Abstract
As recent advances in human genetics have begun to more rapidly identify the individual genes contributing to risk of psychiatric disease, the spotlight now turns to understanding how disruption of these genes alters the brain, and thus behavior. Compared to other tissues, cellular complexity in the brain provides both a substantial challenge and a significant opportunity for systems biology approaches. Current methods are maturing that will allow for finally defining the ‘parts list’ for the functioning mouse and human brains, enabling new approaches to defining how the system goes awry in disorders of the CNS. However, the availability of tissue is certainly a challenge for systems biology of neuroscience, compared to systems biology of other tissues, where biopsy is feasible. This challenge is particularly notable for disorders caused by extremely rare genetic variants. Thus computational and systems biology approaches, as well as precise experimental models by way of genome editing, will play key roles in defining mechanisms for disorders, and their individual symptoms, across varied genetic etiologies. Here, we highlight recent progress in neurogenetics, postmortem genomics, cell-type specific profiling, and precision modeling toward defining mechanisms in disease.
Graphical Abstract
Introduction
The central nervous system(CNS) is the most elaborately specialized of the body’s systems. It is composed of hundreds of morphologically and molecularly definable subtypes of neurons and glia, all organized into complex yet stereotyped collections of circuits, of which the final fundamental role is the production of appropriate behavior. Disorders of the CNS are equally complex, each manifesting as a collection of diagnostically definitive disruptions in behavior. For example, a neurological disorder associated with the aging brain, Parkinson’s disease (PD), results in consistent impairment in initiation of voluntary activity, while Autism Spectrum Disorder (ASD) is characterized by a constellation of symptoms apparent from early childhood that include restricted interests, repetitive behaviors, and deficits in social interactions. For late-onset diseases, decades of postmortem pathological studies have mapped specific behavioral deficits to the degeneration of particular circuits or cell types in the brain, such as loss of dopaminergic inputs to striatal motivation circuits in PD, or atrophy of medial temporal lobe structures for episodic memory deficits in Alzheimer’s disease (AD). From these specific cases, along with studies of other lesions in patients or experimental disruption in animals, the general model has emerged that specific behavioral manifestations of a given disease are mappable to the specific cells or circuits impacted in the disorder. However, for most psychiatric disorders, such as Schizophrenia (SCZ) or ASD, there is no consensus on where the corresponding circuit disruptions are found, though there is a consistent, strong genetic component to their risk. For example, ASD has an estimated heritability of >50%[1,2]. Yet it is clear that the genetic contribution for many CNS disorders is mediated by hundreds of loci, only some of which have been identified. Thus, there is a need to define the mechanisms for CNS disorders at two levels: Which genetic alterations lead to each disorder? And, what are the circuit disruptions that result in unusual behavior or disease? In addition, there is also a need to connect these two levels of inquiry to explain how genetic disruptions alter brain circuits, and thus behavior. Therefore, there is now a conspicuous opportunity for emerging systems-level approaches in both neuroscience and genetics to address these needs.
Genetics of CNS Disorders: Important Advances, New Opportunities
Prior to the modern genomic era, discovery of rare and private (family specific), coding genetic variants had steadily identified a few genes for CNS disorders based on either linkage in large pedigrees, or with genetic association studies for some clearly definable neurological syndromes and developmental disorders(DD)(e.g.[3],[4],[5]). Array based methods since the mid-2000’s allowed for genotyping of millions of polymorphisms across thousands of patients and controls for large scale association studies, but were largely limited to analysis of variation in the genome that was found commonly in the population. This approach rapidly identified a common variant of large effect associated with a retinal neurodegeneration, Macular Degeneration[6], but initially struggled to detect common alleles of smaller effect sizes collectively contributing risk for other common disorders. Given small effect sizes, large samples were needed in order for loci for SCZ[7], Major Depressive Disorder (MDD)[8] and AD[9] (Figure 1), and polygenic risk estimates for several other psychiatric diseases to be identified[10,11]. Since 2012, reduced cost of sequencing, combined with capture reagents, allowed for genome-wide analysis of rare protein-coding variation (exome sequencing), resulting in substantial new gene associations for DDs including ASD, intellectual disability (ID), and childhood epilepsy([12–14] and references therein). Though each associated mutation was incredibly rare or private, a gestalt analysis of mutation rates in these genes in large control databases[15] indicates strong selective pressure on these genes in human populations. Approaches taking advantage of prior information on constraint, and potentially information on brain gene expression[16], are increasing our ability to identify additional causative mutations, even when only observed in a small number of, or even single cases[17,18].
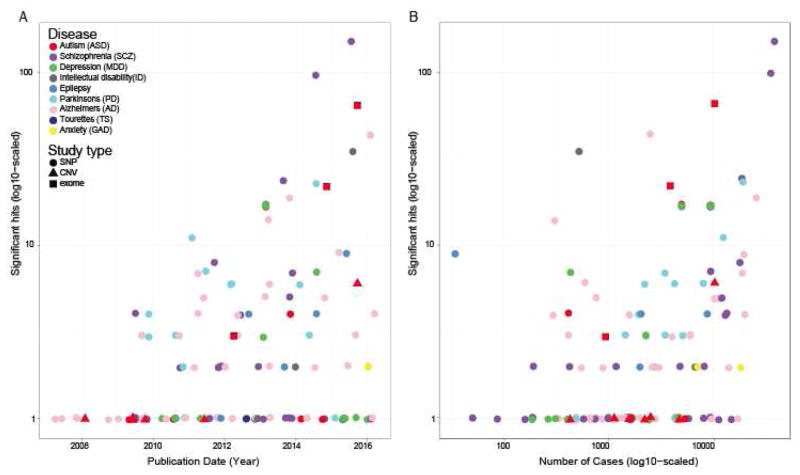
Figure 1. Burgeoning sample numbers have driven identification for loci contributing to several diseases.
A) Number of loci or genes implicated as a function of year. Across multiple neuropsychiatric diseases, common single nucleotide polymorphism (SNP) studies show substantial discovery, particularly for SCZ, but with notable recent gains in AD, PD. Several other adult diseases are beginning to detect some loci. Success in ASD has primarily been in identification of rare de novo variants causing disease (via copy number variation (CNV) analysis and exome sequencing). B) Number of loci discovered is largely a function of accumulating samples sizes. Common SNP data curated from https://www.ebi.ac.uk/gwas/ on 1/25/17, using each disease name as a search term. ASD rare variants manually curated.
These rare variant findings have led to a burgeoning enthusiasm for clinical application of exome sequencing. Indeed, the diagnostic yield in DDs can be seen as one of the early successes of precision medicine. An observational study of 2000 consecutive patients analyzed with clinical whole-exome sequencing reported a diagnosis rate of 25.2% across genetic disorders[19]. As the number of ID-, ASD-, and epilepsy-associated mutations grows, prospective studies are showing that a ‘sequence first’ approach to diagnosis is providing a substantial yield[12,20,21] and thus may become standard. While there is currently an argument to be made that these rare loss-of-function forms of ASD and ID ought to be considered as new, molecularly defined, rare monogenic syndromes(e.g. [22]), perhaps with separate etiology from ‘idiopathic’ ASD, there is also clearly an opportunity to leverage this collection of genes to understand why rare mutations in distinct genes still result in a shared CNS disorder. Likewise, understanding the small effects of the large number of common variant contributions currently discovered for SCZ and MDD will also require leveraging additional information from genomics and neuroscience.
Enter systems biology
Systems biology to understand common variant associations and rare variant mechanisms
The recent discoveries of both common loci and rare genic variants contributing to disease present some distinct and some overlapping challenges. For a common, noncoding genetic association, the first challenge is in defining the relevant gene(s). Noncoding variants are presumed to be acting via regulation of gene expression. However, because of correlation in polymorphism in adjacent regions of the genome (linkage disequilibrium), a variant’s associated region typically includes many noncoding variants, any one(s) of which could alter expression of any gene(s) within hundreds of kilobase pairs. Four parallel systems biology approaches leveraging either genomic annotations, or massively parallel reporter assays(MPRAs, [23]) are currently being applied to this problem(Figure 2, see also review in [24]). First, large descriptive datasets of ‘epigenetic’ marks such as DNA and histone modifications when combined with prediction algorithms (reviewed in [25]) can provide insight into which polymorphisms might be in regulatory DNA sequences. Second, association studies testing for polymorphisms associated with changes in gene expression (expression Quantitative Trait Loci, or eQTLs), have begun to link putative regulatory polymorphisms with corresponding genes[26–28]. Third, regulatory elements such as enhancers have been shown to physically interact with the promoters of their target genes; sequencing based measures of chromatin conformation such as Hi-C have begun to define regulator-gene pairs[29–31], which in turn has enabled further structural predictions from other epigenetic data[32]. These methods thus begin to shape a choreography for the key steps involved in defining the relevant gene for each associated polymorphism, as has been demonstrated for some specific loci (e.g. [33]). However, in each case, these three analytical approaches do not clearly demonstrate that a given variant does have functional consequences. For efficient functional analysis, in a fourth approach, several teams have recently demonstrated that using MPRAs, which couple the available economies of array based oligonucleotide synthesis with high throughput sequencing based readouts, can efficiently query the functional consequences of hundreds or thousands of variants in parallel(Figure 2A,B, reviewed in [34]). However, there is a unique challenge to applying any of these four approaches for diseases of the nervous system: defining the appropriate cell type and time point to assess these genomic features. Gene regulation is a cell type specific phenomenon, and the activity of particular regulatory elements (as assessed by analytical approaches or MPRA) will depend on the cells in which they are measured. Given the diversity of cell types in the nervous system, as well as the possibility of transient regulatory phenomena during development, or in very rare cell populations, there is high potential for false-negative results leveraging only existing datasets. Thus, there is a clear need to expand the genomic annotative approaches to more time windows, brain regions, and cell types, and adapt the MPRAs to cell-type specific measures (Figure 2C). Also, most of the descriptive studies thus far have been relatively cortico-centric[26,35–38]. While the human cortical expansion is fairly remarkable, the pathologic and therapeutic importance of deep brain structures to psychiatric (e.g. the target regions of monoaminergic drugs) and neurological (e.g. the substantia niagra in PD) disorders is well established; thus there is a clear need to deepen the survey outside of the neocortex. It is also likely, and eventually essential, that new single cell analysis approaches may be adapted to these analyses (more below).
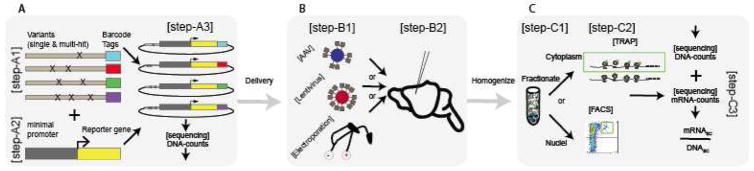
Figure 2. MPRAs in the CNS, current and future directions.
A) Library preparation for MPRA as developed over last few years. [34] A1) Massively parallel oligonucleotide synthesis is used to generate genomic element sequences with specified genomic variants and unique barcode tags. A2) A minimal promoter and reporter gene are inserted between candidate enhancer variants and barcode tags. A3) These are combined in a plasmid such that the barcode for each enhancer is now part of the 3’UTR of the reporter. Traditionally, these are delivered en masse into cell lines where the ratio of RNA barcode counts to DNA barcode counts can provide a measure of activity for each element and variant. B) Recent methods are adapting MPRAs to the mouse brain. The library constructed in A) could be delivered into mouse brain(B2) through packaging into 1) Adeno-associated virus (AAV)[90] or 2) Lentivirus[91] or 3) electroporation directly[92]. C) Given the substantial cellular heterogeneity of the mouse brain, enhancer activity (and potentially variant activity) is likely cell type specific. Provided RNA is harvested using cell type specific methods such as Translating Ribosome Affinity Purification[54,55] or flow sorting of tagged nuclei[87,88] (C2), MPRA should be amenable to cell type specific assessment. Finally, together with Plasmid DNA tag counts, as for in vitro assays, the ratio of mRNA barcode counts to DNA barcode counts can be used to estimate the expression for each element(C3).
In contrast to common variants, for protein-coding variants, the first step in defining the relevant gene is already complete. In addition, this class of variation is eminently modelable in model organisms or induced Pluripotent Stem Cell (iPSC)-derived cells, especially with the advent of efficient genome modification with targeted nucleases[39,40]. From either class of variation, once a gene is defined, there remain two current challenges. The first is to develop experimental approaches that can efficiently scale to systematic functional evaluation of many protein coding variants in parallel, much MPRAs have for non-coding variants, a thorough discussion of which is beyond the scope of this review. The second challenge for both the rare, and eventually the common, variants is to move from a genetic understanding of the disorder to a molecular and cellular one.
Need for a cellular level understanding of psychiatric diseases is underscored by the development of one of the most dramatically effective treatments for a disorder in neuroscience: the reversal of PD symptoms by either supplementation of dopaminergic signaling[41] or deep brain stimulation[42], which predated understanding of the underlying genetics. Rather, knowledge from pathology of the circuit deficits suggested routes for rescuing the behavioral manifestations of the disease. A variety of genetic lesions result in the propensity for nigral neurons to degenerate, creating a common phenotype from diverse genetic causes[43]. The PD case suggests a hypothesis for other diseases: that the specific behavioral disruptions of each are caused by ‘loss-of-function’ circuit mutations. If so, the myriad genetic mutations being discovered in DD, or the common variants highlighted in SCZ, must have shared consequences at the level of cells or circuits. A simple example might be a common loss, or a failure to develop, a key cell type within the circuit. The challenge is to define these disruptions. Systems biology approaches are beginning to be implemented in attempt to both indirectly and directly infer these changes.
From Genes to Cells to Treatments
Leveraging human genetics to define cellular mechanisms
A variety of informatics approaches have been applied to inferring commonalities across the gene sets emerging from psychiatric genetics studies (reviewed recently in [44] for ASD). A typical, and perhaps unsurprising result, is the discovery of disproportionate number of synaptic related genes amongst many of these lists. Gene expression varies widely by tissue and cell type. The ‘selective expression’ hypothesis posits that genes expressed in the disease-afflicted tissue or cell type are more likely to contribute to diseases of that tissue or cell type. While certainly not all disease genes follow this trend, a tool leveraging gene lists to define cell types or tissues can identify a specific tissue for half of traits, even using very crude single nucleotide polymorphism (SNP) to gene mapping[45] (Figure 3A,B). Thus, as synaptic genes are disproportionately brain expressed, it is not unexpected that they will be frequent contributors to genetic variation in behavioral traits. A more focused analysis of gene expression however, could lead to insights as to which cell types or circuits within the brain are mediating the genetic effects of the disorder. For example, recent work has identified low frequency coding variants in Trem2[46,47] and common variants in CD33[9,48,49], as associated with AD. Both of these genes are highly expressed in microglia – a key phagocytic cell type in the brain, with recently discovered roles in supporting synaptic pruning. Thus, the expression of these risk genes has now spotlighted an underappreciated aspect of AD, and suggested a new cell type to target for treatment[50,51]. Systems biology approaches are coming online now to systematically address this question of which cell type(s) in the brain might be disproportionately expressing risk genes as a way of indirectly inferring the circuits disrupted in the disorder[52,53]. However, there are limitations to this approach, as inference requires a knowledge of transcriptomic profiles from each cell type. Pioneering studies in cell type specific profiling had identified profiles for several cell types, but depended on availability of genetically labeled cell types in mice[54–57], clever retrograde labeling strategies[58], or cell surface markers[59,60] coverage was fairly sparse, and many cell types remain undefined. Thus, these analyses were inherently limited. However, new efforts designed to more comprehensively and unbiasedly profile all cells in adult and developing brain, discussed below, could substantially improve the resolution of these tools and provide key benchmark data for these indirect inferential approaches.
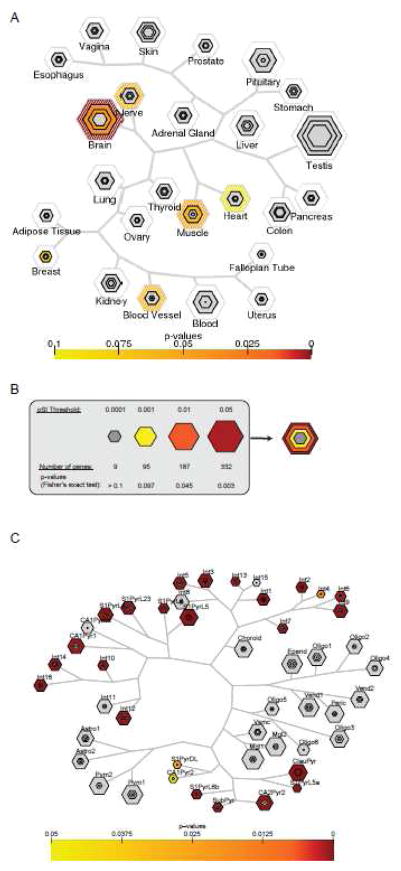
Figure 3. Leveraging expression data can provide both insight into genetics causes and interpretation of transcriptomic consequences.
A) Loci that were implicated in a recent GWA study of body mass index[93] contain genes that are disproportionately expressed in the brain, as shown by Tissue Specific Enrichment Analysis[45]. This suggests that in our current, calorie abundant environment, obesity is mediated in large part by behavior(consumption) rather than by the function of adipose tissues. Some suggestive signal is also seen in muscle. B) Key for a single hexagon plot: For each tissue, the size of the hexagon in is scaled to the number of cell type specific and enriched genes at different cutoffs based on a permuted measure of specificity (pSI). Each hexagon is then color coded by significance of overlap with the GWAS gene list, using Fisher’s exact test p values as shown. As a highly specific list (pSI<.0001) is always included is a less specific list (pSI<.05), all hexagons for the same tissue can be stacked. All tissues tested in a dataset can then be displayed in a single plot as in A, organized by overall similarity of gene expression across tissues. C) Cell Type Specific Enrichment analysis[53] leveraging single cell expression data from somatosensory cortex and hippocampal CA1 region[80] suggests that the decrease in neuronal transcripts previously detected in human postmortem cortical transcriptomics in an individual with autism[64] might represent a loss or dysregulation of a subset of neuronal cell types. Specific interneurons (Int1 & 3) show enrichment at the relatively stringent cutoff of pSI<0.001 (third from outermost hexagon), perhaps suggesting a complete loss of these cell populations. In certain cell subtypes (Int4–7, Int9–10, Int12–13, S1PyrL23, S1PyrL5a, S1PyrL6b, S1PyrDL, SubPyr, CA1Pyr1, CA1Pyr2, CA2Pyr2), the enrichment is seen only in the least stringently enriched transcript lists (outermost hexagon), suggesting a dysregulation of these genes rather than a complete loss of these cell types. For a given cell type, presence of only 2 or 3 hexagons (e.g. Int1) indicates that no transcripts were identified at the most stringent specificity thresholds.
Genomic approaches to the postmortem human brain
Of course, to the extent patient brains are available, more direct approaches to observe cellular deficits are feasible. For end of life diseases, like PD, AD, and amyotrophic lateral sclerosis (ALS), brains are readily available, and decades of pathological studies have created gold standard definitions for each disease in terms of regional and circuit atrophy and specific classes of pathological signatures, and at least for AD, a corresponding inflammatory signature is apparent in postmortem transcriptomic[60,61] and proteomic data. However psychiatric diseases with earlier onset, and that are not directly lethal (ASD, SCZ), provide substantially less opportunity for post mortem pathology to define cellular deficits. This has been gradually changing and thanks to the dedicated efforts of several groups there are beginning to be reasonably sized collections of postmortem tissue from these diseases (e.g. [35,36,62–64]). From these, the community is currently engaged in a discovery phase of ‘transcriptomic’ pathology wherein portions of these brains are being analyzed using genomic approaches to define a molecular pathology indicative of the disorder. These studies are challenged by the variable integrity of tissues from donors, the likely pleiotropy of causes included in each collection, and the difficulty in determining causal changes from those that are a consequence of the course of disease pathology, or that are responses to treatment regimens. Nonetheless, with inclusion of covariates to try to account for sample variation, for at least ASD and SCZ some themes have started to emerge across studies. For ASD studies multiple groups have reported a consistent pattern of gene expression changes. A variety of groups have taken a systems approach to inferring changes in cellularity from this data in ASD (reviewed in [44]) and SCZ (reviewed in [65]). For example, in ASD there does seem to be a consistent cellular signature with loss of some neuronal transcripts from at least the messenger RNA (mRNA) data (Figure 3B,C).
Additional conclusions from across these studies is that the effects are fairly subtle, and direct case-control designs are sometimes less informative than network or module based approaches (see [66] for a thorough review). It is also notable that differences are detectable in all aspects of the transcriptomic signature from coding mRNAs, to large intervening noncoding RNAs (lincRNAs) and microRNAs (miRNAs), to splicing, and epigenetic marks[63,64,67,68], and it is unclear if any one class of this transcriptomic variation is more important than another. One possible explanation is that all of these changes are correlated and reflecting the same underlying biology. One example would be changes in proportion of cellularity: if a particular cell type was reduced or absent in disease then all mRNA, miRNA, lincRNA, splicing, or epigenetic marks associated with that cell type should change in parallel. It is noteworthy though, that neuronal and glial-contained transcripts may have distinct responses to the tissue trauma associated with mortality and dissection, and there appear to be previously unnoticed RNA quality covariates that were confounding recent analyses[69]. In addition, only limited brain regions have been sampled (primarily cortex) and thus if the primary cellular deficits are elsewhere they would not be detected in the current studies.
It is worth noting that transcriptomic measures do have limitations compared to other measures, such as imaging or physiology. As most genomics methods are inherently destructive, they lose information on connectivity and physiology. Thus, if the primary deficit is a miswiring phenomenon, then transcriptomics may not detect it. Likewise, deficits in the electrophysiological performance of cells are only very indirectly measured by their consequences on the transcriptome. In addition, genomics are also only indirectly related to post-translational states such as phosphorylation of proteins or activity of signaling pathways(though advances in proteomics might allow for their assessment in a high-throughput way). Nonetheless, because it is generally the much more scalable than imaging or physiology, transcriptomics could be a powerful tool to dissect CNS disorders in a systematic fashion, provided the alterations can be properly interpreted.
‘Cellomics’ - a new cellular-molecular pathology on the horizon
New advances including microfluidics(e.g. [70]), single tube single cell amplification strategies[71], and droplet based library preparative approaches[72] are now permitting efficient transcriptome analysis at the level of single cells. Both the indirect or direct approaches to trying to define the cellular pathology of CNS disorders will be accelerated by these advances. Most immediately, there is clear enthusiasm from the NIH BRAIN Initiative[73] and other allied sources[74] to definitively generate a complete cellular ontology defining the ‘parts list’ for the human and mouse brains. Consistent with the pioneering targeted work thus far, sequencing thousands of single cells from a few regions of adult brain seems to indicate that transcriptomics can be used to readily cluster mature cells into discrete, separable classes, including previously undefined subclasses of neuronal types[72,75–80]. Expansion of this beginning index to a complete list of cell types, and the transcriptomic profiles that define them, will allow the ‘indirect’ analysis methods developed so far(e.g.[53,81,82]) to be rapidly applied to entire brain, overcoming a substantial limitation in the current analyses.
Eventually, however, as these single cell methods become more robust and cost effective, it will be feasible to directly assess the molecular pathology of postmortem samples at the single cell level. This final ‘cellomic’ pathology will allow for direct quantification of cell numbers of different classes of cells, and allow for analyses that can distinguish between a loss of a cell type from dysregulation within a particular cell type. Cell type-specific transcriptional derangement is typically difficult or impossible to identify from bulk cell transcriptomics. While still cost prohibitive on a large scale, key technical hurdles towards ‘cellomic’ pathology are gradually being overcome. For example, the feasibility of sequencing nuclear RNA for clustering (as intact nuclei are more easily obtained than intact cells) has been established and applied to specific cell types[83–88]. Profiling of methylation status might confer an alternative measure of expression to RNAseq which can provide similar information regarding cell class[63], while potentially overcoming some of the challenges of oversampling abundant transcripts and undersampling rare transcripts in RNAseq. However, it is not clear whether the method also captures dynamic changes as well as traditional RNAseq. Regardless, scalability of droplet based methods are increasing at such a rate it is not unimaginable they may eventually permit cellular level coverage of the entire brain in pathological samples. Previously, it was estimated that a comprehensive cellular survey would cost $20,000 for a brain by 2027[89]. However, with a more rapid decrease in library prep costs, comprehensive cell based molecular pathology might reach this level within a few years.
Clinical heterogeneity and precision modeling
Work in ASD and DD in particular has highlighted the remarkable genetic heterogeneity contributing to these diseases. While roughly 10–20% of cases may be attributable to dominant, loss-of-function mutations, each individual mutation occurs only in a very small number of cases, further limiting access to afflicted tissue. It is also unclear if all mutations will converge on a single downstream cellular pathology, or if there are multiple cellular routes to the same disease, analogous to the clearly multiple genetic routes. Thus, there will be a sustained need for modeling a large variety of precise mutations to define shared and distinct consequences. Advances in genome editing allow for the routine modeling of precise mutations in either rodent or human cell line models, and both have proven successful in recent studies to define possible cellular pathologies and treatments. Current challenges include coping with the genetic heterogeneity outside of the lesioned locus, in both systems, to identify robust consequences of a given genetic deficit. However, the efficiencies associated with single cell analysis highlighted above will also benefit phenotyping in model systems. Overall, defining efficacious treatments will rely on understanding the commonalities and distinctions of disease mechanisms across precise subtypes of disorders.
Highlights.
Large subject numbers are yielding substantial numbers of new loci and gene associations for CNS disorders.
Systematic approaches are maturing for using chromatin annotations and massively parallel functional assays for defining relevant polymorphisms and genes within loci.
Transcriptomic data in both healthy and diseased brains are being leveraged to identify corresponding cellular deficits in CNS disorders.
Advances in single cell profiling methods are poised to contribute substantially to our understanding of disease.
Acknowledgments
We would like to thank D. Conrad, D. King, B. Cohen, and B. Mulvey for thoughtful discussions of the manuscript and assistance with figures.
Funding
We would like to thank the Brain and Behavior Research Foundation (JDD), the Simons Foundation, and the NIH (5R01HG008687, 1R01MH107515, 1R21DA041883, 5R21DA038458, 5U01MH109133 and 9R01MH100027) for support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bourgeron T. Current knowledge on the genetics of autism and propositions for future research. C R Biol. 2016;339:300–307. doi: 10.1016/j.crvi.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, Mahajan M, Manaa D, Pawitan Y, Reichert J, et al. Most genetic risk for autism resides with common variation. Nat Genet. 2014;46:881–885. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacDonald ME, Ambrose CM, Duyao MP, Myers RH, Lin C, Srinidhi L, Barnes G, Taylor SA, James M, Groot N, et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 4.Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CAM, Devon RS, Clair DMS, Muir WJ, Blackwood DHR, et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 5.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 6.Klein RJ, Zeiss C, Chew EY, Tsai J-Y, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, et al. Complement Factor H Polymorphism in Age-Related Macular Degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •7.Ripke S, Neale BM, Corvin A, Walters JT, Farh K-H, Holmans PA, Lee P, Bulik-Sullivan B, Collier DA, Huang H, et al. Biological Insights From 108 Schizophrenia-Associated Genetic Loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. A benchmark work implicating 108 common variant loci in the genetic etiology of Schizophrenia. An exampler of how large collaborative efforts have led to successes in common variant analyses for neurogenetic disorders. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hyde CL, Nagle MW, Tian C, Chen X, Paciga SA, Wendland JR, Tung JY, Hinds DA, Perlis RH, Winslow AR. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet. 2016;48:1031–1036. doi: 10.1038/ng.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambert J-C, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, Jun G, DeStefano AL, Bis JC, Beecham GW, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Consortium C-DG of the PG. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. The Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klei L, Sanders SJ, Murtha MT, Hus V, Lowe JK, Willsey AJ, Moreno-De-Luca D, Yu TW, Fombonne E, Geschwind D, et al. Common genetic variants, acting additively, are a major source of risk for autism. Mol Autism. 2012;3:9. doi: 10.1186/2040-2392-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anazi S, Maddirevula S, Faqeih E, Alsedairy H, Alzahrani F, Shamseldin HE, Patel N, Hashem M, Ibrahim N, Abdulwahab F, et al. Clinical genomics expands the morbid genome of intellectual disability and offers a high diagnostic yield [Internet] Mol Psychiatry. 2016 doi: 10.1038/mp.2016.113. [DOI] [PubMed] [Google Scholar]
- •13.Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, Murtha MT, Bal VH, Bishop SL, Dong S, et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. A benchmark work implicating 71 rare variant loci in the genetic etiology of Autism. A parallel examplar to Ripke et al for how large collaborative efforts have led to successes in rare variant analyses for childhood neurogenetic disorders. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •15.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. This study provides a key resource defining the spectrum of rare variation across the general human population using the largest single collection of exomes to date. Comparing variants discovered in individual neurogenetic studies to this resource allow for better inference in the cases of rare disease causing mutations. See also Samocha 2014 and Wilfert 2016 for similar approaches. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang C, Shen Y. A Cell Type-Specific Expression Signature Predicts Haploinsufficient Autism-Susceptibility Genes. Hum Mutat. 2016 doi: 10.1002/humu.23147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, Kosmicki JA, Rehnström K, Mallick S, Kirby A, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilfert AB, Chao KR, Kaushal M, Jain S, Zöllner S, Adams DR, Conrad DF. Genome-wide significance testing of variation from single case exomes. Nat Genet. 2016;48:1455–1461. doi: 10.1038/ng.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, Ward P, Braxton A, Wang M, Buhay C, et al. Molecular Findings Among Patients Referred for Clinical Whole-Exome Sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helbig KL, Farwell Hagman KD, Shinde DN, Mroske C, Powis Z, Li S, Tang S, Helbig I. Diagnostic exome sequencing provides a molecular diagnosis for a significant proportion of patients with epilepsy. Genet Med. 2016;18:898–905. doi: 10.1038/gim.2015.186. [DOI] [PubMed] [Google Scholar]
- 21.Stark Z, Tan TY, Chong B, Brett GR, Yap P, Walsh M, Yeung A, Peters H, Mordaunt D, Cowie S, et al. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders [Internet] Genet Med. 2016 doi: 10.1038/gim.2016.1. [DOI] [PubMed] [Google Scholar]
- 22.Bernier R, Golzio C, Xiong B, Stessman HA, Coe BP, Penn O, Witherspoon K, Gerdts J, Baker C, Vulto-van Silfhout AT, et al. Disruptive CHD8 Mutations Define a Subtype of Autism Early in Development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melnikov A, Murugan A, Zhang X, Tesileanu T, Wang L, Rogov P, Feizi S, Gnirke A, Callan CG, Kinney JB, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol. 2012;30:271–277. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vockley CM, Barrera A, Reddy TE. Decoding the role of regulatory element polymorphisms in complex disease. Curr Opin Genet Dev. 2017;43:38–45. doi: 10.1016/j.gde.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 25.Whitaker JW, Nguyen TT, Zhu Y, Wildberg A, Wang W. Computational schemes for the prediction and annotation of enhancers from epigenomic assays. Methods. 2015;72:86–94. doi: 10.1016/j.ymeth.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •26.Ardlie KG, Deluca DS, Segrè AV, Sullivan TJ, Young TR, Gelfand ET, Trowbridge CA, Maller JB, Tukiainen T, Lek M, et al. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. The largest resource to date for eQTL analysis in human postmortem tissues, with hundreds of genotyped samples across dozens of tissues. A clear reference for defining which SNPs are associated with expression changes of nearby genes, aiding interpretation of common variant GWAS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, Montgomery GW, Goddard ME, Wray NR, Visscher PM, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- •28.Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, Ruderfer DM, Oh EC, Topol A, Shah HR, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. A large postmortem transcriptome case-control study of 279 individuals with schizophrenia. Demonstrates how gene expression can be used to interpret GWAS results from the 108 loci highlighted in Ripke et al, using eQTL analysis as well as postmortem differntial expression analysis and analysis of cellularity and pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, Wingett SW, Andrews S, Grey W, Ewels PA, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 30.Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •31.Won H, de la Torre-Ubieta L, Stein JL, Parikshak NN, Huang J, Opland CK, Gandal MJ, Sutton GJ, Hormozdiari F, Lu D, et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 2016;538:523–527. doi: 10.1038/nature19847. An elegant application of Hi-C methods for measuring chromatin confirmation to developing human brain. Notably, defines enhancer-promoter interactions for many enhancers containing schizophrenia risk SNPs from Ripke et al, demonstrating how Hi-C information can be used to associate a polymorphism with a specific gene. The study also demonstrates that simple proximity of SNP to gene often performs poorly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whalen S, Truty RM, Pollard KS. Enhancer-promoter interactions are encoded by complex genomic signatures on looping chromatin. Nat Genet. 2016;48:488–496. doi: 10.1038/ng.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, Tooley K, Presumey J, Baum M, Van Doren V, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •34.White MA. Understanding how cis-regulatory function is encoded in DNA sequence using massively parallel reporter assays and designed sequences. Genomics. 2015;106:165–170. doi: 10.1016/j.ygeno.2015.06.003. A thorough review of MPRAs and their applications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.BrainSeq: A Human Brain Genomics Consortium. BrainSeq: Neurogenomics to Drive Novel Target Discovery for Neuropsychiatric Disorders. Neuron. 2015;88:1078–1083. doi: 10.1016/j.neuron.2015.10.047. [DOI] [PubMed] [Google Scholar]
- 36.Gupta S, Ellis SE, Ashar FN, Moes A, Bader JS, Zhan J, West AB, Arking DE. Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism [Internet] Nat Commun. 2014:5. doi: 10.1038/ncomms6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •37.Jaffe AE, Shin J, Collado-Torres L, Leek JT, Tao R, Li C, Gao Y, Jia Y, Maher BJ, Hyde TM, et al. Developmental regulation of human cortex transcription and its clinical relevance at single base resolution. Nat Neurosci. 2015;18:154–161. doi: 10.1038/nn.3898. A human developmental postmortem RNAseq study. Of particular note here is the authors ground-up rethinking of RNAseq analysis, basing it on an agnostic region-based approach rather than depending on a priori gene annotations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller JA, Ding S-L, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Connolly BS, Lang AE. Pharmacological Treatment of Parkinson Disease: A Review. JAMA. 2014;311:1670–1683. doi: 10.1001/jama.2014.3654. [DOI] [PubMed] [Google Scholar]
- 42.Weaver FM, Follett KA, Stern M, Luo P, Harris CL, Hur K, Marks WJ, Rothlind J, Sagher O, Moy C, et al. Randomized trial of deep brain stimulation for Parkinson disease: thirty-six-month outcomes. Neurology. 2012;79:55–65. doi: 10.1212/WNL.0b013e31825dcdc1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gasser T. Usefulness of Genetic Testing in PD and PD Trials: A Balanced Review. J Park Dis. 2015;5:209–215. doi: 10.3233/JPD-140507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kopp N, Climer S, Dougherty JD. Moving from capstones toward cornerstones: successes and challenges in applying systems biology to identify mechanisms of autism spectrum disorders [Internet] Front Genet. 2015:6. doi: 10.3389/fgene.2015.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wells A, Kopp N, Xu X, O’Brien DR, Yang W, Nehorai A, Adair-Kirk TL, Kopan R, Dougherty JD. The anatomical distribution of genetic associations. Nucleic Acids Res. 2015;43:10804–10820. doi: 10.1093/nar/gkv1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JSK, Younkin S, et al. TREM2 Variants in Alzheimer’s Disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert J-C, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naj AC, Jun G, Beecham GW, Wang L-S, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heneka MT, Carson MJ, Khoury JE, Landreth GE, Brosseron F, Feinstein DL, Jacobs AH, Wyss-Coray T, Vitorica J, Ransohoff RM, et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 2015;14:388–405. doi: 10.1016/S1474-4422(15)70016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hong S, Dissing-Olesen L, Stevens B. New insights on the role of microglia in synaptic pruning in health and disease. Curr Opin Neurobiol. 2016;36:128–134. doi: 10.1016/j.conb.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skene NG, Grant SGN. Identification of Vulnerable Cell Types in Major Brain Disorders Using Single Cell Transcriptomes and Expression Weighted Cell Type Enrichment [Internet] Front Neurosci. 2016:10. doi: 10.3389/fnins.2016.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu X, Wells AB, O’Brien DR, Nehorai A, Dougherty JD. Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J Neurosci Off J Soc Neurosci. 2014;34:1420–1431. doi: 10.1523/JNEUROSCI.4488-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suárez-Fariñas M, Schwarz C, Stephan DA, Surmeier DJ, et al. A Translational Profiling Approach for the Molecular Characterization of CNS Cell Types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- 57.Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ, Nelson SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- 58.Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 59.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A Transcriptome Database for Astrocytes, Neurons, and Oligodendrocytes: A New Resource for Understanding Brain Development and Function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang B, Gaiteri C, Bodea L-G, Wang Z, McElwee J, Podtelezhnikov AA, Zhang C, Xie T, Tran L, Dobrin R, et al. Integrated Systems Approach Identifies Genetic Nodes and Networks in Late-Onset Alzheimer’s Disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seyfried NT, Dammer EB, Swarup V, Nandakumar D, Duong DM, Yin L, Deng Q, Nguyen T, Hales CM, Wingo T, et al. A Multi-network Approach Identifies Protein-Specific Co-expression in Asymptomatic and Symptomatic Alzheimer’s Disease. Cell Syst. 2016 doi: 10.1016/j.cels.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jaffe AE, Deep-Soboslay A, Tao R, Hauptman DT, Kaye WH, Arango V, Weinberger DR, Hyde TM, Kleinman JE. Genetic neuropathology of obsessive psychiatric syndromes. Transl Psychiatry. 2014;4:e432. doi: 10.1038/tp.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jaffe AE, Gao Y, Deep-Soboslay A, Tao R, Hyde TM, Weinberger DR, Kleinman JE. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat Neurosci. 2016;19:40–47. doi: 10.1038/nn.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •65.Jaffe AE. Postmortem human brain genomics in neuropsychiatric disorders--how far can we go? Curr Opin Neurobiol. 2016;36:107–111. doi: 10.1016/j.conb.2015.11.002. A well thought through recent perspective on human postmortem transcriptomics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •66.Parikshak NN, Gandal MJ, Geschwind DH. Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat Rev Genet. 2015;16:441–458. doi: 10.1038/nrg3934. A well considered discussion of the applications of network approaches to human postmortem transcriptomic data for neurogenetic disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parikshak NN, Swarup V, Belgard TG, Irimia M, Ramaswami G, Gandal MJ, Hartl C, Leppa V, de la Ubieta LT, Huang J, et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature. 2016;540:423–427. doi: 10.1038/nature20612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu YE, Parikshak NN, Belgard TG, Geschwind DH. Genome-wide, integrative analysis implicates microRNA dysregulation in autism spectrum disorder [Internet] Nat Neurosci. 2016 doi: 10.1038/nn.4373. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jaffe AE, Tao R, Norris A, Kealhofer M, Nellore A, Jia Y, Hyde T, Kleinman J, Straub R, Leek JT, et al. A framework for RNA quality correction in differential expression analysis. bioRxiv. 2016 doi: 10.1101/074245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu AR, Neff NF, Kalisky T, Dalerba P, Treutlein B, Rothenberg ME, Mburu FM, Mantalas GL, Sim S, Clarke MF, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods. 2014;11:41–46. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- •72.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. A landmark paper describing a new method for massively parallel analysis of transcriptomes of thousands of cells in parallel. The dramatic decrease in library-prep costs for each cell are enabling thorough surveys and cataloging of cell types in the brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.17 EU. Pm 6:15: NIH Details Plan for BRAIN Initiative [Internet] Sci AAAS. 2013;2013 [no volume] [Google Scholar]
- 74.The Cell Atlas – Chan Zuckerberg Biohub [Internet] [date unknown], [no volume] [Google Scholar]
- 75.Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Gephart MGH, Barres BA, Quake SR. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci. 2015;112:7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Földy C, Darmanis S, Aoto J, Malenka RC, Quake SR, Südhof TC. Single-cell RNAseq reveals cell adhesion molecule profiles in electrophysiologically defined neurons. Proc Natl Acad Sci. 2016;113:E5222–E5231. doi: 10.1073/pnas.1610155113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fuzik J, Zeisel A, Máté Z, Calvigioni D, Yanagawa Y, Szabó G, Linnarsson S, Harkany T. Integration of electrophysiological recordings with single-cell RNA-seq data identifies neuronal subtypes. Nat Biotechnol. 2016;34:175–183. doi: 10.1038/nbt.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gokce O, Stanley GM, Treutlein B, Neff NF, Camp JG, Malenka RC, Rothwell PE, Fuccillo MV, Südhof TC, Quake SR. Cellular Taxonomy of the Mouse Striatum as Revealed by Single-Cell RNA-Seq. Cell Rep. 2016;16:1126–1137. doi: 10.1016/j.celrep.2016.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci. 2016;19:335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, Manno GL, Juréus A, Marques S, Munguba H, He L, Betsholtz C, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 81.Cahan P, Li H, Morris SA, Lummertz da Rocha E, Daley GQ, Collins JJ. CellNet: Network Biology Applied to Stem Cell Engineering. Cell. 2014;158:903–915. doi: 10.1016/j.cell.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kuhn A, Thu D, Waldvogel HJ, Faull RLM, Luthi-Carter R. Population-specific expression analysis (PSEA) reveals molecular changes in diseased brain. Nat Methods. 2011;8:945–947. doi: 10.1038/nmeth.1710. [DOI] [PubMed] [Google Scholar]
- 83.Grindberg RV, Yee-Greenbaum JL, McConnell MJ, Novotny M, O’Shaughnessy AL, Lambert GM, Araúzo-Bravo MJ, Lee J, Fishman M, Robbins GE, et al. RNA-sequencing from single nuclei. Proc Natl Acad Sci. 2013;110:19802–19807. doi: 10.1073/pnas.1319700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Habib N, Li Y, Heidenreich M, Swiech L, Avraham-Davidi I, Trombetta JJ, Hession C, Zhang F, Regev A. Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science. 2016;353:925–928. doi: 10.1126/science.aad7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lacar B, Linker SB, Jaeger BN, Krishnaswami SR, Barron JJ, Kelder MJE, Parylak SL, Paquola ACM, Venepally P, Novotny M, et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat Commun. 2016;7:11022. doi: 10.1038/ncomms11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung H-L, Chen S, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Reddy AS, O’Brien D, Pisat N, Weichselbaum CT, Sakers K, Lisci M, Dalal JS, Dougherty JD. A Comprehensive Analysis of Cell Type-Specific Nuclear RNA From Neurons and Glia of the Brain. Biol Psychiatry. 2016 doi: 10.1016/j.biopsych.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Matevossian A, Akbarian S. Neuronal nuclei isolation from human postmortem brain tissue. J Vis Exp JoVE. 2008 doi: 10.3791/914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dougherty J. The OMICs: Applications in Neuroscience. OUP; USA: 2014. Cellomics: Characterization of Neural Subtypes by High-Throughput Methods and Transgenic Mouse Models. [Google Scholar]
- 90.Shen SQ, Myers CA, Hughes AEO, Byrne LC, Flannery JG, Corbo JC. Massively parallel cis-regulatory analysis in the mammalian central nervous system. Genome Res. 2016;26:238–255. doi: 10.1101/gr.193789.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maricque BB, Dougherty JD, Cohen BA. A genome-integrated massively parallel reporter assay reveals DNA sequence determinants of cis-regulatory activity in neural cells. Nucleic Acids Res. 2017;45:e16–e16. doi: 10.1093/nar/gkw942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.White MA, Kwasnieski JC, Myers CA, Shen SQ, Corbo JC, Cohen BA. A Simple Grammar Defines Activating and Repressing cis-Regulatory Elements in Photoreceptors. Cell Rep. 2016;17:1247–1254. doi: 10.1016/j.celrep.2016.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]