Abstract
Purpose
Deleterious germline mutations contribute to pancreatic cancer susceptibility and are well documented in families in which multiple members have had pancreatic cancer.
Methods
To define the prevalence of these germline mutations in patients with apparently sporadic pancreatic cancer, we sequenced 32 genes, including known pancreatic cancer susceptibility genes, in DNA prepared from normal tissue obtained from 854 patients with pancreatic ductal adenocarcinoma, 288 patients with other pancreatic and periampullary neoplasms, and 51 patients with non-neoplastic diseases who underwent pancreatic resection at Johns Hopkins Hospital between 2000 and 2015.
Results
Thirty-three (3.9%; 95% CI, 3.0% to 5.8%) of 854 patients with pancreatic cancer had a deleterious germline mutation, 31 (3.5%) of which affected known familial pancreatic cancer susceptibility genes: BRCA2 (12 patients), ATM (10 patients), BRCA1 (3 patients), PALB2 (2 patients), MLH1 (2 patients), CDKN2A (1 patient), and TP53 (1 patient). Patients with these germline mutations were younger than those without (mean ± SD, 60.8 ± 10.6 v 65.1 ± 10.5 years; P = .03). Deleterious germline mutations were also found in BUB1B (1) and BUB3 (1). Only three of these 33 patients had reported a family history of pancreatic cancer, and most did not have a cancer family history to suggest an inherited cancer syndrome. Five (1.7%) of 288 patients with other periampullary neoplasms also had a deleterious germline mutation.
Conclusion
Germline mutations in pancreatic cancer susceptibility genes are commonly identified in patients with pancreatic cancer without a significant family history of cancer. These deleterious pancreatic cancer susceptibility gene mutations, some of which are therapeutically targetable, will be missed if current family history guidelines are the main criteria used to determine the appropriateness of gene testing.
INTRODUCTION
Pancreatic cancer is expected to be the second leading cause of cancer death in the United States by the year 2030.1 Inherited gene mutations are known to contribute to pancreatic cancer in patients with familial pancreatic cancer (defined by the presence of two first-degree relatives with the disease),2 but the extent to which deleterious gene mutations contribute to pancreatic cancer risk in individuals without a family history of pancreatic cancer is not well defined. Identifying inherited susceptibility gene mutations in an individual improves assessment and decisions regarding cancer screening for family members and can guide treatment of patients with pancreatic cancer.
The established familial pancreatic cancer susceptibility genes include the BRCA2, ATM, PALB2, CDKN2A, PRSS1, STK11, MLH1, and MSH23 genes.4-8 The results of whole-genome sequencing of more than 600 individuals with familial pancreatic cancer were recently reported, with analysis focused on the role of low-frequency truncating mutations.2 Deleterious germline mutations in the BRCA2 gene account for the biggest fraction of known familial pancreatic cancer genes (found in approximately 5% to 10% of familial pancreatic cancer families7,9-12), followed by ATM (deleterious mutations found in approximately 2% to 3%).2,4 Deleterious germline mutations involving other genes are less common (each found in approximately ≤ 1% of affected individuals from familial pancreatic cancer kindred). These genes include CDKN2A (deleterious mutations cause familial atypical melanoma mole syndrome),13-17 PALB2 (DNA mismatch–repair genes that cause Lynch syndrome),3 STK11 (Peutz-Jeghers syndrome), and PRSS1 (hereditary recurrent acute pancreatitis).8,18-21 Germline BRCA1 mutations increase the overall risk of developing pancreatic cancer by approximately two- to -four-fold.2,7,22,23 The role of other genes in pancreatic cancer susceptibility is still being evaluated.
The prevalence of germline mutations in individual pancreatic cancer susceptibility genes in patients with apparently sporadic forms of the disease (ie, without a family history of pancreatic cancer) has also been studied.24-27 These studies have primarily focused on BRCA genes. For example, germline BRCA2 mutations are found in a small percentage of patients with apparently sporadic pancreatic cancer,28 with a higher prevalence found in populations with many individuals of Ashkenazi Jewish heritage because of the common 6174delT BRCA2 founder mutation in that population.27,29-33 In one study, germline BRCA mutations were identified in 4.6% of an unselected series of 306 patients with pancreatic cancer from a single center.24
The absence of a significant family history in patients with an established deleterious germline mutation is probably primarily as the result of incomplete penetrance, rather than de novo mutation in the germline. For example, the average lifetime risk of developing pancreatic cancer among BRCA2 gene mutation carriers is estimated to be approximately 5% to 10%.34-36 Notably, these estimates have been determined primarily in families ascertained for breast and/or ovarian cancer and therefore may be an underestimate.
There is considerable potential clinical utility to identifying a germline susceptibility gene in a patient with pancreatic cancer. Mutation carriers with pancreatic cancer may have more options for personalized medicine directed against the genetic drivers of their cancer,37 and their family members may benefit from cancer screening and cancer prevention strategies for pancreatic and extrapancreatic cancers.38-42 Relatives of patients with apparently sporadic pancreatic cancer are at increased risk of mortality from other cancers.43
In this study, we determined the prevalence of germline mutations in known and candidate pancreatic cancer susceptibility genes in a large hospital-based series of patients unselected for their family history of pancreatic cancer. We compared the prevalence of deleterious mutations in these patients with the prevalence in patients who underwent surgery for other periampullary cancers and diseases.
METHODS
Patients and Specimens
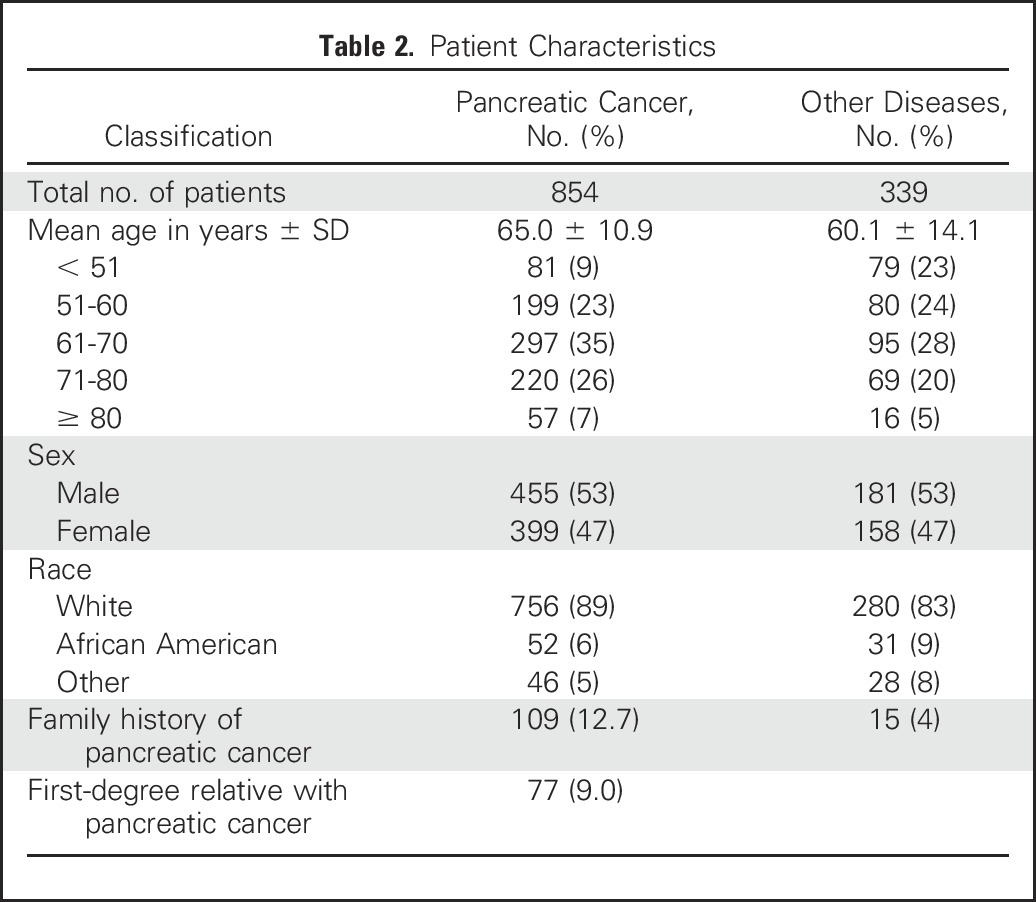
This study included 854 patients with pancreatic ductal adenocarcinoma who were evaluated and treated at the Johns Hopkins Hospital between 2000 and 2015. Patients were enrolled in the study during their preoperative evaluation or during their multidisciplinary clinic visit. Personal and family history information was obtained from the medical record and from the National Familial Pancreas Tumor Registry. To estimate the prevalence of deleterious germline mutations in patients with other periampullary/pancreatic diseases referred to the same clinical services, we included 339 patients who had undergone pancreatic resection for periampullary/biliary pathology other than pancreatic cancer, including 108 with other cancers (duodenal, biliary, gall bladder), 113 with other neoplasms (pancreatic neuroendocrine tumors, GI stromal tumors, carcinoid), 25 with precancerous neoplasms (duodenal, ampullary adenoma), 25 with serous cystadenoma, and 51 with non-neoplastic conditions including 37 with pancreatitis (Table 1); patient demographic data are listed in Table 2.
Table 1.
Pathology and Indications for Pancreatic Resection
Table 2.
Patient Characteristics
All elements of this study were approved by the Johns Hopkins Institutional Review Board, and written informed consent was obtained from all patients.
DNA Extraction
Genomic DNA was extracted from either frozen normal tissue from pancreatic resection specimens (duodenum, spleen, or pancreas) or peripheral blood mononuclear cells using QIAamp DNA Micro Kit (QIAGEN, Hilden, Germany), according to the manufacturer’s instructions. DNA samples were quantified using Quantifiler (Thermo Fisher Scientific, Waltham, MA).
DNA Sequencing
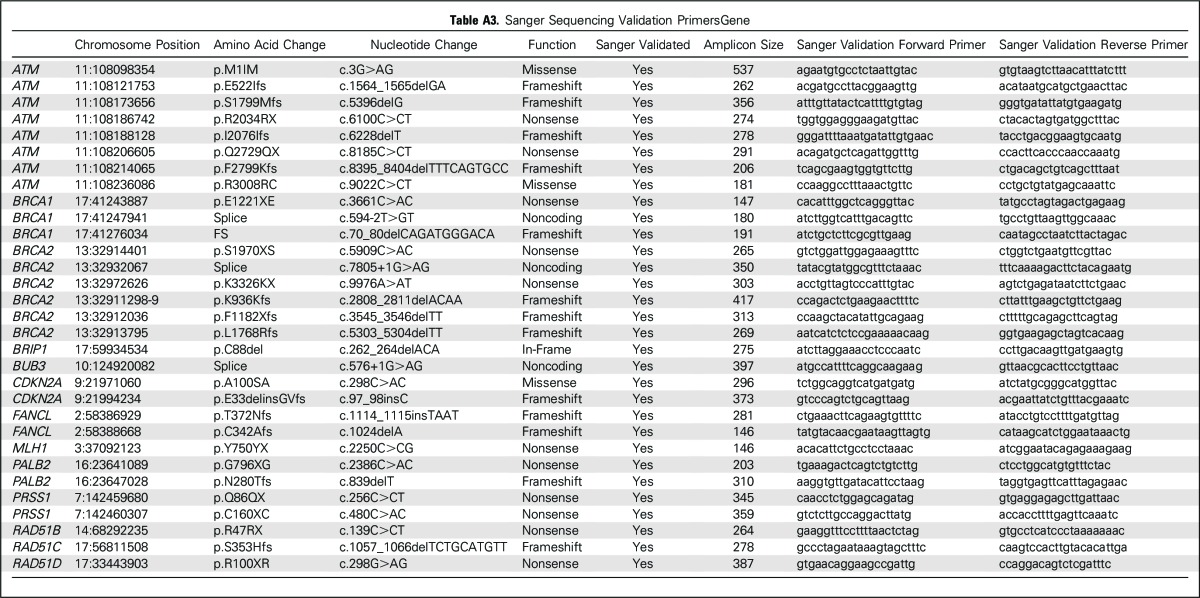
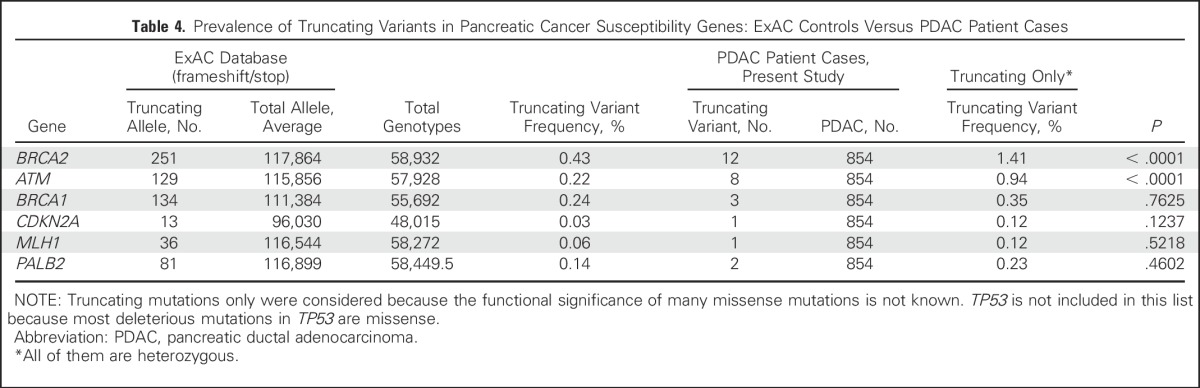
Thirty-two genes (Appendix Table A1 [online only]) were sequenced using an AmpliSeq Custom Panel. Next-generation sequencing was performed using the Ion Proton system (Life Technologies [Life-Tech], Carlsbad, CA), according to the manufacturer’s protocols and as previously described.44 These genes were either known pancreatic cancer susceptibility genes (BRCA2, ATM, PALB2, BRCA1, CDKN2A, MLH1, MSH2, PRSS1, STK11, and TP53), known cancer susceptibility genes (MSH6, PMS2, CDH1, RAD51C, RAD51D, BUB1B, and FANCJ), or candidate pancreatic cancer susceptibility genes (FANCA, FANCC, FANCG, FANCL, ARID1A, RECQL4, XRCC2, XRCC3, ERCC4, TERT, BAP1, BUB1, BUB3, and RNF43). Thus, 20 ng of DNA (10 ng per primer pool) were used for AmpliSeq polymerase chain reaction. After FuPa digestion, P1 adaptor/Xpress barcode ligation, and library clean up (Agencourt AMPure XP Reagent; Beckman Coulter, Brea, CA), libraries were eluted into low Tris-EDTA and quantified (Ion Quantitation Kit; Life-Tech). Libraries underwent emulsion polymerase chain reaction in an Ion OneTouch2 (Life-Tech) for 5 hours; Ion Sphere Particles were then cleaned and enriched in the OneTouch ES (Life-Tech). Enriched Ion Sphere Particles were loaded into P1v3 chips for sequencing (Ion Proton; Life-Tech). The postsequencing raw FASTQ files were launched in NextGENe (version 2.41; SoftGenetics, Chicago, IL) software for alignment to the hg19 human reference genome and single-nucleotide variant calling. Alignments were visually verified using Integrative Genomics Viewer (version 2.3; Broad Institute, Cambridge, MA) and NextGENe Viewer. The functional significance of variants was determined by interrogating ClinVar and PubMed. Variants of unknown significance are listed in Appendix Table A2 (online only). Variants identified as truncating (nonsense, frameshift, splice intervening sequence ± 1 or 2) and deleterious missense variants were validated by Sanger sequencing performed at The Johns Hopkins Synthesis & Sequencing Facility (Applied Biosystems DNA sequencers; Sanger validation primer sequences are listed in Appendix Table A3 [online only]). Deleterious variants were identified in several candidate pancreatic cancer susceptibility genes. To further evaluate these variants, primary pancreatic cancer tissue from three patients was laser-capture microdissected44 and sequenced to evaluate for biallelic inactivation (tumor was not available from the patients with the RAD51D or BUB1B mutation). For genes with deleterious truncating variants (Table 3), we provide the truncating variant data in control subjects from the ExAC database (Broad Institute; Table 4).
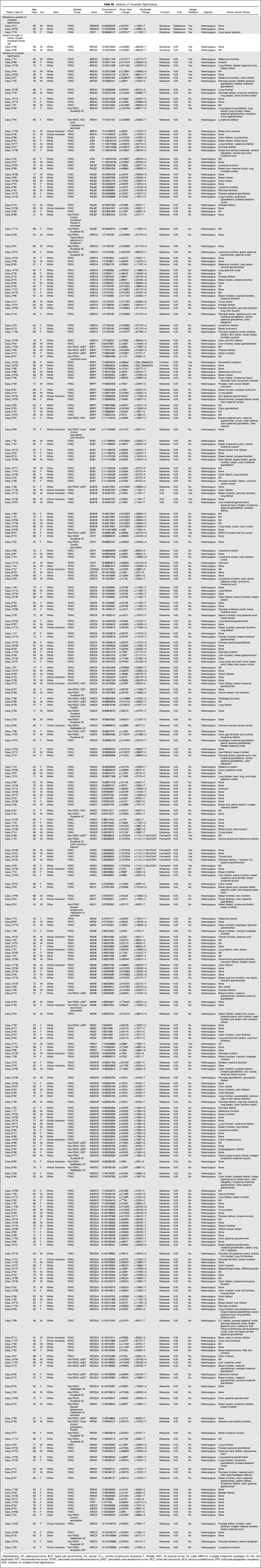
Table 3.
Deleterious Mutations in Pancreatic Cancer Susceptibility Genes
Table 4.
Prevalence of Truncating Variants in Pancreatic Cancer Susceptibility Genes: ExAC Controls Versus PDAC Patient Cases
Statistical Analysis
The mean age of carriers of a deleterious germline variant was compared with noncarriers using the t test. SPSS software was used (version 22; IBM, Armonk, NY). A two-tailed P < .05 was considered statistically significant.
RESULTS
Characteristics of the study population are listed in Table 1 and Table 2. Thirty-three (3.9%) of the 854 patients with pancreatic ductal adenocarcinoma had an identifiable deleterious germline mutation, of which 28 were truncating (Table 3). Thirty-one (3.5%) of these patients had a deleterious mutation in a known pancreatic cancer susceptibility gene. This included 12 patients with deleterious germline BRCA2 mutations, five of whom carried the Ashkenazi Jewish founder BRCA2 mutation (6174delT); ten with ATM; three with BRCA1; two with PALB2; two with MLH1; and one each with a CDKN2A and TP53 mutation. Two patients had a mutation in a candidate pancreatic cancer susceptibility gene (BUB1B and BUB3). Patients with deleterious BUB1B mutations are prone to aneuploidy, chromosomal alterations, and cancer,45 and BUB1B has been identified as a candidate pancreatic cancer susceptibility gene.2 Evidence regarding the role of BUB3 as a cancer susceptibility gene is less clear46,47; both BUB1B and BUB3 regulate mitotic checkpoints and, in mouse models, BUB3 heterozygotes are similarly prone to premature aging and chromosomal instability.48 We sequenced the pancreatic cancer DNA from the patient with the deleterious germline BUB3 variant but did not find evidence of biallelic inactivation.
In addition to the 33 patients with a deleterious mutation in a known or suspected pancreatic cancer susceptibility gene, three patients carried a deleterious mutation in another cancer susceptibility gene (one each involving CDH1, RAD51D, and RAD51B). The significance of these variants for pancreatic cancer susceptibility is not certain (they are listed in Appendix Table A2). RAD51D and RAD51C are ovarian cancer susceptibility genes.49,50 Common variants in RAD51B are associated with increased breast cancer risk.51 Germline CDH1 mutations predispose to hereditary gastric cancer and lobular breast cancer. The CDH1 P373L mutation identified in one patient has been described in a hereditary gastric cancer family.52 We sequenced microdissected pancreatic cancer DNA from the patients with the germline RAD51B and CDH1 mutation, but we did not find evidence of biallelic inactivation of their germline mutated gene in their pancreatic cancer. Overall, there is not sufficient evidence to indicate that RAD51B and CDH1 germline mutations contribute to pancreatic cancer development.
Numerous variants of unknown significance were also identified (listed in Appendix Table A2). In addition to the 33 patients with deleterious mutations, two patients with pancreatic ductal adenocarcinoma carried the BRCA2 polymorphic stop variant p.K3326X. This variant was initially not thought to confer a cancer risk, but is now considered a modifier allele with evidence that carriers have a small increased risk of breast, pancreatic, and other cancers.53-55
The majority (82%) of patients with deleterious germline mutations had a family history of other cancers, but only five mutation carriers (15%) had cancer family histories that suggested a familial cancer syndrome (Table 3). Eighteen of the 33 mutation carriers had a family history of breast cancer reported in a first- or second-degree relative, six had a family history of prostate cancer, and three had a family history of ovarian cancer. Only three (9%) of the 33 individuals with a germline deleterious mutation had a family history of pancreatic cancer. In comparison, among the 818 patients with pancreatic ductal adenocarcinoma without a germline mutation, 117 (14.3%) had a family history of pancreatic cancer identified; for 86 of these patients, it was in a first-degree relative.
Although there are no dedicated pancreatic cancer National Comprehensive Cancer Network (NCCN) guidelines for gene testing, the breast/ovarian cancer NCCN guidelines for gene testing56 include recommendations for when to consider gene testing patients with pancreatic cancer. Candidates for gene testing include individuals 1with a close relative with pancreatic cancer, 2who are of Ashkenazi Jewish descent with pancreatic cancer, and 3with a close blood relative with ovarian cancer or young-onset breast cancer. On the basis of these criteria, five pancreatic cancer cases with the Ashkenazi BRCA2 founder mutation would be eligible for gene testing, as would nine others with a significant cancer family history (first-degree relative with pancreatic cancer, a close blood relative with ovarian cancer or young-onset breast cancer, or multiple close relatives with breast cancer).
The average age ± SD at diagnosis of patients with pancreatic ductal adenocarcinoma identified as having a germline mutation in a known (BRCA2, ATM, CDKN2A, PALB2, MLH1, BRCA1, and TP53) pancreatic cancer susceptibility gene was 60.8 ± 10.6 years, significantly lower than the average age of the patients without an identifiable susceptibility gene mutation (65.1 ± 10.1 years; P = .03).
A significantly smaller percentage (five of 339 patients [1.5%]; P = .02) of patients with diagnoses other than pancreatic ductal adenocarcinoma had an identifiable deleterious germline mutation. All of the nonpancreatic cancer cases with a deleterious germline mutation had another malignancy (five of 238 patients [2.1%]). These included two of the 47 patients with cholangiocarcinoma (one with an ATM mutation and another with a BRIP1 [FANCJ] mutation), one patient who had a renal cell carcinoma with pancreatic metastasis (ATM mutation), one patient with a duodenal GI stromal tumor (RAD51C mutation), and one patient who underwent pancreaticoduodenectomy for a recurrent bleeding duodenal ulcer after radiation therapy for lymphoma (BRCA2 mutation). The significance of the BRIP1 mutation is not clear. Evidence from large studies indicates that carriers of germline BRIP1 mutations are at moderately increased risk of developing ovarian cancer but not breast cancer.57,58
No deleterious mutations were identified in any of the other disease control cases. Pancreatic cancer cases were more likely than disease controls without a malignant neoplasm to have a deleterious mutation (P = .035). We also compared the prevalence of truncating deleterious variants for each pancreatic cancer susceptibility gene in our pancreatic cancer cases with their prevalence in controls in the ExAC database. Germline truncating mutations involving BRCA2 and ATM were significantly more common in pancreatic cancer cases than in ExAC controls (Table 4).
DISCUSSION
We found a significant yield of deleterious germline mutations in pancreatic cancer susceptibility genes in patients with pancreatic cancer without a pancreatic cancer family history. These patients with apparently sporadic pancreatic cancer also often do not have an extensive family history of pancreatic or other cancers that would trigger consideration for germline gene testing. The prevalence of deleterious germline mutations in patients with apparently sporadic pancreatic cancer is approximately half of that reported to date in patients with familial forms of pancreatic cancer.7,27
Family history remains one of the best predictors of future pancreatic cancer risk.59-61 For common cancers such as breast/ovarian and colorectal cancer (where risk assessment, genetic counseling, and gene testing are well established), current guidelines recommend using family history to risk stratify family members to identify those individuals most likely to benefit from gene testing.62 In contrast, our results for pancreatic cancer highlight the limitations of relying solely on current NCCN family history guidelines to determine which patients are most likely to carry a deleterious pancreatic cancer susceptibility gene. Indeed, most deleterious germline mutations found in patients with pancreatic cancer are in those who do not meet the familial criteria for gene testing.
Although the evidence is mostly anecdotal in the context of pancreatic cancer, not only can affected relatives have the opportunity to undertake cancer screening and prevention strategies, but it also would have an impact on the patient’s treatment. Identifying BRCA mutations in patients with pancreatic cancer would provide the opportunity to have personalized therapy with poly (ADP-ribose) polymerase inhibitors or platinums,63 and Lynch syndrome carriers with pancreatic cancer would have the potential to benefit from immunotherapy.64 It is suspected that patients with ATM germline mutations would benefit from radiotherapy to control local disease (assuming that the gene is biallelically inactivated in their cancer)65 and may also be more sensitive to certain chemotherapeutics.
Although a large gene panel was evaluated for this study, our results indicate that gene testing patients with pancreatic cancer should be limited to established pancreatic cancer susceptibility genes (BRCA2, ATM, PALB2, CDKN2A, and BRCA1, and mismatch-repair genes), with testing for PRSS1 and STK11 mutations for families suspected of having the corresponding clinical syndromes. Although using large gene panels to identify cancer susceptibility genes as part of research studies can be informative, early experience with using large gene panels to perform germline gene testing in clinical settings has shown that such panels pose challenges.66 Because of the greater chance of identifying variants of unknown significance, many patients receive inconclusive results, and the potential for misunderstanding and anxiety is significant.
The potential to benefit the few individuals with actionable gene mutations would seem to justify the effort to routinely offer gene testing to all patients with pancreatic ductal adenocarcinoma to identify such cases.39,67 However, offering widespread genetic testing for patients with pancreatic cancer has significant challenges, not the least being that patients should undergo genetic counseling before and after such testing to provide understanding and reassurance and to avoid harm.68 Unfortunately, there are not enough genetic counselors to provide this service. This shortage of genetic counselors applies to other cancers and, as a result, most patients, even those who undergo gene testing for BRCA mutations, do so without genetic counseling.69 The demand for BRCA gene testing has led to alternative approaches. One approach taken in some centers that are sequencing cancer samples is to provide an opt-out option for patients who do not want to know about germline information. Another approach in situations where there is considerable demand, such as for patients with ovarian cancer, is to provide genetic counseling where clinicians (including nurses trained in genetic counseling) provide the service.70 Because of the potential for adverse events when gene testing is performed without adequate genetic counseling, most experts recommend that appropriate counseling71 and testing should only be undertaken by those with the expertise.72 Genetic counseling for patients with pancreatic cancer poses additional challenges. Because pancreatic cancer often progresses rapidly, patients can greatly benefit from optimal selection of their first-line therapy; thus, ideally, counseling and testing would be incorporated into routine patient care so that it can be performed rapidly. Relatives of patients with pancreatic cancer who are also mutation carriers can also benefit from gene testing. Here, the need for genetic counseling is perhaps more important, because the lifetime estimates of developing pancreatic and other cancers for carriers of deleterious mutations need to be better defined, and the benefits of pancreatic screening are still being established.38,41 Because cancer genetics risk assessment is not a routine component of pancreatic cancer care, it would be valuable to undertake studies to determine the benefits and challenges of incorporating risk assessment and gene testing into routine pancreatic cancer practice. Additional studies are also needed to determine how cancer family history and other risk factor information can help refine pancreatic cancer risk in mutation carriers. Furthermore, the pancreatic cancer risk associated with mutations in some pancreatic cancer susceptibility genes (such as ATM and PALB2) is not well defined, and the effectiveness of screening these mutation carriers is not established.
There are some limitations to our study. First, this was a retrospective study where we relied on self-reported family history of pancreatic and other cancers obtained from the medical record, and patient reporting of their familial cancer history is often incomplete. We were also not able to determine if detecting these mutations resulted in clinical benefit to the patients or their families.
In summary, we found that there is a significant yield of deleterious germline mutations in pancreatic cancer susceptibility genes in unselected patients with apparently sporadic pancreatic cancer. Routine gene testing of patients with newly diagnosed pancreatic cancer and their families may yield significant clinical benefits.
Appendix
Table A1.
Targeted Regions of the 32 Genes*
Table A2.
Variants of Uncertain Significance
Table A3.
Sanger Sequencing Validation PrimersGene
Footnotes
Processed as a Rapid Communication manuscript.
Supported by National Institutes of Health grants CA62924, R01CA176828, U01 CA210170, R01CA15482, and K99CA190889 (N.J.R.), Susan Wojcicki and Dennis Troper, and the Rolfe Pancreatic Cancer Foundation.
See accompanying Editorial on page 3375
AUTHOR CONTRIBUTIONS
Conception and design: Jun Yu, Alison P. Klein, Michael Goggins
Collection and assembly of data: Koji Shindo, Jun Yu, Shahriar Fesharakizadeh, Christy Cho, Anne Macgregor-Das, Abdulrehman Siddiqui, P. Dane Witmer, Koji Tamura, Tae Jun Song, Jose Alejandro Navarro Almario, Aaron Brant, Michael Borges, Madeline Ford, Thomas Barkley, Jin He, Matthew J. Weiss, Christopher L. Wolfgang, Ralph H. Hruban, Alison P. Klein, Michael Goggins
Data analysis and interpretation: Koji Shindo, Jun Yu, Masaya Suenaga, Anne Macgregor-Das, Koji Tamura, Nicholas J. Roberts, Alison P. Klein, Michael Goggins
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Deleterious Germline Mutations in Patients With Apparently Sporadic Pancreatic Adenocarcinoma
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO’s conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Koji Shindo
No relationship to disclose
Jun Yu
No relationship to disclose
Masaya Suenaga
No relationship to disclose
Shahriar Fesharakizadeh
No relationship to disclose
Christy Cho
No relationship to disclose
Anne Macgregor-Das
No relationship to disclose
Abdulrehman Siddiqui
Stock or Other Ownership: 22nd Century Group
P. Dane Witmer
No relationship to disclose
Koji Tamura
No relationship to disclose
Tae Jun Song
No relationship to disclose
Jose Alejandro Navarro Almario
No relationship to disclose
Aaron Brant
No relationship to disclose
Michael Borges
No relationship to disclose
Madeline Ford
No relationship to disclose
Thomas Barkley
No relationship to disclose
Jin He
No relationship to disclose
Matthew J. Weiss
No relationship to disclose
Christopher L. Wolfgang
No relationship to disclose
Nicholas J. Roberts
No relationship to disclose
Ralph H. Hruban
Leadership: miDIAGNOSTICS
Patents, Royalties, Other Intellectual Property: Myriad Genetics
Alison P. Klein
Patents, Royalties, Other Intellectual Property: Myriad Genetics
Michael Goggins
Patents, Royalties, Other Intellectual Property: Myriad Genetics
REFERENCES
- 1.Rahib L, Smith BD, Aizenberg R, et al. : Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 74:2913-2921, 2014 [DOI] [PubMed] [Google Scholar]
- 2.Roberts NJ, Norris AL, Petersen GM, et al. : Whole genome sequencing defines the genetic heterogeneity of familial pancreatic cancer. Cancer Discov 6:166-175, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kastrinos F, Mukherjee B, Tayob N, et al. : Risk of pancreatic cancer in families with Lynch syndrome. JAMA 302:1790-1795, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts NJ, Jiao Y, Yu J, et al. : ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov 2:41-46, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein AP: Genetic susceptibility to pancreatic cancer. Mol Carcinog 51:14-24, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lowenfels AB, Maisonneuve P, DiMagno EP, et al. : Hereditary pancreatitis and the risk of pancreatic cancer. J Natl Cancer Inst 89:442-446, 1997 [DOI] [PubMed] [Google Scholar]
- 7.Zhen DB, Rabe KG, Gallinger S, et al: BRCA1, BRCA2, PALB2, and CDKN2A mutations in familial pancreatic cancer: A PACGENE study. Genet Med 17:569-577, 2015 doi:10.1038/gim.2014.153 [DOI] [PMC free article] [PubMed]
- 8.Jones S, Hruban RH, Kamiyama M, et al. : Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science 324:217, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grant RC, Selander I, Connor AA, et al. : Prevalence of germline mutations in cancer predisposition genes in patients with pancreatic cancer. Gastroenterology 148:556-564, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couch FJ, Johnson MR, Rabe KG, et al. : The prevalence of BRCA2 mutations in familial pancreatic cancer. Cancer Epidemiol Biomarkers Prev 16:342-346, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Hahn SA, Greenhalf B, Ellis I, et al. : BRCA2 germline mutations in familial pancreatic carcinoma. J Natl Cancer Inst 95:214-221, 2003 [DOI] [PubMed] [Google Scholar]
- 12.Murphy KM, Brune KA, Griffin C, et al. : Evaluation of candidate genes MAP2K4, MADH4, ACVR1B, and BRCA2 in familial pancreatic cancer: Deleterious BRCA2 mutations in 17%. Cancer Res 62:3789-3793, 2002 [PubMed] [Google Scholar]
- 13.Hussussian CJ, Struewing JP, Goldstein AM, et al. : Germline p16 mutations in familial melanoma. Nat Genet 8:15-21, 1994 [DOI] [PubMed] [Google Scholar]
- 14.Kamb A, Shattuck-Eidens D, Eeles R, et al. : Analysis of the p16 gene (CDKN2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat Genet 8:23-26, 1994 [DOI] [PubMed] [Google Scholar]
- 15.Goldstein AM, Chan M, Harland M, et al. : Features associated with germline CDKN2A mutations: A GenoMEL study of melanoma-prone families from three continents. J Med Genet 44:99-106, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lynch HT, Fusaro RM, Lynch JF, et al. : Pancreatic cancer and the FAMMM syndrome. Fam Cancer 7:103-112, 2008 [DOI] [PubMed] [Google Scholar]
- 17.de Snoo FA, Bishop DT, Bergman W, et al. : Increased risk of cancer other than melanoma in CDKN2A founder mutation (p16-Leiden)-positive melanoma families. Clin Cancer Res 14:7151-7157, 2008 [DOI] [PubMed] [Google Scholar]
- 18. Slater EP, Langer P, Niemczyk E, et al: PALB2 mutations in European familial pancreatic cancer families. Clin Genet 78:490-494, 2010. [DOI] [PubMed]
- 19.Tischkowitz MD, Sabbaghian N, Hamel N, et al. : Analysis of the gene coding for the BRCA2-interacting protein PALB2 in familial and sporadic pancreatic cancer. Gastroenterology 137:1183-1186, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider R, Slater EP, Sina M, et al. : German national case collection for familial pancreatic cancer (FaPaCa): Ten years experience. Fam Cancer 10:323-330, 2011 [DOI] [PubMed] [Google Scholar]
- 21.Harinck F, Kluijt I, van Mil SE, et al. : Routine testing for PALB2 mutations in familial pancreatic cancer families and breast cancer families with pancreatic cancer is not indicated. Eur J Hum Genet 20:577-579, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson D, Easton DF, Breast Cancer Linkage Consortium : Cancer incidence in BRCA1 mutation carriers. J Natl Cancer Inst 94:1358-1365, 2002 [DOI] [PubMed] [Google Scholar]
- 23.Mocci E, Milne RL, Méndez-Villamil EY, et al. : Risk of pancreatic cancer in breast cancer families from the breast cancer family registry. Cancer Epidemiol Biomarkers Prev 22:803-811, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holter S, Borgida A, Dodd A, et al. : Germline BRCA mutations in a large clinic-based cohort of patients with pancreatic adenocarcinoma. J Clin Oncol 33:3124-3129, 2015 [DOI] [PubMed] [Google Scholar]
- 25.Hu C, Hart SN, Bamlet WR, et al. : Prevalence of pathogenic mutations in cancer predisposition genes among pancreatic cancer patients. Cancer Epidemiol Biomarkers Prev 25:207-211, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucas AL, Frado LE, Hwang C, et al. : BRCA1 and BRCA2 germline mutations are frequently demonstrated in both high-risk pancreatic cancer screening and pancreatic cancer cohorts. Cancer 120:1960-1967, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salo-Mullen EE, O’Reilly EM, Kelsen DP, et al. : Identification of germline genetic mutations in patients with pancreatic cancer. Cancer 121:4382-4388, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goggins M, Schutte M, Lu J, et al. : Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res 56:5360-5364, 1996 [PubMed] [Google Scholar]
- 29.Couch FJ, Farid LM, DeShano ML, et al. : BRCA2 germline mutations in male breast cancer cases and breast cancer families. Nat Genet 13:123-125, 1996 [DOI] [PubMed] [Google Scholar]
- 30.Neuhausen S, Gilewski T, Norton L, et al. : Recurrent BRCA2 6174delT mutations in Ashkenazi Jewish women affected by breast cancer. Nat Genet 13:126-128, 1996 [DOI] [PubMed] [Google Scholar]
- 31.Oddoux C, Struewing JP, Clayton CM, et al. : The carrier frequency of the BRCA2 6174delT mutation among Ashkenazi Jewish individuals is approximately 1%. Nat Genet 14:188-190, 1996 [DOI] [PubMed] [Google Scholar]
- 32.Figer A, Irmin L, Geva R, et al. : The rate of the 6174delT founder Jewish mutation in BRCA2 in patients with non-colonic gastrointestinal tract tumours in Israel. Br J Cancer 84:478-481, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ozçelik H, Schmocker B, Di Nicola N, et al. : Germline BRCA2 6174delT mutations in Ashkenazi Jewish pancreatic cancer patients. Nat Genet 16:17-18, 1997 [DOI] [PubMed] [Google Scholar]
- 34.van Asperen CJ, Brohet RM, Meijers-Heijboer EJ, et al. : Cancer risks in BRCA2 families: Estimates for sites other than breast and ovary. J Med Genet 42:711-719, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Struewing JP, Abeliovich D, Peretz T, et al. : The carrier frequency of the BRCA1 185delAG mutation is approximately 1 percent in Ashkenazi Jewish individuals. Nat Genet 11:198-200, 1995 [DOI] [PubMed] [Google Scholar]
- 36.Ferrone CR, Levine DA, Tang LH, et al. : BRCA germline mutations in Jewish patients with pancreatic adenocarcinoma. J Clin Oncol 27:433-438, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knudsen ES, O’Reilly EM, Brody JR, et al. : Genetic diversity of pancreatic ductal adenocarcinoma and opportunities for precision medicine. Gastroenterology 150:48-63, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vasen H, Ibrahim I, Ponce CG, et al. : Benefit of surveillance for pancreatic cancer in high-risk individuals: Outcome of long-term prospective follow-up studies from three European expert centers. J Clin Oncol 34:2010-2019, 2016 [DOI] [PubMed] [Google Scholar]
- 39.Manchanda R, Loggenberg K, Sanderson S, et al. : Population testing for cancer predisposing BRCA1/BRCA2 mutations in the Ashkenazi-Jewish community: A randomized controlled trial. J Natl Cancer Inst 107:379, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Metcalfe K, Lynch HT, Foulkes WD, et al. : Effect of oophorectomy on survival after breast cancer in BRCA1 and BRCA2 mutation carriers. JAMA Oncol 1:306-313, 2015 [DOI] [PubMed] [Google Scholar]
- 41. Canto MI, Hruban RH, Fishman EK, et al: Frequent detection of pancreatic lesions in asymptomatic high-risk individuals. Gastroenterology 142:796-804; quiz e14-e15, 2012. [DOI] [PMC free article] [PubMed]
- 42.Stoffel EM, Mangu PB, Gruber SB, et al. : Hereditary colorectal cancer syndromes: American Society of Clinical Oncology Clinical Practice Guideline endorsement of the familial risk-colorectal cancer: European Society for Medical Oncology Clinical Practice Guidelines. J Clin Oncol 33:209-217, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L, Brune KA, Visvanathan K, et al. : Elevated cancer mortality in the relatives of patients with pancreatic cancer. Cancer Epidemiol Biomarkers Prev 18:2829-2834, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yu J, Sadakari Y, Shindo K, et al: Digital next-generation sequencing identifies low-abundance mutations in pancreatic juice samples collected from the duodenum of patients with pancreatic cancer and intraductal papillary mucinous neoplasms. Gut doi: 10.1136/gutjnl-2015-311166 [epub ahead of print on July 18, 2016] [DOI] [PMC free article] [PubMed]
- 45.Hanks S, Coleman K, Reid S, et al. : Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet 36:1159-1161, 2004 [DOI] [PubMed] [Google Scholar]
- 46.de Voer RM, Geurts van Kessel A, Weren RD, et al. : Germline mutations in the spindle assembly checkpoint genes BUB1 and BUB3 are risk factors for colorectal cancer. Gastroenterology 145:544-547, 2013 [DOI] [PubMed] [Google Scholar]
- 47.Broderick P, Dobbins SE, Chubb D, et al. : Validation of recently proposed colorectal cancer susceptibility gene variants in an analysis of families and patients—A systematic review. Gastroenterology 152:75-77.e4, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kalitsis P, Fowler KJ, Griffiths B, et al. : Increased chromosome instability but not cancer predisposition in haploinsufficient Bub3 mice. Genes Chromosomes Cancer 44:29-36, 2005 [DOI] [PubMed] [Google Scholar]
- 49.Song H, Dicks E, Ramus SJ, et al. : Contribution of germline mutations in the RAD51B, RAD51C, and RAD51D genes to ovarian cancer in the population. J Clin Oncol 33:2901-2907, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Norquist BM, Harrell MI, Brady MF, et al. : Inherited mutations in women with ovarian carcinoma. JAMA Oncol 2:482-490, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orr N, Lemnrau A, Cooke R, et al. : Genome-wide association study identifies a common variant in RAD51B associated with male breast cancer risk. Nat Genet 44:1182-1184, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roviello F, Corso G, Pedrazzani C, et al. : Hereditary diffuse gastric cancer and E-cadherin: Description of the first germline mutation in an Italian family. Eur J Surg Oncol 33:448-451, 2007 [DOI] [PubMed] [Google Scholar]
- 53.Martin ST, Matsubayashi H, Rogers CD, et al. : Increased prevalence of the BRCA2 polymorphic stop codon K3326X among individuals with familial pancreatic cancer. Oncogene 24:3652-3656, 2005 [DOI] [PubMed] [Google Scholar]
- 54.Thompson ER, Gorringe KL, Rowley SM, et al. : Reevaluation of the BRCA2 truncating allele c.9976A > T (p.Lys3326Ter) in a familial breast cancer context. Sci Rep 5:14800, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meeks HD, Song H, Michailidou K, et al. : BRCA2 polymorphic stop codon K3326X and the risk of breast, prostate, and ovarian cancers. J Natl Cancer Inst 108:djv315, 2015. doi:10.1093/jnci/djv315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Provenzale D, Gupta S, Ahnen DJ, et al. : Genetic/familial high-risk assessment: Colorectal version 1.2016, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 14:1010-1030, 2016 [DOI] [PubMed] [Google Scholar]
- 57.Ramus SJ, Song H, Dicks E, et al. : Germline mutations in the BRIP1, BARD1, PALB2, and NBN genes in women with ovarian cancer. J Natl Cancer Inst 107:djv214, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Easton DF, Lesueur F, Decker B, et al. : No evidence that protein truncating variants in BRIP1 are associated with breast cancer risk: Implications for gene panel testing. J Med Genet 53:298-309, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klein AP: Identifying people at a high risk of developing pancreatic cancer. Nat Rev Cancer 13:66-74, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klein AP, Brune KA, Petersen GM, et al. : Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res 64:2634-2638, 2004 [DOI] [PubMed] [Google Scholar]
- 61.Brune KA, Lau B, Palmisano E, et al. : Importance of age of onset in pancreatic cancer kindreds. J Natl Cancer Inst 102:119-126, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nelson HD, Fu R, Goddard K, et al. U.S. Preventive Services Task Force Evidence Syntheses, formerly Systematic Evidence Reviews, Risk Assessment, Genetic Counseling, and Genetic Testing for BRCA-Related Cancer: Systematic Review to Update the U.S. Preventive Services Task Force Recommendation. Rockville, MD, Agency for Healthcare Research and Quality, 2013 [PubMed] [Google Scholar]
- 63.Kaufman B, Shapira-Frommer R, Schmutzler RK, et al. : Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol 33:244-250, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Le DT, Uram JN, Wang H, et al. : PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 372:2509-2520, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ayars M, Eshleman J, Goggins M: Susceptibility of ATM-deficient pancreatic cancer cells to radiation. Cell Cycle 16:991-998, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tung N, Domchek SM, Stadler Z, et al. : Counselling framework for moderate-penetrance cancer-susceptibility mutations. Nat Rev Clin Oncol 13:581-588, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Foulkes WD, Knoppers BM, Turnbull C: Population genetic testing for cancer susceptibility: Founder mutations to genomes. Nat Rev Clin Oncol 13:41-54, 2016 [DOI] [PubMed] [Google Scholar]
- 68.Wolf SM, Branum R, Koenig BA, et al. : Returning a research participant’s genomic results to relatives: Analysis and recommendations. J Law Med Ethics 43:440-463, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Armstrong J, Toscano M, Kotchko N, et al. : Utilization and outcomes of BRCA genetic testing and counseling in a national commercially insured population: The ABOUT study. JAMA Oncol 1:1251-1260, 2015 [DOI] [PubMed] [Google Scholar]
- 70.Percival N, George A, Gyertson J, et al. : The integration of BRCA testing into oncology clinics. Br J Nurs 25:690-694, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kinney AY, Butler KM, Schwartz MD, et al. : Expanding access to BRCA1/2 genetic counseling with telephone delivery: A cluster randomized trial. J Natl Cancer Inst 106:dju328, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bonadies DC, Brierley KL, Barnett RE, et al. : Adverse events in cancer genetic testing: The third case series. Cancer J 20:246-253, 2014 [DOI] [PubMed] [Google Scholar]