Abstract
Objective
Micro RNAs (miRNAs) are a large family of short (∼21-nucleotide) non-coding mRNAs that repress gene expression through degradation of target mRNAs and/or inhibition of their translation. In the mouse, miRNAs play a key role in cumulus cell function and miR-21 is implicated in the regulation of cumulus cell viability. In this study, we asked whether miRNA expression is associated with the number of oocytes retrieved in women undergoing IVF and aimed to identify candidate miRNAs that may play a role in human cumulus cell function.
Design
Experimental study.
Materials and Methods
Pooled cumulus cells were collected from 189 consecutive women undergoing in vitro fertilization-intracytoplasmic sperm injection (IVF-ICSI). Poor responders were identified as patients who produced fewer oocytes than the 25th percentile of their respective age group. MiRNAs were extracted from cumulus cells and miRNA microarray was performed comparing normo-responders (n=3) to poor responders (n=3). Data were analyzed using Partek Genomics Suite software and MATLAB. Expression of miR-21-5p (active strand of miR-21) and miR-21-3p was tested in poor responders (n=21) and non-poor responders (n=29) using quantitative real time polymerase chain reaction (qRT-PCR). Regulation of miR-21-5p and miR-21-3p in KGN cells by estradiol was tested in vitro.
Results
MiRNA microarray analysis showed up-regulation of 16 miRNAs and down-regulation of 88 miRNAs in poor responders compared to the non-poor responders. Notably, miR-21 was significantly up-regulated 5-fold in poor responder samples (p=0.03). qRT-PCR analysis confirmed that miR-21-5p expression was significantly upregulated in poor responder patients (p=0.04), while miR-21-3p expression was significantly lower (p=0.003), suggesting that elevated miR-21-5p expression in cumulus cells is not regulated at the pre-miR-21 level in poor responders. Lastly, we found that both miR-21-5p and miR-21-3p are increased in KGN cells in response to higher doses of estradiol (p<0.05), while their expression is not affected at lower estradiol concentrations.
Conclusion
We found that poor response to IVF is associated with altered miRNA expression in cumulus cells, specifically with elevated expression of miR-21-5p, and that this elevated expression is independent of lower serum estradiol levels in poor responders. Whether miR-21 plays a role in human cumulus cell function and whether miRNA expression in cumulus cells may be used as a biomarker for oocyte or follicular viability remains to be investigated.
Keywords: IVF, miRNA, miR-21, poor ovarian response, cumulus cells
Introduction
The process of oocyte maturation and ovulation is a tightly regulated event requiring the rapid yet highly ordered phenotypic transformation of multiple cell types within the developing follicle. A thorough understanding of this process is particularly important for the successful treatment of infertility, which may require controlled ovarian hyperstimulation (COH) and in-vitro fertilization (IVF). The goal of COH is the recruitment of multiple follicles that may yield fertilizable oocytes of good quality, thus allowing for optimal early embryo development and successful embryo transfer [1]. Individual ovarian response to COH varies widely, ranging from poor to high response to gonadotropins [2]. A poor response to ovarian stimulation typically results in a reduced number of retrieved oocytes [3].
There exists a great diversity of opinions as to the exact definition of a poor ovarian response (POR). In a consensus paper from the European Society of Human Reproduction and Embryology (ESHRE), 24 previous definitions were cited, with the authors concluding that at least two of the following three features must be present: 1) advanced maternal age (≥40 years) or any other risk factor for POR, 2) a previous POR (defined as ≤ 3 oocytes using a conventional stimulation protocol), or 3) an abnormal ovarian reserve test (i.e. antral follicle count <5–7 follicles or anti-mullerian hormone <0.5–1.1 ng/ml). Alternatively, two episodes of POR after maximal stimulation were considered sufficient to define a patient as POR in the absence of advanced maternal age or abnormal ovarian reserve testing [3]. An alternative classification defines poor responders as those patients who produce a lesser number of oocytes than 25% of their respective age group [2].
In any case, the molecular mechanisms responsible for a so-called “poor response” to COH are still largely unknown. A number of germline-specific transcriptional changes have been identified during the process of oocyte maturation [4,5], and we are just beginning to understand the mechanisms by which posttranscriptional alterations in gene expression can also facilitate oocyte development. One process by which such alterations can occur is via changes in miRNA expression, and gonadal-selective miRNAs may play important roles in ovarian development and female fertility.
Micro RNAs (miRNAs) are a large family of short non-coding RNAs that repress expression of target genes via degradation of target mRNAs or inhibition of their translation by interactions with the 3′-untranslated regions (3′-UTRs). MiRNAs regulate essential cellular processes including growth, differentiation, and apoptosis and are thus required for normal mammalian development [6], and their function is conserved across evolution from yeast to mammals [7–11].
MiRNAs are processed from precursor molecules (which correspond either to transcripts of independent miRNA genes or the introns of protein coding mRNAs) and subsequently matured in a two-step process. First, in the nucleus, Drosha catalyzes the processing of pri-miRNAs to ~70-nucleotide pre-miRNAs; then, these pre-miRNAs are transported into the cytoplasm, where they are cleaved by the ribonuclease III endonuclease Dicer. One strand of the resulting ~21-bp miRNA duplex is then preferentially incorporated into the RNA-induced silencing complex (RISC). These single-stranded miRNAs harbor a seven-nucleotide seed sequence which mediates suppression of target transcripts by binding to partially complementary sequences within the 3′-UTR [6].
MiRNAs appear to play a role in granulosa cell (GC) function and thus may be important for follicular signaling and oocyte development. Fiedler et al [12] and Carletti et al [13] demonstrated that 212 known miRNAs are expressed in mouse mural GCs. Of these miRNAs, 13 were regulated by the LH surge, with three in particular (miR-21, miR-132, and miR-212) being highly upregulated by the LH surge [12,13]. Additionally, miR-21 increases in vivo in response to hCG and has been implicated as an antiapoptotic factor in mouse granulosa cells [13]. Disruption of miRNA processing also appears to result in altered ovarian morphology and gene expression. Lei et al [14] generated a granulosa cell-specific Dicer knockout and observed accelerated early follicular recruitment and increased degeneration, as well as alterations in genes involved in follicular development such as Amh1, Cyp17a1, and Cyp19a1. The above studies lend weight to the potential role for miRNAs in follicular development and function.
As miRNAs appear to be important mediators of differentiation, proliferation, and apoptotic events in granulosa cells, we hypothesized that an altered response to gonadotropin stimulation in women with infertility may be associated with altered miRNA expression in somatic follicular cells. We therefore sought to determine whether expression of miRNAs in cumulus cells is altered in women with infertility who demonstrate poor response to COH-IVF.
Materials and Methods
Stimulation protocols and collection of cumulus cells
Pooled cumulus cells were collected from a total of 189 consecutive cycles in women undergoing infertility treatment with IVF-ICSI at Gazi University School of Medicine IVF Center (Ankara, Turkey) between 1/22/2010 and 10/31/2011. Only one cycle from each patient was included in the study. Causes of infertility included diminished ovarian reserve, male factor, tubal factor, anovulation, endometriosis, and unexplained infertility, or a combination of these factors. This study was approved by the Gazi University Institutional Review Board committee (HIC protocol: 131/11.05.2011).
For patients undergoing agonist cycles, treatment was initiated by pituitary suppression with GnRH agonists during the luteal phase of the preceding cycle. Stimulation with gonadotropins was initiated only after downregulation had been achieved (estradiol level <50 pg/ml in the absence of ovarian cysts on transvaginal sonography). For patients undergoing antagonist cycle, treatment with gonadotropins was initiated on cycle day 3 when serum progesterone was <1 ng/ml and transvaginal sonography confirmed absence of ovarian cysts. A GnRH antagonist was added for pituitary suppression after five days of gonadotropin stimulation. Stimulation protocols included 150–300 IU/day of gonadotropins, either recombinant (GonalF, Merck Serono, Darmstadt, Germany) or in combination with human menopausal gonadotropin (hMG, Menopur, Ferrring Pharmaceuticals, Saint-Prex, Switzerland). Patients received hCG (Ovidrel 250 ug, Merck Serono) when two or more follicles > 18mm in diameter were present with an adequate estradiol response. Oocytes were collected 36 hours after hCG injection.
Percentiles of the number of retrieved oocytes were determined separately for four age groups of study population (<35, 35–37, 38–40 and >40). The number of oocytes included in the first quartile, which correspond to percentile values lower than 25%, was considered as indication of poor response to controlled ovarian stimulation.
Retrieved cumulus-oocyte complexes (COCs) were placed in culture medium (G-MOPS, VitroLife) and cumulus cells were dissected from the oocyte mechanically in the absence of hyaluronidase. Cumulus cells from each patient were pooled in a single eppendorf tube. Samples were washed twice with 0.5 ml 1X Phosphate Buffered Saline (PBS) and centrifuged at 1000 × g for 1 minute with the supernatant removed after each wash. After the final wash, the cell pellet was resuspended in 50 μl SideStep™ Lysis and Stabilization Buffer (Stratagene, La Jolla, CA). Samples were stored at −80°C until analysis.
miRNA extraction and microarray analysis
To purify miRNAs from cumulus or KGN cells, Qiagen miRNeasy Mini Kit (cat #217004) and RNeasy MinElute Cleanup Kit (cat # 74204) were used according to the manufacturer’s instructions. MiRNAs extracted from pooled cumulus cell samples of 3 poor and 3 non-poor responder patients were sent to Keck Biotechnology Resource Laboratory at Yale University (New Haven, CT, USA) for microarray analysis using Affymetrix GeneChip miRNA 2.0 Arrays. Sample labeling and hybridization was performed per manufacturer’s instructions. The probe intensities were extracted from microarray scan images and raw data were uploaded to Matlab programming environment (R2011b; The MathWorks, Natick, MA) in .CEL format for background correction, normalization and probe set summarization according to Robust Multichip Average (RMA) algorithm [15]. The relative expressions of miRNAs in cumulus cells were compared across poor and non-poor responders. Differential expression was evaluated in terms of both statistical significance and biological significance. Probesets with a fold change of >|1.5| and a p-value of <0.05 were considered differentially expressed.
Reverse Transcription (RT) and real time polymerase chain reaction (qPCR) for miR-21
To synthesize cDNA, RT master mix was prepared using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Grand Island, NY) according to manufacturer protocol. In brief, 5 μl of isolated total small RNAs was combined with 7 μl RT master mix including RT buffer, dNTPs, MultiScribe reverse transcriptase and RNase inhibitor. RT primer specific for each microRNA was added to a total volume of 15 μl. The reaction was incubated on ice for 5 minutes, followed by reverse transcription in the C1000 Touch Thermal Cycler (Bio-Rad, Hercules, CA) thermal cycler with the following parameters: 16°C/30 min; 42°C/30 min; 85°C/5 min.
Real time PCR (qPCR) was performed using the TaqMan MicroRNA Assay Kits for miR-21-3p, miR-21-5p, let7-f, miR150, and RNU-43 (Applied Biosystems) according to the manufacturer protocol. In brief, 10 μl of TaqMan 2×Universal PCR Master Mix was combined with 7.67 μl of nuclease free water, 1 μl of 20× TaqMan MicroRNA Assay mix, and 1.33 μl of RT reaction in the PCR reaction tube. Amplification was carried out using 40 cycles of PCR in CFX96 Touch Real-Time PCR Detection System (Bio-Rad), with the following program: initial denaturation 95°C/10 min followed by 40 cycles of 95°C/15 sec, 60°C/60 sec. All reactions were run in triplicate. Expression of the target microRNAs was normalized to RNU43 levels, and 2−Δ ΔCt method was used to calculate relative expression levels. Results were reported as a fold change in miR21-5p and -3p gene expression between groups.
In vitro studies
To determine the effect of estradiol on miRNA expression, we performed in vitro experiments using the human granulosa-like tumor cell line (KGN). KGN cells were cultured in αMEM (1:1v/v; Sigma-Aldrich) containing 10% fetal bovine serum (FBS, 10% v/v; Gibco/BRL, Rockville, MD), penicillin (100U/ml), and streptomycin (200μg/ml), in a standard 95% air: 5% CO2 incubator at 37°C. When they reached confluence, cells were passaged with Trypsin-EDTA (0.25%, Sigma-Aldrich), and incubated in serum-free, phenol red-free media (Sigma-Aldrich) for 24h prior to treatment with vehicle, estradiol (E2 10−8 M or 10−7 M, Sigma-Aldrich), for 6h. Following treatments, media were removed, plates were rinsed in PBS and stored at −80°C until analyzed. To measure expression of miRNAs, cells were lysed with QIAzol Lysis Reagent (Qiagen) and miRNA extraction (for miR-21-5p and miR-21-3p) and qRT-PCR was performed as described in the previous section.
Statistical analysis
Statistical analyses were performed using Matlab (R2011b; The MathWorks, Natick, MA) considering two groups of patients, poor responders and non-poor responders, as described above. The significance of the differences between the clinical parameters and qRT-PCR results among the poor and non-poor responder groups was assessed using ANOVA or t-test, when appropriate. Alpha error of <0.05 was considered significant for the comparisons.
Results
Clinical characteristics
The clinical characteristics of the study population are represented in Table 1 by age group and response levels in terms of number of cycles, number of oocytes, the amount of administered FSH and the E2 value on the day of HCG injection (Table 1). A cut-off value for the number of retrieved oocytes was calculated for each age group to define poor ovarian response. These values correspond to first quartile points and were determined as 8, 6, 4 and 3 for age groups of <35, 35–37, 38–40 and >40, respectively. The patients having oocytes less than the cut-off values were classified as poor responders.
Table 1.
Clinical characteristics of study population represented by age groups and ovarian response levels.
| Age group | <35 | 35–37 | 38–40 | >40 | ||||
|---|---|---|---|---|---|---|---|---|
| Response level | Poor | Non-poor | Poor | Non-poor | Poor | Non-poor | Poor | Non-poor |
| Number of cyclesa | 37 | 98 | 7 | 17 | 5 | 14 | 2 | 9 |
| Number of retrieved oocytes*b | 5.8 ± 0.2 | 16.8 ± 0.5 | 4.4 ± 0.4 | 13.9 ± 0.9 | 2.6 ± 0.3 | 11.1 ± 1.2 | 1.5 ± 0.2 | 9.0 ± 1.6 |
| Total FSH administered* | 2984 ± 128 | 2694 ± 97 | 3439 ± 391 | 2827 ± 194 | 3660 ± 501 | 3130 ± 278 | 4000 ± 0 | 3094 ± 425 |
| E2 value on the day of HCG* | 1598 ± 161† | 2205 ± 122† | 1017 ± 176 | 2222 ± 533 | 1076 ± 170 | 1643 ± 355 | 750 ± 249 | 1735 ± 608 |
Numerical values are in the form of mean ± SEM.
Only one cycle from each patient was included in the study.
The number of retrieved oocytes is significantly different among the poor and non-poor responder patients within the <35, 35–37 and >40 age groups. The difference between the oocyte numbers was not significant in >40 age group (p = 0.063).
Significantly different from each other (p < 0.01).
There was no statistical difference in the amount of total gonadotropin used for COH for poor responders, compared to non-poor responders in any of the age groups, although there was a trend toward higher doses for poor responders (Table 1). Serum estradiol levels on the day of HCG injection were lower in poor responders, however, the difference was only statistically significant in the <35 year old age group (Table 1). As the response levels were defined according to the oocyte numbers, the number of retrieved oocytes was also significantly different between the poor and non-poor responder patients within <35, 35–37 and 38–40 age groups. Surprisingly, the difference was not significant in >40 age group probably due to low number of samples with a high variation of oocyte numbers in poor and non-poor responders.
Poor ovarian response is associated with differential miRNA expression in cumulus cells
We first determined whether poor responders differ from non-poor responders in the cumulus cell miRNA expression. MiRNAs were extracted from cumulus cell samples of 6 individual patients including 3 poor and 3 non-poor responders and analyzed using Affymetrix GeneChip miRNA 2.0 Arrays. Table 2 represents clinical characteristics of the patients whose cumulus cell samples were used for microarray analysis. We applied Principal Component Analysis (PCA) to the expression values obtained from microarray analysis of cumulus cells. Samples from both poor and non-poor responder groups are illustrated in Figure 1 in three-dimensional plot across the first three principal components accounting for 88.2% of the total variance in the data. PCA-plot shows a separation between the miRNA expression profiles of poor and non-poor responder groups and high within-group variations (Figure 1). The variation of overall miRNA expressions between poor responder samples is higher compared to the non-poor responder samples. Differential expression analysis revealed up-regulation of 16 miRNAs (Supplemental Table 1) and down-regulation of 88 miRNAs (Supplemental Table 2) in poor responders compared to the non-poor responders. The statistical and biological significance of the differences of expression values between groups are also shown as a volcano plot (Supplemental Figure 1). Notably, miR-21 was significantly up-regulated 5-fold in poor responder samples (p=0.03) with the highest fold change among the all up-regulated miRNAs.
Table 2.
Clinical characteristics of the patients used for microarray analysis.
| Reason for IVF | Age | Number of retrieved oocytes | Total FSH administered (IU) | E2 value on the day of hCG (pg/ml) | |
|---|---|---|---|---|---|
| Poor responders | AMA | 42 | 2 | 4000 | 502 |
| UNEXP. | 24 | 7 | 3500 | 1100 | |
| AMA | 43 | 3 | 4500 | 200 | |
| Non-Poor responders | MALE | 25 | 23 | 2000 | 1800 |
| GENETIC | 33 | 21 | 2025 | 895 | |
| UNEXP. | 29 | 22 | 4000 | 1870 |
AMA, advanced maternal age; UNEXP., unexplained infertility.
Figure 1.
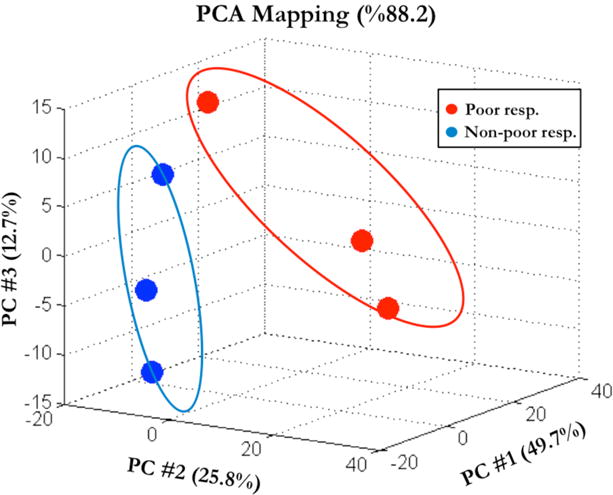
Principal component analysis (PCA) of miRNA expression in cumulus cells of poor responder (red circles) and non-poor responder samples (blue circles). The percentages of variability represented by the first three principal components are displayed across PC#1, 2 and 3 on X, Y and Z axes, respectively.
miR-21 5p is elevated in the cumulus cells of poor responders
Microarray analysis identified miR-21 as a miRNA that is significantly elevated in cumulus cells of women with poor response. As miR-21 was previously implicated as a regulator of granulosa cell function in mouse, we further investigated miR-21 in human cells. First, we tested cumulus cell samples from poor (n=21) and non-poor (n=29) responders for miR-21 5p (active strand of miR-21) expression using qRT-PCR. We found miR-21-5p expression to be significantly upregulated in poor responder patients (p<0.05) (Figure 2).
Figure 2.
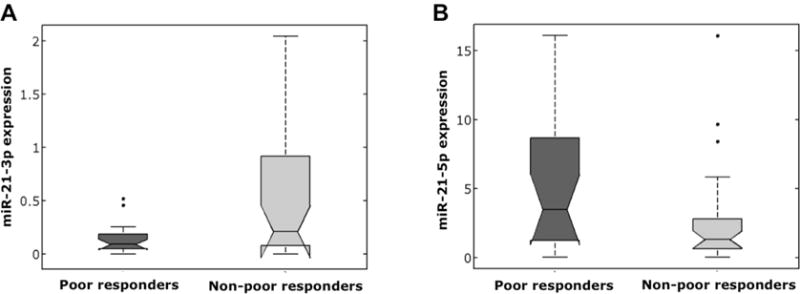
The distributions of miR-21 expressions according to ovarian response are shown as box plots. (A) miR-21 3p expression is significantly lower in poor responder group (p<0.01). (B) miR-21 5p expression is significantly higher in poor responder group (p <0.05)).
The stem–loop precursor of miR-21 (pre-miR-21) contains both MiR-21-5p (the active form implicated for most biological functions, with a sequence 5′-uagcuuaucagacugauguuga-3′), and miR-21-3p (the complementary sequence in the closed loop configuration with the sequence 5′-aacaccagucgaugggcugu-3′) [16,17]. We therefore asked whether the altered miR-21 expression affects the pre-miR-21 with resulting parallel changes in cumulus cell miR-21-5p and miR-21-3p levels, or whether the effect is specific for miR-21-5p. Using qRT-PCR in the same samples, we found miR-21-3p expression to be significantly lower in poor responders (p=0.003) (Figure 2), suggesting that elevated miR-21-5p expression in cumulus cells is not regulated at the pre-miR-21 level in poor responders.
To validate our microarray results, we also tested two additional miRNAs, let7-f and miR150. Consistent with microarray findings, miR150 was significantly downregulated in poor responders (p<0.05) and let7-f was significantly upregulated (p<0.05) (Figure 3).
Figure 3.
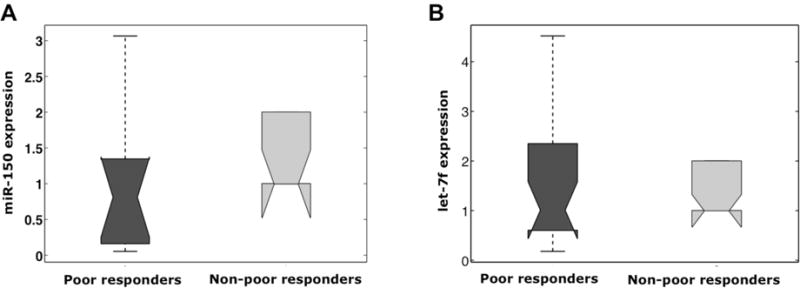
The distribution of miR-150 and let7-f expression according to ovarian response are shown as box plots. (A) miR-150 expression is significantly lower in poor responder group (p<0.05). (B) let7-f expression is significantly higher in poor responder group (p<0.05)
Altered miR-21 expression in poor responders is not associated with maternal age in the assessed patient population
Clinical characteristics of the patients used for qPCR validation of miR-21 expression are given in Table 3. There is no significant difference in the age, total FSH administered and E2 value on the day of hCG between the poor and non-poor responder groups. The only difference was observed in the number of retrieved oocytes as the separation of poor and non-poor responders was based on the oocyte numbers in the entire study population.
Table 3.
Clinical characteristics of the poor responder and non-poor responder patients used for qPCR validation of mir21 expression.
| Patient group | Number of cycles | Age* | Number of retrieved oocytes* | Total FSH administered* | E2 value on the day of HCG* |
|---|---|---|---|---|---|
| Poor responders | 21 | 33.1 ± 1.1 | 3.6 ± 0.4a | 3359 ± 218 | 1254 ± 188 |
| Non-poor responders | 29 | 33.1 ± 1.4 | 15.5 ± 1.2a | 2976 ± 193 | 1733 ± 210 |
Numerical values are in the form of mean ± SEM.
The number of retrieved oocytes is significantly different among the poor and non-poor responder groups (p << 0.01).
In the PCA graph (Figure 1), one of each of the poor and non-poor samples seems slightly different from the other two samples of the same group. The samples that are located relatively apart in the PC space belong to 24 year-old poor responder and 25 year-old non-poor responder patients (Table 2). Since both patients are the youngest of the three patients in their groups, we investigated the association between miR-21 expression and maternal age. The entire qPCR cohort was divided into two age groups as patients younger than 35 (n = 30) and patients 35 or older (n = 20). miR-21 5p and 3p expression were compared among the age groups and no significant difference was found indicating lack of relationship between age and mir21 expression in the study population (Supplemental Figure 2).
Elevated miR-21 expression in poor responders may not be a result of decreased serum estradiol levels in these women
As women with poor response had lower serum estradiol levels, and miR-21expression has previously been shown to be down-regulated by estradiol in MCF-7 cells, we tested whether estradiol affects miR-21 5p or miR-21 3p expression in KGN cells in culture. We found that both miR-21-5p and miR-21-3p are increased in KGN cells in response to higher doses of estradiol (p<0.05), while their expression is not affected at lower estradiol concentrations (Figure 4). While it is not possible to state conclusively that KGN cells and cumulus cells will respond similarly to estradiol, the cell line provides a useful mechanism for studying patterns of miRNA regulation, and these findings suggest that the elevated miR-21-5p expression in cumulus cells of poor responders may be independent of lower serum estradiol levels in these women.
Figure 4.
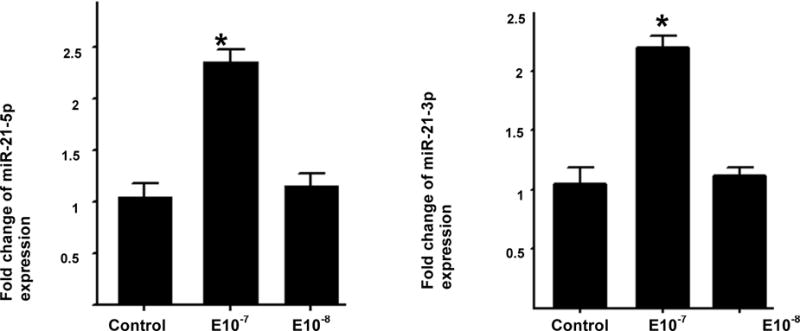
MiR-21 expression in KGN cells in response to estradiol treatment. KGN cells were treated with 10−7 and 10−8 M estradiol, respectively. Raw Ct values were normalized to RNU43 housekeeping snoRNA and relative expression was determined using the 2− Δ ΔCt method. Fold change compared to controls treated with media alone was determined. One-way ANOVA was used to determine statistical differences between groups. Expression of both miR-21-5p (A) and miR-21-3p (B) increased significantly in response to treatment with Estradiol 10−7 M. The results represent mean±SD, *p<0.05 and **p<0.001.
Discussion
Our understanding of the regulation of follicular development and oocyte maturation by miRNAs is rapidly evolving. We now know, for example, that miRNA expression profiles are differentially expressed in MII oocytes and cumulus cells, including genes important for cumulus-oocyte communication [18]. Additionally, miRNA expression profiles in granulosa cells are associated with the maturity of adjacent oocytes, and modulation of these profiles may regulate oocyte maturation [19]. There is little data, however, on miRNA expression patterns in “poor responder” women, who demonstrate a subpar response to controlled ovarian hyperstimulation. Such knowledge has the potential to further the development of therapeutic interventions in fertility treatment. In this study, we demonstrate a significant difference between women with poor and normal responses to controlled ovarian hyperstimulation in terms of miRNA expression. In women with poor response to COH-IVF, 88 miRNAs are downregulated, while 16 miRNAs are upregulated. Specifically, we found that the expression of miR-21, which is highly upregulated by the LH surge [13], is increased in poor responders. We confirmed our findings in a large number of samples using qRT-PCR and determined that the active form of miR-21, miR-21-5p, is specifically elevated in cumulus cells of women with poor response and that this increase is not likely to be mediated by the lower serum estradiol levels observed in poor responders.
A thorough understanding of the genetic and molecular basis of poor ovarian response is imperative in order to design successful therapies. Many autosomal genes identified in animal and human models have been implicated as possible contributors to premature ovarian failure (which is, in turn, associated with the early onset of poor ovarian response). These include Fshr, Fmr1, Foxl2, Foxo3a, Gdf9, Nobox, and Oct4, among others (reviewed in [20]). With respect to poor ovarian response in particular, the most well-studied potential biomarker appears to be the FSH receptor. Variants in the receptor itself as well as abnormal signaling pathways have been implicated in several forms of infertility [2, 20]. Other polymorphisms under investigation include Esr1 and Esr2 [21]. However, while progress has been made, there is still much to learn regarding the molecular pathogenesis of poor ovarian response.
Our findings, which demonstrate differential expression of miR-21 in poor versus non-poor responders, add to a growing body of literature exploring the role of miRNAs in reproductive function. Hong et al [22] generated mice with a targeted deletion of Dicer (which is required for miRNA synthesis) in GCs. These mice demonstrated decreased ovarian weight, fewer large antral follicles, and decreased ovulation rates, suggesting fewer preovulatory follicles in these mice [22]. Conversely, Nagaraja et al [6] demonstrated normal folliculogenesis with increased follicular atresia in female mice, and ultimate sterility, after targeted Dicer deletion in somatic cells of the female reproductive tract. In another mouse model with 80% reduction in Dicer expression, female mice had normal ovulation rates, but corpora lutea function was reduced (as evidenced by decreased progesterone production) and the mice were unable to sustain pregnancies [23]. In any case, disruption of proper miRNA synthesis clearly leads to dysfunctional reproductive outcomes.
MicroRNA 21 is a particularly interesting target of study given that it is highly upregulated by the hCG and the LH surge, and may function as an antiapoptotic factor in GCs [13]. Mir-21 is encoded by the Mir21 gene; it was one of the first mammalian microRNAs identified, and the mature sequence is strongly conserved throughout evolution. The human microRNA-21 gene is located on the plus strand of chromosome 17q23.2 within coding gene TMEM49 (also known as vacuole membrane protein). Despite its location within intronic regions of a coding gene in the direction of transcription, the gene has its own promoter regions [24]. Within the ~3433-nt long, independently transcribed primary miR-21 transcript (pri-miR-21) resides a miR-21 RNA stem-loop precursor (pre-miR-21) [16]. This precursor, found between nucleotides 2445 and 2516, contains both miR21-5p (the active form, derived from the 5′ arm of the precursor miRNA, which is implicated in most biological functions) and miR21-3p (the complimentary sequence in the closed loop configuration, derived from the 3′ arm) [17]. In addition to its roles in reproductive function, aberrant miR-21 expression has also been implicated in multiple other disease states including cancer (where expression of miR-21 has been found to be deregulated in nearly all cancer types) and cardiovascular disease [25].
Notably, in our study, only miR-21-5p was elevated in cumulus cells of poor responders (while miR-21-3p was in fact significantly decreased), suggesting that the increase in miR-21-5p expression is not regulated at the pre-mir-21 level in these poor responder patients. Additionally, we found that both miR-21-5p and miR-21-3p are increased in a granulosa cell line in response to higher doses of estradiol (while expression was not affected at lower estradiol concentrations), suggesting that altered miR-21 expression in poor responders is not a result of decreased serum estradiol levels in these women. The use of the KGN granulosa cell line in these experiments provided a readily accessible system for performing mechanistic experiments to better characterize miR-21 regulation. It is noteworthy that our finding on the effect of estradiol on miR-21 expression differ from those of Wickramasinghe et al [26], who showed that estradiol repressed the expression of miR-21 by activating the estrogen receptor in MCF-7 cells. However, these differences could be explained by cell-type specific effects of estradiol on miR-21. Perhaps estradiol may have both agonist and antagonist effects on miR-21 expression, and exert these effects via both ER-dependent and ER-independent mechanisms.
In this study, we have identified a list of miRNAs with altered expression in cumulus cells of women with a poor response to COH-IVF. We have also identified miR-21-5p as a specific miRNA which is altered in poor responders. Our findings suggest a role for miRNAs in human cumulus cell function and potentially in the pathogenesis of poor ovarian response in women undergoing infertility treatment with COH-IVF. Potential target mRNAs that might be regulated by miR-21 in human cumulus and granulosa cells remain to be identified. In addition, other miRNAs identified as differentially expressed in women with poor response to IVF remain to be validated and characterized.
Supplementary Material
Supplemental Figure 1: Volcano plot of microarray results highlighting differentially expressed miRNAs. Horizontal axes represents the biological significance as log2(ratio) where ratio is the expression fold change between sample groups. Vertical axes, -log10(p-value), denote statistical significance. Significantly up- and down-regulated miRNAs are shown in the upper-right and upper-left regions of the plot, respectively. 16 miRNAs (including miR-21) are up-regulated and 88 miRNAs are down-regulated in poor responders with respect to non-poor responders.
Supplemental Figure 2: The box plot representation of miR-21 expression levels investigated in two groups considering the maternal age: patients under 35, and patients at 35 or higher. (A) miR-21 5p expression represents quite similar distributions in the younger and older age groups; no statistical significance was found between the groups (p = 0.639). (B) The difference between miR-21 3p expressions of the two patient groups was not significant (p = 0.543).
Supplemental Table 1: List of significantly up-regulated miRNAs in the cumulus cell samples of poor responder patients compared to non-poor responders. The miRNAs are listed in descending order based on fold change value.
Supplemental Table 2: List of significantly down-regulated miRNAs in the cumulus cell samples of poor responder patients compared to non-poor responders. The miRNAs are listed in descending order based on absolute fold change value.
Summary sentence.
Poor response to IVF is associated with altered miRNA expression in cumulus cells, specifically with elevated expression of miR-21-5p, an effect which is likely independent of lower serum estradiol levels in poor responders.
References
- 1.Arslan M, Bocca S, Mirkin S, Barroso G, Stadtmauer L, Oehninger S. Controlled ovarian hyperstimulation protocols for in vitro fertilization: two decades of experience after the birth of Elizabeth Carr. Fertil Steril. 2005;84:555–569. doi: 10.1016/j.fertnstert.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 2.Gerasimova T, Thanasoula MN, Zattas D, Seli E, Sakkas D, Lalioti MD. Identification and in vitro characterization of follicle stimulating hormone (FSH) receptor variants associated with abnormal ovarian response to FSH. J Clin Endocrinol Metab. 2010;95:529–536. doi: 10.1210/jc.2009-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferraretti AP, La Marca A, Fauser BC, Tarlatzis B, Nargund G, Gianaroli L, et al. ESHRE consensus on the definition of ‘poor response’ to ovarian stimulation for in vitro fertilization: the Bologna criteria. Hum Reprod. 2011;26:1616–1624. doi: 10.1093/humrep/der092. [DOI] [PubMed] [Google Scholar]
- 4.Espey LL, Richards JS. Temporal and spatial patterns of ovarian gene transcription following an ovulatory dose of gonadotropin in the rat. Biol Reprod. 2002 Dec;67(6):1662–1670. doi: 10.1095/biolreprod.102.005173. [DOI] [PubMed] [Google Scholar]
- 5.Pangas SA, Rajkovic A. Transcriptional regulation of early oogenesis: in search of masters. Hum Reprod Update. 2006;12:65–76. doi: 10.1093/humupd/dmi033. [DOI] [PubMed] [Google Scholar]
- 6.Nagaraja AK, Andreu-Vieyra C, Franco HL, Ma L, Chen R, Han DY, et al. Deletion of Dicer in somatic cells of the female reproductive tract causes sterility. Mol Endocrinol. 2008;22:2336–2352. doi: 10.1210/me.2008-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 8.Brennecke J, Cohen SM. Towards a complete description of the microRNA complement of animal genomes. Genome Biol. 2003;4:228. doi: 10.1186/gb-2003-4-9-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 10.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wienholds E, Koudijs MJ, van Eeden FJ, Cuppen E, Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat Genet. 2003;35:217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 12.Fiedler SD, Carletti MZ, Hong X, Christenson LK. Hormonal regulation of MicroRNA expression in periovulatory mouse mural granulosa cells. Biol Reprod. 2008;79:1030–1037. doi: 10.1095/biolreprod.108.069690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carletti MZ, Fiedler SD, Christenson LK. MicroRNA 21 blocks apoptosis in mouse periovulatory granulosa cells. Biol Reprod. 2010;83:286–295. doi: 10.1095/biolreprod.109.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lei L, Jin S, Gonzalez G, Behringer RR, Woodruff TK. The regulatory role of Dicer in folliculogenesis in mice. Mol Cell Endocrinol. 2010;315:63–73. doi: 10.1016/j.mce.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 16.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartels CL, Tsongalis GJ. MicroRNAs: novel biomarkers for human cancer. Clin Chem. 2009;55:623–631. doi: 10.1373/clinchem.2008.112805. [DOI] [PubMed] [Google Scholar]
- 18.Assou S, Al-edani T, Haouzi D, Phillipe N, Lecellier CH, Piquemal D, et al. Hum Reprod. 2013;28(11):3038–49. doi: 10.1093/humrep/det321. [DOI] [PubMed] [Google Scholar]
- 19.Kim YJ, Ku SY, Kim YY, Liu HC, Chi SW, Kim SH, et al. MicroRNAs transfected into granulosa cells may regulated oocyte meiotic competence during in vitro maturation of mouse follicles. Hum Reprod. 2013;28(11):3050–61. doi: 10.1093/humrep/det338. [DOI] [PubMed] [Google Scholar]
- 20.Skillern A, Rajkovic A. Recent developments in identifying genetic determinants of premature ovarian failure. Sex Dev. 2008;2:228–243. doi: 10.1159/000152039. [DOI] [PubMed] [Google Scholar]
- 21.de Castro F, Moron FJ, Montoro L, Galan JJ, Hernandez DP, Padilla ES, et al. Human controlled ovarian hyperstimulation outcome is a polygenic trait. Pharmacogenetics. 2004;14:285–293. doi: 10.1097/00008571-200405000-00003. [DOI] [PubMed] [Google Scholar]
- 22.Hong X, Luense LJ, McGinnis LK, Nothnick WB, Christenson LK. Dicer1 is essential for female fertility and normal development of the female reproductive system. Endocrinology. 2008;149:6207–6212. doi: 10.1210/en.2008-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otsuka M, Zheng M, Hayashi M, Lee JD, Yoshino O, Lin S, et al. Impaired microRNA processing causes corpus luteum insufficiency and infertility in mice. J Clin Invest. 2008;118:1944–1954. doi: 10.1172/JCI33680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MiRBase: Stem-loop sequence hsa-miR-21. Available at: http://www.mirbase.org/cgi-bin/mirna_entry.pl?acc=MI0000077.
- 25.Kumarswamy R, Volkmann I, Thum T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011;8:706–713. doi: 10.4161/rna.8.5.16154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wickramasinghe NS, Manavalan TT, Dougherty SM, Riggs KA, Li Y, Klinge CM. Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res. 2009;37:2584–2595. doi: 10.1093/nar/gkp117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Volcano plot of microarray results highlighting differentially expressed miRNAs. Horizontal axes represents the biological significance as log2(ratio) where ratio is the expression fold change between sample groups. Vertical axes, -log10(p-value), denote statistical significance. Significantly up- and down-regulated miRNAs are shown in the upper-right and upper-left regions of the plot, respectively. 16 miRNAs (including miR-21) are up-regulated and 88 miRNAs are down-regulated in poor responders with respect to non-poor responders.
Supplemental Figure 2: The box plot representation of miR-21 expression levels investigated in two groups considering the maternal age: patients under 35, and patients at 35 or higher. (A) miR-21 5p expression represents quite similar distributions in the younger and older age groups; no statistical significance was found between the groups (p = 0.639). (B) The difference between miR-21 3p expressions of the two patient groups was not significant (p = 0.543).
Supplemental Table 1: List of significantly up-regulated miRNAs in the cumulus cell samples of poor responder patients compared to non-poor responders. The miRNAs are listed in descending order based on fold change value.
Supplemental Table 2: List of significantly down-regulated miRNAs in the cumulus cell samples of poor responder patients compared to non-poor responders. The miRNAs are listed in descending order based on absolute fold change value.


