Abstract
The conventional technique such as patrilocality suggests some substantial effects on population diversity. With that, this particular study investigated the paternal line, specifically Scientific Working Group on DNA Analysis Methods (SWGDAM)-recommended Y-STR markers, namely, DYS19, DYS385, DYS389I/II, DYS390, DYS391, DYS392, DYS393, DYS438, and DYS439. These markers were tested to compare 184 Orang Asli individuals from 3 tribes found in Peninsular Malaysia. As a result, the haplotype diversity and the discrimination capacity obtained were 0.9987 and 0.9076, respectively. Besides, the most diverse marker was DYS385b, whereas the least was DYS391. Furthermore, the Senoi and Proto-Malay tribes were found to be the most distant, whereas the Senoi and Negrito clans were almost similar to each other. In addition, the analysis of molecular variance analysis revealed 82% of variance within the population, but only 18% of difference between the tribes. Finally, the phylogenetic trees constructed using Neighbour Joining and UPGMA (Unweighted Pair Group Method with Arithmetic Mean) displayed several clusters that were tribe specific. With that, future studies are projected to analyse individuals based on more specific sub-tribes.
Keywords: Y-STR, Orang Asli, Senoi, Negrito, Peninsular Malaysia
Introduction
The 18 sub-tribes of Malaysian aborigines, also known as Orang Asli, residing in Peninsular Malaysia make up 0.5% of the total population or approximately 178 000 individuals.1 However, the Orang Asli is not a homogeneous group, but they are made up of 3 separate tribes, namely, Negrito, Senoi, and Proto-Malay, based on their physical appearances and sociological variances. These groupings, in fact, ease the administrative work at the Department of Orang Asli Development (JAKOA).2 They mostly live together and their social interactions, for instance, marriage and mating with people outside of their tribe, are almost unheard.3 Besides, according to Act 134 stipulated in the Malaysian Federal Constitution, a person is considered as an Orang Asli if one’s parents are members of the Orang Asli ethnic group who adhere to the related laws, beliefs, and rituals. In fact, these tribes differ by a variety of factors, including language and physical differences. According to the work by Bellwood,4 physical variances between the Negrito and the Senoi are mainly due to gene flow and founder effect, instead of local differentiation. Nevertheless, only a handful of studies have compared them genetically via molecular study. Moreover, the related molecular studies that investigated these Orang Asli tribes mostly employed maternal or autosomal markers.5
In addition, cultural practices such as patrilocality, polygamy, and polygyny could have significant impacts on genetic diversity of a population, making studying markers specific to either paternal or maternal line leading to varied conclusions.6 Nevertheless, Y-STR does not undergo recombination because it is placed at a non-recombining region on Y chromosome. Hence, those from similar paternal line share similar Y-STR profile, unless in a case of mutation.7,8 Moreover, Y-STRs are used extensively to deduce population histories, ancestries, as well as for forensic purposes.9 The repeat motif can be classified into mono-, di-, tri-, tetra-, or pentanucleotide based on the presence of nucleotide number in the repeat motif.10 In addition, Ellegren noted that the longer the repeat motifs, the less chance for the occurrence of mutation. However, with increment in the number of repeat, the polymorphism also increases due to mutation.11,12 Hence, this study focused on 11 Y-STR markers, namely, DYS19, DYS385a, DYD385b, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS438, and DYS439. The markers were selected because they were embedded in the Scientific Working Group on DNA Analysis Methods (SWGDAM), as these were the recommended markers for population study worldwide.13 Furthermore, this study looked into the differentiation of 3 tribes of Orang Asli in Peninsular Malaysia using SWGDAM with the recommended Y-STR markers. As for the samples, 3 tribes of Orang Asli in Peninsular Malaysia were chosen, specifically from Pahang, Kelantan, Johor, Perak, and Kedah.
Materials and Methods
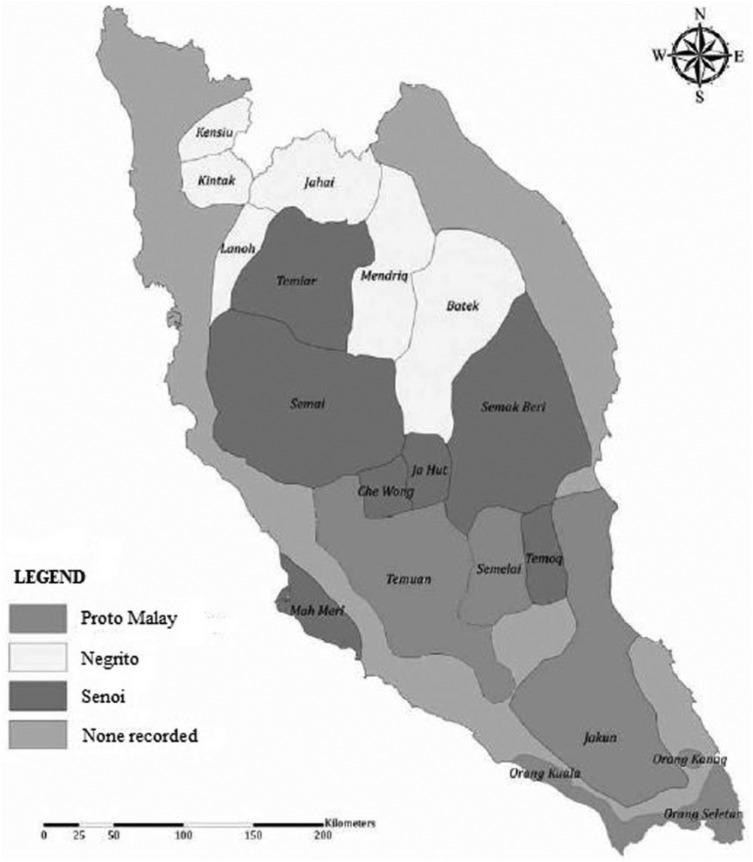
Written approvals from JAKOA, National Medical Research Register (NMRR), and UiTM Research Ethics Committee (REC) were acquired before data collection had begun. Hence, informed consent was obtained from all potential subjects. With that, a total of 184 respondents were chosen from several villages located at Pahang, Kelantan, Johor, Perak, and Kedah. Table 1 presents the names of the villages and the number of samples collected from of the villages. After that, buccal swabs were collected from pure lineage among male individuals with both parents of Orang Asli origin and also above 18 years old at the time of sample collection. Figure 1 shows the distribution pattern of Orang Asli in Peninsular Malaysia.
Table 1.
The locations of sample collection and samples collected.
| No. of samples, n | Tribe | State | Village |
|---|---|---|---|
| 10 | Negrito | Pahang | Kg. Kuala Atok |
| 2 | Senoi | Pahang | Kg. Sungai Tiang |
| 2 | Senoi | Pahang | Kg. Sungai Tiang |
| 8 | Senoi | Pahang | Kg. Sungai Tiang |
| 3 | Negrito | Pahang | Kg. Dedari |
| 2 | Negrito | Pahang | Kg. Teresek |
| 10 | Senoi | Pahang | Kg. Kuching |
| 1 | Senoi | Pahang | Kg. Kuching |
| 7 | Negrito | Pahang | Kg. Gam |
| 10 | Senoi | Kelantan | Kg. Tuel |
| 10 | Senoi | Kelantan | Kg. Hendrop |
| 10 | Negrito | Perak | Kg. Aman Damai |
| 3 | Proto-Malay | Johor | Kg. Mawai |
| 10 | Proto-Malay | Johor | Kg. Layau |
| 8 | Proto-Malay | Johor | Kg. Kempas Menang |
| 4 | Negrito | Perak | Kg. Air Raba |
| 5 | Negrito | Perak | Kg. Air Banun |
| 3 | Negrito | Perak | Kg. Aman Permai |
| 1 | Senoi | Perak | Kg. Aman Permai |
| 10 | Negrito | Kedah | Lubok Legong |
| 10 | Negrito | Perak | Kg. Bukit Asu |
| 10 | Negrito | Perak | Kg. Air Bah |
| 10 | Proto-Malay | Pahang | Kg. Chinta Manis |
| 5 | Proto-Malay | Pahang | Kg. Sungai Yol |
| 3 | Senoi | Pahang | Kg. Sungai Yol |
| 5 | Senoi | Pahang | Kg. Ulu Renggol |
| 5 | Senoi | Pahang | Kg. Sungai Pasu |
| 1 | Proto-Malay | Pahang | Kg. Sungai Pasu |
| 1 | Senoi | Pahang | Kg. Sungai Pasu |
Figure 1.
The locations of Orang Asli in Peninsular Malaysia. Adapted from Kardooni et al.14
Sample collection
Samples for the study were obtained by swabbing both inner sides of the cheeks for 30 seconds using FTA sterile foam applicator. After that, the foam applicator with saliva and buccal cells was pressed onto FTA card and left to dry for at least an hour. This process ensured the integrity of the DNA samples (Whatman, Sigma-Adrich, St. Louis, MO USA).
FTA card washing
FTA card was punched with a micro-puncher to obtain discs with a diameter of 2 mm. The discs were washed using 200 μL FTA reagents and rinsed with 200 μL Tris-Borate-EDTA (TBE) buffer, pH 8.0 before they were dried for an hour at room temperature (Whatman, USA).
Polymerase chain reaction amplification
An amount of 25 μL of polymerase chain reaction (PCR) component mix containing DNA polymerase, KCl, MgCl2, (NH4)2SO4, and nucleotide mix was added to the card. Polymerase chain reaction was performed in T100 Thermal Cycler (Bio-Rad, Berkeley, CA, USA) with an initial denaturation at 95°C for 13 minutes and followed by 30 cycles of denaturation at 95°C for 15 seconds, annealing at 50°C to 60°C in accordance with the markers for 45 seconds, extension at 72°C for 90 seconds, and finally, a final extension for 7 minutes at 72°C. In addition, the primer pairs used for this study were created using the primer sequences retrieved from a paper published by Butler et al15 as tabulated in Table 2.
Table 2.
Information on Y-STR markers applied in this study.
| Marker name | Repeat motif | Allele range | PCR product sizes, bp | GenBank accession |
|---|---|---|---|---|
| DYS19 | TAGA | 10–19 | 214–250 | AC017019 |
| DYS385 a/b | GAAA | 7–28 | 240–304 | AC022486 |
| DYS389I | (TCTG) (TCTA) | 9–17 | 133–169 | AC004617 |
| DYS389II | (TCTG) (TCTA) | 23–34 | 257–289 | AC004617 |
| DYS390 | (TCTA) (TCTG) | 17–28 | 191–227 | AC011289 |
| DYS391 | TCTA | 6–14 | 91–119 | AC011302 |
| DYS392 | TAT | 6–17 | 242–272 | AC011745 |
| DYS393 | AGAT | 9–17 | 108–136 | AC006152 |
| DYS438 | TTTTC | 6–14 | 299–334 | AC002531 |
| DYS439 | GATA | 9–19 | 204–224 | AC002992 |
PCR product detection
Polymerase chain reaction product was detected in 2% agarose gel electrophoresis, pre-stained with ethidium bromide, and run in TBE buffer at 40 minutes, 100 V, and 400 mA. Later, the DNA was visualized under UV transilluminator (Bio-Rad). After that, the sizes of the products were compared with 100-bp ladder for estimation (New England Biolabs Inc., Ipswich, MA, USA).
Fragment analysis
The samples were sent to service provider for fragment analysis (Applied Biosystems, Thermo Fisher Scientific, Forster City, CA, USA). The fragment analysis was completed using ABI Prism 310 Genetic Analyzer. Next, the samples were prepared via denaturation using deionized formamide. In fact, the internal size standard was added to the samples. Later, the samples were introduced into the capillary via electrokinetic injection, where the samples were applied with positive voltage to pull the DNA molecules into the capillary, and each fragment was segregated based on its sizes. The dye molecules on primers were excited using laser source to emit lights of various wavelengths. The light signals were amplified with a photomultiplier to be converted into electronic signals, which were translated into electropherograms for further statistical analysis. After that, the number of repeats was obtained by calculating the number of repeats from a known sample sequence and then comparing the fragment size of the known sample with other samples.18 Moreover, a control sample was incorporated for every analysis to ascertain that the sizes of fragments are indeed reliable. The number of repeats or alleles was recorded to construct the haplotype data.
Statistical analysis
After all the number of repeats for each sample had been obtained, the haplotypes for Orang Asli using Y-STR were developed. Besides, 2 samples with similar haplotype should have similar number of repeats for each marker. In addition, because Y-STR is highly polymorphic, it has been expected to have a large number of haplotypes. However, the allelic frequencies were calculated using simple and direct frequency calculation, where the number of a specific allele found in a sample was divided by the total number of samples. In addition, all the samples were also checked for the presence of private alleles, ie, unique alleles only found in one tribe. Moving on, pairwise population matrix using Nei’s genetic distance, Nei’s genetic identity, and Shannon’s mutual information index had been calculated to observe both similarities and variances between the selected tribes.19 After that, analysis of molecular variance (AMOVA) was calculated using GenAlEx 6.2 with 9999 permutations to increase the level of confidence. Moreover, phylogenetic trees were constructed using Neighbour Joining (NJ) and UPGMA (Unweighted Pair Group Method with Arithmetic Mean) methods based on the pairwise genetic distance of all the samples. These trees were built using freely available software SplitsTree Version 4.8.
Results and Discussion
Haplotype diversity and discrimination capacity
The summary for the number of haplotypes obtained for each sub-tribe, haplotype diversity, and discrimination capacity is depicted in Table 3.
Table 3.
The haplotype diversity and discrimination capacity for each sub-tribe.
| Sub-tribe | No. of individuals | No. of haplotypes | Haplotype diversity | Discrimination capacity |
|---|---|---|---|---|
| Bateq | 24 | 11 | 0.861 | 0.458 |
| Che Wong | 2 | 2 | 0.500 | 1.000 |
| Jahai | 23 | 18 | 0.926 | 0.783 |
| Jakun | 8 | 8 | 0.843 | 0.750 |
| Kanaq | 3 | 3 | 0.667 | 1.000 |
| Kensiu | 12 | 9 | 0.875 | 0.750 |
| Kintak | 10 | 8 | 0.860 | 0.800 |
| Kuala | 10 | 8 | 0.840 | 0.800 |
| Lanoh | 10 | 9 | 0.880 | 0.900 |
| Semai | 11 | 9 | 0.876 | 0.818 |
| Semoq Beri | 18 | 11 | 0.809 | 0.611 |
| Temuan | 21 | 19 | 0.936 | 0.904 |
| Temiar | 33 | 22 | 0.920 | 0.666 |
As a result, a total of 137 haplotypes had been retrieved from 184 samples. From the 137 haplotypes, 13 were found to be shared, whereas 124 were exclusive for 1 individual. In fact, the most common haplotype was shared by 4 individuals (H001), whereas the second most common haplotypes were shared by 3 individuals per haplotype (H005, H097), and 10 other haplotypes were shared by 2 subjects (H003, H012, H021, H023, H041, H095, H103, H123, H130, H160). From all the 137 haplotypes, only 1 haplotype was not tribe specific. Therefore, the findings suggest that the Y-STR data could be used to differentiate tribes of individuals.
Table 3 portrays that the Temuan sub-tribe has the highest value of haplotype diversity, whereas the Che Wong sub-tribe has the smallest value of haplotype diversity. In fact, this finding could be weighed in as the number of sample per sub-tribe had been dissimilar. Next, the discriminatory power or the discrimination capacity for this set of markers had been the highest for Che Wong and Kanaq, in which the set of markers had been able to differentiate the individuals of both sub-tribes at all times.
Furthermore, haplotype diversity was calculated using the haplotype data and resulted in the value of 0.9987, whereas the discrimination capacity was 0.9076. Moreover, the values obtained in this study have been lower than the values of haplotype diversity and discrimination capacity obtained from a study that investigated other major groups in Malaysia, namely, Malays, Chinese, and Indians.20 The haplotype diversity obtained by Chang et al, for the 3 major groups in Malaysia was 0.9996, whereas the discriminatory capacity was 94.6%. These suggest that the Orang Asli population in Peninsular Malaysia are more closely related to each other, in comparison with the other major groups due to geographical isolation. In addition, based on the haplotype diversity value, one can conclude that the 11 markers of Y-STR applied in this study had been able to provide the haplotypes with high diversity. Meanwhile, the discrimination capacity shows that in a given time, the 11 markers used in the experiment did differentiate an Orang Asli individual from another at 90.76% of the time.
Allele frequencies
As for allele frequencies, the most diverse marker was DYS385b with 11 alleles, whereas the least diverse marker was DYS391 with only 4 alleles, which could be due to mutation. Meanwhile, DYS391 displayed lower mutation rate compared with DYS385b. The DYS385 is a multi-locus, where 2 loci can be amplified using a set of primers. Besides,16 claimed that multi-locus markers such as DYS385 and DYS389 have the ability to increase the aspect of polymorphism in selected set of markers, thus increasing its power of discrimination. Furthermore, the locus diversity of all markers had been similar to those from prior studies, except for DYS19 that is lower than the average. Nonetheless, the results fail to concur with high gene diversity of DYS19 in studies of Y-STR conducted in the US, Iran, and Hispanic populations.21–23 As such, more studies have to be conducted with more samples to ascertain whether this is just an exception for the markers or whether Orang Asli does possess lower diversity for DYS19.5 Meanwhile, the other marker with small locus diversity value is DYS392, which is a trinucleotide marker. Nonetheless, in comparison with the surprisingly low value of DYS19, this marker had been expected to exhibit low locus diversity as the mutation rate for this particular marker was not too high.
Genetic distance and AMOVA
The summary of pairwise population values for Nei’s genetic distance, Nei’s genetic identity, and Shannon’s mutual information index is presented in Table 4.
Table 4.
Summary of pairwise population values of Nei’s genetic distance, Nei’s genetic identity, and Shannon’s mutual information index.
| Population 1 | Population 2 | Nei’s genetic distance | Nei’s genetic identity | Shannon’s mutual information index |
|---|---|---|---|---|
| Negrito | Senoi | 0.064 | 0.938 | 0.085 |
| Senoi | Proto-Malay | 0.165 | 0.848 | 0.128 |
| Proto-Malay | Negrito | 0.135 | 0.874 | 0.107 |
From the calculation of Nei’s genetic distance, the Senoi tribe was found to be the most distant from the Proto-Malay but exemplified the highest value of genetic distance when compared with other tribes. Besides, the Senoi and Negrito sub-tribes displayed the closest genetic distance between each other. Genetic identity refers to the value of similarity between 2 groups and it is the opposite to genetic distance. In precise, the pair with the highest genetic distance showed the least value of genetic identity, whereas the pair with the lowest genetic distance had the highest value of genetic identity. Moreover, the value obtained from Nei’s genetic distance was translated in a similar way when Shannon’s mutual information index was determined. Furthermore, based on the Shannon’s mutual information index, the lowest value was between Senoi and Negrito pair, followed by Proto-Malay and Negrito, and finally, Senoi and Proto-Malay.
Although the number of samples for both Senoi and Negrito differed, their diversity values did not vary much due to the characteristic of Y-STR distribution, where the distribution was uniform across various populations, except for several exceptions, such as DYS19, as mentioned earlier.24 Nonetheless, more samples should be incorporated in future study to increase its reliability.
In addition, the summary of AMOVA calculation is presented in Table 5. The calculation was repeated using 99, 999, and 9999 permutations, in which all calculations with those varied permutations resulted in similar findings. The results displayed in this section were obtained after running the AMOVA calculation using 9999 permutations.
Table 5.
Summary of analysis of molecular variance calculation.
| Source | df | SS | MS | Est. var. | % of estimated variances |
|---|---|---|---|---|---|
| Among population | 2 | 1121.244 | 560.622 | 8.741 | 18 |
| Within population | 181 | 7271.337 | 40.173 | 40.173 | 82 |
| Total | 183 | 8392.582 | 48.914 | 100 |
Abbreviations: Est. var., estimated variances; MS, mean sum of squares; SS, sum of squares.
Based on the AMOVA analysis, it was found that from the total variances obtained, 82% derived from the diversities among the population, whereas the remaining 18% reflected the 3 tribes of Orang Asli. In other words, most of the variances found were obtained between individuals in a tribe, whereas the other minor differences were found among groups. Furthermore, the percentage of difference between the individuals of Orang Asli had been lower than the percentage difference of other major groups in Peninsular Malaysia.20 This suggests higher genetic similarities between Orang Asli individuals when compared with the rest of the population.
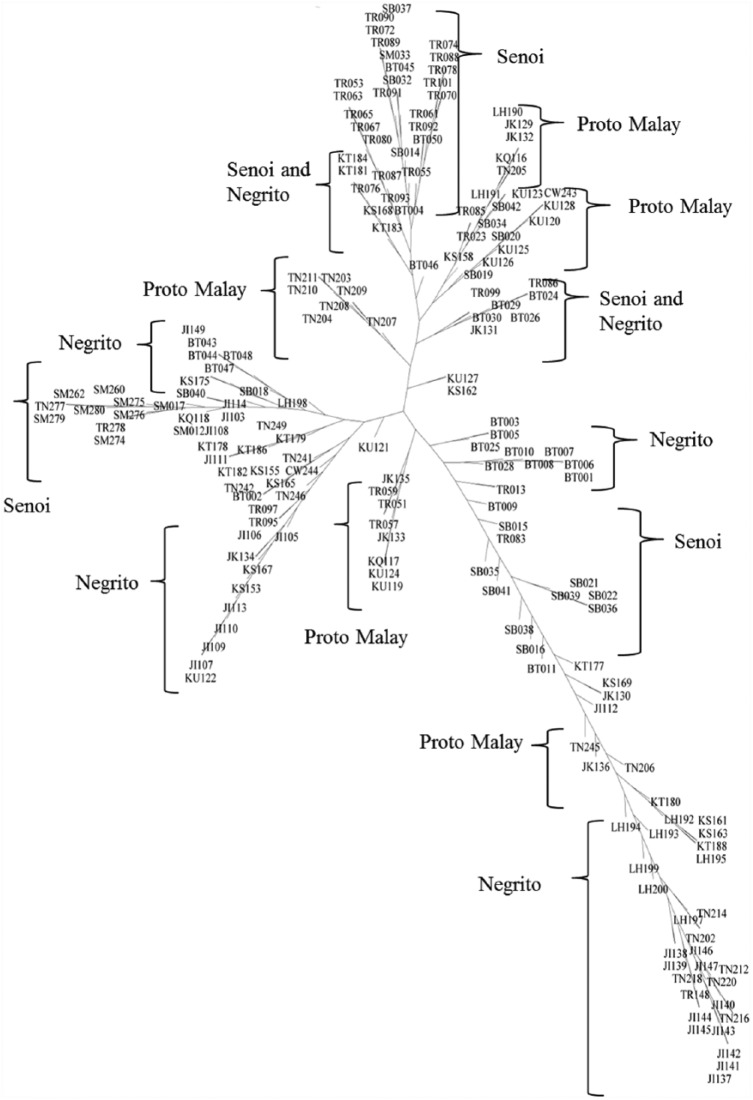
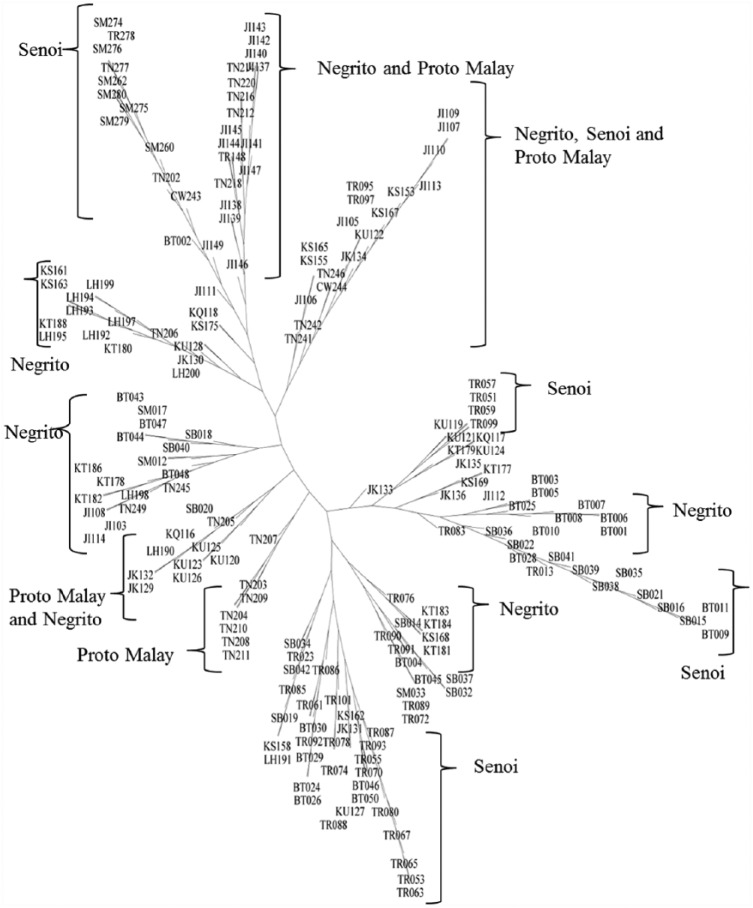
Phylogenetic trees
Phylogenetic trees were constructed based on pairwise differences derived from the subjects using phylogenetic software SplitsTree4. Hence, Figures 2 and 3 illustrate the phylogenetic trees, which were constructed using NJ and UPGMA, respectively. The leaves were labelled with designated names for the samples, including sub-tribe names, following by the order of the samples collected. The sub-tribes and their abbreviations are given in the following: Kintak (KT), Kensiu (KS), Bateq (BT), Jahai (JH), and Lanoh (LH) for Negrito tribe; Semoq Beri (SB), Temiar (TR), Che Wong (CW), Jah Hut (JH), and Semai (SM) for Senoi tribe; and Jakun (JK), Temuan (TN), Kuala (KU), and Kanaq (KQ) for Proto-Malay tribe.
Figure 2.
Neighbour Joining Tree.
Figure 3.
UPGMA (Unweighted Pair Group Method with Arithmetic Mean) tree.
Overall, both NJ and UPGMA generated trees that reflected the traditional method of classification for Orang Asli, where various sub-tribes from the same tribe were clustered into the same split. This suggests close correlation between the sub-tribes from the same tribe. As for NJ trees, 3 clusters represented the Senoi tribe, whereas 4 clusters for Negrito samples, and 5 clusters were made up of Proto-Malay samples. Nevertheless, 2 clusters reflected samples from both Negrito and Senoi. As for genetic calculation, the Negrito and Senoi tribes appeared to have smaller genetic distance between each other, hence explaining the mixture of samples from the 2 tribes in 1 split. Meanwhile, in the UPGMA tree, 4 splits were made up of Senoi samples, whereas another 4 splits represented the Negrito tribe and 1 for Proto-Malay. However, the UPGMA tree exhibited more mixed splits when compared with NJ tree with 2 splits for Proto-Malay and Negrito samples each, whereas 1 for all tribes of Orang Asli. Besides, the Proto-Malay and Negrito tribes showed the second closest genetic distance, right after Negrito and Senoi tribes, which depicts the splits for both tribes.
Conclusions
In conclusion, the SWGDAM recommended Y-STR markers could be used to differentiate Orang Asli based on tribes. Moreover, as insignificant marker allele pattern was not found, it is recommended to get more samples from the population to set a larger haplotype database if Y-STR is used to differentiate the individuals according to tribes. However, the discrimination capacity had been low for it was inadequate to differentiate individuals. In short, the set of markers could be used for population studies, especially those for tribal affinities, but not for forensic purposes. In addition, all the alleles found in both Y-STR and autosomal STR had been in the range of the expected alleles, indicating no unique allele for Orang Asli (YHRD [Y Chromosome Haplotype Reference Database]). Nevertheless, the allele frequency distribution was not always congruent with worldwide databases. Based on the Y-STR, the Senoi and Proto-Malay tribes had been the most distant between each other, followed by Proto-Malay and Negrito, as well as Negrito and Senoi. Meanwhile, according to AMOVA analysis, all the variances found had been mostly caused by the differences between individuals, in comparison with the differences between varied populations. Furthermore, phylogenetic trees were built using Y-STR and displayed clusters that reflected their tribal affinities. Therefore, for those with intention to expand this study, it is highly recommended to incorporate more samples from all other sub-tribes of Orang Asli while also considering other locations to better determine similarities or differences between the sub-tribes. Moreover, to increase the accuracy of the clusters, the samples for future studies should be more pure and not tainted. This can be achieved by selecting samples with known tribal background, which refers to 5 pure generations, when compared with just 3, as employed in this study. Moreover, in upcoming studies, more population data should be embedded while comparing the tribes to detect affinities of Orang Asli in Peninsular Malaysia with other populations worldwide.
Acknowledgments
The authors would like to thank RMI for granting the ethical approval, Jabatan Kemajuan Orang Asli (JAKOA), National Medical Research Register (NMRR), Institute of Research Management and Innovation (IRMI), UiTM, and Universiti Teknologi MARA for their continuous support. They specially thank all volunteers who have given their samples for the study.
Footnotes
Peer review:Five peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1619 words, excluding any confidential comments to the academic editor.
Original Research
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by research grant from the Malaysian Ministry of Higher Education (MOHE) (600-RMI/FRGS TD 5/3 [1/2013]) and this publication was funded by Malaysian Ministry of Higher Education (MOHE) (600-RMI/FRGS 5/3 [46/2015]).
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: FZMY and SSMY conceived and designed the experiments and contributed to the writing of the manuscript. SSMY analysed the data and wrote the first draft of the manuscript. SSMY, FZMY, MM, BMM-Z, and KM agreed with manuscript results and conclusions. SSMY and MM jointly developed the structure and arguments for the paper. FZMY made critical revisions and approved final version. All authors reviewed and approved the final manuscript.
References
- 1. Kardooni R, Fatimah K, Siti Rohani Y, Siti Hajar Y. Traditional knowledge of Orang Asli on forests in Peninsular Malaysia. Indian J Tradit Know. 2014;13:283–291. [Google Scholar]
- 2. Lim LS, Ang KC, Wahid SA, Md-Zain BM. Mitochondrial DNA polymorphism and phylogenetic relationships of Proto Malays in Peninsular Malaysia. J Biol Sci. 2010;10:71–83. [Google Scholar]
- 3. Ang KC, Ngu MS, Reid KP, et al. Skin color variation in Orang Asli tribes of Peninsular Malaysia. PLoS ONE. 2012;7:e42752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bellwood P. Cultural and biological differentiation in Peninsular Malaysia: the last 10,000 years. Asian Perspect. 1993;32:37–60. [Google Scholar]
- 5. Sofia SMY, Farida ZMY, Md-Zain BM. A preliminary study on phylogenetic and population genetic study using DYS19 for Orang Asli in Taman Negara Pahang. Malays Appl Biol. 2015;44:137–139. [Google Scholar]
- 6. Stanyon R, Sazzini M, Luiselli D. Timing the first human migration into eastern Asia. J Biol. 2009;8:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Underhill PA, Shen P, Lin AA, et al. Y chromosome sequence variation and the history of human populations. Nat Genet. 2000;26:358–361. [DOI] [PubMed] [Google Scholar]
- 8. Ge J, Budowle B, Planz JV, Eisenberg AJ, Ballantyne J, Chakraborty R. US forensic Y-chromosome short tandem repeats database. Legal Med. 2010;12:289–295. [DOI] [PubMed] [Google Scholar]
- 9. Bentayebi K, Abada F, Ihzmad H, Amzazi S. Genetic ancestry of a Moroccan population as inferred from autosomal STRs. Meta Gene. 2014;2:427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Powell W, Machray GC, Provan J. Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1996;1:215–222. [Google Scholar]
- 11. Ballantyne KN, Goedbloed M, Fang R, et al. Mutability of Y-chromosomal microsatellites: rates, characteristics, molecular bases, and forensic implications. Am J Hum Genet. 2010;87:341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5:435–445. [DOI] [PubMed] [Google Scholar]
- 13. Willuweit S, Roewer L; for International Forensic Y Chromosome User Group. Y chromosome haplotype reference database (YHRD): update. Forensic Sci Int Genet. 2007;1:83–87. [DOI] [PubMed] [Google Scholar]
- 14. Masron T, Masami F, Ismail N. Orang Asli in Peninsular Malaysia: population, spatial distribution and socio-economic condition. J Ritsumeikan Soc Sci Humanit. 2013;6:75–115. [Google Scholar]
- 15. Butler JM, Schoske R, Vallone PM, Kline MC, Redd AJ, Hammer MF. A novel multiplex for simultaneous amplification of 20 Y-chromosome STR markers. Forensic Sci Int. 2002;129:10–24. [DOI] [PubMed] [Google Scholar]
- 16. Butler JM. Recent development in Y-short tandem repeat and Y-single nucleotide polymorphism analysis. Forensic Sci Rev. 2003;15:91–111. [PubMed] [Google Scholar]
- 17. Schoske R. The design, optimization and testing of Y chromosome short tandem repeat megaplexes. Unpublished Doctoral Dissertation, American University, Washington DC; 2003. [Google Scholar]
- 18. Butler JM, Kline MC, Decker AE. Addressing Y-chromosome short tandem repeat allele nomenclature. J Genetic Geneal. 2008;4:125–148. [Google Scholar]
- 19. Nei M. Genetic distance and molecular phylogeny. In: Ryman N, Utter F, eds. Population Genetics and Fishery Management. Seattle, WA: Washington University Press; 1987:193–223. [Google Scholar]
- 20. Chang YM, Perumal R, Keat PY, Kuehn DLC. Haplotype diversity of 16 Y-chromosomal STRs in three main ethnic populations (Malays, Chinese and Indians) in Malaysia. Forensic Sci Int. 2007;167:70–76. [DOI] [PubMed] [Google Scholar]
- 21. Calderon S, Perez-Benedico D, Mesa L, Guyton D, Rowold DJ, Herrera RJ. Phylogenetic and forensic studies of the Southeast Florida Hispanic population using the next-generation forensic PowerPlex® Y23 STR marker system. Legal Med. 2013;15:289–292. [DOI] [PubMed] [Google Scholar]
- 22. Coble MD, Hill CR, Butler JM. Haplotype data for 23 Y-chromosome markers in four US population groups. Forensic Sci Int Genet. 2013;7:e66–e68. [DOI] [PubMed] [Google Scholar]
- 23. Tabrizi AA, Hedjazi A, Kerachian MA, Honarvar Z, Dadgarmoghaddam M, Raoofian R. Genetic profile of 17 Y-chromosome STR haplotypes in East of Iran. Forensic Sci Int Genet. 2015;14:e6–e7. [DOI] [PubMed] [Google Scholar]
- 24. Agrawal S, Khan F. Reconstructing recent human phylogenies with forensic STR loci: a statistical approach. BMC Genet. 2005;6:47. [DOI] [PMC free article] [PubMed] [Google Scholar]