 Transcriptome profiling reveals that the cellular processes affected by Hg and TCDD in combination could be significantly different from those affected by Hg or TCDD alone.
Transcriptome profiling reveals that the cellular processes affected by Hg and TCDD in combination could be significantly different from those affected by Hg or TCDD alone.
Abstract
Mercury (Hg) and 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) are major environmental contaminants that commonly co-occur in the environment. Both Hg and TCDD are associated with a number of human diseases including cancers. While the individual toxicological effects of Hg and TCDD have been extensively investigated, studies on co-exposure are limited to a few genes and pathways. Therefore, a significant knowledge gap exists in the understanding of the deleterious effects of co-exposure to Hg and TCDD. Due to the prevalence of Hg and TCDD co-contamination in the environment and the major human health hazards they pose, it is important to obtain a fuller understanding of genome-wide effects of Hg and TCDD co-exposure. In this study, by performing a comprehensive transcriptomic analysis of human bronchial epithelial cells (BEAS-2B) exposed to Hg and TCDD individually and in combination, we have uncovered a subset of genes with altered expression only in the co-exposed cells. We also identified the additive as well as antagonistic effects of Hg and TCDD on gene expression. Moreover, we found that co-exposure impacted several biological and disease processes not affected by Hg or TCDD individually. Our studies show that the consequences of Hg and TCDD co-exposure on the transcriptional program and biological processes could be substantially different from single exposures, thus providing new insights into the co-exposure-specific pathogenic processes.
1. Introduction
Environmental exposure to hazardous substances often involves exposure to mixtures of chemicals. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) and heavy metals like mercury (Hg), which accumulate in the atmosphere due to both natural and industrial processes, are among the most common environmental contaminants that co-occur in the environment.1,2 A recent comprehensive study on fish tissues collected from five hundred US lakes and reservoirs showed that 81% and 100% of fish samples were contaminated with dioxins and Hg, respectively.3 Humans can be exposed to Hg and TCDD through the consumption of contaminated fish and water4,5 and inhalation.6,7 In addition, human TCDD exposure can also occur through dermal absorption.7 Exposure to Hg is associated with a number of diseases including pneumonitis, respiratory failure, lung injury, neurological disorders, immune system disorders and cardiovascular toxicity.6,8 TCDD exposure can cause skin lesions, compromised liver function, endocrine disorders and several cancers, including lung cancer, soft-tissue sarcoma and non-Hodgkin lymphoma.9,10 Furthermore, studies have suggested that Hg and TCDD can cross the placenta and affect the developing embryo.11,12 Therefore, exposure to Hg and TCDD is a major human health hazard.
The biological effects of Hg and TCDD individually have been extensively characterized. Multiple animal and in vitro studies have shown a major role for the aryl hydrocarbon receptor (AhR) in mediating TCDD toxicity.13 AhR is a ligand activated transcription factor, which upon binding to TCDD localizes to the nucleus and elevates the transcription of xenobiotic-metabolizing genes, including CYP1A1, CYP1A2 and CYP1B1.14 The increased CYP1A family of enzymes elevates reactive oxygen species (ROS) levels and promotes mitochondrial and endothelial dysfunction, vascular diseases and cancers.15–18 Mercury exposure is associated with lipid and DNA peroxidation resulting in oxidative stress and tissue injury, and causes nuclear localization of nuclear factor erythroid 2-related factor 2 (Nrf2).19,20 Nrf2 nuclear localization elicits an antioxidant response by activating genes such as NAD(P)H quinone dehydrogenase 1 (NQO1) and glutathione S-transferases (GSTs).19 However, the uncontrolled expression of Nrf2 can lead to cellular differentiation, proliferation, chemotherapy resistance and cancers.21
The principal signaling molecules activated by TCDD and Hg are AhR and Nrf2, respectively. Increasing evidence shows extensive cross-talk between these two transcription factors.22,23 Mouse Hep1c1c7 cells exposed to TCDD displayed an AhR-activation-dependent increase in Nrf2 levels.22 Moreover, Hg exposure in Hep1c1c7 cells increased the expression of AhR regulated genes.24 Furthermore, earlier studies have shown that TCDD exposure could activate antioxidant genes through Nrf2 activation25 and inhibit adipogenesis.23 These studies suggest that co-exposure to Hg and TCDD could potentially have a wider impact compared to individual exposures. However, effects due to single chemical exposure have been the focus of a majority of previous toxicological studies.15,20,25 A few studies have shown that the effect of Hg and TCDD co-exposure could differ significantly from individual exposures.26–29 For instance, a recent study showed that Hg modulated the expression of TCDD-induced genes.28 Furthermore, dioxin and Hg co-exposure causes an increase in insulin resistance.30 However, co-exposure studies have focused primarily on investigating the effects on specific genes such as CYP1A1, CYP1A2, CYP1B1, NQO1, GSTA1 and HO-1.26–29 Given the ubiquitous nature of the major transcriptional regulators AhR and Nrf2, it is conceivable that the outcome of co-exposure could be more extensive than the effects on specific genes and pathways and potentially have genome-wide consequences. Moreover, the effects of Hg and TCDD co-exposure could extend beyond AhR and Nrf2-dependent regulation. Due to the prevalence of Hg and TCDD co-contamination and the significant health hazards that they pose, there is a need to obtain a fuller understanding of the effects of co-exposure. In this study, by performing a comprehensive transcriptomic analysis of human bronchial epithelial cells (BEAS-2B) exposed to Hg and TCDD individually and in combination, we have discovered unique transcriptional alterations in the co-exposed cells. Co-exposure resulted in additive or antagonistic effects of Hg and TCDD on the expression of a number of genes. Furthermore, our studies have uncovered pathways and biological processes that are affected only by co-exposure.
2. Materials and methods
2.1. Cell culture and treatments
BEAS-2B was cultured in Dulbecco's Modified Eagle's Medium (Cellgro) supplemented with 1% penicillin streptomycin and 10% fetal bovine serum (Atlanta Biologicals) at 37 °C and 5% CO2. For Hg and TCDD exposures, the cells were treated with different doses of HgCl2 (0, 0.5 and 2.5 μM) and TCDD (0, 2 and 10 nM), either individually or in combination (Hg 0.5 μM/TCDD 2.0 nM; Hg 2.5 μM/TCDD 10 nM) for 3 weeks. The cells were split at 80% confluence. The cell culture medium was changed every 48 h and appropriate amounts of fresh Hg and TCDD were added to the cells. HgCl2 was obtained from Sigma (215465) and 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) was obtained from AccuStandard (D-404S).
2.2. RNA isolation and RNA-Seq
Total RNA was isolated from untreated and Hg and TCDD treated cells using an RNeasy Kit (Qiagen 74104). RNA-Seq libraries were prepared using an Illumina TruSeq RNA Sample Preparation Kit (RS-122-2002) according to the manufacturer's protocol. Sequencing was performed as described earlier31,32 using an Illumina HiSeq 2500 to obtain 50-nucleotide single-end reads. RNA-Seq data analysis was performed using a BioWardrobe Experiment Management System.33 Briefly, the raw Fastq sequence files were aligned to the human genome (hg19) using STAR34 (version 2.4.oj using default parameters; multi hits removed) with a known reference annotation gtf file from RefSeq. Gene expression levels were quantified as Reads Per Kilobase of transcript per Million mapped reads (RPKM) using the BioWardrobe algorithm.33 Genes with RPKM > 1 in at least one experimental condition were considered as expressed.
Differential gene expression was calculated using DESeq2.35 Genes that showed ≥1.5-fold up- or down-regulation between the conditions compared, along with the FDR adjusted p-value <0.1 were considered as differentially expressed and used for further analysis. The RNA-Seq data were deposited in the Gene Expression Omnibus (GEO) under the accession number GSE83886. Quantitative RT-PCR (qRT-PCR) analysis was performed using FastStart Universal SYBR Green Master Mix (Roche) on a 7900HT Fast Real-Time PCR system (Applied Biosystems). PCR was performed with the following conditions: 95 °C for 10 min followed by 40 cycles of 95 °C for 15 seconds and 60 °C for 1 minute. Primer sequences are provided in ESI Table S1.† The statistical significance of all qPCR results was evaluated using the t-test (p < 0.05 (*); p < 0.01 (**); p < 0.001 (***)).
2.3. Identification of canonical pathways and upstream regulators
Differentially expressed gene lists from RNA-Seq analysis were imported into Ingenuity Pathway Analysis (IPA) (QIAGEN Redwood City, http://www.qiagen.com/ingenuity) to identify the enriched canonical pathways. To identify common and unique canonical pathways, the ‘comparison analysis’ option from IPA was used. Fischer's exact test (p < 0.05) was used to identify the enriched canonical pathways. Enrichment scores (Fisher's exact test p-values) along with the z-scores were used for ranking the top canonical pathways.
The matrix derived from the top pathways (–log(p-values)) was visualized as a heat map (Fig. 3) using Java TreeView 3.0.36 Upstream Regulator Analysis (URA)37 was used to identify the upstream regulators potentially involved in generating the gene expression profiles observed in cells exposed to Hg and TCDD. The enrichment scores (Fisher's exact test p-values) along with the z-scores were used for ranking the top upstream regulators.
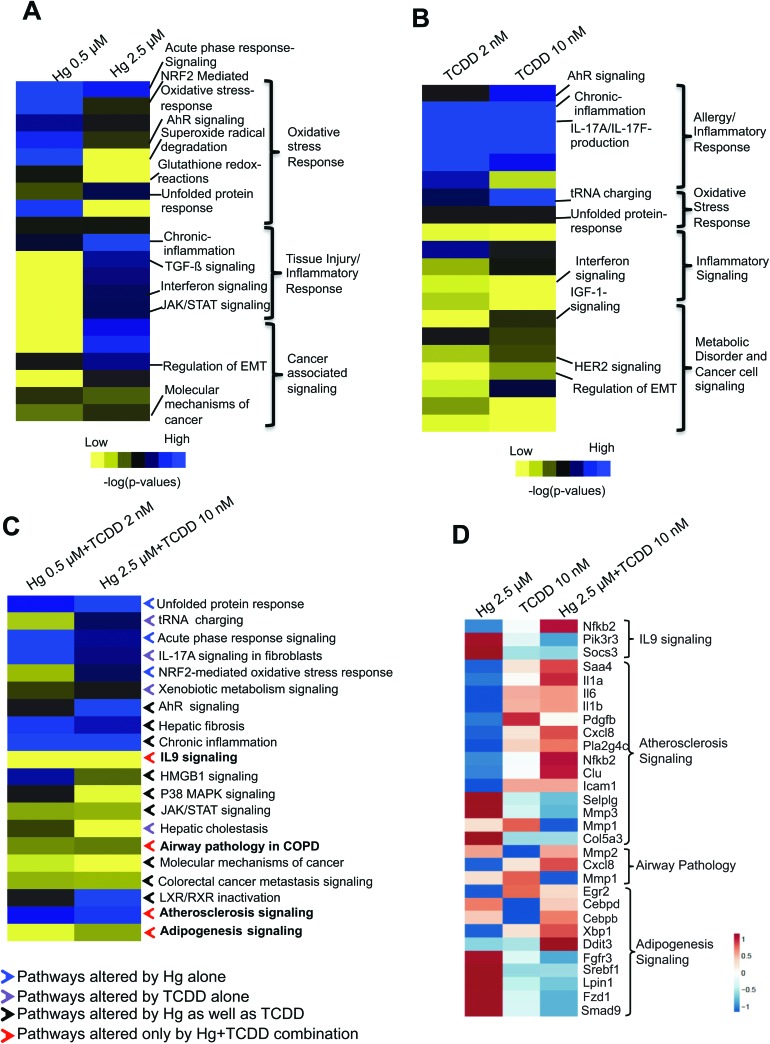
Fig. 3. Pathway enrichment in BEAS-2B cells exposed to Hg or TCDD individually and in combination. Pathways affected by exposure to low- and high-doses of (A) Hg, (B) TCDD and (C) Hg and TCDD combination. The contribution from each chemical is annotated in (C) as colored arrows. Blue arrows: pathways altered in the cells exposed to Hg alone; purple arrows: pathways altered in the cells exposed to TCDD alone; black arrows: pathways altered in the cells individually exposed to Hg as well as TCDD; red arrows: pathways altered only in the cells co-exposed to Hg and TCDD. IPA comparison analysis was performed on the differentially expressed genes (≥1.5 up- or down-regulation) and the pathways are ranked based on the enrichment score (p < 0.05). The pathways were then annotated/classified based on their function and visualized as a heat map using TreeView 3.3. (D) Heat map showing the expression of genes associated with biological processes uniquely enriched by Hg and TCDD co-exposure. The heat map was generated with ClustVis using a fold change matrix of genes in the exposed cells compared to the corresponding control values.
2.4. Identification of Hg and TCDD interactions
k-Means clustering of differentially expressed genes (gene cluster n = 5, array cluster n = 1) (Fig. 2) was performed using Cluster 3.0 and visualized with Java TreeView 3.0.36,38 Unsupervised hierarchical clustering (Fig. 4C, D and 5) was performed using ClustVis.39
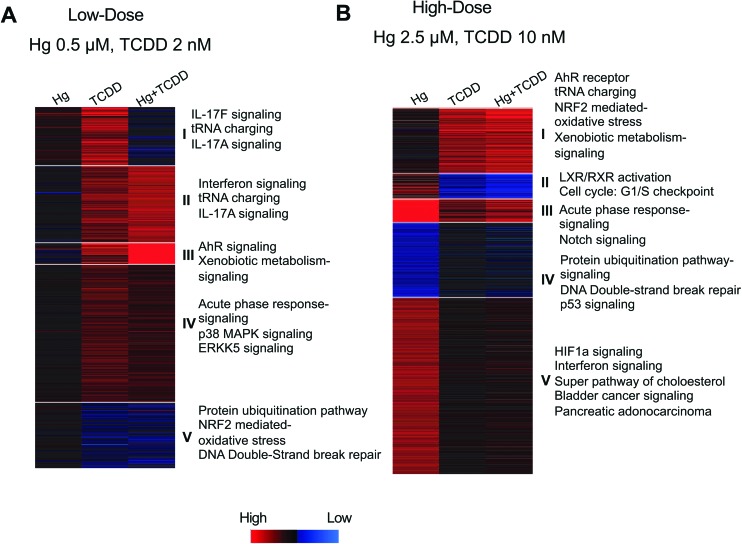
Fig. 2. k-Means clustering analysis showing additive or antagonistic effects of Hg and TCDD on gene expression. Gene cluster analysis was performed using the k-means clustering (gene cluster n = 5, array cluster n = 1) of genes differentially expressed by (A) low-dose Hg; low-dose TCDD; low-dose Hg and TCDD combination. (B) High-dose Hg; high-dose TCDD; high-dose Hg and TCDD combination. Pathway analysis of the genes in each cluster was performed using ingenuity pathway analysis (IPA). Fischer's exact test (p < 0.05) was used for canonical pathway identification. Blue: low expression; red: high expression.
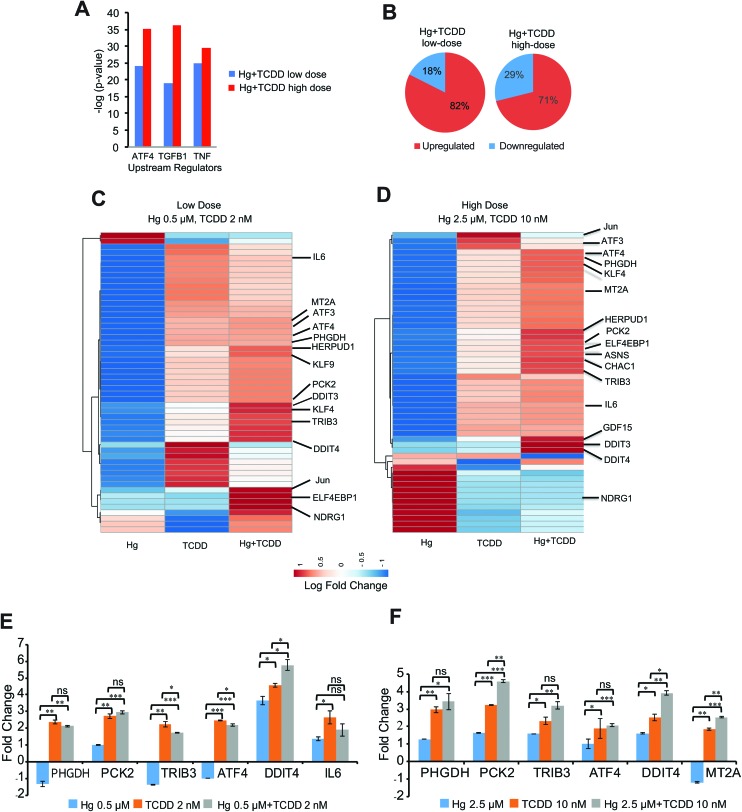
Fig. 4. ATF4 is the top upstream regulator activated by Hg and TCDD co-exposure. (A) Top upstream regulators activated by Hg and TCDD co-exposure. Upstream regulator analysis (URA) was performed on the differentially expressed genes, followed by a comparison analysis of upstream regulators between low- and high-dose Hg and TCDD. Fisher's exact test (p < 0.05) along with the z-scores was used to rank the top upstream regulators. (B) Pie charts showing that higher percentages of genes are upregulated by Hg and TCDD exposure. Unsupervised hierarchical clustering and heat map of the ATF4 target gene fold change following (C) low-dose Hg and TCDD co-exposure and (D) high-dose Hg and TCDD co-exposure. For (C) and (D) heat maps were generated with ClustVis using the log fold change matrix of ATF4 target genes in exposed cells compared to the corresponding control values. (E and F) qPCR validation of RNA-Seq results. mRNA levels of select ATF4 target genes, differentially expressed by low- (E) and high-dose (F) Hg and TCDD exposure shown as fold changes compared to the control. Gapdh was used as an internal control. Error bars represent standard deviations for at least two biological replicates. Statistical significance was evaluated using the t-test (p < 0.05 (*); p < 0.01 (**); p < 0.001 (***)).
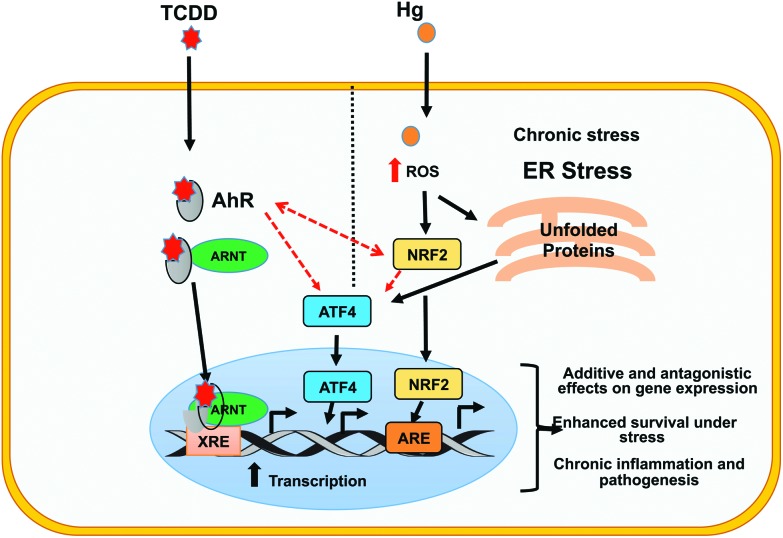
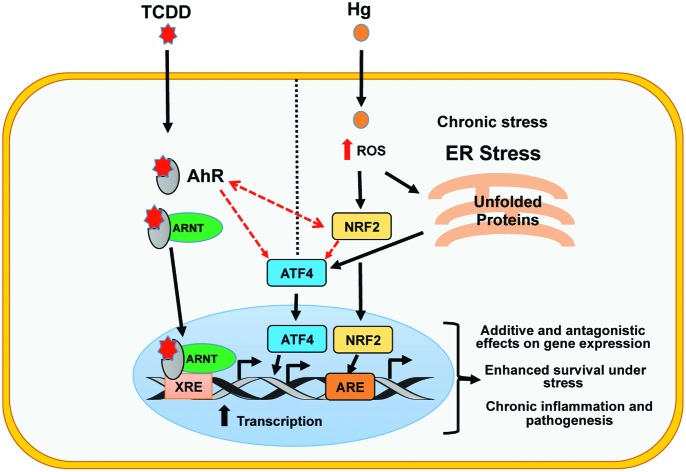
Fig. 5. Proposed model for the generation of a unique gene expression signature by Hg and TCDD co-exposure. Individual exposures to Hg and TCDD activate a number of genes primarily through AhR (TCDD) and Nrf2 (Hg) signaling. Upon Hg and TCDD co-exposure, both the AhR and Nrf2 signaling pathways are further activated, potentially through AhR–Nrf2 cross-talk and the ability of these molecules to regulate each other as well as their ability to activate master regulators such as ATF4. This results in the generation of a co-exposure specific unique transcriptional program through (i) additive interactions, where the expression of target genes is higher in co-exposed cells compared to single exposed cells, (ii) antagonistic interactions, where the effects of one toxicant on the gene expression are negated by the addition of the other toxicant, often resulting in a gene expression fold change that is an average of the fold change induced during the two individual exposures and (iii) differential expression of genes only by co-exposure. Moreover, anti-apoptotic signaling including IL9 signaling is significantly enriched in the co-exposed cells. This could provide the cells with the ability to survive under stress resulting in chronic inflammation and pathogenesis. Red dashed arrows represent interactions between transcription factors.
3. Results
3.1. A subset of genes is differentially expressed only by Hg and TCDD co-exposure
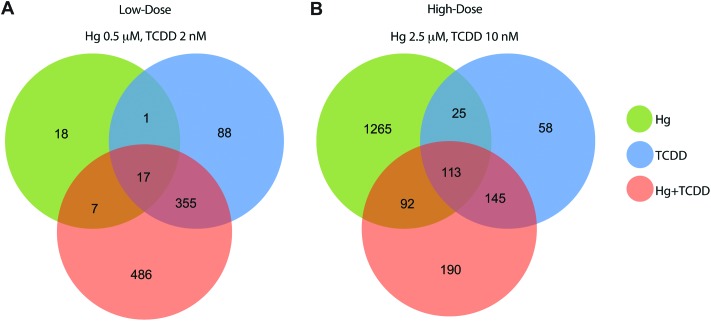
We treated BEAS-2B cells with multiple doses of (i) Hg (0, 0.5 and 2.5 μM); (ii) TCDD (0, 2 and 10 nM) and (iii) Hg and TCDD in combination (Hg 0.5 μM + TCDD 2 nM and Hg 2.5 μM + TCDD 10 nM) for 3 weeks. The doses are well within the range of Hg and TCDD levels in exposed humans,40–42 animals43 and in vitro studies.44,45 Analysis of the differentially expressed genes (RNA-Seq) in the various exposures showed that 50% of the genes (486 out of 972) in the low-dose exposure (Fig. 1A) and 10% of the genes (190 out of 1888) in the high-dose exposure (Fig. 1B) were differentially expressed only in the co-exposed cells.
Fig. 1. Differential gene expression (≥1.5 fold up- or down-regulated compared to control) identified using RNA-Seq analysis in BEAS-2B cells treated with (i) Hg, (ii) TCDD and (iii) Hg and TCDD in combination. (A) Low-dose exposure and (B) high-dose exposure. Co-exposure to Hg and TCDD induces the differential expression of 486 (low-dose) and 190 (high-dose) genes that are not transcriptionally altered by either Hg or TCDD alone.
Surprisingly, the number of Hg and TCDD co-exposure-specific differentially expressed genes was higher in the low-dose exposure (486) (Fig. 1A) compared to high-dose exposure (190) (Fig. 1B), although the total number of differentially expressed genes was substantially higher in the high-dose exposure (Fig. 1B). This suggests that although at low-doses Hg and TCDD individually have minimal effects, when combined, the consequences of exposure could be significantly elevated. Interestingly, over 75% of the genes that were differentially expressed by TCDD alone were differentially expressed by Hg and TCDD combination as well. In contrast, only 55% (in low-dose) and 13% (in high-dose) of the genes that were differentially expressed by Hg were differentially expressed by the Hg and TCDD combination (Fig. 1A and B). These results suggest that the effect of TCDD predominates the gene expression outcome of Hg and TCDD co-exposure at both low- and high-doses.
3.2. Hg and TCDD co-exposure elicits additive or antagonistic effects on gene expression
We next wanted to investigate if Hg and TCDD could have additive or antagonistic effects on gene expression during co-exposure. To accomplish this, we examined the expression levels of each gene that was differentially expressed by either Hg or TCDD individually or in combination. To group the genes based on their expression levels, we performed k-means clustering (gene cluster n = 5, array cluster n = 1) of the differentially expressed genes. If the differential expression fold in the co-exposure was higher than both the individual exposures, we considered the interaction to be additive. If the differential expression fold in the co-exposure was lower than one or both the individual exposures, we considered the interaction antagonistic. As shown in Fig. 2A and B, we could identify antagonistic as well as additive effects of Hg and TCDD in the co-exposed cells. Following this, we examined the pathways associated with each cluster of genes using Ingenuity Pathway Analysis (IPA) (Fig. 2A and B). Aryl hydrocarbon receptor (AhR) signaling, xenobiotic signaling (Fig. 2A: cluster III; B: cluster I), and oxidative stress response (Fig. 2A: cluster V; B: cluster I) were some of the pathways that showed additive effects of Hg and TCDD in the co-exposed cells. In contrast, pathways such as acute phase response (Fig. 2A: cluster IV; B: cluster III), protein ubiquitination (Fig. 2A: cluster V; B: cluster IV) and DNA damage repair processes (Fig. 2A: cluster V; B: cluster IV) showed antagonistic effects of Hg and TCDD.
3.3. Co-exposing cells to Hg and TCDD impacts several biological processes not affected by individual exposures
The additive and antagonistic effects of Hg and TCDD on gene expression (Fig. 2A and B) strongly suggest the possibility of co-exposure-specific effects on the cells. Therefore, we next examined how Hg and TCDD could potentially impact cellular processes individually and in combination. As shown in Fig. 3A, the major biological processes affected in cells exposed to Hg alone include the oxidative stress response, tissue injury/inflammatory response and cancer signaling. Interestingly, we found dose-dependent variations in the processes affected by Hg.
The oxidative stress response was significantly enriched in the cells exposed to low-dose Hg (0.5 μM) (Fig. 3A). In contrast, the cells exposed to high-dose Hg (2.5 μM) displayed a significantly reduced oxidative stress response (Fig. 3A). This suggests that at a high dose, Hg could overwhelm the cellular antioxidant capacity, resulting in a diminished oxidative stress response. In support of this, we found injury, inflammatory response and cancer associated signaling pathways to be highly enriched during high-dose Hg exposure, compared to low-dose exposure (Fig. 3A). TCDD exposure was primarily associated with allergy and autoimmunity-associated chronic inflammation (Fig. 3B). In contrast to Hg, TCDD did not induce significant dose-dependent variations in the affected pathways, with the exception of AhR signaling, which showed a dose-dependent induction (Fig. 3B).
Interestingly, co-exposure to Hg and TCDD resulted in a combined effect on several pathways individually altered by Hg and TCDD (Fig. 3C). Chronic inflammation, Nrf2-mediated oxidative stress response, xenobiotic metabolism and AhR signaling were some of the pathways that were highly enriched by co-exposure, compared to single exposure (Fig. 3C). In addition, stress survival and anti-apoptotic pathways such as an ER stress associated unfolded protein response and tRNA charging were enriched in the co-exposed cells (Fig. 3C). Surprisingly, pathways including IL9 signaling, atherosclerosis signaling and adipogenesis signaling, which were not highly enriched in cells exposed to either Hg or TCDD individually were significantly enriched only in the Hg and TCDD co-exposed cells (Fig. 3C and D). These results suggest that co-exposure to Hg and TCDD produces unique effects on the transcriptional program. Interestingly, we found that there was an enrichment of the anti-apoptotic IL9 signaling in the co-exposed cells, compared to the cells exposed to Hg or TCDD individually (Fig. 3C and D). Along with the enrichment of the oxidative stress response and xenobiotic detoxification process, the potential anti-apoptotic response in Hg and TCDD co-exposed cells suggests enhanced survival ability and resistance to apoptosis, which are hallmarks of cancer cells.
3.4. Activating transcription factor 4 (ATF4) is the top upstream regulator activated by Hg and TCDD co-exposure
We next wanted to identify the key proteins that are potentially responsible for the observed alterations in the transcriptional program of Hg and TCDD co-exposed cells. To accomplish this, we performed Upstream Regulator Analysis (URA).37 URA of the gene expression profiles in cells co-exposed to Hg and TCDD showed ATF4, TGFB1 and TNF as the top upstream regulators (Fig. 4A). The identification of ATF4 as the top upstream regulator is particularly interesting since it has been shown to heterodimerize with several transcriptional co-regulators including chromatin remodeling complexes and influence transcription.46,47 In addition, recent studies have shown the association of ATF4 with the induction of H3K4me3 at specific loci.48 H3K4me3 is a histone modification associated with active gene transcription. Consistent with this, our results show upregulation (≥1.5 fold) of over 70% of the differentially expressed genes in the cells co-exposed to both high- and low-doses of Hg and TCDD (Fig. 4B).
To understand the influence of Hg and TCDD individually and in combination on the ATF4 signaling pathway, we next examined the expression levels of ATF4 target genes (Fig. 4C–F). The exposure of cells to TCDD alone resulted in the upregulation number of ATF4 target genes. In contrast, the exposure of cells to Hg alone downregulated several ATF4 target genes (Fig. 4C–F). However, the fold upregulation of ATF4 target genes was higher in the co-exposed cells compared to TCDD exposed cells, although Hg alone could not activate these genes. For example, stem cell growth, development and differentiation associated transcription factors such as KLF4 and KLF9, which were downregulated by Hg exposure were upregulated by TCDD.
Surprisingly, the levels of upregulation were significantly higher in the co-exposed cells compared to cells exposed to TCDD alone (Fig. 4C–F). A similar phenomenon was observed in the upregulation of ER stress response associated ATF4 target genes DDIT3 and DDIT4 as well. These results suggest that although Hg alone could not upregulate the expression of several ATF4 target genes, when combined with TCDD, it could contribute to the upregulation of gene expression, further suggesting the importance of understanding the combined effects of these co-contaminants. Interestingly, NDRG1, which was highly activated at low-dose Hg and TCDD co-exposure, was repressed at a high dose (Fig. 4C and D). NDRG1 is a tumor suppressor gene and is essential for p53-induced apoptosis.49 Therefore, the loss of NDRG1 expression in cells co-exposed to high-dose Hg and TCDD further suggests a potential resistance to apoptosis in these cells.
4. Discussion
In this study, through a comprehensive analysis of the transcriptomes of cells exposed to Hg and TCDD individually, as well as in combination, we identified several co-exposure specific differentially expressed genes. Interestingly, although the total number of differentially expressed genes was more in the high-dose exposure, low-dose exposure produced more co-exposure-specific differentially expressed genes. This suggests higher tolerability and transcriptional variability in cells co-exposed to low-dose Hg and TCDD. Interestingly, over 70% of genes that were differentially expressed by TCDD alone were differentially expressed by Hg and TCDD co-exposure as well (Fig. 1A). In contrast, only 13.7% of genes differentially expressed by Hg were differentially expressed by co-exposure. This suggests a predominating effect of TCDD on the transcriptional program in the cells co-exposed to Hg and TCDD.
A number of earlier studies have demonstrated the pulmonary toxicity and immunomodulatory roles of TCDD and Hg.50–54 TCDD increased the expression of inflammatory cytokines, mucin 5AC and several matrix metalloproteinases (MMPs) in lungs of mice.55 Exposure to mercury vapour or mercury ions causes direct airway irritation.6 Although both Hg and TCDD are associated with lung injury and inflammation, we surprisingly found a significant enrichment of pathways associated with the ‘airway pathology in COPD’ in the co-exposed cells compared to the single exposed cells (Fig. 3C and D). Earlier studies have implicated the upregulation of C–X–C Motif Chemokine Ligand 8 (CXCL8) and MMP2 in extracellular matrix (ECM) degradation, airway epithelial remodelling and COPD.56,57 Consistent with this, our gene expression analysis shows the upregulation of CXCL8 and MMP2 in the co-exposed cells, compared to single exposed cells. Therefore, our results suggest that the risk of airway tissue remodelling and COPD is higher in Hg and TCDD co-exposed cells, compared to cells exposed to Hg or TCDD alone.
TCDD is a known AhR agonist, which affects a multitude of downstream signaling events.13 On the other hand, Hg primarily induces dose-dependent oxidative stress.20 As shown in Fig. 3A, cells exposed to low-dose Hg exhibited an elevated oxidative stress response and low levels of tissue injury and inflammation. However, at a high Hg dose there is a reduction in the oxidative stress response along with an increase in inflammation and injury. This suggests lower tolerance and increased cellular stress in cells exposed to Hg. In contrast, we did not find significant dose-dependent effects on the pathways altered by cells exposed to both low- and high-doses (5-fold higher) of TCDD. This suggests that compared to Hg, TCDD is better tolerated in the cells in terms of stress and cell survival. Therefore, it is plausible that the better-tolerated TCDD could induce extensive transcriptional alterations, thereby being the primary influencer of the transcriptional program in the co-exposed cells.
Notably, our analyses showed that several processes related to cardiovascular diseases including IL9 signaling, atherosclerosis signaling and adipogenesis signaling were highly enriched in the co-exposed cells compared to cells exposed to either Hg or TCDD individually (Fig. 3C). An examination of the expression levels of the genes involved in these signaling events showed that a number of genes including SAA4, CLU, CXCL8 or IL8, PLA2 and pro-inflammatory cytokines, IL1α, IL1β and IL6 were upregulated in the co-exposed cells (Fig. 3D). Upregulation of these genes is associated with an elevated inflammatory response and can lead to atherosclerosis.58–60 Furthermore, our results show the upregulation of CEBPβ and DDIT3, genes associated with adipocyte differentiation, in the co-exposed cells (Fig. 3D). The upregulation of CEBPβ and DDIT3 has been shown to inhibit adipocyte differentiation.61 In addition, we found the downregulation of LPIN1 and SREBF1 only in the co-exposed cells (Fig. 3D). Since LPIN1 and SREBF1 are known to promote adipocyte differentiation, the down regulation of these genes in co-exposed cells could suggest the inhibition of adipocyte differentiation.62,63 The inhibition of adipocyte differentiation decreases the fat storage capacity and results in the accumulation of triglycerides and cholesterol in other tissues.64 Moreover, the inhibition of adipocyte differentiation could lead to insulin resistance and diabetes. Elevated levels of triglycerides and cholesterol increase the risk of atherosclerosis. Although Hg and TCDD individually are known to be associated with cardiovascular diseases, our results suggest a significantly increased risk of cardiovascular diseases by Hg and TCDD co-exposure. These results suggest that the Hg and TCDD co-exposure presents higher risks for human health compared to single exposures. Although the lung epithelial BEAS-2B cell is not the appropriate cell-type to understand the effects of Hg and TCDD co-exposure on the cardiovascular system or adipocyte differentiation, our results emphasize the importance of co-exposure studies in various cell-types and animal systems to obtain a complete understanding of the exposure to these toxicants.
Our results reveal ATF4 as the major upstream regulator of the transcriptional program in the Hg and TCDD co-exposed cells (Fig. 4A), with most of the ATF4 target genes being upregulated. A closer inspection of the expression levels of ATF4 target genes showed that while TCDD exposure resulted in the activation of ATF4 target genes, Hg exposure resulted in the downregulation of a majority of them. Surprisingly, the fold upregulation in the co-exposed cells was higher than in the TCDD exposed cells (Fig. 4C–F). This indicates that Hg contributed to the upregulation of the ATF4 target genes in the co-exposed cells, although individually it downregulated these genes. These results strongly suggest that co-exposure of cells to Hg and TCDD elicits unique changes to the transcriptional program not seen during individual exposures (Fig. 4 and 5). ATF4 is a member of the ATF/CREB transcription factor family, activated during nutrient deprivation, unfolded protein response (UPR) and or endoplasmic reticulum stress (ER stress).65,66 The activation of AhR by TCDD exposure as well as oxidative stress caused by exposure to Hg can contribute to UPR and ER stress which in turn can cause the upregulation of ATF4 and its downstream target genes. Previous studies have reported the TCDD induced activation of IL-9 as a part of its immunomodulatory role.67,68 Interestingly, AhR has been shown to upregulate IL-9 and ATF4, both of which were found to be activated in asthma, COPD and lung inflammation.69–72 Recent studies have shown ATF4 to be a positive regulator of TLR4-triggered inflammatory cytokine production.73 Moreover, several studies have shown a positive regulatory role of ATF4 in adipocyte differentiation and adipogenesis signalling. In addition, ATF4 and several of its target genes are implicated in atherosclerosis.74–76 Consistent with the known functions of ATF4, our results show the enrichment of processes associated with IL9 signaling, atherosclerosis signalling, airway pathology and adipogenesis signalling in Hg and TCDD co-exposed cells. These results suggest ATF4 upregulation as a key event in the eventual outcome of Hg and TCDD co-exposure.
A recent study has shown that ATF4 binding to the ASNS gene promoter resulted in persistent H3K4me3 increase and a loss of histone H3 indicating the remodelling of the chromatin architecture and activation of the gene expression.48 Moreover, ATF4 has been shown to act as a docking site for several chromatin remodeling enzymes and recruits coactivators (histone acetyl transferases [HATs]) or corepressors (histone deacetylases [HDACs]) directly to promoters and enhancers.46,47 Gene expression studies have shown that ATF4 activation resulted in the induction of several genes associated with chromatin remodeling including H3K27me3 demethylase, JMJD3.47 Interestingly, our coexpression analysis77,78 of the genes differentially expressed by Hg and TCDD co-exposure using MSigDB78 showed enrichment of a gene signature that is similar to the knockdown of EZH2 [p-values, 1.919 × 10–22 (low-dose Hg + TCDD) and 1.002 × 10–22 (high-dose Hg + TCDD)]. EZH2 is a major methyltransferase responsible for the deposition of the repressive histone modification H3K27me3.79 Consistent with this, our results show that a large majority of genes that are differentially expressed by Hg and TCDD co-exposure are upregulated. Taken together, these results suggest the ATF4 mediated reduction of H3K27me3 and increase of H3K4me3 in the co-exposed cells. Therefore, it would be interesting to investigate potential alterations to the epigenetic landscape in the Hg and TCDD co-exposed cells.
5. Conclusions
For examining the transcriptional program of cells co-exposed to Hg and TCDD, we have uncovered the impacted biological pathways and potential disease processes, which could not have been identified by studying Hg and TCDD individually. Consistent with the known functions of Hg and TCDD,20,80 our transcriptome analyses of the co-exposed cells suggest increased oxidative stress and potential anti-apoptotic response. Based on our results, it could be speculated that in the co-exposed cells, the anti-apoptotic effect of TCDD could ensure better cell survival in spite of the increased oxidative stress caused by Hg (Fig. 5). This could potentially have major implications in disease development in the co-exposed cells, since better survival of the cells subjected to increased stress could result in pathogenic processes not seen during Hg or TCDD individual exposures.
Conflict of interest statement
There are no conflicts of interest to declare.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NIH) grants R01ES023174, R01ES024727 and P30ES000260 pilot program to S. C. The authors would like to thank Dr M. Costa for a critical reading of the manuscript. Next-Generation Sequencing was performed at the NYUMC Genome Technology Center, partially supported by the Cancer Center Support Grant, P30CA016087, at the Laura and Isaac Perlmutter Cancer Center.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c6tx00432f
References
- Kim E. J., Oh J. E., Chang Y. S. Sci. Total Environ. 2003;311:177–189. doi: 10.1016/S0048-9697(03)00095-0. [DOI] [PubMed] [Google Scholar]
- Kulkarni P. S., Crespo J. G., Afonso C. A. Environ. Int. 2008;34:139–153. doi: 10.1016/j.envint.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Stahl L. L., Snyder B. D., Olsen A. R., Pitt J. L. Environ. Monit. Assess. 2009;150:3–19. doi: 10.1007/s10661-008-0669-8. [DOI] [PubMed] [Google Scholar]
- Bloom N. S. Can. J. Fish. Aquat. Sci. 1992;49:1010–1017. [Google Scholar]
- Svensson B. G., Nilsson A., Hansson M., Rappe C., Akesson B., Skerfving S. N. Engl. J. Med. 1991;324:8–12. doi: 10.1056/NEJM199101033240102. [DOI] [PubMed] [Google Scholar]
- Rowens B., Guerrero-Betancourt D., Gottlieb C. A., Boyes R. J., Eichenhorn M. S. Chest. 1991;99:185–190. doi: 10.1378/chest.99.1.185. [DOI] [PubMed] [Google Scholar]
- Paustenbach D. J., Wenning R. J., Lau V., Harrington N. W., Rennix D. K., Parsons A. H. J. Toxicol. Environ. Health. 1992;36:103–149. doi: 10.1080/15287399209531628. [DOI] [PubMed] [Google Scholar]
- Holmes P., James K. A., Levy L. S. Sci. Total Environ. 2009;408:171–182. doi: 10.1016/j.scitotenv.2009.09.043. [DOI] [PubMed] [Google Scholar]
- Steenland K., Piacitelli L., Deddens J., Fingerhut M., Chang L. I. J. Natl. Cancer Inst. 1999;91:779–786. doi: 10.1093/jnci/91.9.779. [DOI] [PubMed] [Google Scholar]
- Pelclova D., Urban P., Preiss J., Lukas E., Fenclova Z., Navratil T., Dubska Z., Senholdova Z. Rev. Environ. Health. 2006;21:119–138. doi: 10.1515/reveh.2006.21.2.119. [DOI] [PubMed] [Google Scholar]
- Suzuki G., Nakano M., Nakano S. Biosci., Biotechnol., Biochem. 2005;69:1836–1847. doi: 10.1271/bbb.69.1836. [DOI] [PubMed] [Google Scholar]
- Yang J., Jiang Z., Wang Y., Qureshi I. A., Wu X. D. Ann. Clin. Lab. Sci. 1997;27:135–141. [PubMed] [Google Scholar]
- Mimura J., Fujii-Kuriyama Y. Biochim. Biophys. Acta. 2003;1619:263–268. doi: 10.1016/s0304-4165(02)00485-3. [DOI] [PubMed] [Google Scholar]
- Gu Y. Z., Hogenesch J. B., Bradfield C. A. Annu. Rev. Pharmacol. Toxicol. 2000;40:519–561. doi: 10.1146/annurev.pharmtox.40.1.519. [DOI] [PubMed] [Google Scholar]
- Aly H. A., Domenech O. Toxicol. Lett. 2009;191:79–87. doi: 10.1016/j.toxlet.2009.08.008. [DOI] [PubMed] [Google Scholar]
- Liou G. Y., Storz P. Free Radical Res. 2010;44:479–496. doi: 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yager J. D., Liehr J. G. Annu. Rev. Pharmacol. Toxicol. 1996;36:203–232. doi: 10.1146/annurev.pa.36.040196.001223. [DOI] [PubMed] [Google Scholar]
- Kopf P. G., Huwe J. K., Walker M. K. Cardiovasc. Toxicol. 2008;8:181–193. doi: 10.1007/s12012-008-9027-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar J. W., Niture S. K., Jaiswal A. K. Free Radical Biol. Med. 2009;47:1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund B. O., Miller D. M., Woods J. S. Biochem. Pharmacol. 1993;45:2017–2024. doi: 10.1016/0006-2952(93)90012-l. [DOI] [PubMed] [Google Scholar]
- Sporn M. B., Liby K. T. Nat. Rev. Cancer. 2012;12:564–571. doi: 10.1038/nrc3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao W., Hu L., Scrivens P. J., Batist G. J. Biol. Chem. 2005;280:20340–20348. doi: 10.1074/jbc.M412081200. [DOI] [PubMed] [Google Scholar]
- Shin S., Wakabayashi N., Misra V., Biswal S., Lee G. H., Agoston E. S., Yamamoto M., Kensler T. W. Mol. Cell. Biol. 2007;27:7188–7197. doi: 10.1128/MCB.00915-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korashy H. M., El-Kadi A. O. Toxicology. 2004;201:153–172. doi: 10.1016/j.tox.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Yeager R. L., Reisman S. A., Aleksunes L. M., Klaassen C. D. Toxicol. Sci. 2009;111:238–246. doi: 10.1093/toxsci/kfp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korashy H. M., El-Kadi A. O. Drug Metab. Dispos. 2006;34:152–165. doi: 10.1124/dmd.105.005397. [DOI] [PubMed] [Google Scholar]
- Korashy H. M., El-Kadi A. O. Toxicol. In Vitro. 2008;22:154–158. doi: 10.1016/j.tiv.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Amara I. E., Anwar-Mohamed A., Abdelhamid G., El-Kadi A. O. Food Chem. Toxicol. 2012;50:2325–2334. doi: 10.1016/j.fct.2012.04.028. [DOI] [PubMed] [Google Scholar]
- Amara I. E., El-Kadi A. O. Free Radicals Biol. Med. 2011;51:1675–1685. doi: 10.1016/j.freeradbiomed.2011.07.021. [DOI] [PubMed] [Google Scholar]
- Chang J. W., Chen H. L., Su H. J., Liao P. C., Guo H. R., Lee C. C. J. Hazard. Mater. 2011;185:749–755. doi: 10.1016/j.jhazmat.2010.09.084. [DOI] [PubMed] [Google Scholar]
- Jose C. C., Xu B., Jagannathan L., Trac C., Mallela R. K., Hattori T., Lai D., Koide S., Schones D. E., Cuddapah S. Proc. Natl. Acad. Sci. U. S. A. 2014;111:14631–14636. doi: 10.1073/pnas.1406923111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan L., Jose C. C., Arita A., Kluz T., Sun H., Zhang X., Yao Y., Kartashov A. V., Barski A., Costa M., Cuddapah S. J. Cell. Physiol. 2016;231:1611–1620. doi: 10.1002/jcp.25262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartashov A. V., Barski A. Genome Biol. 2015;16:158. doi: 10.1186/s13059-015-0720-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha A. J. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Kramer A., Green J., Pollard, Jr. J., Tugendreich S. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon M. J., Imoto S., Nolan J., Miyano S. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Metsalu T., Vilo J. Nucleic Acids Res. 2015;43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVito M. J., Birnbaum L. S., Farland W. H., Gasiewicz T. A. Environ. Health Perspect. 1995;103:820–831. doi: 10.1289/ehp.95103820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, Jr. D. G., Needham L. L., Pirkle J. L., Roberts D. W., Bagby J., Garrett W. A., Andrews, Jr. J. S., Falk H., Bernert J. T., Sampson E. J. Arch. Environ. Contam. Toxicol. 1988;17:139–143. doi: 10.1007/BF01056017. [DOI] [PubMed] [Google Scholar]
- Gibb H., O'Leary K. G. Environ. Health Perspect. 2014;122:667–672. doi: 10.1289/ehp.1307864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin M., Jerksell L. G., von Ubisch H. Arch. Environ. Health. 1966;12:33–42. doi: 10.1080/00039896.1966.10664334. [DOI] [PubMed] [Google Scholar]
- Chatterjee S., Ray A., Mukherjee S., Agarwal S., Kundu R., Bhattacharya S. Toxicol. Ind. Health. 2014;30:611–620. doi: 10.1177/0748233712462442. [DOI] [PubMed] [Google Scholar]
- Tsang H., Cheung T. Y., Kodithuwakku S. P., Chai J., Yeung W. S., Wong C. K., Lee K. F. Reprod. Toxicol. 2012;33:60–66. doi: 10.1016/j.reprotox.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Cherasse Y., Maurin A. C., Chaveroux C., Jousse C., Carraro V., Parry L., Deval C., Chambon C., Fafournoux P., Bruhat A. Nucleic Acids Res. 2007;35:5954–5965. doi: 10.1093/nar/gkm642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan J., Fu L., Balasubramanian M. N., Anthony T., Kilberg M. S. J. Biol. Chem. 2012;287:36393–36403. doi: 10.1074/jbc.M112.399600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian M. N., Shan J., Kilberg M. S. Biochem. J. 2013;449:219–229. doi: 10.1042/BJ20120958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein S., Thomas E. K., Herzog B., Westfall M. D., Rocheleau J. V., Jackson, 2nd R. S., Wang M., Liang P. J. Biol. Chem. 2004;279:48930–48940. doi: 10.1074/jbc.M400386200. [DOI] [PubMed] [Google Scholar]
- Li X. M., Peng J., Gu W., Guo X. J. PLoS One. 2016;11:e0150551. doi: 10.1371/journal.pone.0150551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. H., Park S. Y., Lee E. J., Cho Y. H., Park H. S., Hong S. H., Kim W. J. Tuberc. Respir. Dis. 2015;78:99–105. doi: 10.4046/trd.2015.78.2.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkvliet N. I. Biochem. Pharmacol. 2009;77:746–760. doi: 10.1016/j.bcp.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallee T. J. Am. Rev. Respir. Dis. 1969;99:430–436. doi: 10.1164/arrd.1969.99.3.430. [DOI] [PubMed] [Google Scholar]
- Pilones K., Tatum A., Gavalchin J. J. Immunotoxicol. 2009;6:161–170. doi: 10.1080/15476910903084021. [DOI] [PubMed] [Google Scholar]
- Wong P. S., Vogel C. F., Kokosinski K., Matsumura F. Am. J. Respir. Cell Mol. Biol. 2010;42:210–217. doi: 10.1165/rcmb.2008-0228OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovina N., Koutsoukou A., Koulouris N. G. Mediators Inflammation. 2013;2013:413735. doi: 10.1155/2013/413735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churg A., Zhou S., Wright J. L. Eur. Respir. J. 2012;39:197–209. doi: 10.1183/09031936.00121611. [DOI] [PubMed] [Google Scholar]
- Ramji D. P., Davies T. S. Cytokine Growth Factor Rev. 2015;26:673–685. doi: 10.1016/j.cytogfr.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targonska-Stepniak B., Majdan M. Mediators Inflammation. 2014;2014:793628. doi: 10.1155/2014/793628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt-Camejo E., Camejo G., Peilot H., Oorni K., Kovanen P. Circ. Res. 2001;89:298–304. doi: 10.1161/hh1601.095598. [DOI] [PubMed] [Google Scholar]
- Vogel C. F., Sciullo E., Park S., Liedtke C., Trautwein C., Matsumura F. J. Biol. Chem. 2004;279:8886–8894. doi: 10.1074/jbc.M310190200. [DOI] [PubMed] [Google Scholar]
- Ayala-Sumuano J. T., Velez-Delvalle C., Beltran-Langarica A., Marsch-Moreno M., Cerbon-Solorzano J., Kuri-Harcuch W. Sci. Rep. 2011;1:178. doi: 10.1038/srep00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan J., Peterfy M., Reue K. J. Biol. Chem. 2004;279:29558–29564. doi: 10.1074/jbc.M403506200. [DOI] [PubMed] [Google Scholar]
- Guilherme A., Virbasius J. V., Puri V., Czech M. P. Nat. Rev. Mol. Cell Biol. 2008;9:367–377. doi: 10.1038/nrm2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Hai T. J. Biol. Chem. 1997;272:24088–24095. doi: 10.1074/jbc.272.38.24088. [DOI] [PubMed] [Google Scholar]
- Hetz C. Nat. Rev. Mol. Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- Forgacs A. L., Dere E., Angrish M. M., Zacharewski T. R. Toxicol. Sci. 2013;133:54–66. doi: 10.1093/toxsci/kft028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Tsukumo S., Suzuki N., Motohashi H., Yamamoto M., Fujii-Kuriyama Y., Mimura J., Lin T. M., Peterson R. E., Tohyama C., Nohara K. J. Biol. Chem. 2004;279:25204–25210. doi: 10.1074/jbc.M402143200. [DOI] [PubMed] [Google Scholar]
- Wilhelm C., Hirota K., Stieglitz B., Van Snick J., Tolaini M., Lahl K., Sparwasser T., Helmby H., Stockinger B. Nat. Immunol. 2011;12:1071–1077. doi: 10.1038/ni.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch M., Goroll T., Bauer M., Hinz D., Schutze N., Polte T., Kesper D., Simon J. C., Hackermuller J., Lehmann I., Herberth G. Mediators Inflammation. 2014;2014:182549. doi: 10.1155/2014/182549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimbara A., Christodoulopoulos P., Soussi-Gounni A., Olivenstein R., Nakamura Y., Levitt R. C., Nicolaides N. C., Holroyd K. J., Tsicopoulos A., Lafitte J. J., Wallaert B., Hamid Q. A. J. Allergy Clin. Immunol. 2000;105:108–115. doi: 10.1016/s0091-6749(00)90185-4. [DOI] [PubMed] [Google Scholar]
- Steiling K., van den Berge M., Hijazi K., Florido R., Campbell J., Liu G., Xiao J., Zhang X., Duclos G., Drizik E., Si H., Perdomo C., Dumont C., Coxson H. O., Alekseyev Y. O., Sin D., Pare P., Hogg J. C., McWilliams A., Hiemstra P. S., Sterk P. J., Timens W., Chang J. T., Sebastiani P., O'Connor G. T., Bild A. H., Postma D. S., Lam S., Spira A., Lenburg M. E. Am. J. Respir. Crit. Care Med. 2013;187:933–942. doi: 10.1164/rccm.201208-1449OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Bai N., Chang A., Zhang Z., Yin J., Shen W., Tian Y., Xiang R., Liu C. Cell. Mol. Immunol. 2013;10:84–94. doi: 10.1038/cmi.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi S. J., Wang C. Y., Zhang J., Lv Z. P., Li Y. X. Int. J. Clin. Exp. Med. 2015;8:21635–21640. [PMC free article] [PubMed] [Google Scholar]
- Crawford R. R., Prescott E. T., Sylvester C. F., Higdon A. N., Shan J., Kilberg M. S., Mungrue I. N. J. Biol. Chem. 2015;290:15878–15891. doi: 10.1074/jbc.M114.635144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berisha S. Z., Hsu J., Robinet P., Smith J. D. PLoS One. 2013;8:e65003. doi: 10.1371/journal.pone.0065003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V. K., Lindgren C. M., Eriksson K. F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstrale M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., Tamayo P., Spiegelman B., Lander E. S., Hirschhorn J. N., Altshuler D., Groop L. C. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R. S., Zhang Y. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Park S., Matsumura F. Toxicology. 2006;217:139–146. doi: 10.1016/j.tox.2005.09.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.