Abstract
Background:
DNA repair mechanisms are crucial for sustaining DNA integrity and preventing carcinogenesis. The xeroderma pigmentosum group G (XPG), X-ray repair cross complementing group 2 (XRCC2) and RAD51 are candidate genes for DNA repair pathways.
Methods:
We performed a meta-analysis of 26 studies that assessed the impact of XPG Asp1104His, XRCC2 rs3218536 A/G and RAD51 135G/C polymorphisms on colorectal cancer (CRC) risk. This study included 10288 CRC patients and 11885 controls, and odds ratio (OR) with its 95% confidence interval (CI) were used to calculate the strength of association.
Results:
The results of overall meta-analysis suggested an association between the XPG Asp1104His polymorphism and CRC susceptibility in allele (OR=1.06; 95% CI=1.01-1.12) and heterozygote model (OR=1.16; 95%CI=1.02-1.31). In the subgroup analysis based on ethnicity and source of control, we found significantly increased CRC cancer risk in Asians (OR=1.12, 95%CI=1.04-1.21) and in hospital-based (OR=1.22, 95%CI=1.08-1.38) populations. Moreover, the RAD51 135 G/C polymorphism increased the risk of CRC in total using allele (OR=1.21) and recessive models (OR=1.62). However, XRCC2 rs3218536 A/G was not associated with the risk of CRC in total or in subgroups.
Conclusions:
According to the results of our meta-analysis, the XPG Asp1104His and RAD51 135 G/C polymorphisms might influence colorectal cancer risk.
Keywords: XPG, XRCC2, RAD51, colorectal cancer, meta-analysis
Introduction
Colorectal cancer (CRC) is the third most prevalent gastrointestinal tract neoplasm and the fourth major cause of cancer mortality globally (Mao et al., 2013, Sun et al., 2015). CRC development is a multistep process, in that several factors are known to be involved including environmental and genetic alterations (Zhang et al., 2011). The polymorphisms of genes involved in different DNA repair pathways may affect repair of bulky DNA lesions and maintenance of genomic stability, and thus cancer risk (Canbay et al., 2011, Hou et al., 2014). DNA repair protects genome damages caused by oxidative DNA compounds or DNA adducts. DNA repair pathways responsible for fixing DNA damages include base excision repair (BER), nucleotide excision repair (NER) (Lefkofsky et al., 2015) and homologous recombination repair (HRR) (Kiyohara and Yoshimasu, 2007).
NER is one of the key pathways that contributes to UV light-induced DNA damage, and protects a cell against a wide spectrum of structurally unrelated DNA lesions (Sugasawa, 2016). In the inherited disorder, xeroderma pigmentosum, NER deficiency is associated with a 1,000-fold higher occurrence of skin cancer, but also a 20-fold increase in internal tumours highlighting the NER importance in the repair of endogenous DNA damage (Mort et al., 2003; Spivak, 2015). One of the key DNA repair enzymes of the NER pathway is Xeroderma Pigmentosum complementation group G (XPG), which is also known as ERCC5 (excision repair cross-complementation group 5) (Chen et al., 2016). XPG gene is mapped on chromosome 13q22-q33, and consists of 15 exons and 14 introns and is one of the seven XP complementation groups (XPA to XPG). It has reported that a defective XPG results in DNA repair malfunction which leads to genomic instability, gene malfunction and initiation of carcinogenesis (Dworaczek and Xiao, 2007; Sollier et al., 2014).
Accurate repair of double-strand breaks (DSBs) arising during DNA replication or from DNA-damaging agents is essential to conserve genomic stability. HRR is the key pathway for repairing DSBs and the maintenance of genetic stability in mammalians (Griffin, 2002). Throughout the HRR process, a sister chromatid works as a template and the homologous sequence of DNA is aligned. Several numbers of key molecules contribute to the HRR process (Griffin and Thacker, 2004). Recent evidence indicated that RAD51 paralogs (RAD51B, RAD51C, RAD51D, XRCC2, XRCC3) play key roles in the HRR process (Griffin, 2002; Curtin et al., 2009). Moreover, coded by X-ray repair cross complementing group 2 (XRCC2) gene produces the XRCC2 protein that is structurally and functionally associated with RAD51; together with each other they form a fundamental complex required for chromosome segregation and apoptotic response to DSBs (Li et al., 2014). Also over 100-folds of HRR reduction in the XRCC2 deficient hamster cells compared with the parental cells has been observed which highlights the vital role of the XRCC2 protein for the HRR process (Johnson et al., 1999).
Growing evidence has explored the role of common single nucleotide polymorphism (SNP) located in exon 15 of XPG (Asp1104His; dbSNP ID rs17655 G/C) in the etiology of CRC in various populations (Chen et al., 2009; Luo et al., 2014; Steck et al., 2014; Zeng et al., 2015). Additionally, common variants within XRCC2 (R188H, dbSNP ID rs3218536), and RAD51 (135G/C, dbSNP ID rs1801320) have been determined as potential cancer susceptibility loci in recent studies (Curtinet et al., 2009). However, the results of some publications are contradictory (Bigleret al., 2005; Pardini et al., 2008), and some of the individual studies included small sample sizes as well as lack of power to identify a mild gene effect (Mort et al., 2003; Canbay et al., 2011; Krupa et al., 2011; Gil et al., 2012; Nissar et al., 2014; Cetinkunar et al., 2015). Hence, a comprehensive retrieval of the related literature would help obtain a more precise estimation of the association with disease susceptibility. Consequently, we performed a meta-analysis of case-control studies and investigated whether XPG Asp1104His, XRCC2 R188H A/G and RAD51 135G/C polymorphisms are associated with susceptibility to CRC using multiple genetic models.
Materials and Methods
Study assessment
Our literature search included the electronic databases such as PubMed, EMBASE, and MEDICINE. All languages were searched, and inclusive search strategies included the Mesh term and Keywords: (‘XPG’, ‘xeroderma pigmentosum group G’, ‘excision repair cross-complementing group 5’, ‘ERCC5’, ‘RAD51’,’XRCC2‘ or ‘NER’), (‘polymorphism’, ‘variant’ or ‘ ‘mutation’) along with (colorectal’, ‘rectal’, ‘gastrointestinal’, ‘colon cancer’.) through Jan 2, 2017. Eligible studies were selected and evaluated cautiously. Review articles and bibliographies of other relevant studies found were hand-searched to find further qualified studies.
Selection of eligible studies
The articles were filtered by two independent reviewers (M.H, A.R) to assess the appropriateness of the articles selected by using a standardized protocol and data collection form. The following inclusion criteria were used to determine qualified studies: (a) a human case-control study on the association between XPG, XRCC2 and RAD51 SNPs and CRC (b) adequate allele or genotype data needed for assessing an odds ratio (OR) and 95% confidence interval (CI). Exclusion criteria were (a) non-human studies, abstracts only, comments, reviews, editorials or letters, mechanism studies and cohort comprising of a case population; (b) family-based design or sibling pair studies, (c) studies with lack of enough information for data extraction and (d) unpublished data. Discrepancies about inclusion of studies and interpretation of data were solved with conversation.
We used following data information from each study: authors, year of publication, country, ethnicity, source of controls, genotype methods, sample size, allele and genotype frequency distribution and Hardy Weinberg equilibrium (HWE) (Table 1).
Table 1.
Main Characteristics of Studies Included for the Association between the XPG Asp1104His, XRCC2 Rs3218536 A/G and RAD51 135G/C Polymorphisms and Colorectal Cancer
| Study | Year | Country | Ethnicity | Source of controls | Genotype methods | Genotype (case/control) | Allele (case/control) | PHWE | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Asp/Asp | Asp/His | His/His | Asp | His | |||||||
| Sun et al. | 2015 | China | Asian | HB | PCR–RFLP | 890/910 | 216/227 | 476/497 | 198/186 | 908/951 | 872/869 | 0.004 |
| Kabzinski, J., et al. | 2015 | Poland | Caucasian | HB | QPCR | 234/238 | 36/43 | 171/175 | 27/20 | 243/261 | 225/215 | 0.001 |
| Paszkowska-Szczur, K., et al. | 2015 | Poland | Caucasian | PB | MassARRAY | 733/1358 | 429/869 | 272/404 | 32/85 | 1130/2142 | 336/576 | 0.001 |
| Steck, S. E, et al. | 2014 | USA | African | PB | MassARRAY | 224/317 | 65/100 | 120/151 | 39/66 | 250/351 | 198/283 | 0.519 |
| Steck, S. E., et al. | 2014 | USA | Caucasian | PB | MassARRAY | 298/532 | 183/335 | 100/170 | 15/27 | 466/840 | 130/224 | 0.372 |
| Du, H., et al. | 2014 | China | Asian | HB | Taqman Assay | 878/884 | 286/355 | 459/405 | 133/124 | 1031/1115 | 725/653 | 0.622 |
| Liu, D., et al. | 2012 | China | Asian | PB | PCR–RFLP | 1028/1085 | 233/329 | 603/537 | 192/219 | 1069/1195 | 987/975 | 0.996 |
| Gil, J., et al. | 2012 | Poland | Caucasian | PB | PCR–RFLP | 132/100 | 86/64 | 35/31 | 11-May | 207/159 | 57/41 | 0.624 |
| Canbay, E., et al. | 2011 | Turkey | Caucasian | HB | PCR–RFLP | 79/247 | 43/148 | 34/83 | Feb-16 | 120/379 | 38/115 | 0.351 |
| Joshi, A. D., et al | 2009 | USA | Caucasian | FB | Taqman assays | 308/361 | 183/213 | 114/137 | 11-Nov | 480/563 | 136/159 | 0.046 |
| Pardini, B., et al | 2008 | Czech | Caucasian | HB | PCR–RFLP | 532/532 | 334/356 | 177/153 | 21/23 | 845/865 | 219/199 | 0.211 |
| Huang, W. Y., et al. | 2006 | USA | Caucasian | PB | Sequencing | 679/697 | 407/403 | 243/265 | 29/29 | 1057/1071 | 301/323 | 0.073 |
| Bigler, J., et al. | 2005 | USA | Caucasian | PB | Taqman assays | 713/616 | 440/353 | 237/226 | 36/37 | 1117/932 | 309/300 | 0.917 |
| Mort, R., et al. | 2003 | UK | Caucasian | HB | PCR-RFLP | 40/33 | - | - | - | 67/58 | 13/22 | - |
| XRCC2 rs3218536 A/G | Year | Country | Ethnicity | Source | Genotype methods | Total | A/A | G/A | G/G | A | G | PHWE |
| Cetinkunar, S., et al. | 2015 | Turkey | Caucasian | HB | PCR-RFLP | 71/86 | 09-Nov | 30/21 | 32/54 | 48/43 | 94/129 | 0.001 |
| Krupa, R., et al. | 2011 | Poland | Caucasian | HB | PCR-RFLP | 100/100 | 75/84 | 18/14 | 07-Feb | 168/182 | 32/18 | 0.146 |
| Curtin, K., et al. | 2009 | UK & US | Caucasian | PB | Sequencing | 1227/1380 | 1014/1167 | 185/204 | 10-Sep | 2213/2538 | 205/222 | 0.979 |
| Moreno [16] | 2006 | Spain | Caucasian | HB | APEX | 350/316 | 287/265 | 57/45 | 06-Jun | 631/575 | 69/57 | 0.018 |
| Tranah, G. J., et al. | 2004 | USA | Caucasian | PB | TaqMan | 518/522 | 450/441 | A/G+G/G | - | - | - | - |
| Tranah, G. J., et al. | 2004 | USA | Caucasian | PB | TaqMan | 354/688 | 302/582 | A/G+G/G | - | - | - | - |
| RAD51 135 G/C | Year | Country | Ethnicity | Source | Genotyping | Total | G/G | G/C | C/C | G | C | PHWE |
| Cetinkunar, S., et al. | 2015 | Turkey | Caucasian | HB | PCR-RFLP | 71/86 | 39/21 | 21-Nov | Nov-54 | 99/53 | 43/119 | 0.001 |
| Nissar, S., et al. | 2014 | India | Asian | HB | PCR-RFLP | 100/120 | 25/60 | 56/25 | 19/35 | 106/145 | 94/95 | 0.001 |
| Gil, J., et al. | 2012 | Poland | Caucasian | HB | PCR-RFLP | 133/100 | 100/73 | 29/27 | 4/0 | 229/173 | 37/54 | 0.118 |
| Mucha, B., et al. | 2012 | Poland | Caucasian | PB | PCR-RFLP | 200/200 | 161/157 | 34/37 | 05-Jun | 356/351 | 44/49 | 0.048 |
| Romanowicz-Makowska, H., et al. | 2012 | Poland | Caucasian | HB | PCR-RFLP | 320/320 | 51/91 | 56/164 | 213/65 | 158/346 | 482/294 | 0.569 |
| Krupa, R., et al. | 2011 | Poland | Caucasian | HB | PCR-RFLP | 100/100 | 61/36 | 36/35 | Mar-29 | 158/107 | 42/93 | 0.003 |
HWE, Hardy-Weinberg equilibrium; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism;. PB, population-based; HB: hospital-based; FB, Family-based; NHS, Nurses’ Health Study; HPFS, Health Professionals Follow-up Study; APEX, arrayed primer extension
Statistical analyses
The risk of CRC associated with the SNPs were examined for each study by odds ratio (OR) and 95% confidence interval (95% CI). The significance of the summary OR was calculated by the Z-test, and P<0.05 was applied as statistically significant. Five different ORs were computed for XPG Asp1104His: the codominant homozygote (His/His vs. Asp/Asp), codominant heterozygote (Asp/His vs. Asp/Asp), dominant (Asp/His+His/His vs. Asp/Asp), recessive model (His/His vs. Asp/Asp+Asp/His), and allelic comparison (His vs Asp). As for XRCC2 rs3218536 A/G, we used codominant heterozygote (A/G vs. A/A), codominant homozygote (G/G vs. A/A), dominant (G/G + A/G vs. A/A), recessive (G/G vs. A/G+A/A) and allelic comparison (G vs A) to calculate the pooled ORs. For the RAD51 135G/C, the codominant heterozygote (G/C vs. G/G), codominant homozygote (C/C vs. G/G), dominant (C/C + G/C vs. G/G), recessive (C/C vs. G/C + G/G) and allelic comparison (C vs G) were chosen to compute the pooled ORs. A χ2-test-based Q statistic test was done to assess the between-study heterogeneity [24]. We also quantified the effect of heterogeneity by I2 test. Once a significant Q test (P > 0.05) or I2 < 50% indicated homogeneity across studies, the fixed effects model was utilized [25]; otherwise the random effects model was used [26]. Then, we performed stratification analyses on ethnicity (Asian, Caucasian or African) and source of control (Population-based or PB, Hospital-based or HB and family-based or FB). Analysis of sensitivity was performed to assess the stability of the results. Potential publication bias was examined using Begg’s funnel plot. All analyses were performed using the Cochrane Collaboration RevMan 5.3. HWE was calculated for each study using an internet-based HWE calculator (http://ihg.gsf.de/csgi-bin/hw/hwa1.pl).
Results
Characteristics of studies
After preliminary search with duplicates discarded, a total of 412 records of publications were yielded. Following the predefined inclusion and exclusion criteria, eventually 26 case-control studies were included in this meta-analysis (details in Figure 1). These 26 studies included a total of 22173 subjects (10,288 cases and 11,885 controls), and examined the impact of XPG Asp1104His, XRCC2 rs3218536 A/G and RAD51 135G/C polymorphisms on CRC risk. Fourteen studies comprising of 6,728 cases and 7,877 controls assessed the impact of XPG Asp1104His polymorphism on CRC (Mort et al., 2003; Bigler et al., 2005; Huang et al., 2006; Pardini, Naccarati et al. 2008, Joshi, Corral et al. 2009, Canbay et al., 2011; Gil et al., 2012; Liu et al., 2012; Du et al., 2014; Li et al., 2014; Steck et al., 2014; Kabzinski et al., 2015; Paszkowska-Szczur et al., 2015; Sun et al., 2015). OF these, 10 were Caucasians, 3 were Asians and one was African. After stratification of studies according to the source of control, 7 studies were stratified as PB and 6 were HB and one was a FB study.
Figure 1.
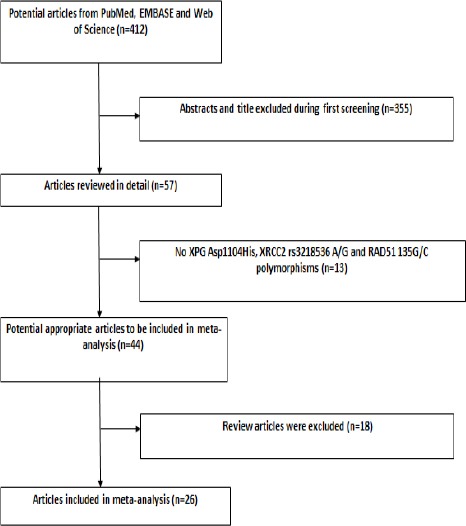
Flow Diagram of Included Studies for This Meta-Analysis
Six studies including 2620 cases and 3092 controls examined the association of XRCC2 rs3218536 A/G and CRC. OF these 6 studies, 3 were HB and 3 were PB studies, but as for ethnicity all were Caucasians. With respect to RAD51 135G/C polymorphism in CRC, 6 studies met the inclusion criteria which included 940 cases and 926 controls. Of these, 5 studies were Caucasians and 5 were HB studies. Baseline characteristics of the included studies for XPG Asp1104His, XRCC2 rs3218536 A/G and RAD51 135G/C polymorphisms on CRC are shown in Table 1.
XPG Asp1104His polymorphism and CRC
The associations of CRC risks with XPG Asp1104His polymorphism were indicated in table 2. At allelic level, the pooled analysis showed that the His vs Asp allele was associated with increased risk of CRC in total studies with the overall OR of 1.06 (OR= 1.06; 95% CI =1.01-1.12; P=0.02) as indicated in fig 2. The subgroup analysis indicated that the variant His allele is a risk factor for CRC in Asians (OR= 1.12; 95% CI = 1.04-1.21’ P=0.002). However, it was not associated with the CRC risk in either of Caucasians or Africans (P>0.05).
Table 2.
Main Results of Pooled ORs for the XPG Asp1104His, XRCC2 Rs3218536 A/G And RAD51 135G/C Polymorphisms in Colorectal Cancer
| Variables | Noa | Allele | Codominant (eterozygous) | Codominant (homozygous) | Dominant | Recessive |
|---|---|---|---|---|---|---|
| His vs Asp | His/Asp vs Asp/Asp | His/His vs Asp/Asp | His/Asp+His/His vs Asp/Asp | His/His vs His/Asp+Asp/Asp | ||
| XPG Asp1104His | OR (95 % CI) | OR (95 % CI) | OR (95 % CI) | OR (95 % CI) | OR (95 % CI) | |
| Total | 14 | 1.06(1.01-1.12) | 1.16(1.02-1.31) | 1.09(0.97-1.23) | 1.12(0.99-1.25) | 0.98(0.88-1.09) |
| P/Ph b/I2 (%) | 0.02/0.22/21 | 0.02/0.002/61 | 0.15/0.50/0 | 0.06/0.005/58 | 0.65/0.54/0 | |
| Ethnicity | ||||||
| Asian | 3 | 1.12(1.04-1.21) | 1.31(1.01-1.70) | 1.22(1.05-1.43) | 1.29(1.05-1.60) | 1.02(0.90-1.17) |
| P/Ph/I2 (%) | 0.002/0.38/0 | 0.04/0.01/78 | 0.01/0.69/0 | 0.02/0.04/69 | 0.73/37/0 | |
| Caucasian | 10 | 1.01(0.94-1.09) | 1.08(0.98-1.19) | 0.93(0.76-1.14) | 1.04(0.95-1.14) | 0.91(0.75-1.12) |
| P/Ph/I2 (%) | 0.70/0.32/13 | 0.10/0.06/47 | 0.50/0.65/0 | 0.35/0.11/39 | 0.37/0.50/0 | |
| Source of controls | ||||||
| Population-based | 7 | 1.04(0.97-1.11) | 1.14(0.93-1.39) | 1.01(0.85-1.20) | 1.01(0.76-1.34) | 0.88(0.75-1.02) |
| P/Ph/I2 (%) | 0.33/0.19/31 | 0.21/0.001/75 | 0.95/0.29/18 | 0.95/0.001/88 | 0.09/0.72/0 | |
| Hospital-based | 6 | 1.11(1.02-1.20) | 1.22(1.08-1.38) | 1.19(0.99-1.42) | 1.21(1.08-1.37) | 1.09(0.93-1.28) |
| P/Ph/I2 (%) | 0.01/0.31/16 | 0.001/0.29/20 | 0.06/0.46/0 | 0.001/0.42/0 | 0.28/0.55/0 | |
| XRCC2 rs3218536 A/G | ||||||
| Total | 6 | G vs A | AG vs AA | GG vs AA | AG+GG vs AA | GG vs AA+AG |
| OR (95 % CI) | 1.06 [0.90, 1.24] | 1.10 [0.92, 1.33] | 1.18 [0.69, 2.00] | 1.02 [0.89, 1.18] | 0.83 [0.54, 1.30] | |
| P/Ph b/I2 (%) | 0.49/0.06/60 | 0.29/0.67/0 | 0.54/0.35/9 | 0.76/0.51/0 | 0.42/0.07/57 | |
| Population-based | 3 | |||||
| OR (95 % CI) | 0.99 [0.82, 1.19] | 1.07 [0.86, 1.32] | 0.99 [0.51, 1.92] | 1.05 [0.86, 1.29] | 0.67 [0.40, 1.13] | |
| P/Ph/I2 (%) | 0.91/0.07/69 | 0.55/0.34/0 | 0.97/0.40/0 | 0.63/0.93/0 | 0.13/0.09/65 | |
| Hospital-based | 3 | |||||
| OR (95 % CI) | 1.28 [0.94, 1.76] | 1.23 [0.85, 1.78] | 1.60 [0.66, 3.89] | 1.27 [0.90, 1.80] | 1.54 [0.64, 3.75] | |
| P/Ph/I2 (%) | 0.12/0.13/57 | 0.28/0.64/0 | 0.30/0.15/52 | 0.18/0.30/7 | 0.34/0.16/50 | |
| RAD51 135 G/C | ||||||
| Total | 6 | C vs G | GC vs GG | CC vs GG | CC+GC vs GG | CC vs GC+GG |
| OR (95 % CI) | 1.21 [1.05, 1.39] | 0.98 [0.77, 1.24] | 1.28 [0.98, 1.67] | 1.06 [0.87, 1.31] | 1.62 [1.30, 2.02] | |
| P/Ph b/I2 (%) | 0.001/0.01/97 | 0.85/0.01/85 | 0.07/0.01/95 | 0.55/0.01/91 | 0.001/0.01/97 | |
, Number of comparisons;
, P value of Q test for heterogeneity test;
c, Random model was used when P value for heterogeneity test was below 0.05; otherwise, fixed model was used
Figure 2.
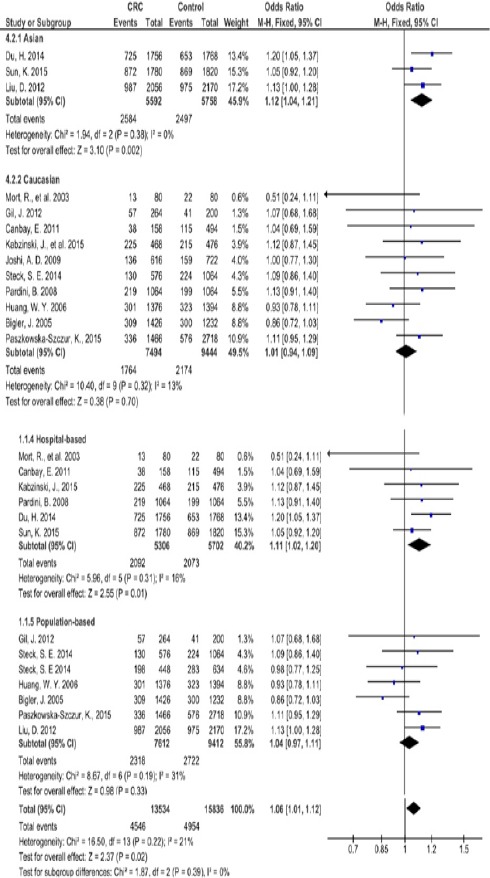
Forest Plot of the Risk of Colorectal Cancer Associated with XPG Asp1104His Polymorphism in Allele Comparison
At genotypic level and using the codominant model, the pooled evidence suggested that His/Asp vs Asp/Asp heterozygote genotype distribution between groups was different and the association was statistically significant in total with the pooled OR of 1.16 (95%CI= 1.02-1.31; P=0.02). Similarly, this genotype was a risk factor for CRC in Asians with the OR of 1.31 (95%CI= 1.01-1.70; P=0.04) but not in Caucasians or Africans (fig 3).
Figure 3.
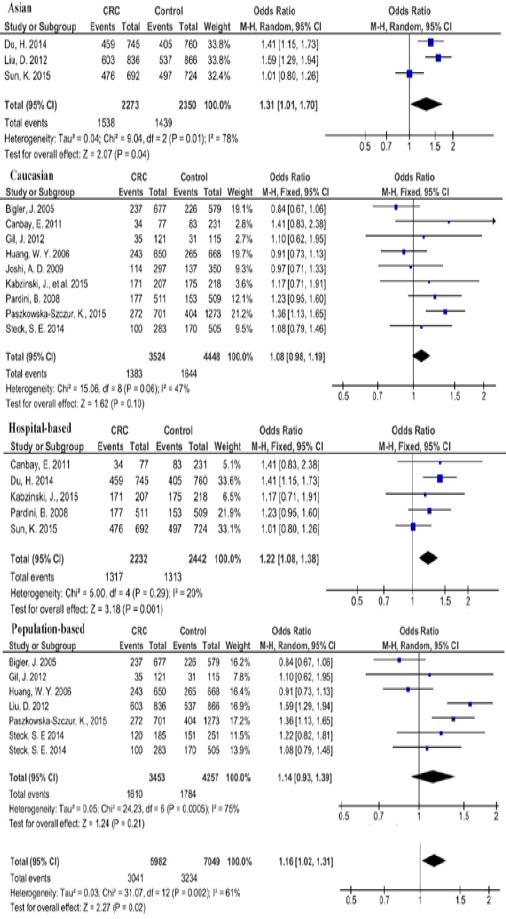
Forest Plot of the Risk of Colorectal Cancer Associated with XPG Asp1104His Polymorphism in Codominant (Heterozygote) Comparison
In contrast, the homozygote genotype His/His vs Asp/Asp in codominant model was not associated with CRC in total (P=0.15). However, the His/His genotype was a risk factor for Asians with OR of 1.22 (95%CI= 1.05-1.43; P=0.01).
In dominant model, the His/Asp+His/His vs Asp/Asp genotype was not correlated with susceptibility to CRC in total (P=0.33) as well as in Caucasians (P=0.35). In Asians, however, His/Asp+His/His was a risk factor for CRC with the pooled OR of 1.29 (P=0.02; 95%CI= 1.05-1.60).
In recessive model, the general difference between groups for His/His vs His/Asp+Asp/Asp was not associated with the risk of CRC either in total (P=0.54) or Caucasians (P=0.50) or Asians (P=0.73).
In the subgroup analysis by source of control, the XPG Asp1104His polymorphism had statistically significant association with elevated CRC risk under allele His vs Asp (P=0.01; OR=1.11, 95% CI=1.02-1.20), codominant heterozygote His/Asp vs Asp/Asp (P=0.001; OR=1.22, 95% CI=1.08-1.38) and dominant His/Asp+His/His vs Asp/Asp (P=0.001; OR=1.21, 95% CI=1.08-1.37) in the HB subgroup.
XRCC2 rs3218536 A/G polymorphism and CRC
As shown in table 2, no significant association was found between XRCC2 rs3218536 A/G polymorphism and CRC using different genetic models. In allelic comparison, the distribution of G vs A allele was not different between cases and controls (P=0.49) in total or in PB (P=0.91) or HB (P=0.12) subgroups. Similar results were found for the polymorphism in total using codominant heterozygote (P=0.29), homozygote (0.54), dominant (0.76) or recessive genetic model (P=0.83). After stratification based on source of controls, no significant association was found either in PB or HB subgroups using different genetic models (P>0.05). As for ethnicity, no stratification was done because all studies belonged to the Caucasian populations.
RAD51 135 G/C polymorphism and CRC
Our pooled evidence revealed that the RAD51 135 G/C polymorphism was a risk factor for CRC in total using allele or recessive models. At allelic level, the C vs G allele was associated with increased risk of CRC with the OR of 1.21 (P=0.001; 95%CI=1.05-1.39). Using the recessive genetic model, a significant relationship between CC vs GC+GG polymorphism and CRC was observed in total (P=0.001; OR=1.62; 95%CI=1.30-2.02). For this polymorphism, no stratification based on ethnicity or sources of controls was performed due to lack of enough data for subgroups.
Heterogeneity and sensitivity analyses
We found heterogeneity in the codominant model for XPG His/Asp genotype using codominant heterozygote in overall (Ph=0.002; I2=61%), and in Asians (Ph=0.01; I2=78%) as well as in PB subgroup (Ph=0.001; I2=75%). Similarly, a heterogeneity among total studies in dominant model for His/Asp+His/His genotype (Ph=0.005; I2=58%), as well as in Asians (Ph=0.04; I2=69%) and in PB subgroup (Ph=0.001; I2=88%). For the RAD51 135 G/C, a significant heterogeneity was observed for all genetic models (Ph<0.05; I2>50%); however, no heterogeneity was found for the XRCC2 rs3218536 A/G whether in total or PB/HB subgroups (Ph>0.05) as demonstrated in Table 2. Sensitivity analysis was performed according to heterogeneity. Due to significant heterogeneity across some studies, individual studies were sequentially omitted to identify the heterogeneity source by sensitivity analysis. The results showed that no individual study influenced the pooled OR values for XPG Asp1104His, XRCC2 rs3218536 A/G and RAD51 135G/C polymorphisms.
Publication bias
The funnel plots were used to evaluate the potential publication bias of included studies under each comparison model. The shape of the funnel plot did not reveal any obvious asymmetry for 3 studied polymorphisms.
Discussion
In this meta-analysis, we investigated the potential genetic association between XPG Asp1104His, XRCC2 rs3218536 A/G and RAD51 135G/C polymorphisms and CRC susceptibility. Using a meta-analytic approach, we synthesized 14 studies from 6 different countries for XPG Asp1104His variation including 6728 cases and 7877 controls. We found that XPG Asp1104His gene polymorphism was a risk factor for CRC in overall population in allele and codominant model. Besides, subgroup analysis stratified by ethnicity and source of control indicated that XPG Asp1104His polymorphism was associated with CRC susceptibility in Asians and HB subgroups.
The association between XPG Asp1104H is polymorphism and CRC has extensively been studied but the results have been inconsistent (Kiyohara and Yoshimasu, 2007; He et al., 2014). Our pooled evidence supports the findings of Du et al., (2014), Liu et al., (2012) and Paszkowska-Szczur et al., (2015). Du et al., (2014) found a significant increased CRC risk for the His vs Asp allele (or C vs G) (OR=1.20), and genotypes under the codominant (OR=1.41) and dominant models (OR of 1.39). They also performed a meta-analysis on the association of the SNP with CRC risk on five studies with a total of 2649 CRC cases and 2848 controls included. In their meta-analysis, the association between XPG rs17655 and CRC risk was replicated under the codominant (His/His: OR=1.24) and dominant model (His/His+Asp/His: OR = 1.35). Their finding for dominant model showed lack of relationship between rs17655 and CRC which does not support our pooled results for this model. Additionally, Paszkowska-Szczur K et al., (2015) reported that XPG Asp1104His heterozygote His/Asp genotype was a CRC risk factor in a polish population, and was associated with 1.36-fold higher risk of CRC supporting our pooled findings (OR=1.16).
The XPG Asp1104His (rs17655 G/C) gene variation is the most commonly studied XPG polymorphism located in the XPG C-terminus, which is essential for its interaction with other members of the NER pathway, such as XPB, XPD and TFIIH subunits. The XPG rs17655 G/C polymorphism causes the replacement of Asp amino acid to His which may influence these protein–protein interactions; however, no functional study has been reported to date. Despite lack of functional studies for XPG rs17655 G/C, this SNP has been reported to contribute to a poorer overall survival (OS) in patients with different cancers, e.g. gastric cancer (Li et al., 2014), cutaneous melanoma (Schrama et al., 2011), squamous cell carcinoma of the oropharynx (SCCOP) (Song et al., 2013) and CRC (Liuet al., 2012; Sun et al., 2015). In CRC, Liu et al., (2012) demonstrated that XPG Asp1104His variant genotypes under dominant and codominant (heterozygote) models were associated with increased risk of CRC.
With respect to RAD51 135G/C polymorphism our pooled revealed that this genetic variation is associated with increased risk of CRC using allele and recessive models. According to our findings, individuals carrying the C vs G variant or CC genotype vs GC+GG of RAD51 135G/C were predisposed to 1.21 or 1.62-fold increased risk of CRC, respectively. In line with our findings, Romanowicz-Makowska et al., (2012) indicated that the variant 135C allele of RAD51 increased the CRC risk in a polish population with the OR of 3.59. Additionally, a recent meta-analysis (Kong et al., 2015) on six studies suggested that RAD51 G135C is associated with increased head and neck cancer (HNC) risk in allele comparison (OR=1.21) which supports our findings (OR=1.21). Another comprehensive meta-analysis (Zhao et al., 2014) indicated that the RAD51 G135C significantly increased the risk of overall cancers using homozygote, recessive and allele models. However, they found no significant association between RAD51 and CRC in all models. A meta-analysis by Cheng et al., (2014) for RAD51 G135C on four types of common cancers revealed that there was no relationship between this variation and CRC risk. Concerning XRCC2 rs3218536 A/G polymorphism, we observed no association between this variation and the risk of total cancers or CRC using all models.
Some limitations of this meta-analysis should be acknowledged. First, a common limitation of meta-analysis was heterogeneity. In our study, there was a considerable heterogeneity of studies for the dominant and codominant models of the XPG rs17655 G/C polymorphism in the overall population. However, after performing the analyses by ethnicity and source of control, the heterogeneity disappeared in Caucasian and hospital-based groups. These results propose that the heterogeneity may somewhat result from ethnicity or lacking of adequate data, hence large studies with subgroup analysis are required. Moreover, considerable inherent heterogeneity existed among different studies for RAD51 135G/C, which was confirmed by significant statistical heterogeneity we obtained. However, we detected no significant heterogeneity when three case-control studies Romanowicz-Makowska et al., (2012), Cetinkunar et al., (2015) and Krupa et al., (2011) in Table 1) were excluded, which implied the likelihood of the removed studies being the origins of heterogeneity. Second, the small sample size in some subgroups reduced the statistical power to examine the association between XRCC2 rs3218536 A/G and RAD51 135G/C and CRC with great confidence, especially in the Asians or PB subgroups. Third, our meta-analysis synthesized only published literatures, considering the fact that some pertinent important but unpublished studies were missed. Thus despite of its limitation, our meta-analysis is valuable to be interpreted with caution.
In conclusion, our meta-analysis suggested that the XPG Asp1104His and RAD51 135 G/C polymorphisms were risk factors for the pathogenesis of CRC in overall population. Besides, subgroup analysis stratified by ethnicity and source of control indicated that XPG Asp1104His polymorphism was associated with CRC susceptibility both in Asians or HB population. Further well designed studies with larger sample size on different ethnic groups are needed to confirm the risk identified in our meta-analysis.
Declaration of interest
The authors declare that there is no conflict of interests.
Funding
This study received no funding from any organization.
Acknowledgments
A.R and EEN extracted the data, MH contributed to data analysis; EEN wrote the paper.
References
- Bigler J, Ulrich CM, Kawashima T, Whitton J, Potter JD. DNA repair polymorphisms and risk of colorectal adenomatous or hyperplastic polyps. Cancer Epidemiol Biomarkers Prev. 2005;14:2501–8. doi: 10.1158/1055-9965.EPI-05-0270. [DOI] [PubMed] [Google Scholar]
- Canbay E, Cakmakoglu B, Zeybek U, et al. Association of APE1 and hOGG1 polymorphisms with colorectal cancer risk in a Turkish population. Curr Med Res Opin. 2011;27:1295–1302. doi: 10.1185/03007995.2011.573544. [DOI] [PubMed] [Google Scholar]
- Cetinkunar S, Gok I, Celep RB, et al. The effect of polymorphism in DNA repair genes RAD51 and XRCC2 in colorectal cancer in Turkish population. Int J Clin Exp Med. 2015;8:2649–55. [PMC free article] [PubMed] [Google Scholar]
- Chen J, Xie F, Chen K, et al. ERCC5 promoter polymorphisms at -763 and +25 predict the response to oxaliplatin-based chemotherapy in patients with advanced colorectal cancer. Cancer Biol Ther. 2009;8:1424–30. doi: 10.4161/cbt.8.14.8889. [DOI] [PubMed] [Google Scholar]
- Chen YZ, Guo F, Sun HW, et al. Association between XPG polymorphisms and stomach cancer susceptibility in a Chinese population. J Cell Mol Med. 2016;20:903–8. doi: 10.1111/jcmm.12773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D, Shi H, Zhang K, Yi L, Zhen G. RAD51 Gene 135G/C polymorphism and the risk of four types of common cancers: a meta-analysis. Diagn Pathol. 2014;9:18. doi: 10.1186/1746-1596-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin K, Lin WY, George R, et al. Genetic variants in XRCC2: new insights into colorectal cancer tumorigenesis. Cancer Epidemiol Biomarkers Prev. 2009;18:2476–84. doi: 10.1158/1055-9965.EPI-09-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Zhang X, Duet M, et al. Association study between XPG Asp1104His polymorphism and colorectal cancer risk in a Chinese population. Sci Rep. 2014;4:6700. doi: 10.1038/srep06700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworaczek H, Xiao W. Xeroderma pigmentosum: a glimpse into nucleotide excision repair, genetic instability, and cancer. Crit Rev Oncog. 2007;13:159–77. doi: 10.1615/critrevoncog.v13.i2.20. [DOI] [PubMed] [Google Scholar]
- Gil J, Ramsey D, Stembalska A, et al. The C/A polymorphism in intron 11 of the XPC gene plays a crucial role in the modulation of an individual's susceptibility to sporadic colorectal cancer. Mol Biol Rep. 2012;39:527–34. doi: 10.1007/s11033-011-0767-5. [DOI] [PubMed] [Google Scholar]
- Griffin CS. Aneuploidy, centrosome activity and chromosome instability in cells deficient in homologous recombination repair. Mutat Res. 2002;504:149–55. doi: 10.1016/s0027-5107(02)00088-x. [DOI] [PubMed] [Google Scholar]
- Griffin CS, Thacker J. The role of homologous recombination repair in the formation of chromosome aberrations. Cytogenet Genome Res. 2004;104:21–7. doi: 10.1159/000077462. [DOI] [PubMed] [Google Scholar]
- He XF, Liu LR, Wei W, et al. Association between the XPG Asp1104His and XPF Arg415Gln polymorphisms and risk of cancer: a meta-analysis. PLoS One. 2014;9:e88490. doi: 10.1371/journal.pone.0088490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou R, Liu Y, Feng Y, et al. Association of single nucleotide polymorphisms of ERCC1 and XPF with colorectal cancer risk and interaction with tobacco use. Gene. 2014;548:1–5. doi: 10.1016/j.gene.2014.05.025. [DOI] [PubMed] [Google Scholar]
- Huang WY, Berndt SI, Kang D, et al. Nucleotide excision repair gene polymorphisms and risk of advanced colorectal adenoma: XPC polymorphisms modify smoking-related risk. Cancer Epidemiol Biomarkers Prev. 2006;15:306–311. doi: 10.1158/1055-9965.EPI-05-0751. [DOI] [PubMed] [Google Scholar]
- Johnson RD, Liu N, Jasin M. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature. 1999;401:397–99. doi: 10.1038/43932. [DOI] [PubMed] [Google Scholar]
- Joshi AD, Corral R, Siegmund KD, et al. Red meat and poultry intake, polymorphisms in the nucleotide excision repair and mismatch repair pathways and colorectal cancer risk. Carcinogenesis. 2009;30:472–9. doi: 10.1093/carcin/bgn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabzinski J, Przybylowska K, Dziki L, Dziki A, Majsterek I. An association of selected ERCC2 and ERCC5 genes polymorphisms, the level of oxidative DNA damage and its repair efficiency with a risk of colorectal cancer in Polish population. Cancer Biomark. 2015;15:413–23. doi: 10.3233/CBM-150488. [DOI] [PubMed] [Google Scholar]
- Kiyohara C, Yoshimasu K. Genetic polymorphisms in the nucleotide excision repair pathway and lung cancer risk: a meta-analysis. Int J Med Sc. 2007;4:59–71. doi: 10.7150/ijms.4.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F, Wu J, Hu L, Du Y, Pan Y. Association between RAD51 polymorphisms and susceptibility of head and neck cancer: a meta-analysis. Int J Clin Exp Med. 2015;8:6412–19. [PMC free article] [PubMed] [Google Scholar]
- Krupa R, Sliwinski T, Wisniewska-Jarosinska M, et al. Polymorphisms in RAD51, XRCC2 and XRCC3 genes of the homologous recombination repair in colorectal cancer-a case control study. Mol Biol Rep. 2011;38:2849–54. doi: 10.1007/s11033-010-0430-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkofsky HB, Veloso A, Ljungman M. Transcriptional and post-transcriptional regulation of nucleotide excision repair genes in human cells. Mutat Res. 2015;776:9–15. doi: 10.1016/j.mrfmmm.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XB, Luo H, Huang J, et al. XRCC2 gene polymorphisms and its protein are associated with colorectal cancer susceptibility in Chinese Han population. Med Oncol. 2014;31:245. doi: 10.1007/s12032-014-0245-8. [DOI] [PubMed] [Google Scholar]
- Li Y, Liu Z, Liu H, et al. Potentially functional variants in the core nucleotide excision repair genes predict survival in Japanese gastric cancer patients. Carcinogenesis. 2014;35:2031–8. doi: 10.1093/carcin/bgu142. [DOI] [PubMed] [Google Scholar]
- Liu D, Wu HZ, Zhang YN, et al. DNA repair genes XPC, XPG polymorphisms: relation to the risk of colorectal carcinoma and therapeutic outcome with Oxaliplatin-based adjuvant chemotherapy. Mol Carcinog. 2012;51:83–93. doi: 10.1002/mc.21862. [DOI] [PubMed] [Google Scholar]
- Luo JF, Yan RC, Zou L. XPG Asp1104His polymorphism and gastrointestinal cancers risk: a meta-analysis. Int J Clin Exp Med. 2014;7:4174–82. [PMC free article] [PubMed] [Google Scholar]
- Mao D, Zhang Y, Lu H, Fu X. Association between X-ray repair cross-complementing group 1 Arg194Trp polymorphism and colorectal cancer risk. Tumour Biol. 2013;34:2529–38. doi: 10.1007/s13277-013-0760-9. [DOI] [PubMed] [Google Scholar]
- Mort R, Mo L, McEwan C, Melton DW. Lack of involvement of nucleotide excision repair gene polymorphisms in colorectal cancer. Br J Cancer. 2003;89:333–7. doi: 10.1038/sj.bjc.6601061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissar S, Baba SM, Akhtar T, et al. RAD51 G135C gene polymorphism and risk of colorectal cancer in Kashmir. Eur J Cancer Prev. 2014;23:264–68. doi: 10.1097/CEJ.0000000000000049. [DOI] [PubMed] [Google Scholar]
- Pardini B, Naccarati A, Novotny B, et al. DNA repair genetic polymorphisms and risk of colorectal cancer in the Czech Republic. Mutat Res. 2008;638:146–53. doi: 10.1016/j.mrfmmm.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Paszkowska-Szczur K, Scott RJ, Gorski B, et al. “Polymorphisms in nucleotide excision repair genes and susceptibility to colorectal cancer in the Polish population. Mol Biol Rep. 2015;42:755–64. doi: 10.1007/s11033-014-3824-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowicz-Makowska H, Samulak D, Michalska M, et al. RAD51 gene polymorphisms and sporadic colorectal cancer risk in Poland. Pol J Pathol. 2012;63:193–8. doi: 10.5114/pjp.2012.31505. [DOI] [PubMed] [Google Scholar]
- Schrama D, Scherer D, Schneider M, et al. ERCC5 p.Asp1104His and ERCC2 p.Lys751Gln polymorphisms are independent prognostic factors for the clinical course of melanoma. J Invest Dermatol. 2011;131:1280–90. doi: 10.1038/jid.2011.35. [DOI] [PubMed] [Google Scholar]
- Sollier J, Stork CT, Garcia-Rubio ML, et al. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell. 2014;56:777–85. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Sturgis EM, Jin L, et al. Variants in nucleotide excision repair core genes and susceptibility to recurrence of squamous cell carcinoma of the oropharynx. Int J Cancer. 2013;133:695–704. doi: 10.1002/ijc.28051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivak G. Nucleotide excision repair in humans. DNA Repair (Amst) 2015;36:13–8. doi: 10.1016/j.dnarep.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steck SE, Butler LM, Keku T, et al. Nucleotide excision repair gene polymorphisms, meat intake and colon cancer risk. Mutat Res. 2014;762:24–31. doi: 10.1016/j.mrfmmm.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K. Molecular mechanisms of DNA damage recognition for mammalian nucleotide excision repair. DNA Repair (Amst) 2016;44:110–7. doi: 10.1016/j.dnarep.2016.05.015. [DOI] [PubMed] [Google Scholar]
- Sun K, Gong A, Liang P. Predictive impact of genetic polymorphisms in DNA repair genes on susceptibility and therapeutic outcomes to colorectal cancer patients. Tumour Biol. 2015;36:1549–59. doi: 10.1007/s13277-014-2721-3. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Wei L, Wang YJ, Liu C. Genetic association between ERCC5 rs17655 polymorphism and colorectal cancer risk: Evidencebased on a meta-analysis. Asian Pac J Cancer Prev. 2015;16:5565–71. doi: 10.7314/apjcp.2015.16.13.5565. [DOI] [PubMed] [Google Scholar]
- Zhang Y B, He S, Pan YQ, et al. Association of OGG1 Ser326Cys polymorphism with colorectal cancer risk: a meta-analysis. Int J Colorectal Dis. 2011;26:1525–30. doi: 10.1007/s00384-011-1258-9. [DOI] [PubMed] [Google Scholar]
- Zhao M, Chen P, Dong Y, Zhu X, Zhang X. Relationship between Rad51 G135C and G172T variants and the susceptibility to cancer: a meta-analysis involving 54 case-control studies. PLoS One. 2014;9:e87259. doi: 10.1371/journal.pone.0087259. [DOI] [PMC free article] [PubMed] [Google Scholar]


