Summary
Tumor microenvironments are a driver of resistance to anti-cancer drugs. Dissecting cell-microenvironment interactions into tractable units of study presents a challenge. Here, we assess the impact of hundreds of tumor-inspired microenvironments in parallel on lapatinib responses in four cancer cell lines. Combinations of ECM and soluble factors were printed on stiffness-tunable substrata to generate a collection of controlled microenvironments in which to explore cell-based functional responses. Proliferation, HER2 protein expression and phosphorylation, and morphology were measured in single cells. Using dimension reduction and linear modeling, the effects of microenvironment constituents were identified, and then validated empirically. Each of the cell lines exhibits unique microenvironment-response patterns. Fibronectin, type IV collagen, and matrix rigidity are significant regulators of lapatinib resistance in HER2-amplified breast cancer cells. Small molecule inhibitors were identified that could attenuate microenvironment-imposed resistance. Thus, we demonstrate a strategy to identify resistance- and sensitivity-driving microenvironments to improve efficacy of anti-cancer therapeutics.
Keywords: Microenvironment, microenvironment microarray, MEMA, HER2, lapatinib, verteporfin, AZD0530, YAP, fibronectin, breast cancer, drug resistance
Graphical Abstract
Introduction
Chemotherapy for cancers are often associated with problems of toxicity and resistance. Pathway targeted therapies have proven efficacious with less toxicity, but durability of the therapeutic effects remains a barrier. Resistance to targeted compounds arises from signaling redundancies in the targeted pathways and genotype selection, presumably due to the intra-tumor heterogeneity of cancer cells, and both have been intensively investigated (Logue and Morrison, 2012, Sun and Yu, 2015). The tumor microenvironment, which is less well studied, also modulates drug responses.
Microenvironment is the sum of cell-cell, cell-extracellular matrix (ECM), cell-soluble factor interactions, as well as the physical and geometric characteristics of the tissue (Bissell and Labarge, 2005). Dynamic and reciprocal communication between cells and microenvironments plays a pivotal role in regulating tissue-specific functions, as well as enabling tumor genesis and metastasis (Xu et al., 2009, Quail and Joyce, 2013). Stromal gene expression and morphological features are important as prognostic factors in a number of tumor types (Beck et al., 2011, Finak et al., 2008), and gene signatures derived from tumor cells, as well as from tumor stroma, independently predict drug efficacy (Klijn et al., 2015, Farmer et al., 2009). Tumor microenvironments modulate efficacy of both cytotoxic agents and targeted therapies (Weaver et al., 2002, Weigelt et al., 2010, Allen and Louise Jones, 2011). Even changes in the physical properties of tumors, such as increased rigidity, are associated with malignancy and drug resistance (Pickup et al., 2014). Delineating of impact of tumor microenvironment constituents is fundamental to understanding the mechanisms of drug resistance.
There are relatively few examples of efficient experimental systems for dissecting cell-microenvironment interactions. Inhibitory effects of HER2-targeted drugs were shown to vary in breast cancer cells cultured in three-dimensional Matrigel (3D) compared to two-dimensional (2D) tissue culture plastic (TCP) environments (Weigelt et al., 2010). Even in this relatively straightforward example, it remains a challenge to identify which microenvironment constituents or characteristics governed the different responses to anti-cancer agents. Matrigel is deceptively complex, consisting of hundreds of components (Hughes et al., 2010). We previously used HCC1569, a HER2-amplified breast cancer cell line that is a model for resistance to HER2-targeted therapeutics, together with lapatinib (Lap), a HER2-targeted small molecule, to show that the Hippo pathway mediated stiffness-dependent resistance to Lap (Lin et al., 2015). HCC1569 paired with Lap exhibits a particularly wide dynamic range of proliferation inhibitory response compared to other combinations of HER2 amplified cell lines with HER2-targeted drugs (Weigelt et al., 2010), thus it embodies a useful system to probe the impacts of microenvironment constituents on responses to targeted therapeutics.
Here, we utilized microenvironment microarrays (MEMA) (LaBarge et al., 2009, Lin et al., 2012) to assess simultaneously the impact of molecular composition of microenvironment in addition to matrix rigidity on Lap responses in HCC1569 and three other cancer cell lines that represent different cancer types, each with differing HER2 protein levels, and sensitivity to Lap. We demonstrate that a continuum of microenvironment-imposed drug response phenotypes is achievable in ostensibly identical cancer cells. Dimension reduction and generalized linear models enabled decoupling of individual effects and synergistic pairings of microenvironment constituents, which were validated experimentally. We demonstrate a combinatorial cell-based approach to understanding the functional impact of tumor microenvironment components on drug responses.
Results
Isolating effects of individual microenvironment components and pairwise synergies
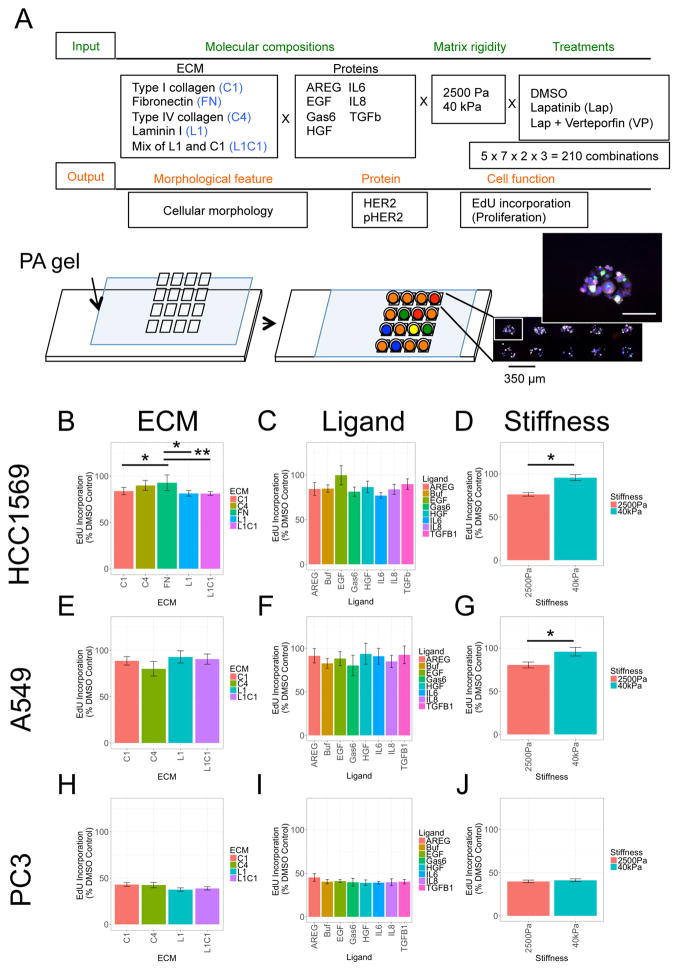
To study the impacts of microenvironments on the inhibitory effects of Lap, cancer cell lines were grown on microenvironment microarrays (MEMA). Four cancer cell lines were examined. HER2-amplified HCC1569, and HER2-negative BT549 breast cancer cells. A549 non-small cell lung cancer (NSCLC) cell line, which has genomic amplification of EGFR and HER2 and was used to show that NSCLC could benefit from lapatinib treatment (Diaz et al., 2010). PC3 prostate cancer cell line that was used to show that radiation resistant prostate cancers express HER2, and may benefit from HER2 targeted therapy (Andersson et al., 2015). 60 replicate spots were printed for each microenvironment composed of pairwise combinations of four ECM and seven ligands on either 2500 Pa or 40 kPa elastic modulus substrata. Proliferation, HER2 expression and phosphorylation, and cell morphology were measured in each cell (Fig 1A, examples of immunofluorescence in Fig S1). The ECM consists of proteins that are enriched in breast stroma or basement membrane, and the soluble factors were chosen for having been implicated in breast cancer progression or therapeutic resistance (Quail and Joyce, 2013, Pickup et al., 2014). Laminin-111 (L1) and type IV collagen (C4) are the major ECM components of basement membrane; and fibronectin (FN) and type I collagen (C1) are major ECM components of stroma. The effects of these ECM as well as other ligands that are related to drug resistance (Jones et al., 2016) were assessed. 2500 Pa is relevant to the stiffness of breast tumors (Tilghman et al., 2010), and 40 kPa is much stiffer and represents a culture environment closer to TCP. Empirically we found that 40 kPa polyacrylamide gels and TCP, which are >3 GPa, elicit similar proliferation rates and activation of YAP in mammary epithelial cell lines (Lin et al., 2015). Cells were cultured on MEMA for 48hr treated with Lap alone (1.5 μM), Lap with verteporfin (VP, 2 μg/mL), or with DMSO. VP, an inhibitor of the Yes associated protein (YAP), was included because it prevents stiffness-induced resistance to Lap (Lin et al., 2015). In all, functional impacts on cell behavior were assessed on 210 unique microenvironments in parallel.
Figure 1. Isolating the effects of individual components of combinatorial microenvironments on lapatinib responses.
(A) Experimental design illustrative diagram and a representative image of cells on a MEMA spot. Scale bars, 100 μm. (B–J) Bar graphs of GLM analyses showing the mean EdU incorporation across all microenvironments that contain the indicated component, focusing on either (B, E, H) ECM, (C, F, I) ligand, or (D, G, J) stiffness. The cell lines considered were (B–D) HCC1569, (E–G) A549, and (H–J) PC3. Data are relative incorporation of EdU expressed as a percentage of DMSO-treated cells, presented as mean ± SEM (n = 60 for each microenvironment). Individually, ECM and stiffness were identified as significant drivers of lapatinib response in HCC1569 and A549.
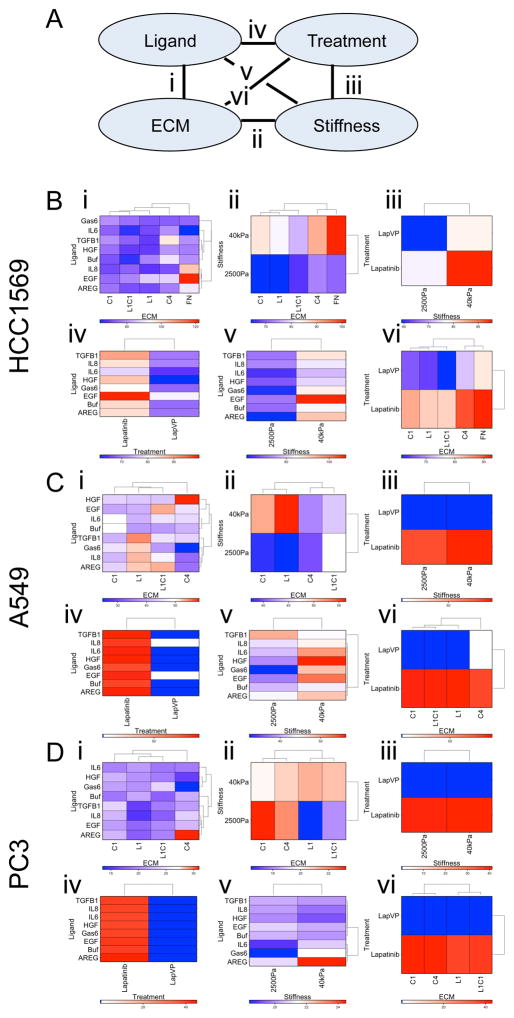
Generalized linear model (GLM) was applied to decouple effects of multiple components and determine whether some single components exert particularly strong effects on the drug response phenotypes (Lin, 2017, Ranga et al., 2014). BT549 cells are completely resistant to Lap because they do not express HER2 (Fig S2A, S2B). In all of the remaining cell lines single soluble ligands were not significantly correlated with differential responses to Lap, as determined by changes in EdU incorporation (Fig 1C, 1F, and 1I). In HCC1569 cells, FN and C4 showed significant correlation with Lap resistance compared to C1 and L1 (Fig 1B), and higher matrix rigidity also was associated with resistance (Fig 1D), but only a trend in the data leaning towards resistance to Lap was detected on EGF-containing microenvironments (Fig 1C). PC3 and A549 cells had reproducibly poor attachment to FN-containing MEMA spots, thus FN features were omitted from analyses for those lines. In A549, neither individual ECM (Fig 1E) or ligands (Fig 1F) were significantly correlated with Lap responses; but, high matrix rigidity was significantly correlated with Lap resistance (Fig 1G). In PC3, neither the molecular components, nor rigidity, were correlated with Lap responses (Fig 1H–J). The pairwise interactions of molecular components were examined systematically by GLM (Fig 2A). Whereas single ligands had relatively minor impact, combining them with ECM or stiffness had a synergistic effect. HCC1569 showed higher resistance to Lap in FN-rich microenvironments with EGF (Fig 2B, interaction i), A549 showed higher resistance to Lap in C4-rich microenvironments with HGF (Fig 2C, interaction i), and PC3 showed higher resistance to Lap in C4-rich microenvironments with AREG (Fig 2D, interaction i). Treatment with VP and Lap showed strong inhibition of proliferation in all lines (Fig 2B–2D interactions iii, iv, and vi; Fig S2C, S2D). The responses of each cell line to the array of different microenvironments that they were exposed to were unique, like functional drug-response fingerprints.
Figure 2. Identification of synergistic interacting microenvironment components.
(A) A diagram showing each of the pair-wise interactions that were tested (i–vi) between individual components of microenvironments. (B–D) Clustered heat maps represent z-scores of EdU-incorporation in all the microenvironments that contain both of the indicated components including: (i) an ECM with a ligand, (ii) stiffness with an ECM, (iii) drug treatment with stiffness, (iv) drug treatment with ligand, (v) stiffness with a ligand, and (vi) drug treatment with an ECM. These pair-wise interactions were tested in (B) HCC1569, (C) A549, and (D) PC3. Each cell line showed resistance (red) or sensitivity (blue) to lapatinib in unique combinations of ECM and soluble ligands (n = 60 for each microenvironment).
Cell morphology imposed by combinatorial microenvironments co-organizes with drug response phenotypes
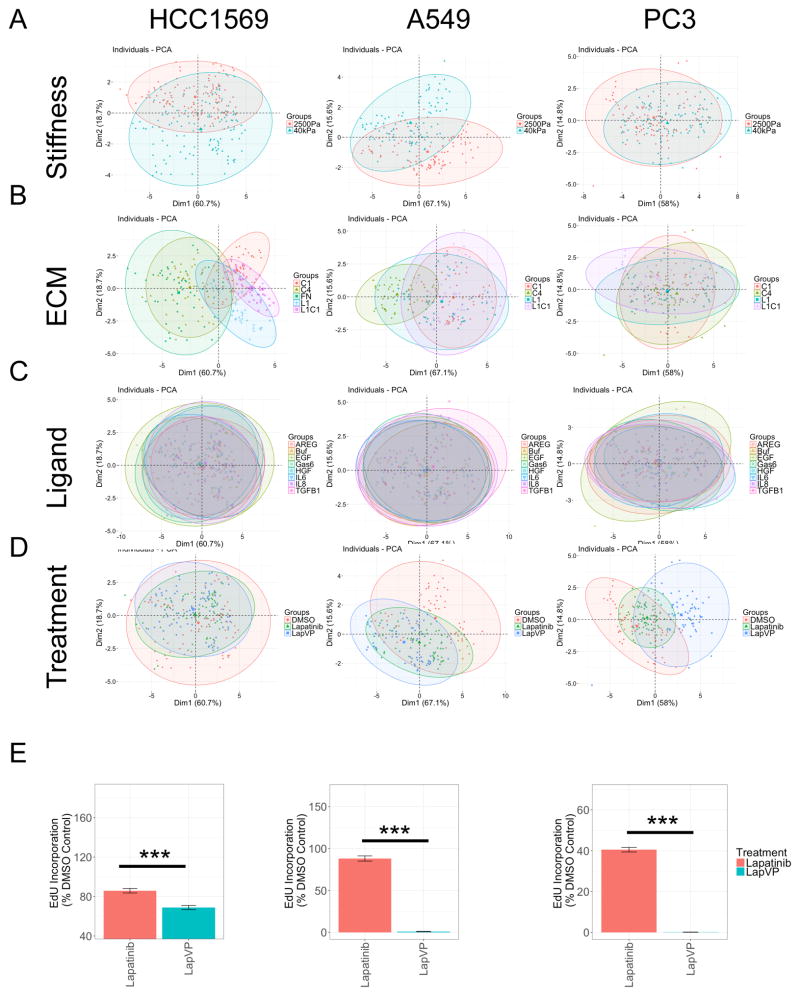
Morphological features of the cells were quantified because cell shape is regulated by mechanical and chemical properties of microenvironments, and is directly related to mechanisms that modulate behaviors such as motility and proliferation (Halder et al., 2012, Katz et al., 2000, Ulrich et al., 2009). We utilized principle component analysis (PCA) and t-distributed stochastic neighbor embedding (t-SNE), to better visualize the high-dimensional data (Amir et al., 2013). PCA is applied to high dimension cytometry data using linear transformation, and t-SNE more faithfully captures the nonlinear relationships in biological data sets (Amir et al., 2013). Cell area, compactness, eccentricity, extent, form factor, major axis length, and solidity of both nucleus and cell body were used to generate relationships in the PCA and t-SNE plots. Each dot in the PCA and t-SNE represents the composite phenotype begotten by a given microenvironment. PCA shows that different microenvironments are well distinguished based on matrix rigidity (Fig 3A) and ECM (Fig 3B) for HCC1569 and A549. Ligands were not well separated, suggesting minimal contributions to altered cell morphology (Fig 3C). Treatment with Lap, or Lap with VP prominently altered morphological features compared to DMSO in A549 and PC3, but less so in HCC1569 (Fig 3D). PCA was validated with t-SNE. Phenotypes formed clusters according to matrix rigidity and treatments in all cell lines (Fig S3A, S3B). t-SNE revealed that all cell lines in Lap with VP had distinctive morphology (Fig S3A), and that response groups in HCC1569 and A549 were clustered based on ECM (Fig S3C). Ligands showed minimal effect across all lines (Fig S3D). VP with Lap showed strong inhibition of proliferation in all cell lines in a majority of conditions (Fig 3E). PCA and t-SNE arrived at similar conclusions, that matrix rigidity and certain ECM significantly influenced cellular morphology and drug response. The particular combinations were specific to each cell line. When compared to GLM, which used EdU incorporation as the major variable, these dimension-reduction methods demonstrate that morphology alone captures a as much useful information as some specific molecular markers regarding responses to drugs and microenvironments.
Figure 3. Cell morphologies imposed by combinatorial microenvironments co-organize with drug-response phenotypes.
(A–D) The impact of microenvironment on eight different morphometric measurements in HCC1569, A549, and PC3 cells are visualized with PCA. Each data point is one microenvironment composition and the major variants are derived from morphological features. Colors represent (A) matrix rigidity, 2500 Pa (red) and 40 kPa (green), respectively, (B) ECM, (C) ligand, and (D) drug treatments in HCC1569, A549, and PC3. (E) Bar graphs represent mean±SEM EdU incorporated as a percentage of DMSO-treated cells (i.e. drug response) in HCC1569, A549, and PC3 treated with lapatinib alone or lapatinib with verteporfin, averaged across all microenvironments (n = 60 for each microenvironment).
Microenvironments modulate HER2 activity
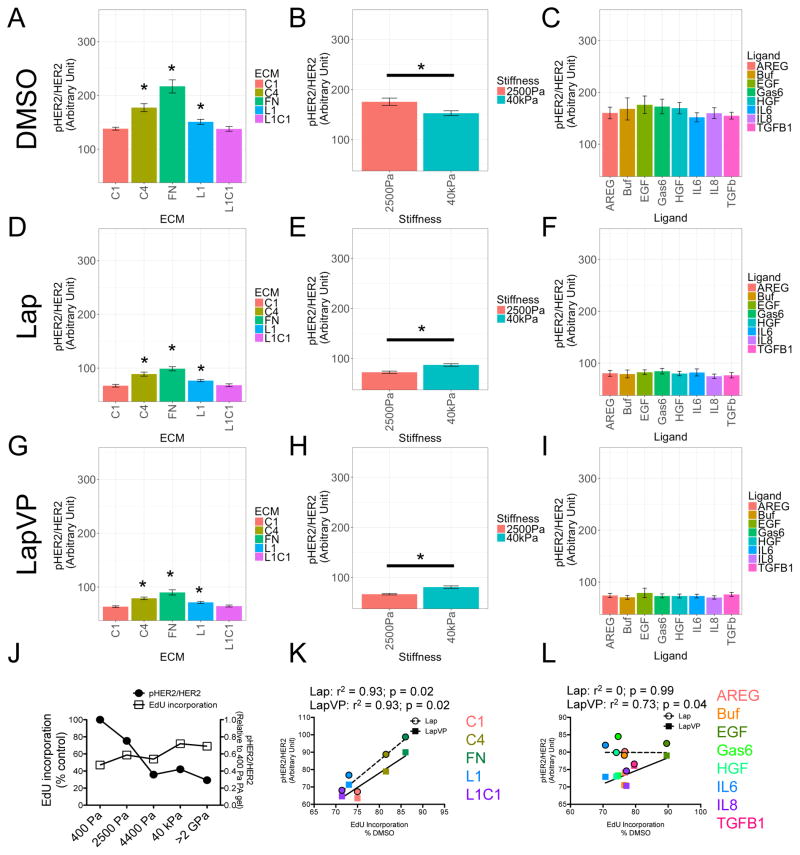
Phosphorylation of HER2 is sensitive to changes in microenvironment (Weigelt et al., 2010, Breslin and O’Driscoll, 2016) and overexpression of HER2 is predictive of response to Lap (Rusnak et al., 2007). Because HCC1569 generally showed more microenvironment-dependent resistance to Lap in all conditions compared to the other cell lines, we focused our attention on understanding the basis of that resistance. Microenvironment did impose trends in HER2 expression and phosphorylation in A549 and PC3 that were not significant (Fig S4), whereas HCC1569 exhibited strong microenvironment-dependent changes in HER2/pHER2 ratios (Fig 4). GLM isolated effects of the different microenvironment components on HER2 phosphorylation. FN and C4 significantly increased the ratio of pHER2 (Fig 4A), and a rigid substrate was associated with lower pHER2 ratio in DMSO treated cells compared to Lap treated (Fig 4B). Ligands caused minimal shifts in pHER2/HER2 ratios (Fig 4C). Addition of Lap, or Lap with VP, significantly decreased the pHER2 ratios; but adhesion to FN or C4 (Fig 4D and 4G) and rigid substrata (Fig 4E and 4H), sustained higher pHER2 ratios in both drug combinations, and effects of the printed ligands were minimal (Fig 4F and 4I). HER2 phosphorylation ratio decreased as matrix rigidity increased and was inversely correlated with Lap resistance (Fig 4J). By applying a linear fit we observed the microenvironment components that associated with higher pHER2/HER2 ratios, C4 and FN, were correlated with greater resistance to the anti-proliferative effects of Lap (Lap: r2 = 0.93, p = 0.02; LapVP: r2 = 0.93, p = 0.02) (Fig 4K), whereas ligands-associated pHER2/HER2 ratios showed less correlation with resistance (Lap: r2 = 0, p = 0.99; LapVP: r2 = 0.73, p = 0.04) (Fig 4L). Thus microenvironment components that alter pHER2/HER2 ratios were identified with GLM, and microenvironments that sustained activated HER2 were correlated with resistance to Lap.
Figure 4. Microenvironments modulate ratios of pHER2 and total HER2, which correlates with lapatinib resistance.
(A–I) Bar graphs represent the ratio of pHER2/HER2 in HCC1569 modulated by (A, D, and G) ECM, (B, E, and H) stiffness, and (C, F, and I) ligand. Data are analyzed by GLM in (A, B, and C) DMSO, (D, E, and F) lapatinib, and (G, H, and I) lapatinib and verteporfin. (J) Line graph represents the ratio of pHER2/HER2 and lapatinib responses of HCC1569 in different matrix rigidity determined by FACS (n = 3, 10000 cells/condition per experiment). (K and L) Line graph represents the correlation between pHER2/HER2 ratio of HCC1569 modulated by (K) ECM, and (L) ligand; versus EdU incorporation determined on MEMA. r2 and p value are calculated from Pearson correlation coefficient. All bar graph data are represented as mean ± SEM (n = 60 for each microenvironment).
Adhesion to fibronectin activates YAP translocation
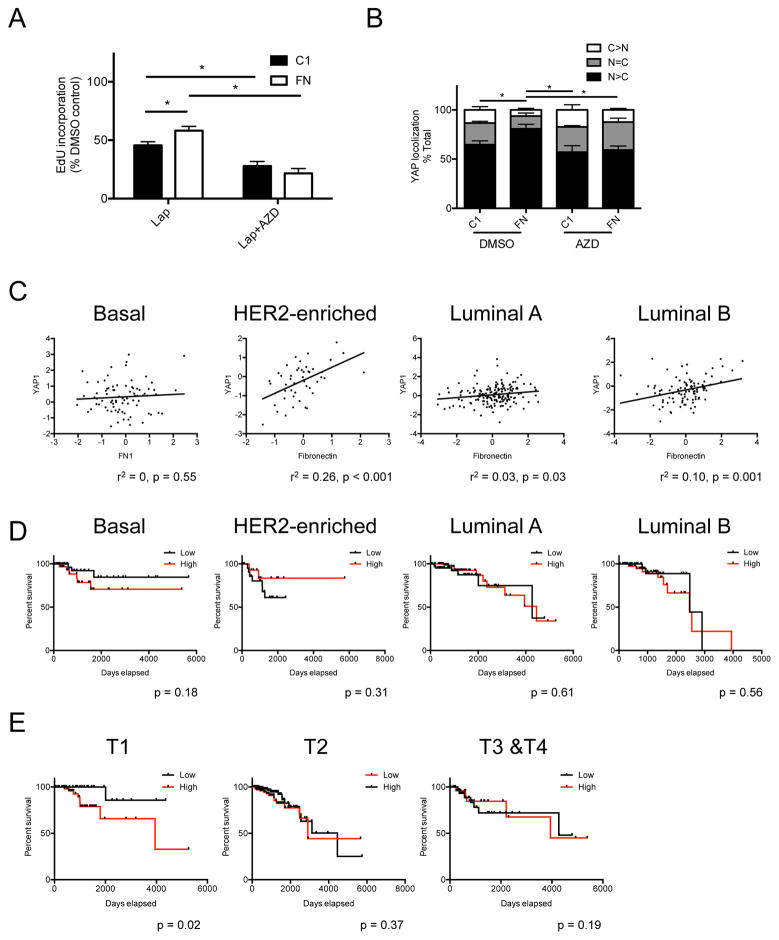
To our surprise, GLM suggested that FN, independent of matrix rigidity, correlated strongly with Lap resistance. Adhesion of MCF10A to FN was shown to activate FAK-Src-YAP (Kim and Gumbiner, 2015), thus we hypothesized that adhesion to FN independently conferred Lap resistance via a Src-YAP-dependent pathway. Based on IC50 analysis (Fig S5A), 1 μM of Src inhibitor, AZD0530, was used because it did not inhibit EdU incorporation in HCC1569 on either C1 or FN-coated coverslips (Fig S5B). Cells grown on FN, compared to C1, were significantly more resistant to Lap, but AZD0530 combined with Lap inhibited proliferation, eliminating the FN-dependent Lap resistance (Fig 5A). Adhesion to FN caused more nuclear translocation of YAP, whereas Src inhibition prevented FN-induced YAP translocation (Fig 5B, and examples of YAP immunofluorescence in Fig S5C). Protein expression data from The Cancer Genome Atlas (TCGA) (The Cancer Genome Atlas Research et al., 2013) showed that FN and YAP protein levels are correlated in HER2-enriched (r2 = 0.26, p < 0.001), luminal A (r2 = 0.03, p = 0.03), and luminal B subtypes (r2 = 0.1, p = 0.001); but no correlation was observed in the basal subtype (r2 = 0, p = 0.55) (Fig 5C). FN protein levels were not associated with significant changes in survival rate (Fig 5D), but high FN levels were correlated with poor survival in cancers diagnosed in early T1 pathological stages (p = 0.02) (Fig 5E). Overall, these MEMA experiments identify putative tumor microenvironments that can better protect some tumor cells compared to other microenvironments. Our cell-based functional results suggested FN in rigid microenvironments conferred Lap resistance, and inhibition of Src or YAP may be of some benefit to eliminate this source of microenvironment-dependent resistance.
Figure 5. Adhesion to fibronectin increases nuclear YAP translocation and confers lapatinib resistance in HCC1569.
(A) Bar graph represents drug responses of HCC1569 cultured on type I collagen (C1) or fibronectin (FN) coated glass coverslips, and treated with lapatinib or lapatinib plus verteporfin. Data are represented as mean ± SEM (n = 3; 500 cells/condition per experiment). (B) Bar graph represents the distribution of YAP (C>N, higher in cytoplasm; N=C, equally between nucleus and cytoplasm; N>C, higher in nucleus) of HCC1569 cultured on C1 or FN, treated with or without AZD0530. Data are represented as mean ± SEM (n = 3; 100 cells/condition per experiment). (C) Scatter plots represent the correlation of protein levels between fibronectin and YAP in basal, HER2-enriched, luminal A, and luminal-B subtypes of breast cancers. r2 and p value are calculated from Pearson correlation coefficient. (D) Kaplan-Meier plots represent the survival rate affected by protein level of fibronectin in basal, HER2-enriched, luminal A, and luminal-B subtypes of breast cancer. p values are calculated from Log-rank (Mantel-Cox) test. (E) Kaplan-Meier plots represent the survival rate affected by protein level of fibronectin in T1, T2, and T3–T4 pathological stages of breast cancer. p values are calculated from Log-rank (Mantel-Cox) test.
Discussion
A continuum of drug response phenotypes is achievable in isogenic cells by making changes to the cellular microenvironment. Here we functionally examined the impact of Lap on four cell lines grown on 210 combinatorial microenvironments. Dimension reduction and visualization, and GLM were used to better define the relationship between responses to Lap and the mechanical and molecular properties of the microenvironment. In addition to matrix rigidity, specific ECM, e.g. FN and C4, had strong impacts on drug responses in HCC1569, which is a model for cancers that are resistant HER2-targeted drugs. The effects of soluble ligands that were printed with ECM had relatively minor effects on Lap responses, although when considered in specific pairwise contexts they exerted modulatory effects. Resistance to Lap correlated with microenvironment-dependent HER2 activation in HCC1569, but it was not an absolute relationship across other cell lines. By identifying specific microenvironments that sustained high HER2 phosphorylation ratios, e.g. FN and matrix rigidity, we were able to attenuate the Lap resistance with addition of VP and AZD0530. Interestingly, when we evaluated responses to the drugs in all three cell lines based only on seven morphometric factors, the conclusions were identical to the analyses that used protein and proliferation measurements. Cell morphology is an emergent property of complex cellular systems, and our analyses reveal much utility in using morphology as a marker of drug and microenvironment responses. Solid tumor microenvironments are infamously heterogeneous, which is thought to be a contributing factor to efficacy and durability of drugs responses. Here we demonstrate a pre-clinical strategy to profile drug activity as it relates to microenvironment context, in way that enables identification of the operative drivers of resistance and sensitivity. This information can then be used to identify potential combined therapeutic approaches to improve drug efficacy and durability.
The ratio of pHER2/HER2 in individual cells was measured in this study, because HER2 protein levels are reportedly more predictive of a response compared to gene amplification (Nuciforo et al., 2016). Taking HCC1569 as an example, our results are consistent with reports that HER2-targeted drug efficacy is correlated with the level of pHER2 inhibition (Konecny et al., 2006). Both HER2 phosphorylation (Hudelist et al., 2006) and total HER2 levels (Konecny et al., 2006) allegedly predict responses to HER2 targeted therapy, but their predictive power of efficacy is inconsistent (Ginestier et al., 2007). Alteration of HER2 expression modulates drug efficacy, for example, long-term exposure to trastuzumab reduced HER2 expression and induced trastuzumab resistance (Burnett et al., 2015). The empirical combinatorial approach we used here reveals that different microenvironments will significantly impact HER2 modification and expression, which may explain the lack of consistency among previous reports. Rigid, FN and C4-rich microenvironments that sustained high pHER2/HER2 ratio after drug treatment were correlated with drug resistance. Matrix rigidity is correlated with breast malignancy and drug resistance (Levental et al., 2009, Lin et al., 2015). Higher levels of stromal FN is correlated with poor patient survival and up-regulation of EMT markers (Ioachim et al., 2002, Park and Schwarzbauer, 2014), and C4 increases proliferation of a number of cancer cells (Öhlund et al., 2013). All of the mechanisms by which microenvironments can sustain activated HER2 are not clear, and further study is needed to understand and to mitigate the various sources of microenvironment-imposed drug resistance.
Adhesion to FN caused nuclear YAP translocation independent of matrix rigidity, which was correlated with Lap resistance and could be abrogated by a small molecule Src inhibitor. YAP, one of the transcriptional co-factors in the Hippo pathway, plays an important role in transducing microenvironment mechanical information from the plasma membrane into the nucleus (Halder et al., 2012). YAP activation is associated with tumorigenesis and malignancy in a number of cancer types (e.g. breast, colon, lung, liver, and mesothelioma) (Moroishi et al., 2015). Targeting YAP is an attractive strategy to overcoming drug resistance in some cases. Indeed, small molecules have been identified, in addition to VP, that inhibit nuclear translocation of YAP and increased sensitivity of chemotherapies to breast cancers (Oku et al., 2015). VP, a FDA approved photosensitizer for photodynamic therapy of age-related macular degeneration (Michels and Schmidt-Erfurth, 2001), has shown potential as an anti-tumor therapy in multiple cancer types (Feng et al., 2016, Donohue et al., 2013, Zhang et al., 2015). Multiple anti-tumor mechanisms of VP have been proposed including: inducing necrosis (Huggett et al., 2014), proteotoxicity independent of YAP (Zhang et al., 2015), and disrupting the interaction of YAP-TEAD to down-regulate transcriptional targets of YAP (Brodowska et al., 2014, Liu-Chittenden et al., 2012). VP can overcome resistance to chemotherapy and HER2-targeted therapy via disruption of YAP-TEAD (Pan et al., 2016, Lin et al., 2015). We further examined the effect of VP to modulate stiffness-dependent Lap resistance. In the HCC1569, Lap treatment with VP together showed a greater inhibitory effect on proliferation, compared to Lap alone, in most microenvironments. Interestingly, using the same concentration of VP, proliferation in A549 and PC3 were strongly inhibited across all conditions. A549 and PC3 cells were used by others as models for lung and prostate carcinomas that might benefit from HER2-targeted therapy, our data indicate potential therapeutic benefit from VP treatment in addition, however, the mechanisms need further investigation and in vivo validation.
Our results here showed that FN increased Lap resistance via an Src-YAP related pathway. The dataset from TCGA suggested that FN protein levels are not directly associated with poor survival rate in the different subtypes of breast cancer. However, a higher level of FN protein was associated with poor survival rate in the earliest pathological stages of cancers, suggesting that tumor microenvironment may have different impacts in different stages of progression. Our analyses of the TCGA data did not reveal robust associations between the MEMA-identified microenvironments that promoted resistance to Lap. However, the TCGA data effectively treat tumors as homogeneous masses, and thus important regional differences are not detectable by the types of analyses we conducted on the TCGA dataset. Reports have connected the expression of stromal FN to breast cancer metastasis, prognostic significance (Fernandez-Garcia et al., 2014), and chemotherapeutic resistance (Yuan et al., 2015). That the relationship between FN expression in patient tumors and survival was not entirely strong challenges our own prediction from the MEMA experiments. This reflects a general challenge with understanding the resolution to which we must understand tumor ecologies. There may well be nests of resistance-supportive microenvironments that may not be readily detected with the types of traditional approaches that homogenize tumors and tissues for high dimensional sequencing and proteomics analyses.
Tissue microenvironments are known to impose and maintain cell lineage specificity, and elements of tumor microenvironments in specific act as regulators of malignant progression and drug response (LaBarge et al., 2009, Pickup et al., 2014). Effective ways are needed to functionally explore the cell-microenvironment relationship in a manner that is akin to high-dimensional omics technologies. Here we demonstrate the proof-of-principal of a way forward to understanding the totality of drug responses that can be elicited within tumor microenvironments by measuring the responses of isogenic cells across numerous microenvironment conditions in parallel. These data suggest that a comprehensive understanding of how ECM modulates drug responses will impact design of therapeutic strategies. Co-targeting microenvironment-regulated pathways, such as by Src or YAP inhibition, revealed potential therapeutic benefit for Lap treatments, which merits in vivo validation and follow-up. The impact of microenvironment should be taken into consideration during the earliest stages of preclinical design. Platforms like MEMA can be applied to identify microenvironments that contribute to drug resistance.
Experimental Procedures
Cell culture and drug treatment
HCC1569 and BT549 breast cancer cells, and androgen-independent prostate PC3 cancer cells (American Type Culture Collection; Manassas, VA, USA) were maintained in RPMI1640 (Invitrogen, Carlsbad, CA, USA). NSCLC A549 cancer cells was maintained in DMEM (Invitrogen, Carlsbad, CA, USA) supplemented with 10 % fetal bovine serum (FBS; Gemini Bio-Products, West Sacramento, CA, USA) USA), and 1% Penicillin/Streptomycin/Glutamine (P/S/Glu) (Invitrogen, Carlsbad, CA, USA).
For drug treatments, both in validation and MEMA experiments, cells were cultured as above for 2 hr after initial adhesion, and were then treated with Lap (1.5 μM, LC Laboratories, Woburn, MA, USA) in media supplemented with 1 % FBS and 1 % P/S/Glu for additional 48 hr. Other pharmaceutical modulators were concurrently added: i.e. Verteporfin (VP, Sigma-Aldrich, St. Louis, MO, USA) was added at 2 μg/mL; and AZD0530 (Selleckchem, Houston, TX, USA) was added at 1 μM. All cell lines were authenticated by DNA Diagnostics Center.
MEMA fabrication and cell culture on MEMA
Microenvironment microarray (MEMA, formerly MEArray (LaBarge etal. 2009)) were fabricated according to (Lin et al., 2012). Briefly, microscope slides were coated with 2500 Pa and 40 kPa polyacrylamide gels based on an adopted protocol from (Tse and Engler, 2010). Combinatorial microenvironments were prepared in printing buffer (100mM Tris-acetate/20%glycerol/0.05%Triton 100X at pH5.2) in 384-well plates. The ECM molecules used were: type I collagen (C1, 100μg/mL), type IV collagen (C4, 100μg/mL), laminin-111 (L1, 100 μg/mL), fibronectin (FN, 100 μg/mL), and a mixture of type I collagen together with laminin-111 (L1C1, 100 μg/mL). The recombinant purified proteins used were, amphiregulin (AREG, 5 μg/mL), epidermal growth factor (EGF, 1 μg/mL), growth arrest-specific gene 6 (GAS6, 1 μg/mL), hepatocyte growth factor (HGF, 0.5 μg/mL), interleukin-6 (IL-6, 2 μg/mL), interleukin-8 (IL-8, μg/mL), and transforming growth factor beta (TGFβ, 1 μg/mL). Arrays were printed with a SpotBot III microarrayer (Array-it). 30 replicate features for each unique microenvironment were printed/array, total of 1920, spaced 350μm apart. MEMA are stored at 4°C for short-term, or −80 °C if over one month. Before use slides are rehydrated in 4 mL of PBS with 1% P/S/Glu and 0.1% fungizone for 60 min. PBS was replaced with complete medium supplemented with 10% FBS and 1% P/S/Glu. 500 μL of cells at 106 cells/mL were added on each slide for 10 min of initial adhesion, and 3 mL of complete medium were then added for another 2hr adhesion. Unbound cells were washed away and remaining adherent cells were treated with media containing drug for 48 hr at 37 °C, 5 % CO2. 10 μM EdU was added 4 hr prior to the experimental endpoint.
Immunofluorescence staining
For MEMA, cells on the slides were fixed in methanol/acetone (1:1) at −20 °C for 20 min, whereas for coverslip-based experiments, cells were fixed in paraformaldehyde (4%) at room temperature for 10 min. Cells were then blocked with PBS, 5 % normal goat serum, and 0.1% Triton X-100 at room temperature for 30 min, and then incubated with primary antibodies: anti-HER2-647 (1:100; Biolegend), anti-pHER2 (1:100; Cell Signaling Technology) overnight at 4 °C. pHER2 primary antibodies was visualized with fluorescent Alexa-568 secondary antibodies raised in goats (1:500; Invitrogen) together with Hoechst 33342 (1:2000; Invitrogen), incubated at room temperature for 2 hr. Images were acquired with an epifluorescence microscope. Image analyses were conducted with CellProfiler. For quantification of YAP/TAZ localization, the ratios of median fluorescence intensity in the cytoplasmic (C) and nuclear (N) compartments of segmented cells were used. The cutoffs ratios were used to establish three classes: C > N (X < 1), N = C (1 < X < 1.2) and N > C (X > 1.2)
MEMA analysis
Single-cell resolution image data was obtained by marker-based watershed segmentation. Morphological features, fluorescence intensity of DAPI, HER2, pHER, and EdU were quantified. Morphological features included cell area, compactness, eccentricity, extent, form factor, major axis length, and solidity. Image analyses were performed with CellProfiler (Lamprecht et al., 2007). Further data analyses were conducted and final reports were generated with R (R Core Team, 2015) and packages (Hadley Wickham and Romain Francois, 2016, Helios De Rosario-Martinez, 2015, Paul C. Boutros, 2015, Simon Urbanek and Jeffrey Horner, 2015, Erich Neuwirth, 2014, Alboukadel Kassambara, 2015). The R package, phia, was used for GLM to test the significance and decouple the impact of single microenvironment factors as well as pairwise interactions among factors to drug responses (Helios De Rosario-Martinez, 2015, Ranga et al., 2014). For dimension reduction and better visualization, PCA analyses with R package, factoextra, as well as t-SNE analyses with ViSNE, were performed with all morphological features as input (Alboukadel Kassambara, 2015, Amir et al., 2013). See supplemental information for details of CellProfiler pipeline.
Coverslip cell culture
For coating proteins on the coverslip surface, 12 mm diameter coverslips were placed in 24-well plates, 100 μg/mL of fibronectin (FN) or type I collagen (C1) were applied to each coverslip and incubated at 37 °C for 2 hr. Before cell culture, coverslips were washed with PBS containing 1% P/S/Glu and 0.1% fungizone at room temperature for 30 min.
Flow Cytometry
Cells were collected via EDTA-PBS (0.4% EDTA) treatment on ice. After washing with PBS, cells were blocked with PBS containing 2% bovine serum albumin, 5% normal goat serum, and 5 mM EDTA on ice for 30 min. Cells were incubated with the primary antibody: anti-HER2-647 (1:100; Biolegend, San Diego, CA), anti-pHER2 (1:200; Cell Signaling Technology, Beverly, MA) on ice for 30 min, washed with PBS, and then treated with the Alexa-555 secondary antibody to pHER2 (1:400, I Invitrogen, Carlsbad, CA) on ice for 15 min. After two PBS washes, the level of pHER2 and HER2 were measured with a FACSCalibur (Becton-Dickinson, San Jose, CA).
Proliferation assay
5-ethynyl-2′-deoxyuridine (EdU) incorporation and clicking reaction were performed according to the manufacture’s protocol (Invitrogen, Carlsbad, CA). Nuclei were stained with Hoechst 33342 (Invitrogen, Carlsbad, CA). Drug response values are expressed as a percentage of DMSO-treated cells.
TCGA Database
All data were obtained from the The Cancer Genome Atlas (TCGA) breast cancer online portal (https://tcga-data.nci.nih.gov/docs/publications/brca_2012/; Cancer Genome Atlas, 2012. The following file was used: for reverse-phase protein array expression data: “rppaData-403Samp-171Ab-Trimmed.txt”.
Statistical analysis
GraphPad Prism 6.0 for Mac and R were used for all statistical analysis(GraphPad Software, 2016, R Core Team, 2015). Variance of the means was confirmed by ANOVA. For multiple comparisons, Dunnette’s t-tests were used for groups more than 5, and fisher’s least significant difference (LSD) tests were used for groups less than 5. To compare two distributions, student t-tests were performed. Log-rank (Mantel-Cox) tests were performed for Kaplan-Meier plots. Microenvironments that impose phenotypes statistically different from the control were detected associate with a p value. Significance was established when *p < 0.05, **p < 0.01, and ***p < 0.001.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award number P30CA033572, strategic recruitment funds from Beckman Research Institute at City of Hope, and by Laboratory Directed Research and Development funding from the Lawrence Berkeley National Laboratory, contract DE-AC02-05CH11231, to MAL. NIH U54HG008100 to JG and MAL, and a Komen Foundation grant to JG. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Author Contributions
Conceptualization, CL, and MAL.; Methodology, CL and MAL; Investigation, CL; Formal Analysis, CL, and TJ; Writing Original Draft, CL and MAL; Writing Review & Editing, CL, TJ, JG, MAL; Funding Acquisition, MAL and JG
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- KASSAMBARA ALBOUKADEL. R package version 1.0.3. 2015. factoextra: Extract and visualize the results of multivariate data analyses. [Google Scholar]
- ALLEN M, LOUISE JONES J. Jekyll and Hyde: the role of the microenvironment on the progression of cancer. J Pathol. 2011;223:162–76. doi: 10.1002/path.2803. [DOI] [PubMed] [Google Scholar]
- AMIR EAD, DAVIS KL, TADMOR MD, SIMONDS EF, LEVINE JH, BENDALL SC, SHENFELD DK, KRISHNASWAMY S, NOLAN GP, PE’ER D. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotech. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ANDERSSON J, ROSESTEDT M, ORLOVA A. Imaging of HER2 may improve the outcome of external irradiation therapy for prostate cancer patients. Oncology Letters. 2015;9:950–954. doi: 10.3892/ol.2014.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BECK AH, SANGOI AR, LEUNG S, MARINELLI RJ, NIELSEN TO, VAN DE VIJVER MJ, WEST RB, VAN DE RIJN M, KOLLER D. Systematic Analysis of Breast Cancer Morphology Uncovers Stromal Features Associated with Survival. Science Translational Medicine. 2011;3:108ra113–108ra113. doi: 10.1126/scitranslmed.3002564. [DOI] [PubMed] [Google Scholar]
- BISSELL MJ, LABARGE MA. Context, tissue plasticity, and cancer: are tumor stem cells also regulated by the microenvironment? Cancer Cell. 2005;7:17–23. doi: 10.1016/j.ccr.2004.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRESLIN S, O’DRISCOLL L. The relevance of using 3D cell cultures, in addition to 2D monolayer cultures, when evaluating breast cancer drug sensitivity and resistance. Oncotarget. 2016 doi: 10.18632/oncotarget.9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRODOWSKA K, AL-MOUJAHED A, MARMALIDOU A, MEYER ZU HORSTE M, CICHY J, MILLER JW, GRAGOUDAS E, VAVVAS DG. The clinically used photosensitizer Verteporfin (VP) inhibits YAP-TEAD and human retinoblastoma cell growth in vitro without light activation. Experimental Eye Research. 2014;124:67–73. doi: 10.1016/j.exer.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BURNETT JP, KORKAYA H, OUZOUNOVA MD, JIANG H, CONLEY SJ, NEWMAN BW, SUN L, CONNARN JN, CHEN CS, ZHANG N, WICHA MS, SUN D. Trastuzumab resistance induces EMT to transform HER2+ PTEN− to a triple negative breast cancer that requires unique treatment options. Scientific Reports. 2015;5:15821. doi: 10.1038/srep15821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DIAZ R, NGUEWA PA, PARRONDO R, PEREZ-STABLE C, MANRIQUE I, REDRADO M, CATENA R, COLLANTES M, PEÑUELAS I, DÍAZ-GONZÁLEZ JA, CALVO A. Antitumor and antiangiogenic effect of the dual EGFR and HER-2 tyrosine kinase inhibitor lapatinib in a lung cancer model. BMC Cancer. 2010;10:188. doi: 10.1186/1471-2407-10-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DONOHUE E, THOMAS A, MAURER N, MANISALI I, ZEISSER-LABOUEBE M, ZISMAN N, ANDERSON HJ, NG SSW, WEBB M, BALLY M, ROBERGE M. The Autophagy Inhibitor Verteporfin Moderately Enhances the Antitumor Activity of Gemcitabine in a Pancreatic Ductal Adenocarcinoma Model. Journal of Cancer. 2013;4:585–596. doi: 10.7150/jca.7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEUWIRTH ERICH. R package version 1.1–2. 2014. RColorBrewer: ColorBrewer Palettes. [Google Scholar]
- FARMER P, BONNEFOI H, ANDERLE P, CAMERON D, WIRAPATI P, BECETTE V, ANDRE S, PICCART M, CAMPONE M, BRAIN E, MACGROGAN G, PETIT T, JASSEM J, BIBEAU F, BLOT E, BOGAERTS J, AGUET M, BERGH J, IGGO R, DELORENZI M. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med. 2009;15:68–74. doi: 10.1038/nm.1908. [DOI] [PubMed] [Google Scholar]
- FENG J, GOU J, JIA J, YI T, CUI T, LI Z. Verteporfin, a suppressor of YAP–TEAD complex, presents promising antitumor properties on ovarian cancer. OncoTargets and therapy. 2016;9:5371–5381. doi: 10.2147/OTT.S109979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FERNANDEZ-GARCIA B, EIRO N, MARIN L, GONZALEZ-REYES S, GONZALEZ LO, LAMELAS ML, VIZOSO FJ. Expression and prognostic significance of fibronectin and matrix metalloproteases in breast cancer metastasis. Histopathology. 2014;64:512–22. doi: 10.1111/his.12300. [DOI] [PubMed] [Google Scholar]
- FINAK G, BERTOS N, PEPIN F, SADEKOVA S, SOULEIMANOVA M, ZHAO H, CHEN H, OMEROGLU G, METERISSIAN S, OMEROGLU A, HALLETT M, PARK M. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- GINESTIER C, ADELAIDE J, GONCALVES A, REPELLINI L, SIRCOULOMB F, LETESSIER A, FINETTI P, GENEIX J, CHARAFE-JAUFFRET E, BERTUCCI F, JACQUEMIER J, VIENS P, BIRNBAUM D. ERBB2 phosphorylation and trastuzumab sensitivity of breast cancer cell lines. Oncogene. 2007;26:7163–7169. doi: 10.1038/sj.onc.1210528. [DOI] [PubMed] [Google Scholar]
- GRAPHPAD SOFTWARE 2016. La Jolla California, USA.
- WICKHAM HADLEY, FRANCOIS ROMAIN. R package version 0.5.0. 2016. dplyr: A Grammar of Data Manipulation. [Google Scholar]
- HALDER G, DUPONT S, PICCOLO S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat Rev Mol Cell Biol. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- DE ROSARIO-MARTINEZ HELIOS. R package version 0.2–1. 2015. phia: Post-Hoc Interaction Analysis. [Google Scholar]
- HUDELIST G, KÖSTLER WJ, CZERWENKA K, KUBISTA E, ATTEMS J, MÜLLER R, GSCHWANTLER-KAULICH D, MANAVI M, HUBER I, HOSCHÜTZKY H, ZIELINSKI CC, SINGER CF. Her-2/neu and EGFR tyrosine kinase activation predict the efficacy of trastuzumab-based therapy in patients with metastatic breast cancer. International Journal of Cancer. 2006;118:1126–1134. doi: 10.1002/ijc.21492. [DOI] [PubMed] [Google Scholar]
- HUGGETT MT, JERMYN M, GILLAMS A, ILLING R, MOSSE S, NOVELLI M, KENT E, BOWN SG, HASAN T, POGUE BW, PEREIRA SP. Phase I/II study of verteporfin photodynamic therapy in locally advanced pancreatic cancer. Br J Cancer. 2014;110:1698–704. doi: 10.1038/bjc.2014.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUGHES CS, POSTOVIT LM, LAJOIE GA. Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics. 2010;10:1886–90. doi: 10.1002/pmic.200900758. [DOI] [PubMed] [Google Scholar]
- IOACHIM E, CHARCHANTI A, BRIASOULIS E, KARAVASILIS V, TSANOU H, ARVANITIS DL, AGNANTIS NJ, PAVLIDIS N. Immunohistochemical expression of extracellular matrix components tenascin, fibronectin, collagen type IV and laminin in breast cancer: their prognostic value and role in tumour invasion and progression. Eur J Cancer. 2002;38:2362–70. doi: 10.1016/s0959-8049(02)00210-1. [DOI] [PubMed] [Google Scholar]
- JONES VS, HUANG RY, CHEN LP, CHEN ZS, FU L, HUANG RP. Cytokines in cancer drug resistance: Cues to new therapeutic strategies. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 2016;1865:255–265. doi: 10.1016/j.bbcan.2016.03.005. [DOI] [PubMed] [Google Scholar]
- KATZ BZ, ZAMIR E, BERSHADSKY A, KAM Z, YAMADA KM, GEIGER B. Physical State of the Extracellular Matrix Regulates the Structure and Molecular Composition of Cell-Matrix Adhesions. Molecular Biology of the Cell. 2000;11:1047–1060. doi: 10.1091/mbc.11.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM NG, GUMBINER BM. Adhesion to fibronectin regulates Hippo signaling via the FAK–Src–PI3K pathway. The Journal of Cell Biology. 2015;210:503–515. doi: 10.1083/jcb.201501025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLIJN C, DURINCK S, STAWISKI EW, HAVERTY PM, JIANG Z, LIU H, DEGENHARDT J, MAYBA O, GNAD F, LIU J, PAU G, REEDER J, CAO Y, MUKHYALA K, SELVARAJ SK, YU M, ZYNDA GJ, BRAUER MJ, WU TD, GENTLEMAN RC, MANNING G, YAUCH RL, BOURGON R, STOKOE D, MODRUSAN Z, NEVE RM, DE SAUVAGE FJ, SETTLEMAN J, SESHAGIRI S, ZHANG Z. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotech. 2015;33:306–312. doi: 10.1038/nbt.3080. [DOI] [PubMed] [Google Scholar]
- KONECNY GE, PEGRAM MD, VENKATESAN N, FINN R, YANG G, RAHMEH M, UNTCH M, RUSNAK DW, SPEHAR G, MULLIN RJ, KEITH BR, GILMER TM, BERGER M, PODRATZ KC, SLAMON DJ. Activity of the dual kinase inhibitor lapatinib (GW572016) against HER-2-overexpressing and trastuzumab-treated breast cancer cells. Cancer Res. 2006;66:1630–9. doi: 10.1158/0008-5472.CAN-05-1182. [DOI] [PubMed] [Google Scholar]
- LABARGE MA, NELSON CM, VILLADSEN R, FRIDRIKSDOTTIR A, RUTH JR, STAMPFER MR, PETERSEN OW, BISSELL MJ. Human mammary progenitor cell fate decisions are products of interactions with combinatorial microenvironments. Integr Biol. 2009;1:70–9. doi: 10.1039/b816472j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAMPRECHT MR, SABATINI DM, CARPENTER AE. CellProfiler: free, versatile software for automated biological image analysis. Biotechniques. 2007;42:71–5. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- LEVENTAL KR, YU H, KASS L, LAKINS JN, EGEBLAD M, ERLER JT, FONG SFT, CSISZAR K, GIACCIA A, WENINGER W, YAMAUCHI M, GASSER DL, WEAVER VM. Matrix Crosslinking Forces Tumor Progression by Enhancing Integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIN C, LABARGE MA. The influence of tissue architecture on drug response: anti-cancer drug development in high-dimensional combinatorial microenvironment platforms. In: LARS A, AKSLEN RSW, editors. Biomarkers of the Tumor Microenvironment. Springer; 2017. [Google Scholar]
- LIN CH, LEE JK, LABARGE MA. Fabrication and Use of MicroEnvironment microArrays (MEArrays) J Vis Exp. 2012 doi: 10.3791/4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIN CH, PELISSIER FA, ZHANG H, LAKINS J, WEAVER VM, PARK C, LABARGE MA. Microenvironment rigidity modulates responses to the HER2 receptor tyrosine kinase inhibitor lapatinib via YAP and TAZ transcription factors. Mol Biol Cell. 2015;26:3946–53. doi: 10.1091/mbc.E15-07-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU-CHITTENDEN Y, HUANG B, SHIM JS, CHEN Q, LEE SJ, ANDERS RA, LIU JO, PAN D. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–5. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOGUE JS, MORRISON DK. Complexity in the signaling network: insights from the use of targeted inhibitors in cancer therapy. Genes Dev. 2012;26:641–50. doi: 10.1101/gad.186965.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MICHELS S, SCHMIDT-ERFURTH U. Photodynamic therapy with verteporfin: a new treatment in ophthalmology. Semin Ophthalmol. 2001;16:201–6. doi: 10.1076/soph.16.4.201.10298. [DOI] [PubMed] [Google Scholar]
- MOROISHI T, HANSEN CG, GUAN KL. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15:73–79. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NUCIFORO P, THYPARAMBIL S, AURA C, GARRIDO-CASTRO A, VILARO M, PEG V, JIMENEZ J, VICARIO R, CECCHI F, HOOS W, BURROWS J, HEMBROUGH T, FERRERES JC, PEREZ-GARCIA J, ARRIBAS J, CORTES J, SCALTRITI M. High HER2 protein levels correlate with increased survival in breast cancer patients treated with anti-HER2 therapy. Molecular Oncology. 2016;10:138–147. doi: 10.1016/j.molonc.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ÖHLUND D, FRANKLIN O, LUNDBERG E, LUNDIN C, SUND M. Type IV collagen stimulates pancreatic cancer cell proliferation, migration, and inhibits apoptosis through an autocrine loop. BMC Cancer. 2013;13:154. doi: 10.1186/1471-2407-13-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OKU Y, NISHIYA N, SHITO T, YAMAMOTO R, YAMAMOTO Y, OYAMA C, UEHARA Y. Small molecules inhibiting the nuclear localization of YAP/TAZ for chemotherapeutics and chemosensitizers against breast cancers. FEBS Open Bio. 2015;5:542–549. doi: 10.1016/j.fob.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAN W, WANG Q, ZHANG Y, ZHANG N, QIN J, LI W, WANG J, WU F, CAO L, XU G. Verteporfin can Reverse the Paclitaxel Resistance Induced by YAP Over-Expression in HCT-8/T Cells without Photoactivation through Inhibiting YAP Expression. Cellular Physiology and Biochemistry. 2016;39:481–490. doi: 10.1159/000445640. [DOI] [PubMed] [Google Scholar]
- PARK J, SCHWARZBAUER JE. Mammary epithelial cell interactions with fibronectin stimulate epithelial-mesenchymal transition. Oncogene. 2014;33:1649–1657. doi: 10.1038/onc.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOUTROS PAULC. R package version 5.3.4. 2015. BoutrosLab.plotting.general: Functions to Create Publication-Quality Plots. [Google Scholar]
- PICKUP MW, MOUW JK, WEAVER VM. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014;15:1243–53. doi: 10.15252/embr.201439246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- QUAIL DF, JOYCE JA. Microenvironmental regulation of tumor progression and metastasis. Nature medicine. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R CORE TEAM. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2015. [Google Scholar]
- RANGA A, GOBAA S, OKAWA Y, MOSIEWICZ K, NEGRO A, LUTOLF MP. 3D niche microarrays for systems-level analyses of cell fate. Nat Commun. 2014:5. doi: 10.1038/ncomms5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RUSNAK DW, ALLIGOOD KJ, MULLIN RJ, SPEHAR GM, ARENAS-ELLIOTT C, MARTIN AM, DEGENHARDT Y, RUDOLPH SK, HAWS TF, JR, HUDSON-CURTIS BL, GILMER TM. Assessment of epidermal growth factor receptor (EGFR, ErbB1) and HER2 (ErbB2) protein expression levels and response to lapatinib (Tykerb, GW572016) in an expanded panel of human normal and tumour cell lines. Cell Prolif. 2007;40:580–94. doi: 10.1111/j.1365-2184.2007.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- URBANEK SIMON, HORNER JEFFREY. R package version 1.5–9. 2015. Cairo: R graphics device using cairo graphics library for creating high-quality bitmap (PNG, JPEG, TIFF), vector (PDF, SVG, PostScript) and display (X11 and Win32) output. [Google Scholar]
- SUN XX, YU Q. Intra-tumor heterogeneity of cancer cells and its implications for cancer treatment. Acta Pharmacologica Sinica. 2015;36:1219–1227. doi: 10.1038/aps.2015.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THE CANCER GENOME ATLAS RESEARCH N. WEINSTEIN JN, COLLISSON EA, MILLS GB, SHAW KRM, OZENBERGER BA, ELLROTT K, SHMULEVICH I, SANDER C, STUART JM. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TILGHMAN RW, COWAN CR, MIH JD, KORYAKINA Y, GIOELI D, SLACKDAVIS JK, BLACKMAN BR, TSCHUMPERLIN DJ, PARSONS JT. Matrix Rigidity Regulates Cancer Cell Growth and Cellular Phenotype. PLOS ONE. 2010;5:e12905. doi: 10.1371/journal.pone.0012905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSE JR, ENGLER AJ. Preparation of hydrogel substrates with tunable mechanical properties. Curr Protoc Cell Biol. 2010;Chapter 10(Unit 10):16. doi: 10.1002/0471143030.cb1016s47. [DOI] [PubMed] [Google Scholar]
- ULRICH TA, DE JUAN PARDO EM, KUMAR S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer research. 2009;69:4167–4174. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEAVER VM, LELIEVRE S, LAKINS JN, CHRENEK MA, JONES JC, GIANCOTTI F, WERB Z, BISSELL MJ. beta4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell. 2002;2:205–16. doi: 10.1016/s1535-6108(02)00125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEIGELT B, LO AT, PARK CC, GRAY JW, BISSELL MJ. HER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on the 3D microenvironment. Breast Cancer Res Treat. 2010;122:35–43. doi: 10.1007/s10549-009-0502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XU R, BOUDREAU A, BISSELL MJ. Tissue architecture and function: dynamic reciprocity via extra- and intra-cellular matrices. Cancer metastasis reviews. 2009;28:167–176. doi: 10.1007/s10555-008-9178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YUAN J, LIU M, YANG L, TU G, ZHU Q, CHEN M, CHENG H, LUO H, FU W, LI Z, YANG G. Acquisition of epithelial-mesenchymal transition phenotype in the tamoxifen-resistant breast cancer cell: a new role for G protein-coupled estrogen receptor in mediating tamoxifen resistance through cancer-associated fibroblast-derived fibronectin and β1-integrin signaling pathway in tumor cells. Breast Cancer Research: BCR. 2015;17:69. doi: 10.1186/s13058-015-0579-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG H, RAMAKRISHNAN SK, TRINER D, CENTOFANTI B, MAITRA D, GYŐRFFY B, SEBOLT-LEOPOLD JS, DAME MK, VARANI J, BRENNER DE, FEARON ER, OMARY MB, SHAH YM. Tumor-selective proteotoxicity of verteporfin inhibits colon cancer progression independently of YAP1. Science Signaling. 2015;8:ra98–ra98. doi: 10.1126/scisignal.aac5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.