Abstract
DNA-modified particles are used extensively for applications in sensing, material science, and molecular biology. The performance of such DNA-modified particles is greatly dependent on the degree of surface coverage, but existing methods for quantitation can only be employed for certain particle compositions and/or conjugation chemistries. We have developed a simple and broadly applicable exonuclease III (Exo III) digestion assay based on the cleavage of phosphodiester bonds—a universal feature of DNA-modified particles—to accurately quantify DNA probe surface coverage on diverse, commonly used particles of different compositions, conjugation chemistries, and sizes. Our assay utilizes particle-conjugated, fluorophore-labeled probes that incorporate two abasic sites; these probes are hybridized to a complementary DNA (cDNA) strand, and quantitation is achieved via cleavage and digestion of surface-bound probe DNA via Exo III’s apurinic endonucleolytic and exonucleolytic activities. The presence of the two abasic sites in the probe greatly speeds up the enzymatic reaction without altering the packing density of the probes on the particles. Probe digestion releases a signal-generating fluorophore and liberates the intact cDNA strand to start a new cycle of hybridization and digestion, until all fluorophore tags have been released. Since the molar ratio of fluorophore to immobilized DNA is 1:1, DNA surface coverage can be determined accurately based on the complete release of fluorophores. Our method delivers accurate, rapid, and reproducible quantitation of thiolated DNA on the surface of gold nanoparticles, and also performs equally well with other conjugation chemistries, substrates, and particle sizes, and thus offers a broadly useful assay for quantitation of DNA surface coverage.
Graphical abstract
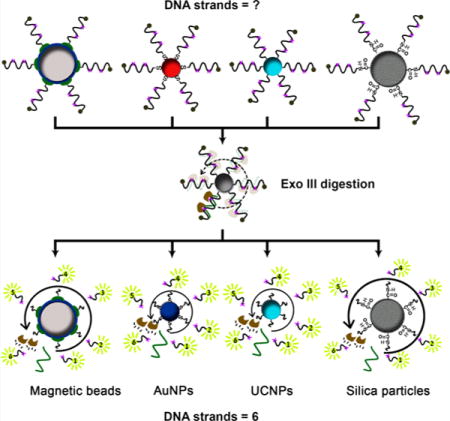
INTRODUCTION
DNA-modified nano- and microparticles can specifically recognize a variety of targets, including nucleic acids, proteins, peptides, small molecules, and metal ions.1 Accordingly, such particles have proven extraordinarily useful for bioimaging, bioseparation, diagnostic assays, drug targeting, nanotherapeutics, and nanomaterial assembly.2–6 Different conjugation chemistries have been used to immobilize DNA strands onto various particle substrates, including metallic nanoparticles, semiconductive quantum dots (QDs), inorganic upconversion nanoparticles (UCNPs), silica microspheres, and magnetic beads.7–15 In particular, alkanethiol adsorption has been employed for attaching thiolated DNA onto gold nanoparticles (AuNPs).7,16 The lanthanide–phosphate interaction has been used to conjugate unmodified DNAs onto lanthanide-doped hydrophobic UCNPs.9 The streptavidin (SA)–biotin interaction is often utilized to attach biotinylated DNA onto SA-coated magnetic beads,17,18 QDs,13 and UCNPs.19 In addition, when analyte detection or sample separation is being performed at elevated temperatures, amino-modified DNA has been covalently conjugated onto carboxylated magnetic11,14 or silica15 particles. Such covalently bound DNA probes offer greater stability for assays that employ heat-induced dehybridization,20,21 as high temperatures can disrupt SA–biotin interaction.18
The extent of DNA surface coverage can profoundly influence DNA hybridization efficiency,22–24 enzyme activity,22,25–29 assay sensitivity,30 cellular uptake efficiency,31 and thermostability of DNA nanoparticle-assembled superlattices.11,32 Many studies have shown that high surface coverage is associated with steric hindrance, electrostatic repulsive interactions, and elevated surface salt concentration, whereas low surface coverage can result in nonspecific binding of oligonucleotides to the particle surface. Both scenarios can greatly reduce DNA hydridization efficiency and enzyme activity for surface-bound DNA probes,33,34 and strategies for accurately quantifying the degree of surface coverage are critical for research applications based on DNA-conjugated particles.
Radioisotope labeling has been used to determine the number of conjugated oligonucleotides on a variety of surfaces.12,35–38 Briefly, probe DNAs are labeled at their 5′ end with a radioisotope such as 32P with T4 polynucleotide kinase before conjugation.12,35 Quantitation is then achieved by direct measurement of the radioactivity of radiolabeled DNA-modified particles using a liquid scintillation counter or a Geiger counter.12,35 Although 32P can be easily incorporated into the phosphate groups of dNTPs to generate highly sensitive, radiolabeled probes,36 the short half-life of 32P means that the labeled probes should be used within a week of preparation, leading to increased costs. Meanwhile, radioactive hazards are also a concern requiring personal protection and separate waste disposal procedures.
Radioisotope methods have become less popular as a variety of fluorescence-based assays have been developed for accurate quantitation of DNA surface coverage on various particle surfaces. Fluorophore-labeled DNA is strongly quenched when bound to particles such as AuNPs and magnetic beads, but this fluorescence is fully recovered once the DNA is detached from the surface. For thiolated, DNA-modified AuNPs, surface coverage is typically determined by measuring the fluorescence of oligonucleotides liberated via ligand displacement with mercaptoethanol33 or dithiothreitol (DTT).39 The standard DTT displacement protocol usually requires overnight incubation of DNA-modified AuNPs with high concentrations (∼0.5 M) of DTT at room temperature.39 Although time-consuming, the DTT displacement-based assay is recognized as a reliable “gold standard” for quantifying surface coverage on AuNPs irrespective of DNA length or sequence. Recently, several label-free methods utilizing PCR, OliGreen, and unimolecular beacon have been developed to characterize DNA surface coverage.40–42 However, these methods are only compatible with thiol-coupled DNA-AuNPs. Fluorescent assays are also available for accurate determination of surface coverage on SA-coated inorganic particles such as QDs, UCNPs, or magnetic beads. In this scenario, the interaction between biotinylated probe DNAs and SA-coated beads is disrupted in a mixture of ethylenediaminetetraacetic acid (EDTA) and formamide via high-temperature treatment (90 °C for 10 min).17 The eluted biotinylated DNAs can then be fluorescently measured. However, this method is not applicable to other conjugation chemistries, such as covalent linkage of thiolated DNA to aminated particles12 or of amino-labeled DNA to carboxylated magnetic or silica substrates.8,14,15,35
Alternatively, dissolution of particle substrates can ensure full release of the attached oligonucleotides for accurate quantification of DNA surface coverage. This is typically achieved with fluorophore labeling33,37 rather than absorbance measurement at 260 nm, which suffers two important impediments. First, such measurements are not sensitive enough to accurately quantify DNA at low levels of surface coverage (∼nM).43 Second, the chemicals used for AuNP dissolution, such as ferrocyanide and ferricyanide, exhibit strong absorbance at 260 nm,44 and this can interfere with accurate quantitation. Dissolution-based determination of DNA surface coverage has been reported for only two substrates: AuNPs33 and metal–organic framework (MOF) nanoparticles.37 A mixture of KCN and K3Fe(CN)6 was used to dissolve DNA-modified AuNPs for several minutes, after which the absorbance of the CN−-released fluorescein (FAM)-labeled DNAs was measured at 494 nm.33 DNA-modified MOF nanoparticles can be dissolved by incubating in NaOH for 18 h under mechanical shaking, with the absorbance of the released TAMRA-labeled DNAs measured at 566 nm.37 Such methods are constrained to specific substrates, however, due to the limited availability of dissolution chemistries.33,37 Thus, there is presently no simple and fast general approach that can be employed for rapidly and accurately quantifying DNA surface coverage across the large spectrum of commonly used particle compositions and conjugation chemistries.
To address this need, we have developed a generalizable method based on the chemical cleavage of phosphodiester bonds—a feature shared by all DNA-modified particles—that consistently achieves rapid and accurate quantitation based on fluorophore release, regardless of the conjugation chemistry employed, particle substrate, or size. Our assay utilizes exonuclease III (Exo III) to achieve cleavage and digestion at abasic sites incorporated into surface-bound, fluorophore-labeled probe DNAs that are hybridized with a complementary DNA (cDNA) strand. Hybridization of the cDNA prevents the surface-bound probes from wrapping around the particle surface, rendering them accessible to Exo III for digestion and enabling accurate quantitation. The presence of the abasic sites in the probe is not strictly essential, but greatly speeds up the process of enzyme digestion, and we have experimentally demonstrated that these abasic sites do not alter the packing density of probes on particle surface relative to probes lacking these sites. As an initial proof-of-concept, we have demonstrated complete fluorophore release from AuNP-conjugated, FAM-labeled DNA using our Exo III-based assay after 60 min at room temperature. The results were in very good agreement with those from an overnight DTT displacement assay for both high and low DNA surface coverage conditions. Our assay also achieved similar sensitivity to this “gold standard” DTT displacement approach. More importantly, our Exo III-based assay proved equally effective for quantifying surface coverage of DNA conjugated to other particles, including magnetic beads (1 μm), QDs (18 nm), silica microspheres (1 μm), and UCNPs (25 or 76 nm). We further demonstrated that our method is effective with probes conjugated via streptavidin–biotin interaction, covalent linkage, or electrostatic interaction. In most instances, we observed far more efficient fluorophore release with our Exo III-based assay relative to existing assays. Our assay therefore delivers valuable new analytical capabilities for accurate DNA quantitation, greatly reducing the measurement time needed for DNA-conjugated AuNPs while also offering a general-purpose assay that can readily be applied to various conjugation chemistries, substrates, and sizes.
RESULTS
We first employed our Exo III-based assay with FAM-labeled probe DNA-modified AuNPs. Exo III has 3′-to-5′ exonuclease activity and processes nucleic acids in a sequence-independent fashion.45 We first exploited the exonucleolytic activity of Exo III to catalyze the stepwise removal of mononucleotides from 3′-hydroxyl termini of duplex DNA as a means to accurately determine DNA surface coverage on AuNPs. The procedure for the Exo III-based assay is illustrated in Figure 1.
Figure 1.
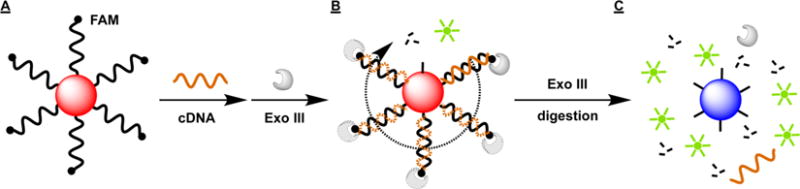
Scheme of the Exo III-based assay for quantifying surface coverage of probe DNA on AuNPs. Fluorophore-labeled, AuNP-coupled DNAs (A) are hybridized with a complementary cDNA and subsequently undergo exonucleolytic digestion by Exo III (B). This releases fluorophores and intact cDNA strands, allowing multiple rounds of digestion until all fluorophores are released (C). The resulting aggregation of the probe-free AuNPs produces a visible red-to-blue color change.
These AuNP-conjugated, FAM-labeled probes (Figure 1A) are hybridized to a complementary DNA (cDNA) strand (Supporting Information (SI), Table S1, cDNA-8A) to form a perfectly matched duplex, and quantitation is achieved via Exo III digestion of surface-bound probe DNAs (Figure 1B). Digestion releases a signal-generating fluorophore and liberates the intact cDNA strand to start anew until all probes have been digested from the particle surface, generating an AuNP aggregation-induced visible color shift from red to blue (Figure 1C). Since the molar ratio of FAM to immobilized probe DNA is 1:1, DNA surface coverage can be determined accurately based on the complete release of fluorophores after enzyme digestion.
We designed a 47-nt probe DNA with the following sequence: 5′-poly(T6)-ACCACATCATCCATATAACTGAAAGCCAAACAGTTTTTTTT-3′. The 5′ poly(T6) acts as a flexible linker to improve cDNA hybridization. We synthesized the probe with a 5′ thiol group and a 3′ FAM-label (SI, Table S1, SH probe) and conjugated it onto AuNPs via thiol-gold chemistry39 at a ratio of 300:1. After the modification, we found that 70% of the DNA was unconjugated and remained in the supernatant (data not shown). This large excess confirms that these conditions ensure saturated DNA loading of the AuNPs, and indicate that coverage would not meaningfully increase at even higher DNA:AuNP ratios. These DNA-modified AuNPs are stable in reaction buffer, and the resulting solution is red in color due to the strong electrostatic repulsion of DNA-modified nanoparticles. We hybridized the modified AuNPs with cDNA-8A, and then incubated with Exo III. Digestion of the surface-bound probes causes the probe-free AuNPs to become unstable in the reaction buffer, leading to salt-induced aggregation that gives rise to a red-to-blue color change. We tracked this response by performing time-dependent measurements of UV absorbance (Figure 2A), specifically monitoring the ratio of absorbance at 650 to 520 nm (A650/A520) (Figure 2B) that has served as an indicator of AuNP aggregation.46 We observed that aggregation occurred gradually, with complete digestion of surface-bound probe DNAs achieved after 255 min. We also measured the time-dependent release of the fluorophore from AuNP-conjugated SH probes. We found that 75% of the FAM molecules were released in the first 60 min, and fluorescence intensity subsequently achieved saturation after 180 min (Figure 2C). As expected, the fluorophore was released before the complete degradation of immobilized probe DNAs on AuNP.
Figure 2.
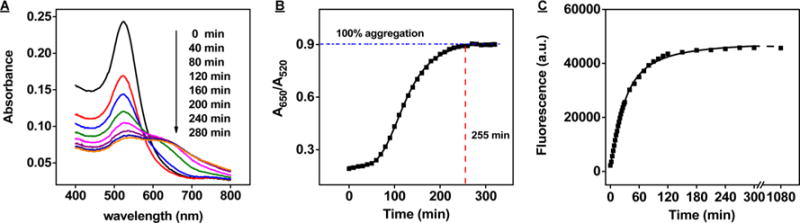
Exo III digestion of DNA-conjugated AuNPs modified at a DNA:AuNP ratio of 300. Time-dependent absorbance spectra (A) and absorbance ratio (A650/A520) (B) and fluorescence intensity (C) of the SH probe-modified AuNPs in Exo III-based assay. The A650/A520 of SH probe DNA-modified AuNPs reached saturation after 255 min.
To test if our Exo III-based assay can quantify AuNP-conjugated probe DNA in both high and low surface-coverage scenarios, we prepared DNA-modified AuNPs at SH probe DNA:AuNP ratios of 60, 80, 120, 150, 200, and 300 and characterized their surface coverage under the conditions described above. After a 255 min incubation, we separated the supernatant from the AuNP precipitate and measured its fluorescence. We confirmed that Exo III was able to cleave the 3′ FAM-modified nucleotide from the probe, thereby releasing it into solution. The fluorescence intensity increased in proportion to the amount of DNA used for the modification (Figure 3A and SI, Figure S1), and we used a standard curve generated from unconjugated FAM-labeled, SH probe DNA with Exo III and cDNA under the same experimental conditions (Figure 3B) to calculate DNA surface coverage. We determined that AuNPs modified with DNA:AuNP ratios of 60, 80, 120, 150, 200, and 300 respectively displayed 38 ± 1, 44 ± 1, 53 ± 1, 65 ± 1, 69 ± 1, and 79 ± 2 oligonucleotides per particle, equivalent to surface coverage of 12.1 ± 0.3, 13.8 ± 0.2, 16.7 ± 0.2, 20.4 ± 0.3, 21.7 ± 0.3, and 24.7 ± 0.6 pmol/cm2, respectively (Figure 3C). To evaluate the accuracy of our Exo III-based assay, we performed a DTT displacement assay39 with the same set of samples. We mixed each batch of SH probe DNA-modified AuNPs with an equal volume of 1.0 M DTT and incubated the mixture at room temperature for 12 h, after which we centrifuged the samples and measured the fluorescence of the collected supernatant (Figure 3A, SI, Figure S2). We used the constructed calibration curve (Figure 3B) to determine that the surface coverage measurements obtained via DTT displacement were in very good agreement with the values obtained via Exo III digestion (Figure 3C).
Figure 3.
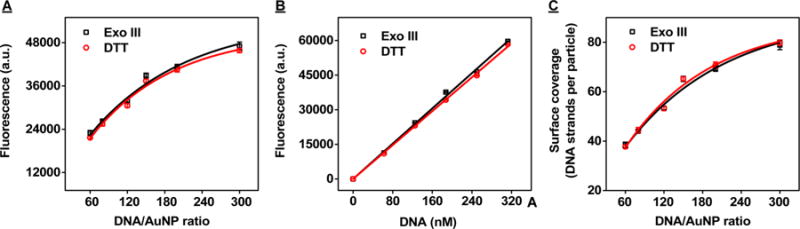
Comparison of the Exo III-based and the DTT displacement assays with SH probe DNA-conjugated AuNPs prepared at different DNA:AuNP ratios. (A) Fluorescence intensities of supernatants after Exo III digestion (black) and DTT displacement (red). (B) Standard calibration curve obtained with known concentrations of FAM-labeled, thiolated probe DNA after Exo III digestion (black) and DTT displacement (red). (C) Surface coverage of SH probe DNA-modified AuNPs as characterized by Exo III digestion (black) and DTT displacement (red). Error bars show standard deviations obtained from three measurements.
In addition to 3′-to-5′ exonuclease activity, Exo III has also been reported to have apurinic (AP) endonuclease activity that digests propanyl abasic sites inserted into DNA strands.47 We predicted that the enzymatic digestion in our Exo III-based assay could be profoundly accelerated by inserting propanyl abasic sites47 into the DNA probe, thereby exploiting both activities of Exo III. Specifically, we utilized Exo III’s endonuclease activity to generate internal nicks at the abasic sites (marked as X in the probe sequence), with subsequent exonucleolytic digestion at the newly created nicks considerably accelerating the assay process. We inserted two internal propanyl abasic sites into our SH probe DNA, producing the SH-2X probe (SI, Table S1, SH-2X probe). The SH-2X probe DNA was then attached onto AuNPs via thiol linkage (Figure 4A). When the immobilized probe hybridizes to the cDNA strand (SI, Table S1, cDNA), it forms a duplex with an 8-nt sticky end (Figure 4B). The two duplexed abasic sites provide binding sites for Exo III, which endonucleolytically cleaves the phosphodiester bond and generates two nicks with hydroxyl groups at the 3′ end (Figure 4C). This converts the SH-2X probe into three nicked duplex fragments (12, 11, and 8 bp). The FAM-labeled 8-bp strand (Tm = 21.9 °C) readily dissociates from the cDNA at room temperature, while the 11- and 12-bp fragments (Tm = 27.8 and 44.5 °C, respectively) remain hybridized to the cDNA. Exo III subsequently initiates 3′-to-5′ exonucleolytic digestion of the SH-2X probe DNA at these newly formed 3′-hydroxyl termini. When the probe is degraded, liberating the fluorophore, the intact cDNA is released (Figure 4D) to start a new cycle of DNA hybridization and Exo III digestion (Figure 4E). Once all of the attached probes have been sheared from the surface, we use the resulting fluorescence to accurately determine DNA surface coverage (Figure 4F).
Figure 4.
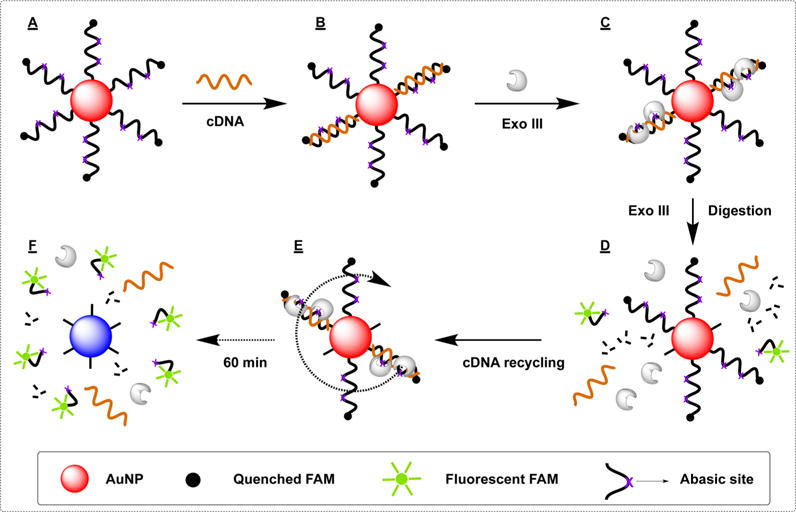
Schematic illustration of our Exo III-based assay for quantifying the surface coverage of SH-2X probe DNA conjugated with AuNPs. AuNPs are modified with DNA probes containing two abasic sites (A). These are hybridized to cDNA (B) and then incubated with Exo III (C). The enzyme introduces internal nicks at the abasic sites, and then performs exonucleolytic digestion of the remaining probe fragments (D). The cDNA is released intact for further rounds of digestion (E), until all probes have been stripped away and the released fluorophores can be quantified to assess surface coverage (F).
We experimentally confirmed that the insertion of abasic sites accelerated DNA probe digestion and that the number of abasic sites strongly influences the reaction rate. We immobilized two versions of our probe DNA containing either one or two abasic sites (Figure 5A and SI, Table S1, SH-1X and -2X probes) onto AuNPs with a saturating DNA:AuNP ratio of 300. We determined that the surface coverage for these two new probes was essentially identical to that of the original SH-probe DNA (79 ± 2 oligonucleotides/particle), with surface densities of 77 ± 3 and 79 ± 3 oligonucleotides/particle for the SH-1X and -2X probes, respectively. We then performed Exo III digestion of these three batches of modified AuNPs and recorded the absorbance change at 520 and 650 nm. The time course of aggregation confirmed that the SH-2X probe DNA produced a much more rapid reaction (SI, Figure S3A) relative to the SH (Figure 2A) or SH-1X probes (SI, Figure S3B). The reaction times required for complete aggregation of the AuNPs were 60, 230, and 255 min for AuNPs modified with SH-2X, -1X probes and SH probe, respectively (Figure 5B). Clearly, the presence of two abasic sites in the 2X probe DNA enables far more rapid digestion. Compared with the SH probe, the SH-1X probe exhibited only moderately faster digestion, presumably because the 20-bp 3′ duplexed fragment (Tm = 56.9 °C) formed by cleavage at the 5′ abasic site is very stable, which prevents the cDNA from releasing at room temperature and thereby delays further digestion steps. We therefore used the SH-2X probe DNA for all subsequent experiments with our Exo III-based assay.
Figure 5.
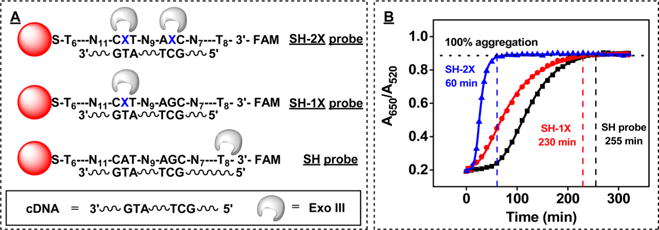
Effect of the number of abasic sites on the rate of digestion for DNA-modified AuNPs in the Exo III-based assay. (A) Scheme of Exo III digestion for cDNA duplexes formed with AuNP-conjugated SH-2X, SH-1X, and SH probes. (B) Time-dependent absorbance changes (A650/A520) of AuNPs modified with SH-2X, SH-1X, and SH probe DNA.
Previous studies have shown that the AP activity of Exo III is marginally influenced by the presence of different bases at positions complementary to abasic sites.48 We conducted experiments to test for this effect under our assay conditions. We found that cDNAs with C or T opposite the abasic site (SI, Table S1, cDNA-C and -T) contributed to more efficient digestion by Exo III relative to those with A or G opposite the abasic site (SI, Table S1, cDNA-A and -G) (SI, Figure S4) at low enzyme concentrations (0.004 U/μL), but the difference became negligible and did not affect the assay’s efficiency when 0.2 U/μL of Exo III was used (SI, Figure S5).
To verify that all fluorophore labels had been released from the particle surface at the point of AuNP aggregation, we measured the time-dependent release of the fluorophore from AuNP-conjugated SH-2X probes by monitoring the change in FAM intensity. Most FAM molecules were released in the first 15 min of the reaction, with 75% of the FAM released into solution (SI, Figure S6). Fluorescence intensity subsequently increased slowly, achieving saturation after 60 min, with no further increase after 18 h. Clearly, the complete release of fluorophores is consistent with the aggregation of the AuNPs, indicating that Exo III can achieve entire digestion of surface-bound 2X-probe within 1 h.
Digestion of surface-bound probes requires the formation of a duplex with cDNA, since Exo III is inactive on single-stranded DNA. When hybridized with the AuNP-immobilized DNA probe, the 3′ end of the cDNA is close to the surface of the AuNPs. We believe that the 3′-to-5′ exonuclease activity of Exo III is greatly inhibited by steric hindrance near the particle surface. We have previously shown that cDNA concentrations as low as 2 nM can result in complete removal of all probe DNA from AuNPs, resulting in aggregation,49 demonstrating that low concentrations of cDNA can be recycled efficiently. However, it is not necessary for the cDNA to be recycled as long as we have sufficient cDNA in the sample. We further investigated the effect of cDNA concentration on Exo III efficiency. We observed that the initial reaction rate of Exo III increased with increasing concentrations of cDNA until reaching a plateau at 200 nM (SI, Figure S7). Further increases in the cDNA concentration up to 500 nM yielded only a slightly higher reaction rate. We therefore used 200 nM cDNA for subsequent experiments. We also examined the effect of Exo III concentration, and found that increasing the amount of Exo III greatly increased the initial reaction rate (SI, Figure S8). 0.2 U/μL of Exo III was used for subsequent experiments to achieve a rapid reaction.
We confirmed the accuracy of the Exo III-based assay utilizing SH-2X probe modified AuNPs under both low and high surface coverage conditions. We conjugated SH-2X probe DNA with AuNPs at DNA:AuNP ratios of 60, 80, 120, 150, 200, and 300 under the same modification conditions described above and characterized the surface coverage of the six batches of AuNPs with the Exo III-based (SI, Figure S9) and DTT displacement (SI, Figure S10) assays. We found that the measurements in high and low surface coverage scenarios obtained via both methods were in very good agreement (SI, Figure S11). However, the Exo III-based assay can be performed in a far shorter time relative to the DTT displacement assay, especially when using abasic site containing probes (1 h vs 12 h). Since the surface coverage values of the SH-2X probe (SI, Figure S11) were essentially identical to those obtained with the SH probe (Figure 3), we conclude that the inclusion of the abasic sites speeds up the enzymatic reaction without altering either the DNA packing density on the particle surface or the accuracy of quantitation.
The Exo III-based assay also demonstrated a comparable limit of quantitation (LOQ) to the DTT displacement assay. We performed Exo III digestion using SH-2X probe DNA-modified AuNPs with maximum surface coverage at particle concentrations ranging from 31 fM to 3.1 nM, equivalent to a concentration of SH-2X probe DNA ranging from 2.5 pM to 250 nM. Our results demonstrated good linearity, with a LOQ of 34.6 pM probe DNA (8.3 × 108 oligonucleotides) (Figure 6). This demonstrates the high sensitivity of the Exo III-based assay; based on the small testing volume of our assay (40 μL), only 1.4 fmol DNA—or a subfemtomole quantity of AuNPs—is required for accurate quantitation. We observed a similar LOQ (26.3 pM, 6.3 × 108 oligonucleotides) for the DTT displacement-based assay using the same set of samples.
Figure 6.
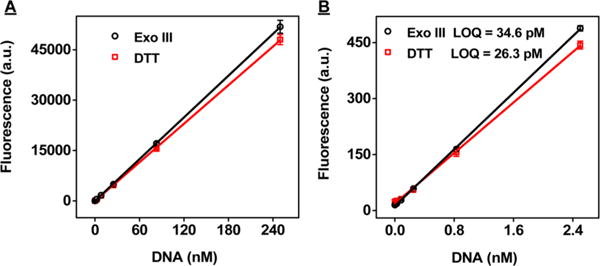
Sensitivity of the Exo III-based (black) and DTT displacement (red) assays. (A) Fluorescence intensities at 520 nm for supernatants from different DNA concentrations of SH-2X probe DNA-modified AuNPs (79 oligonucleotides per particle) after performing both assays. (B) We determined the limit of quantitation (LOQ) for both assays based on a signal-to-noise ratio of 10 (ref 50). Error bars show standard deviations obtained from three measurements.
Our Exo III-based assay achieves rapid and accurate determination of DNA surface coverage due to the high efficiency of Exo III and increased enzyme accessibility resulting from cDNA hybridization. DNase I digestion offers an alternative nuclease-based method51,52 that cleaves fluoro-phore-labeled DNA from gold surfaces for subsequent quantification. DNase I is an endonuclease that nonspecifically cleaves single- and double-stranded DNA to release di-, tri-, and oligonucleotide products with 5′-phosphorylated and 3′-hydroxylated ends.53 To compare the digestion efficiency of Exo III and DNase I, we monitored the time course for digestion of SH-2X probe DNA-modified AuNPs with maximum surface coverage with 80 nM Exo III (0.2 U/μL) or 80 nM DNase I (0.125 U/μL) as reported optimized condition51 in the presence of 200 nM cDNA. After 60 min, Exo III achieved 100% probe digestion whereas DNase I digested only 42% of the probe DNA (Figure 7A). We further compared the capacity of the DNase I-based assay to quantify DNA at different levels of surface coverage relative to Exo III. We mixed six batches of SH-2X probe DNA-modified AuNPs displaying various DNA surface coverage with DNase I and incubated the mixture at room temperature for 16 h, then collected the supernatants to obtain fluorescence spectra (SI, Figure S12A). We used a standard curve established with different concentrations of unconjugated FAM-labeled, thiolated 2X probe DNA and 0.125 U/μL of DNase I to calculate the DNA surface coverage on these modified AuNPs (SI, Figure S12B). The performance of the DNase I-based assay was generally inferior to that of the Exo III-based assay, with estimates of surface coverage for all six batches of DNA-modified AuNPs that were significantly smaller than those obtained by our Exo III-based assay (Figure 7B and SI, Table S2).
Figure 7.
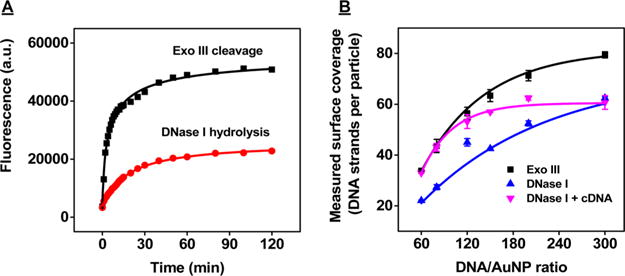
Reaction kinetics for Exo III digestion and DNase I hydrolysis of SH-2X probe DNA-modified AuNPs. (A) Time-dependent fluorescence changes for AuNPs modified with 79 oligonucleotides per particle upon addition of either 80 nM Exo III (0.2 U/μL) or DNase I (0.125 U/μL) in the presence of 200 nM cDNA. (B) Surface coverage measurements obtained for DNA-modified AuNPs with different amounts of SH-2X probe DNA, as characterized by Exo III digestion with 200 nM cDNA (black) or DNase I hydrolysis with (pink) or without 200 nM cDNA (blue). Error bars show standard deviations from three measurements.
This incomplete digestion by DNase I could be attributable to two reasons. First, DNase I activity is highly dependent on salt concentration, and digestion can be inhibited when DNA surface coverage is high due to an elevated local salt concentration.26 In contrast, Exo III is robust against high local salt concentrations. Furthermore, the wrapping of immobilized single-stranded probe DNAs around particles54 typically reduces the accessibility of the probes to enzymes,55,56 impairing accurate quantitation with DNase I—especially when DNA surface coverage is low. The persistence length of ssDNA is around 8 nm (∼24 nucleotides),57 and this lack of stiffness makes it easy for the probe to bind to the particle surface. When cDNA binds with the probe to form a duplex, however, the persistence length of dsDNA increases to 50 nm (∼150 nucleotides),58 which is much longer than our probe-cDNA duplex (14 nm/41 nucleotides). Thus, the duplexed probe would be predicted to not bend and wrap around the curved particle surface, and should extend perpendicularly to the AuNP59 regardless of the degree of surface coverage.
We predicted that duplex formation between surface-bound probe DNA and cDNA should minimize surface adsorption in the DNase I-based assay and thus enhance the accuracy of the measurements. To confirm this, we incubated our SH-2X probe DNA-modified AuNPs with 0.125 U/μL of DNase I and 200 nM of cDNA for 16 h. The supernatants were collected and recorded (SI, Figure S13A). We used a standard curve (SI, Figure S13B) established with different concentrations of unconjugated SH-2X probe DNA, 200 nM of cDNA, and 0.125 U/μL of DNase I to calculate DNA surface coverage on these modified AuNPs. As expected, the presence of the cDNA improved the performance of the DNase I assay, yielding measurements that were in good agreement with our Exo III-based assay for AuNPs with relatively low surface coverage (Figure 7B and SI, Table S2). This suggests that the duplexed probe DNAs provide more access to DNase I, thereby facilitating complete digestion. However, the DNase I-based assay still produced lower measurements when DNA surface coverage was greater than 56 oligonucleotides/particle (Figure 7B and SI, Table S2), presumably due to inhibition of DNase I by high local salt concentrations at the surface.26
To demonstrate the generalizability of our Exo III-based method for determining DNA surface coverage on various substrates with different conjugation chemistries and sizes, we first quantified surface coverage of biotinylated 2X probe DNA (SI, Table S1, biotin-2X probe) on SA-coated magnetic beads (MB-SA, 1 μm diameter). We modified the beads with equal volumes of 5 μM biotinylated DNA in binding buffer, and then washed and separated the DNA-modified beads from unbound DNA using a magnetic particle concentrator. Using the same Exo III digestion procedure described above, we could readily obtain complete fluorophore release after 60 min, with no measurable increase of fluorescence after an overnight reaction (Figure 8A). Based on the corresponding standard curve (SI, Figure S14), we measured an average (3.4 ± 0.1) × 105 DNA molecules per MB-SA, equivalent to a surface coverage of 18.0 ± 0.6 pmol/cm2. Heat-treated elution in a mixture of EDTA and formamide has been used to measure DNA surface coverage under such conditions by disrupting SA-biotin binding.17 Using this approach, we obtained a value of 16.1 ± 0.6 pmol/cm2 (SI, Figure S15A and C). This is 10% less than the Exo III-based measurement, indicating that not all of the biotinylated DNA was successfully dissociated under such conditions. Employing the same conjugation method, we attached the biotin-2X probe DNA onto SA-coated quantum dots (QD-SA, 18 nm diameter) and performed Exo III digestion with the DNA-modified QDs. We used the established standard curve (SI, Figure S16) to calculate surface coverage on the DNA-modified QDs after 60 min (Figure 8B), measuring an average 34 ± 1 oligonucleotides per QD-SA, equivalent to a surface coverage of 5.6 ± 0.1 pmol/cm2. We obtained a comparable value using the heating-based elution assay, finding an average of 32 ± 1 oligonucleotides per QD-SA (5.3 ± 0.1 pmol/cm2) (SI, Figure S17A,B).
Figure 8.
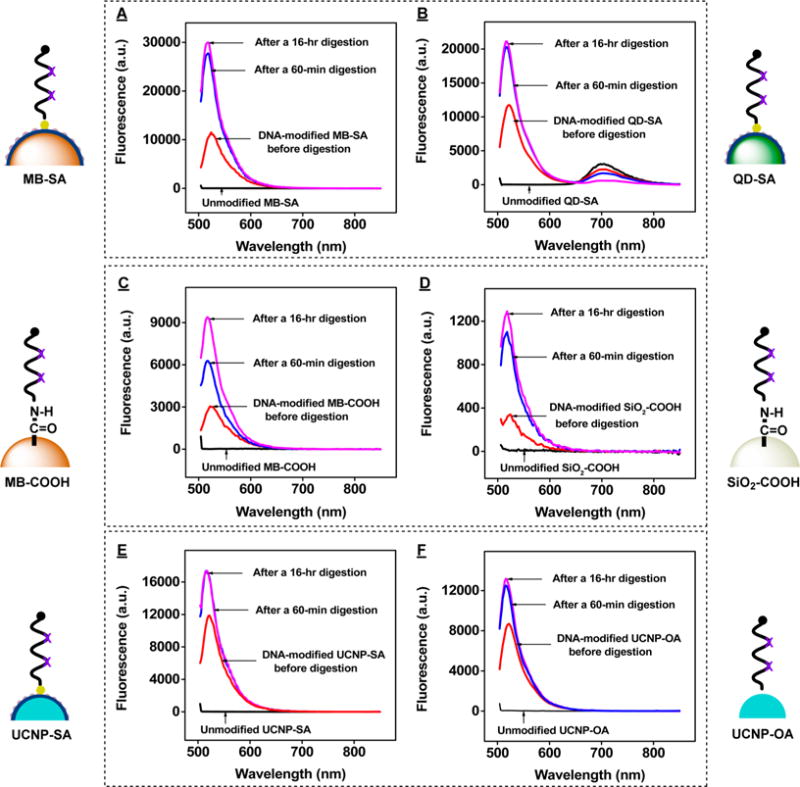
Exo III-based quantification of DNA surface coverage on various particles. Fluorescence spectra of unmodified beads (black), undigested modified beads (red), and supernatants from Exo III-treated, DNA-modified (A) streptavidin-coated magnetic beads (MB-SA), (B) streptavidin-coated quantum dots (QD-SA), (C) carboxylated magnetic beads (MB-COOH), (D) carboxylated silica microspheres (SiO2-COOH), (E) streptavidin-coated UCNPs (UCNP-SA), and (F) oleic acid-capped UCNPs (UCNP-OA). Assays were conducted for 1 h (blue) or 16 h (magenta) with 200 nM cDNA and 0.2 U/μL Exo III.
Our method was equally successful in quantifying amino-labeled DNA (SI, Table S1, NH2-2X probe) covalently conjugated onto carboxylated magnetic beads (MB-COOH, 1 μm diameter) and silica microspheres (SiO2-COOH, 1 μm diameter). We used ethyl(dimethylaminopropyl) carbodiimide (EDC) to activate the carboxylic acid groups on the surface of MB-COOH or SiO2-COOH in MES buffer, and incubated with NH2-2X probe DNA. The DNA-modified magnetic beads were collected with a magnetic particle concentrator, and the DNA-modified silica microspheres were separated from unbound DNA by several rounds of centrifugation and resuspension. We mixed the modified beads with 0.2 U/μL of Exo III and 200 nM of cDNA and incubated for either 1 or 16 h. After centrifuging the samples, we collected the supernatants and obtained the sample fluorescence. Based on a standard curve (SI, Figure S18) established with different concentrations of unconjugated NH2-2X probe DNA, 200 nM of cDNA, and 0.2 U/μL of Exo III, we determined that the surface coverage was (1.1 ± 0.1) × 105 DNA strands per magnetic bead (Figure 8C) and (7.4 ± 0.2) × 103 DNA strands per silica microsphere (Figure 8D), equivalent to surface coverage of 5.8 ± 0.3 and 0.39 ± 0.01 pmol/cm2, respectively. We noted that overnight incubation was required to achieve complete digestion with covalently conjugated DNA particles. The reason for this is still unclear, but one possibility is that nonspecific adsorption of Exo III onto the surface of these nonprotein-coated microparticles60,61 greatly reduces the enzyme’s ability to access the conjugated oligonucleotides. This limitation aside, it is important to note that our Exo III-based assay represents the first published fluorescence method capable of accurately quantifying coverage for such covalently conjugated DNA probes.
Finally, we demonstrated successful determination of DNA surface coverage on UCNPs. DNA-modified UCNPs have gained considerable attention in the field of DNA-based bionanotechnology because of their distinctive properties, such as exceptional photostability, suppression of autofluorescence, and low in vitro and in vivo toxicity.62,63 DNA can be attached to UCNPs through either SA-biotin binding or direct adsorption. Therefore, we prepared 2X probe DNA-modified UCNPs by attaching biotin-2X probe DNA onto SA-coated UCNPs (UCNP-SA, 76 nm diameter)19 or directly conjugating 5′-unmodified 2X probe DNA onto oleic acid-capped UCNPs (UCNP-OA, 25 nm diameter)9 via the electrostatic interaction between phosphoric acid groups on the DNA and lanthanide ions on the UCNPs.64 Employing the Exo III-based assay, we readily obtained complete fluorescence recovery after 60 min, with no measurable increase of fluorescence after an overnight reaction (Figure 8E,F). Based on the corresponding standard curves (SI, Figures S14 and S19), we measured an average of 30 ± 1 DNA molecules per UCNP-SA and an average of 19 ± 1 DNA molecules per UCNP-OA, equivalent to a surface coverage of 0.27 ± 0.01 pmol/cm2 and 1.6 ± 0.1 pmol/cm2, respectively. We obtained a comparable value using the heating-based elution assay, finding an average of 25 ± 1 DNA molecules per UCNP-SA, equivalent to a surface coverage of 0.22 ± 0.01 pmol/cm2 (SI, Figure S15B,C).
CONCLUSION
We have demonstrated a general-purpose method for the quantitation of DNA surface coverage that targets the phosphodiester bonds of DNA probes—a feature shared by all DNA-modified particles. Our system exploits the distinct dual enzymatic activities of Exo III (with the assistance of a hybridized cDNA) to achieve efficient probe digestion and complete fluorophore release. The resulting fluorescence serves as a reporter for accurately quantifying DNA conjugated onto particles of diverse sizes and compositions via a variety of different chemistries.
Using DNA-modified AuNPs as an initial proof-of-concept, we demonstrated that our Exo III-based method can deliver accurate and reproducible quantitation for both high- and low-surface-coverage scenarios, with results closely comparable to the “gold standard” DTT-displacement assay, yet much faster. Importantly, our method generated essentially identical measurements from AuNP conjugation experiments with probes that either contain or lack abasic sites, and both probes achieve equivalent coverage on the AuNP surface after the modification. However, the addition of abasic sites to the probe significantly accelerates enzymatic digestion. Note that we obtained equivalent surface coverage measurements for both unmodified and abasic site-containing probes, indicating that the effects of any alterations in cDNA hybridization in the latter scenario are minimal in terms of assay performance. Additionally, our assay proved effective with other particle substrates, such as QDs, UCNPs, magnetic beads, and silica microspheres, delivering accurate quantitation of both noncovalently and covalently bound DNAs with results that were comparable to or superior to other standard assays (SI, Table S3). Although the fluorophore-labeled DNA probes and excess amount of complementary DNA are required to complete the characterization of DNA surface coverage in the Exo III-based assay, we believe that our Exo III-based assay has the potential to provide simple and accurate measurement of surface coverage for virtually any class of DNA-particle conjugates.
EXPERIMENTAL SECTION
Quantitation of DNA Surface Coverage on AuNPs via Exo III-Based Assay
1.25 μL of SH-2X probe DNA-modified AuNPs (final concentration 3.1 nM) was mixed with 36.75 μL of reaction buffer (20 mM Tris-Ac, 50 mM KAc, 25 mM NaCl, 20 mM CaCl2, 3 mM MgCl2, 1 × BSA, pH 7.9), 2 μL of 4 μM cDNA and 3 μL of 2.7 U/μL Exo III (both diluted with reaction buffer). After 60 min incubation at room temperature, the solution was centrifuged at 21,130 rcf × 10 min to remove the AuNP precipitates, after which 40 μL of supernatant was transferred into a 384-well microplate to record fluorescence intensities of fluorescein label using a Tecan Infinite M1000 PRO with λex = 495 nm and λem = 520 nm. To establish the standard calibration curve, 38 μL of reaction buffer containing known concentrations of SH-2X probe DNA was mixed with 2 μL of 4 μM cDNA and 3 μL of Exo III (2.7 U/μL) to obtain probe concentrations ranging from 0 to 250 nM. These samples underwent the same incubation and centrifugation process as the modified AuNP samples, with 40 μL of each solution used for measurement. To investigate the effect of cDNA concentration on the reaction time, SH-2X probe DNA-modified AuNPs with a surface coverage of 79 oligonucleotides/particle were mixed with various concentrations of cDNA (0, 10, 20, 50, 100, 200, and 500 nM) and Exo III (0.01 U/μL) in 40 μL reaction buffer. The mixtures were loaded into a 384-well microplate and the time-dependent fluorescence changes (λex = 495 nm and λem = 520 nm) were recorded. The initial reaction rate was determined by the slope of the exponential fitting curve at 0 min.
Quantitation of DNA Surface Coverage on Different Particles via Exo III-Based Assay
1.25 μL of 2X probe DNA-modified particles (final concentration 3.1 nM) were mixed with 36.75 μL of reaction buffer, 2 μL of 4 μM cDNA, and 3 μL of 2.7 U/μL Exo III (both diluted with reaction buffer). After incubation for 60 min or 16 h at room temperature, the solution was centrifuged at 21,130 rcf × 10 min to remove the particles, after which 40 μL of supernatant was transferred into a 384-well microplate to record the fluorescence spectra from 504 to 850 nm (λex = 495 nm). To establish the standard calibration curve, 38 μL of reaction buffer containing known concentrations of biotin-2X probe DNA (for SA-coated magnetic beads, QDs or UCNPs), NH2-2X probe DNA (for carboxylated magnetic or silica beads), or 5′-unmodified-2X probe DNA (for oleic acid-capped UCNPs) was mixed with 2 μL of 4 μM cDNA and 3 μL of 2.7 U/μL Exo III to obtain probe solutions ranging from 0 to 250 nM. These samples underwent the same incubation and centrifugation process as the DNA-modified AuNP samples, with 40 μL of each solution used for fluorescence measurement. Since DNA-modified QD-SA particles cannot be completely removed by centrifugation and DNA-modified QD-SA particles did not significantly quench FAM fluorescence, we directly collected the fluorescence spectra from 504 to 850 nm (λex = 495 nm) from samples of DNA-modified QD-SA particles. The same amount of unmodified QD-SA particles was added for calibration before Exo III digestion.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health – National Institute on Drug Abuse [R15DA036821], and National Institute of Justice, Office of Justice Programs, U.S. Department of Justice [2013-DN-BX-K032 and 2015-R2-CX-0034]. The opinions, findings, and conclusions or recommendations expressed in this publication/program/exhibition are those of the author(s) and do not necessarily reflect those of the Department of Justice. The authors thank Dr. Lele Li and Professor Yi Lu for their advice on synthesis of UCNPs, and Nitya Sai Reddy Satyavolu for collecting the TEM images for UCNPs.
Footnotes
ORCID
Yi Xiao: 0000-0001-7278-9811
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bioconjchem.6b00660.
Materials; instrumentation; detailed methods; DNA sequences and surface coverage, time-dependent absorbance and fluorescence intensity, reaction rates, calibration curves, fluorescence spectra, TEM imaging (PDF)
Notes
The authors declare no competing financial interest.
References
- 1.Liu JW, Cao ZH, Lu Y. Functional nucleic acid sensors. Chem Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nutiu R, Li YF. In vitro selection of structure-switching signaling aptamers. Angew Chem, Int Ed. 2005;44:1061–1065. doi: 10.1002/anie.200461848. [DOI] [PubMed] [Google Scholar]
- 3.Cutler JI, Auyeung E, Mirkin CA. Spherical nucleic acids. J Am Chem Soc. 2012;134:1376–1391. doi: 10.1021/ja209351u. [DOI] [PubMed] [Google Scholar]
- 4.Rosi NL, Mirkin CA. Nanostructures in biodiagnostics. Chem Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 5.Smith JE, Wang L, Tan WH. Bioconjugated silica-coated nanoparticles for bioseparation and bioanalysis. TrAC, Trends Anal Chem. 2006;25:848–855. [Google Scholar]
- 6.Lee JH, Yigit MV, Mazumdar D, Lu Y. Molecular diagnostic and drug delivery agents based on aptamer-nanomaterial conjugates. Adv Drug Delivery Rev. 2010;62:592–605. doi: 10.1016/j.addr.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 8.Xue C, Chen XD, Hurst SJ, Mirkin CA. Self-assembled monolayer mediated silica coating of silver triangular nanoprisms. Adv Mater. 2007;19:4071–4074. [Google Scholar]
- 9.Li LL, Wu PW, Hwang K, Lu Y. An exceptionally simple strategy for DNA-functionalized up-conversion nanoparticles as biocompatible agents for nanoassembly, DNA delivery, and imaging. J Am Chem Soc. 2013;135:2411–2414. doi: 10.1021/ja310432u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell GP, Mirkin CA, Letsinger RL. Programmed assembly of DNA functionalized quantum dots. J Am Chem Soc. 1999;121:8122–8123. [Google Scholar]
- 11.Cutler JI, Zheng D, Xu XY, Giljohann DA, Mirkin CA. Polyvalent oligonucleotide iron oxide nanoparticle “click” conjugates. Nano Lett. 2010;10:1477–1480. doi: 10.1021/nl100477m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strömberg M, Gunnarsson K, Johansson H, Nilsson M, Svedlindh P, Strømme M. Interbead interactions within oligonucleotide functionalized ferrofluids suitable for magnetic biosensor applications. J Phys D: Appl Phys. 2007;40:1320–1330. [Google Scholar]
- 13.Zhang CY, Yeh HC, Kuroki MT, Wang TH. Single-quantum-dot-based DNA nanosensor. Nat Mater. 2005;4:826–831. doi: 10.1038/nmat1508. [DOI] [PubMed] [Google Scholar]
- 14.Parham NJ, Picard FJ, Peytavi R, Gagnon M, Seyrig G, Gagne PA, Boissinot M, Bergeron MG. Specific magnetic bead based capture of genomic DNA from clinical samples: application to the detection of group B streptococci in vaginal/anal swabs. Clin Chem. 2007;53:1570–1576. doi: 10.1373/clinchem.2007.091389. [DOI] [PubMed] [Google Scholar]
- 15.Cheng XG, Basuray S, Senapati S, Chang HC. Identification and separation of DNA-hybridized nanocolloids by taylor cone harmonics. Electrophoresis. 2009;30:3236–3241. doi: 10.1002/elps.200900159. [DOI] [PubMed] [Google Scholar]
- 16.Alivisatos AP, Johnsson KP, Peng XG, Wilson TE, Loweth CJ, Bruchez MP, Schultz PG. Organization of “nanocrystal molecules” using DNA. Nature. 1996;382:609–611. doi: 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]
- 17.Tong X, Smith LM. Solid-phase method for the purification of DNA sequencing reactions. Anal Chem. 1992;64:2672–2677. doi: 10.1021/ac00046a004. [DOI] [PubMed] [Google Scholar]
- 18.Holmberg A, Blomstergren A, Nord O, Lukacs M, Lundeberg J, Uhlén M. The biotin-streptavidin interaction can be reversibly broken using water at elevated temperatures. Electrophoresis. 2005;26:501–510. doi: 10.1002/elps.200410070. [DOI] [PubMed] [Google Scholar]
- 19.Hwang SH, Im SG, Sung H, Soo SH, Cong VT, Lee DH, Jun SS, Oh HB. Upconversion nanoparticle-based fÖrster resonance energy transfer for detecting the IS6110 sequence of mycobacterium tuberculosis complex in sputum. Biosens Bioelectron. 2014;53:112–116. doi: 10.1016/j.bios.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 20.Xu F, Li P, Ming X, Yang D, Xu HY, Wu XL, Shah NP, Wei H. Detection of cronobacter species in powdered infant formula by probe-magnetic separation PCR. J Dairy Sci. 2014;97:6067–6075. doi: 10.3168/jds.2014-8287. [DOI] [PubMed] [Google Scholar]
- 21.Rogers PH, Michel E, Bauer CA, Vanderet S, Hansen D, Roberts BK, Calvez A, Crews JB, Lau KO, Wood A, et al. Selective, controllable, and reversible aggregation of polystyrene latex microspheres via DNA hybridization. Langmuir. 2005;21:5562–5569. doi: 10.1021/la046790y. [DOI] [PubMed] [Google Scholar]
- 22.Nicewarner Peña SR, Raina S, Goodrich GP, Fedoroff NV, Keating CD. Hybridization and enzymatic extension of Au nanoparticle-bound oligonucleotides. J Am Chem Soc. 2002;124:7314–7323. doi: 10.1021/ja0177915. [DOI] [PubMed] [Google Scholar]
- 23.Herne TM, Tarlov MJ. Characterization of DNA probes immobilized on gold surfaces. J Am Chem Soc. 1997;119:8916–8920. [Google Scholar]
- 24.Lytton-Jean AKR, Mirkin CA. A Thermodynamic investigation into the binding properties of DNA functionalized gold nanoparticle probes and molecular fluorophore probes. J Am Chem Soc. 2005;127:12754–12755. doi: 10.1021/ja052255o. [DOI] [PubMed] [Google Scholar]
- 25.Wang ZX, Kanaras AG, Bates AD, Cosstick R, Brust M. Enzymatic DNA processing on gold nanoparticles. J Mater Chem. 2004;14:578–580. [Google Scholar]
- 26.Seferos DS, Prigodich AE, Giljohann DA, Patel PC, Mirkin CA. Polyvalent DNA nanoparticle conjugates stabilize nucleic acids. Nano Lett. 2009;9:308–311. doi: 10.1021/nl802958f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin WJ, Yung LYL. Efficient manipulation of nanoparticle-bound DNA via restriction endonuclease. Biomacromolecules. 2006;7:3047–3051. doi: 10.1021/bm060517o. [DOI] [PubMed] [Google Scholar]
- 28.Palanisamy R, Connolly AR, Trau M. Considerations of solid-phase DNA amplification. Bioconjugate Chem. 2010;21:690–695. doi: 10.1021/bc900491s. [DOI] [PubMed] [Google Scholar]
- 29.Prigodich AE, Alhasan AH, Mirkin CA. Selective enhancement of ncleases by polyvalent DNA-functionalized gold nanoparticles. J Am Chem Soc. 2011;133:2120–2123. doi: 10.1021/ja110833r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strömberg M, Zardán Gómez de la Torre T, Göransson J, Gunnarsson K, Nilsson M, Strømme M, Svedlindh P. Microscopic mechanisms influencing the volume amplified magnetic nanobead detection assay. Biosens Bioelectron. 2008;24:696–703. doi: 10.1016/j.bios.2008.06.043. [DOI] [PubMed] [Google Scholar]
- 31.Giljohann DA, Seferos DS, Patel PC, Millstone JE, Rosi NL, Mirkin CA. Oligonucleotide loading determines cellular uptake of DNA-modified gold nanoparticles. Nano Lett. 2007;7:3818–3821. doi: 10.1021/nl072471q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin RC, Wu G, Li Z, Mirkin CA, Schatz GC. What controls the melting properties of DNA-linked gold nanoparticle assemblies? J Am Chem Soc. 2003;125(6):1643–1654. doi: 10.1021/ja021096v. [DOI] [PubMed] [Google Scholar]
- 33.Demers LM, Mirkin CA, Mucic RC, Reynolds RA, III, Letsinger RL, Elghanian R, Viswanadham G. A fluorescence-based method for determining the surface coverage and hybridization efficiency of thiol-capped oligonucleotides bound to gold thin films and nanoparticles. Anal Chem. 2000;72:5535–5541. doi: 10.1021/ac0006627. [DOI] [PubMed] [Google Scholar]
- 34.Henry MR, Wilkins Stevens P, Sun J, Kelso DM. Real-time measurements of DNA hybridization on micro-particles with fluorescence resonance energy transfer. Anal Biochem. 1999;276:204–214. doi: 10.1006/abio.1999.4344. [DOI] [PubMed] [Google Scholar]
- 35.Dugas V, Depret G, Chevalier Y, Nesme X, Souteyrand É. Immobilization of single-stranded DNA fragments to solid surfaces and their repeatable specific hybridization: covalent binding or adsorption? Sens Actuators, B. 2004;101:112–121. [Google Scholar]
- 36.Urdea MS, Warner BD, Running JA, Stempien M, Clyne J, Horn TA. Comparison of non-radioisotopic hybridization assay methods using fluorescent, chemiluminescent and enzyme labeled synthetic oligodeoxyribonucleotide probes. Nucleic Acids Res. 1988;16:4937–4956. doi: 10.1093/nar/16.11.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morris W, Briley WE, Auyeung E, Cabezas MD, Mirkin CA. Nucleic acid–metal organic framework (MOF) nanoparticle conjugates. J Am Chem Soc. 2014;136:7261–7264. doi: 10.1021/ja503215w. [DOI] [PubMed] [Google Scholar]
- 38.Álvarez M, Carrascosa LG, Moreno M, Calle A, Zaballos Á, Lechuga LM, Martínez-A C, Tamayo J. Nanomechanics of the formation of DNA self-assembled monolayers and hybridization on microcantilevers. Langmuir. 2004;20:9663–9668. doi: 10.1021/la0489559. [DOI] [PubMed] [Google Scholar]
- 39.Hurst SJ, Lytton-Jean AKR, Mirkin CA. Maximizing DNA loading on a range of gold nanoparticle sizes. Anal Chem. 2006;78:8313–8318. doi: 10.1021/ac0613582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim EY, Stanton J, Vega RA, Kunstman KJ, Mirkin CA, Wolinsky SM. A real-time PCR-based method for determining the surface coverage of thiol-capped oligonucleotides bound onto gold nanoparticles. Nucleic Acids Res. 2006;34:e54. doi: 10.1093/nar/gkl147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu CW, Huang CC, Chang HT. Control over surface DNA density on gold nanoparticles allows selective and sensitive detection of mercury(II) Langmuir. 2008;24:8346–8350. doi: 10.1021/la800589m. [DOI] [PubMed] [Google Scholar]
- 42.Paliwoda RE, Li F, Reid MS, Lin YW, Le XC. Sequential strand displacement beacon for detection of DNA coverage on functionalized gold nanoparticles. Anal Chem. 2014;86:6138–6143. doi: 10.1021/ac501341t. [DOI] [PubMed] [Google Scholar]
- 43.Heaton PA. Quantification of total DNA by spectroscopy. In: Saunders GC, Helen P, editors. Analytical Molecular Biology: Quality and Validation. Chapter 4. Royal Society of Chemistry; Cambridge: 1999. pp. 47–57. [Google Scholar]
- 44.Chakrabarti MH, Roberts EPL. Analysis of mixtures of ferrocyanide and ferricyanide using UV-Visible spectroscopy for characterisation of a novel redox flow battery. J Chem Soc Pak. 2008;30:817–823. [Google Scholar]
- 45.Rogers SG, Weiss B. Exonuclease III of Escherichia coli K-12, an AP endonuclease. Methods Enzymol. 1980;65:201–211. doi: 10.1016/s0076-6879(80)65028-9. [DOI] [PubMed] [Google Scholar]
- 46.Link S, El-Sayed MA. Size and temperature dependence of the plasmon absorption of colloidal gold nanoparticles. J Phys Chem B. 1999;103:4212–4217. [Google Scholar]
- 47.Takeuchi M, Lillis R, Demple B, Takeshita M. Interactions of Escherichia coli endonuclease IV and exonuclease III with abasic sites in DNA. J Biol Chem. 1994;269:21907–21914. [PubMed] [Google Scholar]
- 48.Shida T, Noda M, Sekiguchi J. Cleavage of single-and double-stranded DNAs containing an abasic residue by Escherichia coli exonuclease III (AP endonuclease VI) Nucleic Acids Res. 1996;24:4572–4576. doi: 10.1093/nar/24.22.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu S, Liang PP, Yu HX, Xu XW, Liu Y, Lou XH, Xiao Y. Amplified single base-pair mismatch detection via aggregation of exonuclease-sheared gold nanoparticles. Anal Chem. 2014;86:3461–3467. doi: 10.1021/ac4040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Armbruster DA, Pry T. Limit of blank, limit of detection and limit of quantitation. Clin Biochem Rev. 2008;29:S49–52. [PMC free article] [PubMed] [Google Scholar]
- 51.McKenzie F, Steven V, Ingram A, Graham D. Quantitation of biomolecules conjugated to nanoparticles by enzyme hydrolysis. Chem Commun. 2009;1:2872–2874. doi: 10.1039/b823057a. [DOI] [PubMed] [Google Scholar]
- 52.Pal S, Kim MJ, Song JM. Quantitation of surface coverage of oligonucleotides bound to chip surfaces: A fluorescence-based approach using alkaline phosphatase digestion. Lab Chip. 2008;8:1332–1341. doi: 10.1039/b804066d. [DOI] [PubMed] [Google Scholar]
- 53.Vanecko S, Laskowski M. Studies of the specificity of deoxyribonuclease I. III. hydrolysis of chains carrying a monoesterified phosphate on carbon 5′. J Biol Chem. 1961;236:3312–3316. [PubMed] [Google Scholar]
- 54.Parak WJ, Pellegrino T, Micheel CM, Gerion D, Williams SC, Alivisatos AP. Conformation of oligonucleotides attached to gold nanocrystals probed by gel electrophoresis. Nano Lett. 2003;3:33–36. [Google Scholar]
- 55.He XX, Wang K, Tan WH, Liu B, Lin X, He CM, Li D, Huang SS, Li J. Bioconjugated nanoparticles for DNA protection from cleavage. J Am Chem Soc. 2003;125:7168–7169. doi: 10.1021/ja034450d. [DOI] [PubMed] [Google Scholar]
- 56.Lu CH, Li J, Lin MH, Wang YW, Yang HH, Chen X, Chen GN. Amplified aptamer-based assay through catalytic recycling of the analyte. Angew Chem, Int Ed. 2010;49:8454–8457. doi: 10.1002/anie.201002822. [DOI] [PubMed] [Google Scholar]
- 57.Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 58.Tinland B, Pluen A, Sturm J, Weill G. Persistence length of single-stranded DNA. Macromolecules. 1997;30:5763–5765. [Google Scholar]
- 59.Anne A, Bouchardon A, Moiroux J. 3′-Ferrocene-labeled oligonucleotide chains end-tethered to gold electrode surfaces: novel model systems for exploring flexibility of short DNA using cyclic voltammetry. J Am Chem Soc. 2003;125:1112–1113. doi: 10.1021/ja028640k. [DOI] [PubMed] [Google Scholar]
- 60.Kondo A, Higashitani K. Adsorption of model proteins with wide variation in molecular properties on colloidal particles. J Colloid Interface Sci. 1992;150:344–351. [Google Scholar]
- 61.Safarik I, Safarikova M. Magnetic techniques for the isolation and purification of proteins and peptides. Biomagn Res Technol. 2004;2:7. doi: 10.1186/1477-044X-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou J, Liu Z, Li FY. Upconversion nanophosphors for small-animal imaging. Chem Soc Rev. 2012;41:1323–1349. doi: 10.1039/c1cs15187h. [DOI] [PubMed] [Google Scholar]
- 63.Wang F, Liu XG. Recent advances in the chemistry of lanthanide-doped upconversion nanocrystals. Chem Soc Rev. 2009;38:976–989. doi: 10.1039/b809132n. [DOI] [PubMed] [Google Scholar]
- 64.Costa D, Burrows HD, Da Graca̧ Miguel M. Changes in hydration of lanthanide ions on binding to DNA in aqueous solution. Langmuir. 2005;21:10492–10496. doi: 10.1021/la051493u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


