the resulting 40 clades (including singletons), 30 lacked a sequenced reference genome. The reconstructed genomes of these clades form the basis for revealing their metabolic potential in the rumen microbiome.
Figure 2.
Phylogeny and CAZome profiles of MAGs. A phylogenomic tree of the 99 moose rumen MAGs (a). Triangles represent genus-level clades based on average amino-acid identity levels of encoded proteins. For Bacteroidetes, family Prevotellaceae and Rikenellaceae are indicated, as well as classifications based on 16S genes for selected MAGs. Included reference genomes are from isolates except the Cyanobacteria/Melainabacteria genomes (Di Rienzi et al., 2013), the MGS00153 Alphaproteobacteria genome (Laczny et al., 2016) and the SRM-1 Bacteroidetes genome (Pope et al., 2012) that were assembled from metagenomes and the SR1 (Campbell et al., 2013) and TM7 genomes (Podar et al., 2007) that were sequenced from single cells. Heatmaps showing counts of selected GHs involved in plant degradation (b) and of cohesins (COHs), dockerins (DOCs) and polysaccharide utilizing loci (PULs) (c) in the MAGs. Relative abundances of the MAGs in the different moose samples (d), expressed as average coverage depth per million read pairs.
The 10 most frequently occurring GH families in the MAGs were also the most common in the metagenome data set overall (Table 1), again indicating that the collection of MAGs are representative of the moose rumen prokaryotic community. In the following sections, we will describe the functional potential of the different groups of MAGs, with emphasis on carbohydrate-degrading capabilities.
Genomes of several novel strains of well-studied fibrolytic rumen species
Three MAGs clustered closely with F. succinogenes strain S85 (Figure 2), a fibrolytic bovine rumen species (Montgomery et al., 1988). Genomic analyses have revealed specialization in the use of cellulose as the sole energy source (Suen et al., 2011b) and several cellulases have been characterized in this species (Brumm et al., 2011). These MAGs may therefore be important for the deconstruction of cellulose in moose rumen. The average number of 16S rRNA encoding reads from phylum Fibrobacteres was low (Figure 1c), which could explain why the presence of Fibrobacteres has not been previously highlighted for moose rumen (Ishaq and Wright, 2012, 2014). The Fibrobacter MAGs contained the highest numbers of putative cellulase genes of the moose rumen MAGs. These were distributed in families GH5, GH9, GH44 and GH45 (Figure 2b) and additionally GH8, GH74 and GH94 (Supplementary Table S3). One GH45 gene product in F. succinogenes S85 has been experimentally verified to be an endoglucanase (Seon Park et al., 2007). Nine GH45 modules were identified in the MAGs of moose rumen and these were restricted to the Fibrobacter-like ones. As a comparison, 19 bacterial GH45 sequences were present in the CAZy database at the time of submission. The GH5 sequences in the Fibrobacter-like MAGs were classified into subfamilies GH5_2, GH5_4, GH5_10 and GH5_37 (Supplementary Table S3). Most of these are probably targeting cellulose, whereas enzymes in subfamily GH5_10 are reported as β-mannanases (Aspeborg et al., 2012). The GH5_37 coding gene was restricted to one of the Fibrobacter MAGs. This is a subfamily actually not present in the F. succinogenes S85 genome. Ten GH5 members found in the Fibrobacteres-like MAGs could not be assigned to any subfamily and may represent enzymes with new substrate or product specificities. F. succinogenes is also known as a major pectin fermenter in the rumen (Sun et al., 2008; Puniya et al., 2015). Accordingly, a considerable number of GH, PL and CE sequences associated with pectin degradation was found in the moose Fibrobacter MAGs. Xylanases (GH10, GH11) and β-mannanases of family GH26 were also observed, suggesting that the Fibrobacter MAGs are equipped with a portfolio of enzymes involved in the hydrolysis of most plant cell wall polysaccharides of woody tissues.
Various Ruminococcus species of the Firmicutes phylum have been identified as primary degraders of plant material in the rumen of cow (Flint, 2008). One MAG clustered with the fibrolytic and cellulosome-producing species R. flavefaciens (Kirby et al., 1997; Ding et al., 2001; Berg Miller et al., 2009; Figure 2a). The numerous identified cohesin and dockerin modules in this MAG (Figure 2c) suggest that this strain is a typical cellulosomal bacterium belonging to the Clostridiales family. Both cohesins of type I and II were present and dockerin modules were found in combination with various CAZymes and CBMs (Supplementary Table S3). Recently, comparative genomics between several strains of R. flavefaciens and R. albus identified species-specific cellulosome architectures (Dassa et al., 2014). The inventory of cellulases (GH5 and GH9) and dockerin modules in the Ruminococcus MAG exhibited higher levels of homology to R. flavefaciens than R. albus. The GH5 members in the Ruminococcus MAG were classified into subfamilies GH5_1 and GH5_4 (Supplementary Table S3) and it was the only genome in the moose rumen data set containing GH48 modules (Figure 2). Clostridiales GH48 enzymes have been biochemically characterized as either endoglucanases or cellobiohydrolases (Kirby et al., 1997; Ding et al., 2001). Another putative endoglucanase gene was identified that belonged to the small GH124 family. The predicted GH124 enzyme is a dockerin-containing protein and thus a putative cellulosome component. The only characterized GH124 gene product has been suggested to target the interface between crystalline and amorphous cellulose and was shown to act in synergy with the major cellulosome exo-acting cellulase from GH48 (Brás et al., 2011). Likewise, there may be a synergy between GH124 and GH48 enzymes in the moose rumen Ruminococcus MAG. Moreover, an expansin module connected to a predicted cellulose-binding CBM63 was present in the MAG, suggesting a role in cellulose degradation. Further analysis of the Rumincococcus MAG revealed that CBM13 was frequently present in modular CAZymes in combination with catalytic modules putatively involved in pectin degradation, including families PL1, PL11, GH43, GH53 and CE12. Identified members of GH42, GH51, GH95, GH105 and PL9 may also be involved in deconstructing pectins, hence it appears that the sequenced moose rumen Ruminococcus species has high capacity for liberation of sugars from pectins. Although xylanases were under-represented in the MAG, both xylanase and β-mannanase genes were predicted together with dockerins implying that hemicellulases are part of the cellulosome complex in this Ruminococcus bacterium. The observed CAZyme repertoire of the Ruminococcus-like MAG suggests a bacterium closely related to R. flavefaciens but with unique features that may reflect adaptation to the moose diet.
Novel diversity and presence of dockerins in Bacteroidetes
Phylogenomic analyses of the 68 Bacteroidetes MAGs revealed extensive diversity; 20 genus-level clades were formed. Seven of these grouped with reference genomes of family Prevotellaceae, two with family Rikenellaceae, while the other 12 were Bacteroidales that could not be determined to the family level (Figure 2a, Supplementary Figure S2 for a more detailed tree and Supplementary Table S3 for detailed taxonomy). Among Prevotellaceae MAGs, P. ruminicola strain 23 was grouped inside one genus-level clade of 18 MAGs, and hence the MAGs within this clade are referred to as Prevotella. For the Rikenellaceae, one genus-level clade of MAGs grouped with a Bacteroidales MAG reconstructed from Svalbard reindeer rumen (SRM-1) reported to constitute 11% of the reindeer rumen community (Pope et al., 2012). Remarkably, no reference genomes were found for the remaining 18 Bacteroidetes genus-level clades. However, based on 16S rRNA genes of the MAGs (due to difficulties of assembling/binning 16S genes, these were only found in a subset of MAGs; Supplementary Table S3), several of the clades represent 16S sequence clusters of uncultivated organisms in the SILVA database (Figure 2a): ‘UCG-001’ (105_bin7), ‘RF16’ (106_bin29), ‘RC9’ (105_bin84 and 106_bin73), and ‘BS11’ (106_bin71). Clusters ‘RF16’ and ‘BS11’ are abundant members of the core rumen microbiome (Henderson et al., 2015) and ‘RC9’ was previously shown to be abundant in moose (Ishaq and Wright, 2014) and turned out to be the same as SRM-1 above. The reconstructed genomes for the previously uncharacterized Bacteroidetes clades have the potential to significantly advance our understanding of the rumen ecosystem and will serve as reference genomes for future metaomics studies of rumen microbiomes.
Several Bacteroidetes species have been identified as capable of utilizing various plant and host glycans (Koropatkin et al., 2012; Naas et al., 2014). The ecological niche and substrate usage of a particular Bacteroidetes member can be predicted not only from the CAZyme profile but also from existing PULs, where each PUL may contain a complete enzymatic machinery for degrading a specific glycan structure. Most of the 68 Bacteroidetes MAGs contained large numbers of GH genes that were candidates for plant polysaccharide degradation, and PULs were observed in 64 of these genomes (Figure 2c; Supplementary Table S4). The number of PULs per genome varied between one and 38, with the highest number found in two Prevotella MAGs. In some predicted PULs, no putative CAZymes were present but proteins of unknown function exhibited remote similarity to CAZymes (Figures 3a and b; Supplementary Table S4), thus forming interesting targets for enzyme discovery. Clades within Bacteroidetes display distinct cellulolytic potential (Figure 2b). The highest number of GH5 genes was noted in the cluster of Prevotella MAGs. However some genomes in the same clade lacked cellulases. In addition, some of the GH5 genes encode enzymes belonging to subfamilies that do not carry cellulase activity, such as the xylanase subfamily GH5_21 and the β-mannanase subfamily GH5_7 (Supplementary Table S3). Several PULs contained members of subfamily GH5_46. Although GH5_46 genes are frequently found in genomes isolated from ruminant guts (Aspeborg et al., 2012), the function of enzymes within this subfamily is currently limited to activity on carboxymethylcellulose.
Figure 3.
Different examples of gene organizations in a subset of the PULs identified in MAGs of phylum Bacteroidetes. PULs absent from CAZymes that rather contained genes encoding proteins of unknown function (a, b). PULs exhibiting similar organization and CAZyme content as experimentally characterized PULs, where (c) is similar to xylan-degrading PULs in B. ovatus ATCC 8483 (Rogowski et al., 2015) and (d) is similar to starch-degrading PULs in B. thetaiotaomicron VPI-5482 (Foley et al., 2016). A PUL with GH5 and GH26 genes (e). PULs exhibiting combinations of GH9 genes with various other GH-encoding genes (f). A PUL shared in four Prevotellaceae MAGs that harbored dockerin-like modules (g). PULs, containing dockerin modules, that are likely involved in pectin or starch breakdown (h). The red horizontal bars indicate contig starts or ends.
The rumen bacterium P. ruminicola has been suggested to represent an exclusively hemicellulolytic species (Purushe et al., 2010). Indeed, the CAZomes of the Prevotella MAGs revealed that most of them are able to deconstruct both xylans and mannans, but GH and PL families involved in starch, cellulose and pectin degradation were also identified (Figure 2b, Supplementary Table S3). Consistently, PULs predicted for breakdown of xylan, mannan, starch and pectin were identified (Figures 3c and d; Supplementary Table S4). For instance, in the reconstructed genome 106_bin108, predicted PULs 2 and 8 were expected to degrade xylan and predicted PUL 12 β-mannan (Supplementary Table S4). In the three Prevotella MAGs with the greatest number of PULs, 102_bin60, 104_bin27 and 103_bin120, many GH28 polygalacturonase genes were observed. Furthermore, several of the predicted PULs were populated with genes coding for pectin-active enzymes. According to our results, the PULs and CAZomes of uncultured species of Prevotella discovered in the current study suggest generalists utilizing pectins, starch, cellulose and hemicelluloses as carbon sources.
Recent investigations of ruminal guts have claimed a fibrolytic role of uncharacterized clades within Bacteroidetes (Naas et al., 2014; Mackenzie et al., 2015). A versatile PUL, including three GH5 genes and one GH26 gene, from the uncultured Bacteroidales phylotype SRM-1 has been demonstrated to target both soluble cellulose derivatives and hemicellulose (Mackenzie et al., 2015). The group of moose rumen MAGs related to SRM-1 (‘RC9’) display a CAZyme content that suggests saccharolytic activity against mannan, xylan and pectin (Figure 2b). For three of the four MAGs, PULs were identified but none were identical to the one characterized in SRM-1. Nonetheless, PUL variants containing GH5 and GH26 genes were observed (Figure 3e). Six Rikenellaceae MAGs form a sister clade to the clade including SRM-1 (also ‘RC9’). These MAGs feature extensive arrays of GHs (Figure 2b), including genes encoding GH5_7 and GH26 β-mannanases and GH5_4 and GH9 cellulases (Supplementary Table S3). Two of the MAGs (104_bin41 and 106_bin73) were especially enriched in GH2, GH3 and GH43 genes, indicative of a high activity on oligosaccharides and side chains. Moreover, >30 PULs were predicted in each of these two MAGs, and several of these displayed GH9 genes in combinations with various other GH-encoding genes (Figure 3f). The only Bacteroidetes MAG possessing an expansin module was Prevotellaceae MAG 103_bin60 (Figure 2a; Supplementary Table S3). The expansin-bearing protein also contained a putative cellulose-binding CBM63. Further analyses of this MAG identified two GH5_4 genes, and one putative GH94 cellobiose phosphorylase that collectively may be involved in cellulose degradation. However, the overall CAZyme profile implies that cellulose is likely not the main target for the organism as the reconstructed genome has an extensive portfolio of hemicellulolytic genes, for example, four GH10 xylanases, one GH5_21 xylanase, two GH115 glucuronidases, five GH26 β-mannanases and one β-mannanase belonging to subfamily GH5_7. Moreover, regarding the MAGs assigned to the phylum Bacteroidetes, one GH12 gene was found in the Prevotellaceae MAG 101B_bin22 located in a clade consisting of seven MAGs with potentially competent plant polysaccharide-degrading enzymes (Figure 2b). GH12 contains endoglucanases, xyloglucanases and licheninases. Furthermore, all GH74 (endoglucanases, xyloglucanases) and five out of the six GH5_13 (unknown activity) genes were found in MAGs placed in this clade. Thus this Prevotellaceae clade, which is lacking previously sequenced genomes but belong to uncultivated cluster ‘UCG-001’ according to the 16S analysis, seems to have an important role in plant cell wall deconstruction in the moose rumen.
Several Prevotellaceae MAGs featured dockerin modules (Figure 2c). These were sometimes encoded by the same gene as GH and PL modules (Supplementary Table S3) and sometimes even found in genes organized in PULs. Dockerins are components of cellulosome and amylosome structures, but dockerin-containing proteins unrelated to these structures have also been identified (Bayer et al., 1999, 2004; Peer et al., 2009; Ze et al., 2015). Although a handful of dockerins have been identified in Bacteroidetes genomes previously (Peer et al., 2009), the genomes analyzed in this study have substantial number of dockerin modules (up to 40). The observed dockerins were, in many cases, combined with GHs, such as GH5, GH98 and GH128 (Supplementary Table S3). In addition, dockerins were identified within PULs. To our knowledge, this is the first reported example of PULs containing dockerins, despite the analysis of approximately 600 Bacteroidetes genomes (unpublished data). Five MAGs possessed a single PUL that harbored dockerins (104_bin3, 105_bin10, 105_bin129, 106_bin111 and 106_bin81), two MAGs had two dockerin-containing PULs (101B_bin22 and 102_bin22) and in one MAG dockerins were identified in eight PULs (105_bin7). These dockerin modules were part of proteins consisting of catalytic modules not only from families GH13, GH30, GH43, PL1, PL11 and CE8 but also from non-classified GHs. Notably, one PUL was found with the same gene organization across four genomes (Figure 3g). Most of the identified dockerin-carrying PULs are likely involved in pectin or starch breakdown (Figure 3h). In the cellulosome, dockerins are interacting with cohesins, and multiple cohesin modules are signatures of the scaffoldin component of the cellulosome (Bayer et al., 2004). Initial CAZyme profiling did not indicate the presence of cohesins in any of the MAGs affiliated with Bacteroidetes (Figure 2c). However, additional searches against NCBI’s conserved domain database (Marchler-Bauer et al., 2014) gave weak hits (e-values⩽10−4) to cohesin modules in seven of the Bacteroidetes MAGs that harbored dockerin modules (but, notably, in none of the Bacteroidetes MAGs that lacked dockerins) and also indications of genes with two cohesin modules in the same gene in two MAGs (data not shown).
After the original submission of this study, a reanalysis of the bovine rumen metagenome was published that revealed dockerins being present on contigs assigned to Bacteroidetes (Bensoussan et al., 2017). Thus the presence of dockerins in Bacteroidetes genomes is not restricted to the moose rumen. To investigate whether dockerins can also be found in PULs in bovine Bacteroidetes, we here conducted another reanalysis of the bovine rumen data by Hess et al. (2011). We identified 1796 PULs on the metagenome contigs. Eight of these PULs encoded in total nine proteins appended to a dockerin (DOC1) module. Five of these proteins were attached to GH modules (from families GH5, GH43, GH13 and GH53). This supports our finding of dockerins in PULs from moose Bacteroidetes and shows that this situation is probably common among ruminants. The function of the Bacteroidetes dockerins, however, is presently unclear given the apparent scarcity (or possibly absence) of cohesins in Bacteroidetes.
The presence of Treponema may reflect a pectin-rich diet
Treponema has been estimated to account for <2.4% of the bovine rumen bacterial community (Sikorová et al., 2010), but as described above, our results suggest that species of this genus are more abundant in the moose rumen. All MAGs of phylum Spirochaetes were of genus Treponema and resembled closely with Treponema saccharophilum, a pectinolytic bacterium isolated from bovine rumen (Paster and Canale-Parola, 1985). It was recently shown that a switch to a pectin-rich diet in cow (Bos taurus) favored the growth of T. saccharophilum but not of other treponemes (Liu et al., 2014). Three of the identified MAGs harbored genes encoding pectin-degrading enzymes in families GH28, GH105, PL1 and PL9, whereas the CAZome of the fourth (106_bin66) suggests capability to degrade other plant polysaccharides, such as xylan (GH10, GH30_1, GH120) and cellulose (GH5_2, GH94; Supplementary Table S3). The presence of two GH5_2 endoglucanases and one cellulose-binding CBM63 module in the putative pectinolytic MAG 106_bin54 (Supplementary Table S3) suggests that this Treponema may also degrade cellulose in addition to pectin. Genes for starch-degrading enzymes were also identified in the four reconstructed Treponema genomes, hence the CAZyme analyses indicate a role for treponemes in plant polysaccharide depolymerization and particularly breakdown of pectin in the moose rumen.
MAGs with limited plant-carbohydrate-degrading ability
Except for the Ruminococcus MAG, only a small number of CAZymes predicted to be involved in plant biomass degradation were identified in reconstructed Firmicutes genomes. Two MAGs that clustered with Butyrivibrio fibrisolvens (Figure 2a) possessed one gene each for a GH5_2 endoglucanase and for a putative GH94 cellobiose phosphorylase. Moreover, a GH10 xylanase was predicted in one of these MAGs, whereas the other contained a GH5_44 gene (Supplementary Table S3). Thus these MAGs seem to have a, perhaps limited, plant-carbohydrate-degradative ability. In MAG 102_bin14, with Clostridiales genomosp. BVAB3 UP119-5 as the closest related reference genome (Figure 2a), one GH10 xylanase and one GH94 cellobiose phosphorylase (Supplementary Table S3) are indicative of a capacity to degrade xylan and cellulose. The five Mitsuokella-clade MAGs contained a high number of GH1 genes (Figure 2b), which could indicate that they utilize cellobiose. However, the presence of other GH and CE families in these MAGs rather indicates a role in breakdown of starch, peptidoglycan and fructans. MAGs from other phyla contained few CAZymes predicted for structural plant-carbohydrate degradation. However, a GH26 β-mannanase gene was predicted in each of the two candidate phylum SR1 genomes, and a GH8 gene was identified in one of the Gammaproteobacteria MAGs (105_bin31). Two MAGs (106_bin52 and 102_bin56) branched closest with the Firmicute Erysipelotrix in the phylogenetic tree, while 16S analysis classified them to the SILVA ‘RF9’ cluster (named ‘RF39’ in Greengenes) of uncultivated organisms of class Mollicutes within the Tenericutes phylum. The placement of Mollicutes within the Firmicutes agrees with earlier phylogenetic analysis, and their taxonomic placement has been a matter of debate (Skennerton et al., 2016). CAZyme identification in the Mollicutes MAGs (Figure 2a) suggests that these bacteria can hydrolyze xylan, starch/glycogen and peptidoglycan. The predicted enzymatic systems of the remaining MAGs, classified into candidate phylum TM7, Cyanobacteria/Melainabacteria, Alphaproteobacteria and Synergistetes, suggest ability to mainly digest starch/glycogen and peptidoglycan.
Archaea of moose rumen are associated with methane production
One archaeal genome was recovered that was highly similar (77.35% average amino-acid identity in >1000 proteins) to that of the rumen methanogen Methanobrevibacter ruminantium strain M1 (Leahy et al., 2010), previously linked to high-fiber/low-energy diets in various ruminants, including moose (Zhou et al., 2010; Ishaq et al., 2015). This MAG contained methanogenesis-associated genes such as methylenetetrahydromethanopterin reductase, CoB–CoM heterodisulfide reductase subunits, methyl-coenzyme M reductases, tetrahydromethanopterin S-methyltransferase subunits and tungsten formylmethanofuran dehydrogenase subunits. Along with Methanobrevibacter, the 16S rRNA analysis revealed the presence of the genus Methanosphaera, but in lower abundance (average ratio 3.5:1), as previously observed in moose (Ishaq et al., 2015). The presence of Methanosphaera is compatible with pectin in the diet, as they utilize methanol (a byproduct of pectin fermentation; Facey et al., 2012; Ishaq et al., 2015). Although methanogens are not directly involved in plant fiber digestion, cooperation with lignocellulose-degrading microbes has been suggested to improve the fiber-degrading capacity of the latter (Wei et al., 2016).
Conclusions
Shotgun metagenomic sequencing of moose rumen contents in combination with binning yielded a high number of MAGs that were characterized based on their CAZyme repertoires, providing comprehensive insights into the complex mechanisms of plant-polysaccharide degradation in this anaerobic environment.
So what were the main findings? First of all, an unusually high prevalence of Spirochaetes for a ruminal environment. Second, a number of enriched pectin-degrading CAZyme families as compared with microbial communities of cow and reindeer rumen. Third, multiple Bacteroidetes clades with fibrolytic potential lacking previously sequenced genomes, and fourth, a large number of predicted dockerins encoded in Bacteroidetes genomes, often in combination with CAZyme modules and sometimes encoded inside PULs, which has never been discovered before. The current study provides reference genomes for a large number of rumen prokaryotic clades previously lacking characterized genomes as well as moose rumen versions of genomes of the well-known fibrolytic species such as F. succinogenes and R. flavefaciens isolated from bovine rumen, genomes that can be used as references in future metaomics studies. Findings of the present investigation may lead to a better understanding of various strategies for plant matter enzymatic digestion, and the described cellulases, hemicellulases and pectinases may, after experimental characterizations, find a future role in biomass-conversion applications. In addition, the numerous PUL genes with unknown functions may lead to the development of novel enzyme cocktails.
Acknowledgments
Roger Bergström is thanked for initial discussions of the project. We also thank Laura Lebrun for essential work in DNA extractions and Hussein Shokry for conducting a pilot study. This study was supported by The Swedish Research Council Formas (grants 213-2012-1513 and 213-2014-176), European Union’s Seventh Framework Program (FP/2007/2013) and European Research Council (ERC Grant Agreement 322820).
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ et al. (2014). Binning metagenomic contigs by coverage and composition. Nat Methods 11: 1144–1146. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Aspeborg H, Coutinho PM, Wang Y, Brumer H 3rd, Henrissat B. (2012). Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol Biol 12: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer EA, Belaich J-P, Shoham Y, Lamed R. (2004). The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol 58: 521–554. [DOI] [PubMed] [Google Scholar]
- Bayer EA, Coutinho PM, Henrissat B. (1999). Cellulosome-like sequences in Archaeoglobus fulgidus: an enigmatic vestige of cohesin and dockerin domains. FEBS Lett 463: 277–280. [DOI] [PubMed] [Google Scholar]
- Bengtsson-Palme J, Hartmann M, Eriksson KM, Pal C, Thorell K, Larsson DGJ et al. (2015). metaxa2: improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol Ecol Resour 15: 1403–1414. [DOI] [PubMed] [Google Scholar]
- Bensoussan L, Moraïs S, Dassa B, Friedman N, Henrissat B, Lombard V et al. (2017). Broad phylogeny and functionality of cellulosomal components in the bovine rumen microbiome. Environ Microbiol 19: 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg Miller ME, Antonopoulos DA, Rincon MT, Band M, Bari A, Akraiko T et al. (2009). Diversity and strain specificity of plant cell wall degrading enzymes revealed by the draft genome of Ruminococcus flavefaciens FD-1. PLoS One 4: e6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergström R, Hjeljord O. (1987). Moose and vegetation interactions in northwestern Europe and Poland. Swedish Wildlife Research (Sweden) http://agris.fao.org/agris-search/search.do?recordID=SE8811428.
- Bjursell MK, Martens EC, Gordon JI. (2006). Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J Biol Chem 281: 36269–36279. [DOI] [PubMed] [Google Scholar]
- Boisvert S, Laviolette F, Corbeil J. (2010). Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol 17: 1519–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brás JLA, Cartmell A, Carvalho ALM, Verzé G, Bayer EA, Vazana Y et al. (2011). Structural insights into a unique cellulase fold and mechanism of cellulose hydrolysis. Proc Natl Acad Sci USA 108: 5237–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumm P, Mead D, Boyum J, Drinkwater C, Deneke J, Gowda K et al. (2011). Functional annotation of Fibrobacter succinogenes S85 carbohydrate active enzymes. Appl Biochem Biotechnol 163: 649–657. [DOI] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH. (2015). Fast and sensitive protein alignment using DIAMOND. Nat Methods 12: 59–60. [DOI] [PubMed] [Google Scholar]
- Campbell JH, O’Donoghue P, Campbell AG, Schwientek P, Sczyrba A, Woyke T et al. (2013). UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota. Proc Natl Acad Sci USA 110: 5540–5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederlund G, Ljungqvist H, Markgren G, Stalfelt F. (1980). Foods of Moose and Roe-deer at Grimsö in Central Sweden. Results of Rumen Contents Analyses. Swed Wildl Res 11: 170–247. [Google Scholar]
- Cuskin F, Lowe EC, Temple MJ, Zhu Y, Cameron EA, Pudlo NA et al. (2015). Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature 517: 165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassa B, Borovok I, Ruimy-Israeli V, Lamed R, Flint HJ, Duncan SH et al. (2014). Rumen cellulosomics: divergent fiber-degrading strategies revealed by comparative genome-wide analysis of six ruminococcal strains. PLoS One 9: e99221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SY, Rincon MT, Lamed R, Martin JC, McCrae SI, Aurilia V et al. (2001). Cellulosomal scaffoldin-like proteins from Ruminococcus flavefaciens. J Bacteriol 183: 1945–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzi SC, Sharon I, Wrighton KC, Koren O, Hug LA. (2013). The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. Elife 2: e01102. http://elifesciences.org/content/2/e01102.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan C-J, Feng J-X. (2010). Mining metagenomes for novel cellulase genes. Biotechnol Lett 32: 1765–1775. [DOI] [PubMed] [Google Scholar]
- Facey HV, Northwood KS, Wright AD. (2012). Molecular diversity of methanogens in fecal samples from captive Sumatran orangutans (Pongo abelii. Am J Primatol 74: 408–413. [DOI] [PubMed] [Google Scholar]
- Flint HJ. (2008) Cellulase systems of anaerobic microorganisms from the rumen and large intestineIn: Biomass Recalcitrance. Blackwell Publishing Ltd.: Oxford, UK, pp 393–406. [Google Scholar]
- Foley MH, Cockburn DW, Koropatkin NM. (2016). The Sus operon: a model system for starch uptake by the human gut Bacteroidetes. Cell Mol Life Sci 73: 2603–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbeck R, Damásio ARL, Gonçalves TA, Machado CB, Paixão DAA, Wolf LD et al. (2014). Development of hemicellulolytic enzyme mixtures for plant biomass deconstruction on target biotechnological applications. Appl Microbiol Biotechnol 98: 8513–8525. [DOI] [PubMed] [Google Scholar]
- Henderson G, Cox F, Ganesh S, Jonker A, Young W et alGlobal Rumen Census Collaborators. (2015). Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci Rep 5: 14567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlemann DPR, Lundin D, Labrenz M, Jürgens K, Zheng Z, Aspeborg H et al. (2013). Metagenomic de novo assembly of an aquatic representative of the verrucomicrobial class Spartobacteria. MBio 4: e00569-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess M, Sczyrba A, Egan R, Kim T-W, Chokhawala H, Schroth G et al. (2011). Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331: 463–467. [DOI] [PubMed] [Google Scholar]
- Hofmann RR. (1989). Evolutionary steps of ecophysiological adaptation and diversification of ruminants: a comparative view of their digestive system. Oecologia 78: 443–457. [DOI] [PubMed] [Google Scholar]
- Hugerth LW, Larsson J, Alneberg J, Lindh MV, Legrand C, Pinhassi J et al. (2015). Metagenome-assembled genomes uncover a global brackish microbiome. Genome Biol 14: 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Mitra S, Ruscheweyh H-J, Weber N, Schuster SC. (2011). Integrative analysis of environmental sequences using MEGAN4. Genome Res 21: 1552–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishaq SL, Sundset MA, Crouse J, Wright A-DG. (2015). High-throughput DNA sequencing of the moose rumen from different geographical locations reveals a core ruminal methanogenic archaeal diversity and a differential ciliate protozoal diversity. Microb Genom 1: e000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishaq SL, Wright A. (2012). Insight into the bacterial gut microbiome of the North American moose. BMC Microbiol 12: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishaq SL, Wright A-D. (2014). High-throughput DNA sequencing of the ruminal bacteria from moose (Alces alces in Vermont, Alaska, and Norway. Microb Ecol 68: 185–195. [DOI] [PubMed] [Google Scholar]
- Joshi NA, Fass JN. (2011). Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ Files (version 1.33) https://github.com/najoshi/sickle.
- Kirby J, Martin JC, Daniel AS, Flint HJ. (1997). Dockerin-like sequences in cellulases and xylanases from the rumen cellulolytic bacterium Ruminococcus flavefaciens. FEMS Microbiol Lett 149: 213–219. [DOI] [PubMed] [Google Scholar]
- Kishi LT, de Jesus RB, Pavani CD, Lemos EGM, de Souza JAM. (2015). Metagenomic assembly and draft genome sequence of an uncharacterized Prevotella sp. from nelore rumen. Genome Announc 3: e00723–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin NM, Cameron EA, Martens EC. (2012). How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol 10: 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laczny CC, Muller EEL, Heintz-Buschart A, Herold M, Lebrun LA, Hogan A et al. (2016). Identification, recovery, and refinement of hitherto undescribed population-level genomes from the human gastrointestinal tract. Front Microbiol 7: 884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K, Hallin P, Rødland EA, Staerfeldt H-H, Rognes T, Ussery DW. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35: 3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy SC, Kelly WJ, Altermann E, Ronimus RS, Yeoman CJ, Pacheco DM et al. (2010). The genome sequence of the rumen methanogen Methanobrevibacter ruminantium reveals new possibilities for controlling ruminant methane emissions. PLoS One 5: e8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS et al. (2008). Evolution of mammals and their gut microbes. Science 320: 1647–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang J-K, Zhu W, Pu Y-Y, Guan L-L, Liu J-X. (2014). Monitoring the rumen pectinolytic bacteria Treponema saccharophilum using real-time PCR. FEMS Microbiol Ecol 87: 576–585. [DOI] [PubMed] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42: D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie AK, Naas AE, Kracun SK, Schückel J, Fangel JU, Agger JW et al. (2015). A polysaccharide utilization locus from an uncultured bacteroidetes phylotype suggests ecological adaptation and substrate versatility. Appl Environ Microbiol 81: 187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY et al. (2014). CDD: NCBI’s conserved domain database. Nucleic Acids Res 43: D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanram S, Amat D, Choudhary J, Arora A, Nain L. (2013). Novel perspectives for evolving enzyme cocktails for lignocellulose hydrolysis in biorefineries. Sust Chem Process 1: 15. [Google Scholar]
- Montgomery L, Flesher B, Stahl D. (1988). Transfer of Bacteroides succinogenes (Hungate) to Fibrobacter gen. nov. as Fibrobacter succinogenes comb. nov. and description of Fibrobacter intestinalis sp. nov. Int J Syst Evol Microbiol 38: 430–435. [Google Scholar]
- Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L et al. (2011). Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332: 970–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas AE, Mackenzie AK, Mravec J, Schückel J, Willats WGT, Eijsink VGH et al. (2014). Do rumen Bacteroidetes utilize an alternative mechanism for cellulose degradation? MBio 5: e01401–e01414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25: 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Canale-Parola E. (1985). Treponema saccharophilum sp. nov., a large pectinolytic spirochete from the bovine rumen. Appl Environ Microbiol 50: 212–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel DD, Patel AK, Parmar NR, Shah TM, Patel JB, Pandya PR et al. (2014). Microbial and Carbohydrate Active Enzyme profile of buffalo rumen metagenome and their alteration in response to variation in the diet. Gene 545: 88–94. [DOI] [PubMed] [Google Scholar]
- Peer A, Smith SP, Bayer EA, Lamed R, Borovok I. (2009). Noncellulosomal cohesin- and dockerin-like modules in the three domains of life. FEMS Microbiol Lett 291: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar M, Abulencia CB, Walcher M, Hutchison D, Zengler K, Garcia JA et al. (2007). Targeted access to the genomes of low-abundance organisms in complex microbial communities. Appl Environ Microbiol 73: 3205–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope PB, Denman SE, Jones M, Tringe SG, Barry K, Malfatti SA et al. (2010). Adaptation to herbivory by the Tammar wallaby includes bacterial and glycoside hydrolase profiles different from other herbivores. Proc Natl Acad Sci USA 107: 14793–14798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope PB, Mackenzie AK, Gregor I, Smith W, Sundset MA, McHardy AC. (2012). Metagenomics of the Svalbard reindeer rumen microbiome reveals abundance of polysaccharide utilization loci. PLoS One 7: e38571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glöckner FO. (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28: 1823–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puniya AK, Singh R, Kamra DN (eds). (2015) Rumen Microbiology: From Evolution to Revolution. Springer: New Delhi, India. [Google Scholar]
- Purushe J, Fouts DE, Morrison M, White BA, Mackie RI et al North American Consortium for Rumen Bacteria. (2010). Comparative genome analysis of Prevotella ruminicola and Prevotella bryantii: insights into their environmental niche. Microb Ecol 60: 721–729. [DOI] [PubMed] [Google Scholar]
- Ransom-Jones E, Jones DL, Edwards A, McDonald JE. (2014). Distribution and diversity of members of the bacterial phylum Fibrobacteres in environments where cellulose degradation occurs. Syst Appl Microbiol 37: 502–509. [DOI] [PubMed] [Google Scholar]
- Rodriguez-R LM, Konstantinos KT. (2014). Bypassing cultivation to identify bacterial species. Microbe Magazine9 https://www.microbemagazine.org/index.php?option=com_content&view=article&id=6634:bypassing-cultivation-to-identify-bacterial-species&catid=1313.
- Rogowski A, Briggs JA, Mortimer JC, Tryfona T, Terrapon N, Lowe EC et al. (2015). Glycan complexity dictates microbial resource allocation in the large intestine. Nat Commun 6: 7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roume H, Muller EEL, Cordes T, Renaut J, Hiller K, Wilmes P. (2013). A biomolecular isolation framework for eco-systems biology. ISME J 7: 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30: 2068–2069. [DOI] [PubMed] [Google Scholar]
- Segata N, Börnigen D, Morgan XC, Huttenhower C. (2013). PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun 4: 2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seon Park J, Russell JB, Wilson DB. (2007). Characterization of a family 45 glycosyl hydrolase from Fibrobacter succinogenes S85. Anaerobe 13: 83–88. [DOI] [PubMed] [Google Scholar]
- Sikorová L, Piknová M, Javorský P, Guczyńska W, Kasperowicz A, Michałowski T et al. (2010). Variability of treponemes in the rumen of ruminants. Folia Microbiol 55: 376–378. [DOI] [PubMed] [Google Scholar]
- Skennerton CT, Haroon MF, Briegel A, Shi J, Jensen GJ, Tyson GW et al. (2016). Phylogenomic analysis of Candidatus ‘Izimaplasma’ species: free-living representatives from a Tenericutes clade found in methane seeps. ISME J 10: 2679–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen G, Stevenson DM, Bruce DC, Chertkov O, Copeland A, Cheng J-F et al. (2011. a). Complete genome of the cellulolytic ruminal bacterium Ruminococcus albus 7. J Bacteriol 193: 5574–5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen G, Weimer PJ, Stevenson DM, Aylward FO, Boyum J, Deneke J et al. (2011. b). The complete genome sequence of Fibrobacter succinogenes S85 reveals a cellulolytic and metabolic specialist. PLoS One 6: e18814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XZ, Joblin KN, Andrew IG, Hoskin SO, Harris PJ. (2008). Degradation of forage chicory by ruminal fibrolytic bacteria. J Appl Microbiol 105: 1286–1297. [DOI] [PubMed] [Google Scholar]
- Terrapon N, Henrissat B. (2014). How do gut microbes break down dietary fiber? Trends Biochem Sci 39: 156–158. [DOI] [PubMed] [Google Scholar]
- Terrapon N, Lombard V, Gilbert HJ, Henrissat B. (2015). Automatic prediction of polysaccharide utilization loci in Bacteroidetes species. Bioinformatics 31: 647–655. [DOI] [PubMed] [Google Scholar]
- Wallace RJ, Rooke JA, McKain N, Duthie C-A, Hyslop JJ, Ross DW et al. (2015). The rumen microbial metagenome associated with high methane production in cattle. BMC Genomics 16: 839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke F, Luginbühl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT et al. (2007). Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450: 560–565. [DOI] [PubMed] [Google Scholar]
- Wei YQ, Long RJ, Yang H, Yang HJ, Shen XH, Shi RF et al. (2016). Fiber degradation potential of natural co-cultures of Neocallimastix frontalis and Methanobrevibacter ruminantium isolated from yaks (Bos grunniens grazing on the Qinghai Tibetan Plateau. Anaerobe 39: 158–164. [DOI] [PubMed] [Google Scholar]
- Weimer PJ. (2013) The ruminant animal as a natural biomass-conversion platform and a source of bioconversion agentsBiological Concerstion of Biomass for Fuels and Chemicals: Exploration from Natural Utilization Systems 10: 248.
- Wilson DB. (2009). Evidence for a novel mechanism of microbial cellulose degradation. Cellulose 16: 723–727. [Google Scholar]
- Wright A-DG. (2015) Rumen Protozoa In: Rumen Microbiology: From Evolution to Revolution. Springer: New Delhi, India, pp 113–120. [Google Scholar]
- Ze X, Ben David Y, Laverde-Gomez JA, Dassa B, Sheridan PO, Duncan SH et al. (2015). Unique organization of extracellular amylases into amylosomes in the resistant starch-utilizing human colonic firmicutes bacterium Ruminococcus bromii. MBio 6: e01058-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Hernandez-Sanabria E, Guan LL. (2010). Characterization of variation in rumen methanogenic communities under different dietary and host feed efficiency conditions, as determined by PCR-denaturing gradient gel electrophoresis analysis. Appl Environ Microbiol 76: 3776–3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.