Abstract
We have identified an aminoacyl-tRNA synthetase/tRNA pair for the efficient and site-specific incorporation of a cyclobutene-containing amino acid into proteins in response to amber nonsense codon. Fast and fluorescent labeling of purified proteins and intact proteins in live cells was demonstrated using the inverse electron demand Diels-Alder reaction with a tetrazine.
The increasing demand for bioorthogonal protein labelling has driven the search for new functional groups that can react with their partner reagents with fast reaction rate, high selectivity, and good biocompatibility.1–6 The inverse electron demand Diels–Alder (IEDDA) reaction between a tetrazine and a strained alkene has been widely explored for bioorthogonal labelling of biomolecules.7–19 The second-order rate constants for the rate-limiting [4+2] cycloaddition forming the first step of this reaction span nine orders of magnitude, depending on the nature of the two reactants and reaction conditions.1, 20–21 The broad application of the tetrazine/alkene cycloaddition in biological systems stems from several unique features: (1) the reaction rate is often increased in the presence of water;8, 22 (2) rapid cycloaddition does not require a metal catalyst; and (3) nitrogen gas is the only by-product.
Medium-sized strained alkenes, predominantly trans-cyclooctene and norbornene, have been extensively explored as IEDDA substrates.7–19 In recent years, cyclopropene, a less bulky strained alkene, has also been successfully applied as a robust bioorthogonal probe.23–27 Curiously, cyclobutenes, a class of small and strained alkenes known to be reactive towards IEDDA reactions,28–30 have received much less attention, with most efforts involving derivatives of the Reppe anhydride, a bicyclic derivative of cyclooctatetraene.31 Monocyclic cyclobutenes, such as those discussed here, have surprisingly good thermal stability,29–30 and, in contrast to cyclopropenes,25 have good chemical stability even in the absence of additional substituents on the alkene.29–30 Preliminary work in one of our labs had demonstrated that the rate of IEDDA cycloaddition of a cyclobutene-containing fatty acid (3,6-di(pyridin-2-yl)-1,2,4,5-tetrazine; k = 1.21±0.12 M−1 s−1)30 is faster than that observed with the majority of cyclopropenes24 and comparable to the most reactive cyclopropenes (k = 2.78±0.37 M−1 s−1).25 In this article, we report an efficient synthesis of a cyclobutene-containing unnatural amino acid (CbK; Scheme 2) that is applied as a chemical reporter for protein labelling in live cells. This amino acid was genetically incorporated into proteins in response to amber nonsense codon. While protein labelling through cysteine or lysine by alkylation or amide bond formation reactions are widely employed,32–33 the selectivity of this approach is limited due to the possibility of containing more than one such residues in the protein of interest. In contrast, the unnatural amino acid mutagenesis approach allows for site-specific conjugation of proteins with a chemical probe.
Scheme 2.
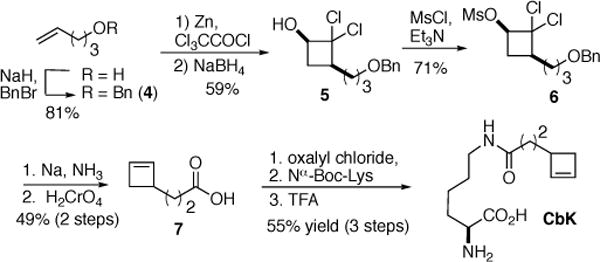
Synthesis of unnatural amino acid CbK containing short chain cyclobutene moiety.
Given the limited data reported for IEDDA reaction of cyclobutenes, the planned ligation was initially investigated using a previously reported cyclobutene (1, Scheme 1).29–30 In contrast to the reaction of tetrazines with cyclopropenes or N-acylazetines,22 we observed no skeletal rearrangement, instead isolating an approximately 1:1 mixture of inseparable 1,4-dihydro-1,2-diazene tautomers 2a and 2b (see ESI-1, Scheme 2 for mechanism). Measurement of kinetics under pseudo-first order conditions (see ESI) revealed bimolecular rate constants lower than those reported for trans-cyclooctene, considerably larger than reported for norbornenes or simple alkenes, and comparable to those recently described for cyclopropenes.1, 34–35 Due to the presence of the tautomers, the cycloadduct(s) were fully characterized following oxidative aromatization to the 1,2-pyrazine (3).
Scheme 1.
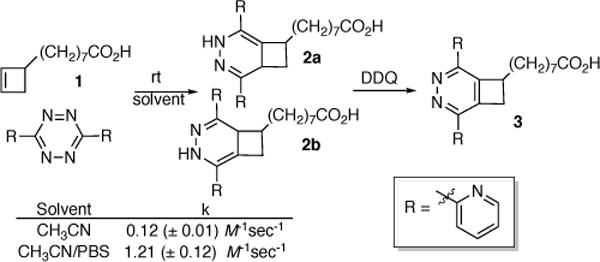
Model of cyclobutene/tetrazine cycloaddition
In order to apply the cyclobutene-tetrazine reaction to protein labelling, a lysine-derived unnatural amino acid containing cyclobutene moiety (CbK; Scheme 2) was synthesized from 3-(2-cyclobutene-1-yl)propanoic acid (7) which was prepared by a variant of a route previously described for cyclobuteneoctanoic acid 1.29 Benzyl ether 4, available from protection of 4-penten-1-ol, underwent [2+2] cycloaddition with dichloroketene; in situ reduction of the intermediate dichloroketone furnished a stable dichlorocyclobutanol (5). Conversion to the corresponding methanesulfonate was followed by a dissolving metal reduction, which achieved both introduction of the cyclobutene and deprotection of the benzyl ether. The volatile cyclobutenyl propanol (6) was not easily isolated and was instead generally carried on to acid 7. The corresponding acid chloride reacted with Nα-protected lysine to furnish modified amino acid CbK.
To site-specifically encode CbK into proteins, a mutant library of Methanosarcina barkeri pyrrolysyl-tRNA synthetase (PylRS) was generated. According to the crystal structures of PylRS,36–38 four amino acid residues involved in recognition of pyrrolysine (Leu270, Tyr271, Leu274, and Cys313) were randomized using site-saturation mutagenesis. In addition, a Tyr349Phe mutation was introduced to all PlyRS mutants by site-directed mutagenesis, since this mutation was previously shown to increase aminoacylation efficiency.27 After one round of negative and one round of positive selection using the previously reported method,39 a PylRS mutant able to incorporate CbK into proteins was identified. This mutant (designated as CbKRS) has the following two mutations, Leu274Met and Cys313Ala.
To further determine the efficiency and fidelity of CbK incorporation by the evolved CbKRS mutant, a fluorescence-based assay was conducted using a GFP mutant (sfGFP-N149TAG) that contains an amber nonsense codon at position Asn149.40 Protein expression was carried out in LB medium supplemented with and without 1 mM CbK. Fluorescence analysis of E. coli cultures showed that full-length sfGFP protein was produced only in the presence of CbK (Fig. 2). In a large-scale expression of sfGFP-N149TAG (50 ml cell culture), 35 mg/L of sfGFP-N149CbK was obtained after purification using affinity chromatography (Fig. 5 of ESI-1). Mass spectrometry analyses of the in-gel digested protein fragment (LEYNFNSH-CbK-VYITADK; Fig. 6 of ESI-1) confirmed site specific incorporation of CbK at position 149 of sfGFP (expected, 684.40 Da, z = 3 and 1026.10 Da, z = 2; observed 684.35 Da, z = 3 and 1026.02 Da, z = 2).
Fig. 2.
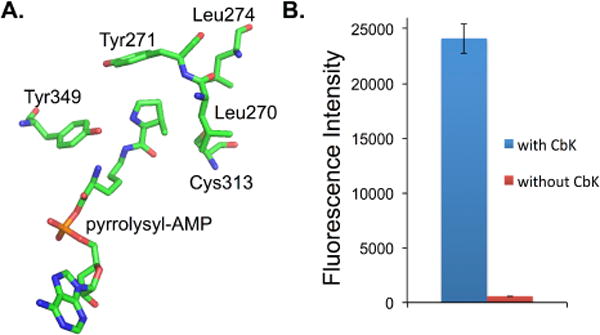
Genetic incorporation of CbK. (A) Crystal structure of the PylRS in complex with pyrrolysyl-AMP. The structure is derived from PDB 2ZIM; (B) Fluorescence readings of cells expressing CbKRS and sfGFP-Asn149TAG mutant. The expressions were conducted either in the presence or in the absence of 1.0 mM CbK. Fluorescence intensity was normalized to cell growth.
With the purified sfGFP-N149CbK in hand, we explored the inverse Diels-Alder reaction between the recombinant sfGFP-N149CbK and a commonly used tetrazine scaffold40–41 containing a fluorescein conjugate (Fl-Tet; Fig. 1). In a typical reaction, 1 μL of Fl-Tet in DMSO (varied stock concentrations to reach the indicated final concentrations in Fig. 3) was added to 9 μL of protein (final concentration, 0.24 mg/mL) in PBS buffer. The reaction mixture was incubated at room temperature with gentle agitation on a platform rocker. The reaction was stopped by the addition of a large excess of 5-norbonene-2-methanol (1 μL; 100 mM) in DMSO. The quenched reaction was subsequently mixed with SDS-PAGE loading buffer and heated at 95 °C for 15 minutes to denature the protein. Therefore, only the fluorescein-labelled sfGFP protein was expected to be fluorescent under UV irradiation. We first examined the labelling of the sfGFP-N149CbK mutant with varied concentrations of Fl-Tet in PBS buffer following an 80 min reaction. As shown in Fig. 3A, fluorescence could be detected at a Fl-Tet concentration of 10 μM or above. The best result was obtained when 100 μM of Fl-Tet was used (Fig. 3A). Control experiments using wild-type sfGFP (sfGFP-wt) and Fl-Tet, or sfGFP-N149CbK only did not show detectable fluorescence (Fig. 3A). These results demonstrate that the unnatural cyclobutene moiety of CbK is biocompatible and orthogonal to functional groups in natural amino acids. Next, we conducted a time dependence labelling study using 50 μM of Fl-Tet. Significant fluorescence was detected from a 5 min reaction (Fig. 3B), indicating that the reaction is very fast as evidenced by our kinetic study. The fluorescence intensity increased gradually in a time-dependent manner. No fluorescence was observed in control experiments when either sfGFP-wt was used or Fl-Tet was omitted. We also conducted mass spectrometry analysis of the product from the 80 min reaction. Expected masses were observed (Fig. 8 of ESI-1; a hydrolysis of thiourea to urea linkage of the labelling reagent, Fl-Tet, was observed). Taken together, the results demonstrate that the cyclobutene/tetrazine IEDDA bioconjugation reaction can be used to efficiently and selectively label a purified protein.
Fig. 1.
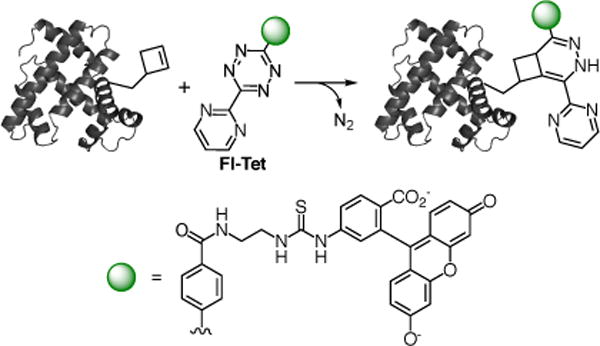
Fluorescent protein labelling with a genetically encoded unnatural amino acid containing a cyclobutene moiety.
Fig. 3.
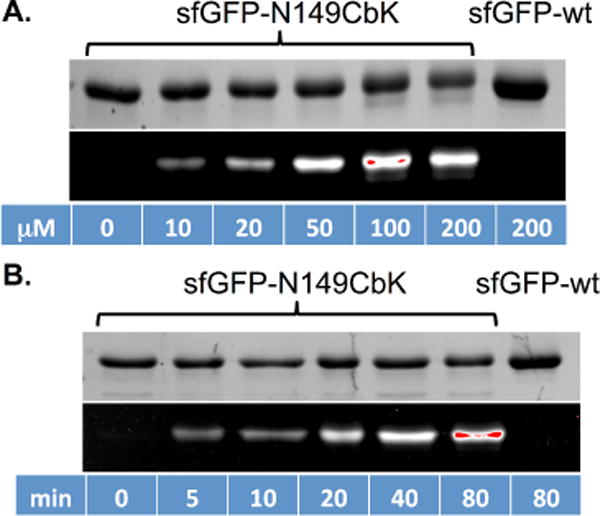
In vitro protein labelling of sfGFP variants with cyclobutene-tetrazine reaction. Following labeling reactions, protein samples were denatured by heating, then analyzed by SDS-PAGE. The top panel in each figure shows Coomassie blue stained gel and the bottom panel shows the fluorescent image of the same gel before Coomassie blue treatment. (A) Labeling of sfGFP-N149CbK protein with varied concentrations of Fl-Tet for 80 minutes; (B) Reaction progress of sfGFP-N149CbK protein labeling with 50 μM of Fl-Tet. Wild-type sfGFP was included in both experiments as the control. The fluorescence intensities of these images were quantified and presented in Fig. 7 of ESI. The kobs was estimated as 0.37 M−1 s−1.
We have also demonstrated the labeling of an outer membrane protein OmpX in live E. coli cells. Plasmid pOmpX-TAG was constructed to encode an OmpX mutant containing a seven amino acid insertion (Ala-Ala-Ala-X-Ala-Ala-Ala; X denotes an amber mutation) between the two extracellular residues Ser76 and Ser77.41 The protein expression was conducted in E. coli in the presence of CbKRS, the amber suppressor tRNA (tRNAPyl), and 1 mM of CbK. Cells containing the mutant OmpX protein (OmpX-CbK) were collected, washed three times, and resuspended in PBS buffer. The labeling experiment was initiated with the addition of Fl-Tet (2 μL; 5 mM) to 100 μL of cell suspension to a final concentration of 100 μM. The reaction mixture was incubated for 1 hour at room temperature with gentle agitation on a platform rocker. Cells were then collected, washed three times with PBS buffer, and imaged under a microscope. As shown in Fig. 4A, labeling of cells containing the mutant protein resulted in strong fluorescent signals which co-localized nicely with cells. As a control, when E. coli cells expressing wild-type OmpX protein in the presence of CbK was used in the labeling experiment under the same conditions, no detectable fluorescence was observed. This confirmed that the observed fluorescence in Fig. 4A resulted from labelled OmpX-CbK protein and not from free CbK. This above data suggested the bioorthogonal reaction between cyclobutene and tetrazine is applicable in live cell labeling. While the present work only demonstrated protein labeling in bacteria, the CbKRS-tRNAPyl pair should support the incorporation of CbK into proteins for labeling experiments in mammalian cells. It is known that the PylRS-derived system can be used to incorporate unnatural amino acids in bacterial, yeast, and mammalian cells.42–44
Fig. 4.
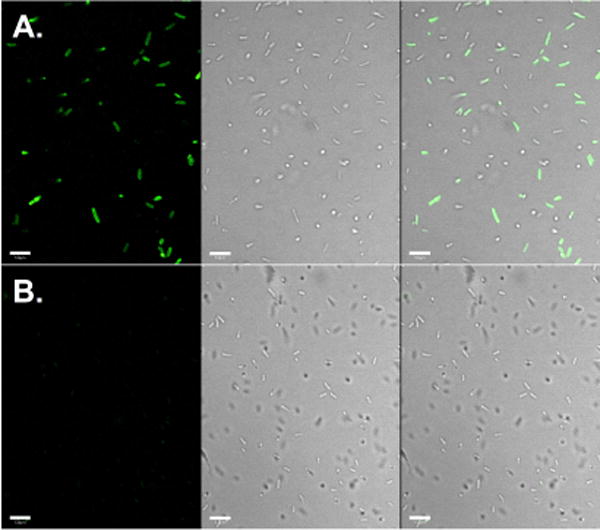
Selective labeling of E. coli cells expressing OmpX-CbK. (A) OmpX-F28CbK mutant that was expressed in the presence of CbK; (B) Wild-type OmpX that was expressed in the presence of CbK. For all images, the left panel shows fluorescent images of E. coli, the middle panel shows bright-field images of the same E. coli cells, and the right panel shows composite images of bright-field and fluorescent images. Scale bars, 5 μm.
In summary, we have synthesized CbK, a cyclobutene-derived unnatural amino acid, and identified a PylRS mutant (CbKRS) for the site-specific incorporation of CbK into proteins in response to amber nonsense codon in E. coli. The bioorthogonal reaction between the strained cyclobutene and a tetrazine achieves efficient protein labeling both in vitro and in vivo. Cyclobutenes offer an excellent structural scaffold in terms of stability and limited size; their addition to the family of reactive IEDDA dienophiles should augment current efforts to expand the chemical toolbox for the study of biological systems.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Health (grant 1R01AI111862 to J.G.), National Science Foundation (grant 1553041 to J.G., and EPS-1004094 to P.D.), and the Life Sciences Interdisciplinary Research Program (Univ. of Nebraska-Lincoln). Portions of this research were performed in facilities renovated with support from the NIH (RR016544). The authors would like to acknowledge assistance from the following individuals at the University of Nebraska-Lincoln: Prof. Ron Cerny (Nebraska Center for Mass Spectrometry, protein mass spectrometry); Dr. Sophie Alvarez (Proteomics and Metabolomics Facility); Prof. Martha Morton (Chemistry Research Instrumentation Facility, kinetics); Prof. David Berkowitz (Department of Chemistry, dissolving metal reduction); Dr. You Zhou (Center for Biotechnology Microscopy facility, fluorescence imaging).
Footnotes
Electronic Supplementary Information (ESI) available: Full experimental details and additional data. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Patterson DM, Nazarova LA, Prescher JA. ACS Chem Biol. 2014;9:592–605. doi: 10.1021/cb400828a. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt MJ, Summerer D. ChemBioChem. 2012;13:1553–1557. doi: 10.1002/cbic.201200321. [DOI] [PubMed] [Google Scholar]
- 3.Shieh P, Bertozzi CR. Org Biomol Chem. 2014;12:9307–9320. doi: 10.1039/c4ob01632g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lang K, Chin JW. ACS Chem Biol. 2014;9:16–20. doi: 10.1021/cb4009292. [DOI] [PubMed] [Google Scholar]
- 5.Ramil CP, Lin Q. Chem Commun. 2013;49:11007–11022. doi: 10.1039/c3cc44272a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruchez MP. Curr Opin Chem Biol. 2015;27:18–23. doi: 10.1016/j.cbpa.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knall AC, Slugovc C. Chem Soc Rev. 2013;42:5131–5142. doi: 10.1039/c3cs60049a. [DOI] [PubMed] [Google Scholar]
- 8.Blackman ML, Royzen M, Fox JM. J Am Chem Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devaraj NK, Weissleder R, Hilderbrand SA. Bioconjugate Chem. 2008;19:2297–2299. doi: 10.1021/bc8004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Royzen M, Yap GPA, Fox JM. J Am Chem Soc. 2008;130:3760–3761. doi: 10.1021/ja8001919. [DOI] [PubMed] [Google Scholar]
- 11.Devaraj NK, Hilderbrand S, Upadhyay R, Mazitschek R, Weissleder R. Angew Chem, Int Ed. 2010;49:2869–2872. doi: 10.1002/anie.200906120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor MT, Blackman ML, Dmitrenko O, Fox JM. J Am Chem Soc. 2011;133:9646–9649. doi: 10.1021/ja201844c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lang K, Davis L, Wallace S, Mahesh M, Cox DJ, Blackman ML, Fox JM, Chin JW. J Am Chem Soc. 2012;134:10317–10320. doi: 10.1021/ja302832g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang Y, Mackey JL, Lopez SA, Liu F, Houk KN. J Am Chem Soc. 2012;134:17904–17907. doi: 10.1021/ja309241e. [DOI] [PubMed] [Google Scholar]
- 15.Liu DS, Tangpeerachaikul A, Selvaraj R, Taylor MT, Fox JM, Ting AY. J Am Chem Soc. 2012;134:792–795. doi: 10.1021/ja209325n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seitchik JL, Peeler JC, Taylor MT, Blackman ML, Rhoads TW, Cooley RB, Refakis C, Fox JM, Mehl RA. J Am Chem Soc. 2012;134:2898–2901. doi: 10.1021/ja2109745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lang K, Davis L, Torres-Kolbus J, Chou C, Deiters A, Chin JW. Nat Chem. 2012;4:298–304. doi: 10.1038/nchem.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plass T, Milles S, Koehler C, Szymanski J, Mueller R, Wiessler M, Schultz C, Lemke EA. Angew Chem, Int Ed. 2012;51:4166–4170. doi: 10.1002/anie.201108231. [DOI] [PubMed] [Google Scholar]
- 19.Borrmann A, Milles S, Plass T, Dommerholt J, Verkade JMM, Wiessler M, Schultz C, van Hest JCM, van Delft FL, Lemke EA. ChemBioChem. 2012;13:2094–2099. doi: 10.1002/cbic.201200407. [DOI] [PubMed] [Google Scholar]
- 20.Wong CH, Zimmerman SC. Chem Commun. 2013;49:1679–1695. doi: 10.1039/c2cc37316e. [DOI] [PubMed] [Google Scholar]
- 21.Wu H, Devaraj NK. Top Curr Chem. 2015;374:1–22. doi: 10.1007/s41061-015-0005-z. [DOI] [PubMed] [Google Scholar]
- 22.Wijnen JW, Zavarise S, Engberts JBFN, Charton M. J Org Chem. 1996;61:2001–2005. [Google Scholar]
- 23.Yang J, Seckute J, Cole CM, Devaraj NK. Angew Chem, Int Ed. 2012;51:7476–7479. doi: 10.1002/anie.201202122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patterson DM, Nazarova LA, Xie B, Kamber DN, Prescher JA. J Am Chem Soc. 2012;134:18638–18643. doi: 10.1021/ja3060436. [DOI] [PubMed] [Google Scholar]
- 25.Kamber DN, Nazarova LA, Liang Y, Lopez SA, Patterson DM, Shih HW, Houk KN, Prescher JA. J Am Chem Soc. 2013;135:13680–13683. doi: 10.1021/ja407737d. [DOI] [PubMed] [Google Scholar]
- 26.Yu Z, Pan Y, Wang Z, Wang J, Lin Q. Angew Chem, Int Ed. 2012;51:10600–10604. doi: 10.1002/anie.201205352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elliott TS, Townsley FM, Bianco A, Ernst RJ, Sachdeva A, Elsaesser SJ, Davis L, Lang K, Pisa R, Greiss S, Lilley KS, Chin JW. Nat Biotechnol. 2014;32:465–472. doi: 10.1038/nbt.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoerner S, Uth C, Avrutina O, Frauendorf H, Wiessler M, Kolmar H. Chem Commun. 2015;51:11130–11133. doi: 10.1039/c5cc03434e. [DOI] [PubMed] [Google Scholar]
- 29.Sittiwong W, Zinniel DK, Fenton RJ, Marshall DD, Story CB, Kim B, Lee JY, Powers R, Barletta RG, Dussault PH. ChemMedChem. 2014;9:1838–1849. doi: 10.1002/cmdc.201402067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sittiwong W. Ph D Thesis. University of Nebraska - Lincoln; 2014. [Google Scholar]
- 31.Wiessler M, Waldeck W, Kliem C, Pipkorn R, Braun K. Int J Med Sci. 2010;7:19–28. doi: 10.7150/ijms.7.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carrico IS. Chemical Society Reviews. 2008;37:1423–1431. doi: 10.1039/b703364h. [DOI] [PubMed] [Google Scholar]
- 33.Chalker JM, Bernardes GJL, Lin YA, Davis BG. Chem–Asian J. 2009;4:630–640. doi: 10.1002/asia.200800427. [DOI] [PubMed] [Google Scholar]
- 34.Wu HX, Devaraj NK. Topics Curr Chem. 2016;374 doi: 10.1007/s41061-015-0005-z. [DOI] [PubMed] [Google Scholar]
- 35.Mayer S, Lang K. Synthesis. 2017;49:830–848. [Google Scholar]
- 36.Ambrogelly A, Gundllapalli S, Herring S, Polycarpo C, Frauer C, Söll D. Proc Natl Acad Sci U S A. 2007;104:3141–3146. doi: 10.1073/pnas.0611634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yanagisawa T, Ishii R, Fukunaga R, Kobayashi T, Sakamoto K, Yokoyama S. J Mol Biol. 2008;378:634–652. doi: 10.1016/j.jmb.2008.02.045. [DOI] [PubMed] [Google Scholar]
- 38.Nozawa K, O’Donoghue P, Gundllapalli S, Araiso Y, Ishitani R, Umehara T, Söll D, Nureki O. Nature. 2009;457:1163–1167. doi: 10.1038/nature07611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang N, Ju T, Niu W, Guo J. ACS Synth Biol. 2014;4:207–212. doi: 10.1021/sb500195w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shang X, Song X, Faller C, Lai R, Li H, Cerny R, Niu W, Guo J. Chem Sci. 2017;8:1141–1145. doi: 10.1039/c6sc03635j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee YJ, Kurra Y, Yang Y, Torres-Kolbus J, Deiters A, Liu WR. Chem Commun. 2014;50:13085–13088. doi: 10.1039/c4cc06435f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen PR, Groff D, Guo J, Ou W, Cellitti S, Geierstanger BH, Schultz PG. Angew Chem, Int Ed. 2009;48:4052–4055. doi: 10.1002/anie.200900683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hancock SM, Uprety R, Deiters A, Chin JW. J Am Chem Soc. 2010;132:14819–14824. doi: 10.1021/ja104609m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niu W, Schultz PG, Guo J. ACS Chem Biol. 2013;8:1640–1645. doi: 10.1021/cb4001662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


