Abstract
Previous studies have suggested that overexpression of the oncogenic protein epithelial membrane protein-2 (EMP2) correlates with endometrial carcinoma progression and ultimately poor survival from disease. To understand the role of EMP2 in the etiology of disease, gene analysis was performed to show transcripts that are reciprocally regulated by EMP2 levels. In particular, EMP2 expression correlates with and helps regulate the expression of several cancer stem cell associated markers including aldehyde dehydrogenase 1 (ALDH1). ALDH expression significantly promotes tumor initiation and correlates with the levels of EMP2 expression in both patient samples and tumor cell lines. As therapy against CSCs in endometrial cancer is lacking, the ability of anti-EMP2 IgG1 therapy to reduce primary and secondary tumor formation using xenograft HEC1A models was determined. Anti-EMP2 IgG1 reduced the expression and activity of ALDH and correspondingly reduced both primary and secondary tumor load. Our results collectively suggest that anti-EMP2 therapy may be a novel method of reducing endometrial cancer stem cells.
Introduction
In developed countries, endometrial cancer is among the most commonly diagnosed gynecologic malignancy1, 2. According to recent cancer statistics, endometrial cancer remains among the leading cause for new cancer cases and deaths in women in the United States3 with studies estimating that 1 in 38 woman will be diagnosed with the disease in her lifetime. Although endometrial cancer is typically identified early, 15% to 20% of patients with presumed localized disease recur with advancement to metastasis4.
Cancer stem cells (CSCs), small subset of cells capable of self-renewal and clonal expansion are responsible for initiating and driving tumor growth, have emerged as a central hypothesis for treatment failure in cancer5–10. CSCs are typically resistant to chemotherapy and radiation, and it is believed that standard chemotherapy can promote or inadvertently select for these cells11–13. CSCs have been documented in multiple cancer types including those that originate within the prostate, colon, ovary, and breast, and recent studies have shown that these cells exist in endometrial cancer as well14, 15. However, there is still debate on the specific markers that identify CSCs in endometrial cancer.
Epithelial membrane protein-2 (EMP2), a tetraspan protein from the GAS-3/PMP22 family, is found in both endometrioid and serous endometrial cancers. Mechanistically, EMP2 regulates integrin-FAK activation driving both tumor migration as well as HIF-1α mediated angiogenesis16, 17, and interestingly, these are both pathways linked into the formation of cancer stem cells18, 19. Growing evidence in endometrial cancer suggests that EMP2 is an oncogenic protein whose expression directly contributes to tumor initiation and growth, and within patient samples increased EMP2 correlates with increased lymphovascular invasion as well as poor survival17, 20, 21.
In order to characterize the potential functions of EMP2 in driving CSCs in endometrial cancers, a comparative genomic analysis of endometrial cancer cells with ectopic overexpression versus knockdown of EMP2 was performed relative to a vector control. EMP2 expression directly correlated with induction of a number of cancer stem cell associated genes including ALDH1a. Further analysis revealed co-expression of ALDH and EMP2 in cell lines derived from endometrial cancers and patient tumors, and these cells exhibited a higher tumor initiation capacity than those lacking ALDH expression. As we have previously shown that anti-EMP2 antibodies improve endometrial cancer survival using mouse xenograft models, we extended the utility of this therapy to determine its effectiveness in reducing tumor re-initiation. In this paper, we reveal that targeting of EMP2 may be a novel therapeutic target for endometrial cancer through the specific reduction of tumor initiating cells.
Results
EMP2 expression correlates with cancer stem cell marker expression
To further our understanding of the etiology of EMP2 in cancer, differential expression in HEC1A cells with modulated EMP2 levels was determined using an Affymetrix U133 2.0 Plus array. Using the criteria where the average fold change between the groups was greater or equal to 2 yielded a set of 997 genes that were modified by EMP2 overexpression (HEC1A/EMP2) and 224 genes that were altered by shRNA knockdown (HEC1A/sh KD) compared to control (HEC1A/VC; Figure 1). Genes that were reciprocally regulated between the shRNA knockdown and overexpression were identified, and the intersection of the two lists consisted of 109 genes including EMP2 (Figure 1B; Supplementary Table 1). Using Ingenuity Pathways Analysis software, modulation of EMP2 expression enriches for genes involved in a number of biological processes involved in cancer, cellular movement, cellular development, cell death and survival, and the top 20 genes altered in either direction are shown (Supplementary Table 2). The most striking differences were the up-regulation of cancer stem cell associated genes, in particular the expression of ALDH1a. Quantitative PCR of four discriminator genes was performed, and similar to the results determined through Ingenuity, reciprocal regulation of Wnt3a, Wnt5a, DUSP4, and ALDH1 by EMP2 was confirmed (Figure 1C).
Figure 1. Variations of cancer related genes in EMP2 over-expressing and knock-down cell lines.
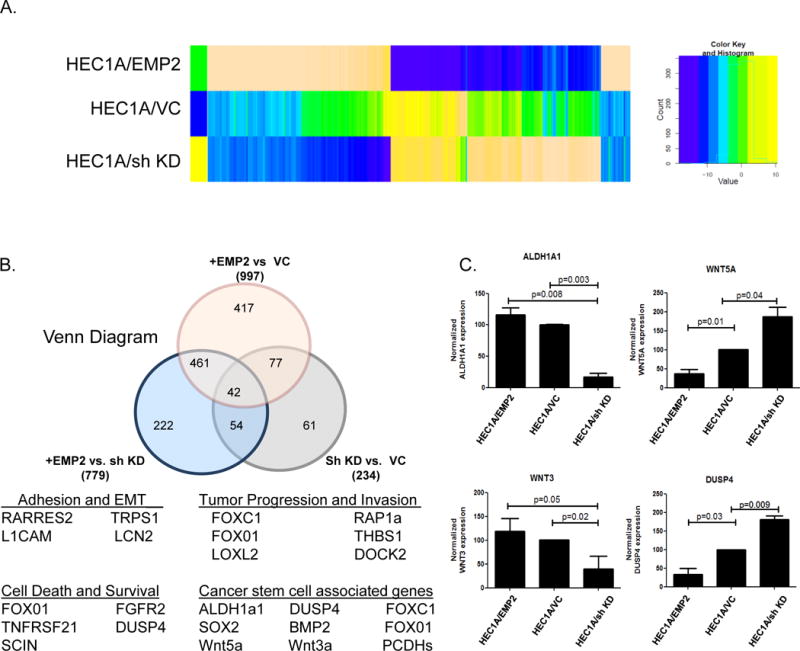
A. The heatmap shows the fold change of gene expression (rows) compared to control cell lines in EMP2 over-expressing (EMP2) and shRNA knock-down (sh KD) cell lines (columns). These cells were compared to the lentiviral vector control cells (VC). The dendrogram shows the hierarchical clustering of rows using Euclidean distance. The heatmap was plotted using the heatmap.2 function in gplots R package (v2.13.0). B. Top, Venn diagram of the intersection between genes differentially regulated by EMP2 expression (fold change > 2 fold for each comparison). Below, Hierarchical cluster analysis was used to classify differentially expressed genes according to their function by means of GCOS 1.2 software. Select gene lists are included. C. Validation of select mRNA differences using real time PCR comparing HEC1A/EMP2 and HEC1A/sh KD. Experiments were performed three times with the average ± SEM presented. Groups were analyzed using Student’s t test.
EMP2 promotes ALDH1 expression and activity
Aldehyde dehydrogenases (ALDH) are a group of detoxifying enzymes involved in the metabolism of intracellular aldehydes22, 23. They are thought to play a role in early stem cell differentiation through oxidation of retinol to retinoic acid. To translate the mRNA changes in ALDH1 to functional activity, cells stably expressing an EMP2 shRNA knockdown, or vector control were assessed by flow cytometry using the ALDEFLUOR assay under normoxic conditions. Knockdown of EMP2 significantly reduced ALDH activity compared to the vector control (Figure 2A).
Figure 2. EMP2 promotes ALDH1 expression in HEC1A cells.
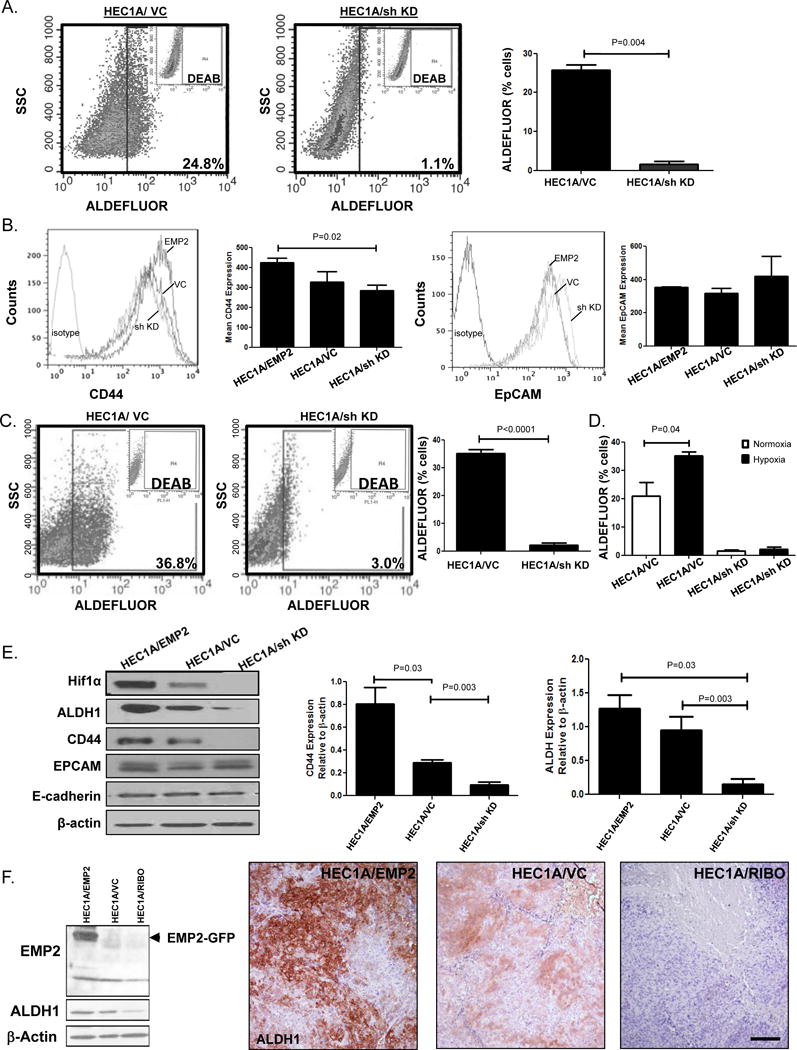
A. ALDH activity was measured in HEC1A/VC and HEC1A/sh KD cells under normoxic conditions. Right, results from three representative experiments are expressed as the mean average positive cells ± SEM. B. CD44 expression (left) or EpCAM (right) was measured using flow cytometry in HEC1A/EMP2, HEC1A/VC and HEC1A/sh KD cells. Right, graph represents compiled data from 3 independent experiments ± SEM. C. ALDH activity was measured in HEC1A/VC and HEC1A/sh KD cells under hypoxic conditions. Right, Results from three representative experiments are expressed as the mean average positive cells ± SEM. D. Comparison of ALDH activity in HEC1A/VC and HEC1A/sh KD cells under hypoxic and normoxic conditions. E. HEC1A cells with modified EMP2 levels were probed for HIF1α, EMP2, ALDH1, CD44, EpCAM, E-cadherin, and β-Actin protein expression using Western blot analysis. Right, semi-quantitative analysis of CD44 and ALDH1 expression relative to β-Actin from three representative blots ± SEM. F. HEC1A cells were engineered to express a ribozyme which has been shown to cleave the mRNA of EMP2. ALDH1 expression correlated with EMP2 levels (left). Right, HEC1A tumors with modified EMP2 levels were injected s.c. into BALB/c nude mice, and tumor growth monitored for 30 days. Tumors were then harvested and ALDH1 expression measured using IHC. Magnification, 200X. Scalebar, 200μM.
Given the potential regulation of ALDH by EMP2, we broadened our scope to examine a number of other endometrial cancer stem cell associated proteins such as CD44, CD133, and EpCAM24–28. CD133 expression was undetectable on HEC1A cells (data not shown). However, while CD44 surface expression showed some dependence on EMP2 levels, no alternation in EpCAM expression was detectable by flow cytometry among the engineered HEC1A cell lines (Figure 2B).
Hypoxia has been shown to drive the cancer stem cell phenotype29–31. Given that EMP2 helps to promote or stabilize HIF-1α expression16, we examined HEC1A cells with modified levels of EMP2 under hypoxic conditions. HEC1A/VC cells showed a statistically significant increase in ALDH activity under hypoxic conditions compared to the normoxia (P=0.04). However, even under hypoxic conditions, knockdown of EMP2 resulted in significantly less ALDH activity compared to the control (Figure 2C). As previously reported and observed here as well, ALDH activity increased under hypoxic conditions (Figure 2D). However, no significant increase in ALDH activity was observed in HEC1A/sh KD cells under hypoxia. This suggested that knockdown of EMP2 was sufficient to suppress ALDH activity in HEC1A cells.
To confirm and expand these results, a number of other cancer stem cell markers were evaluated under hypoxic conditions using western blot analysis. EMP2 overexpression promoted HIF-1α, ALDH1, and CD44 expression with reciprocal changes under knockdown conditions (Figure 2E). In contrast, no differences were observed in E-cadherin or EpCAM expression, and even under hypoxic conditions, CD133 expression was below detection using western blot analysis (data not shown).
Previous experiments have shown that EMP2 expression correlates with increased tumor initiation and growth in HEC1A cells in vivo17. However, these experiments were performed using a ribozyme as an alternate method to reduce EMP2 levels by specific mRNA cleavage32. To confirm that the reduction in ALDH1 was not dependent on a particular method of EMP2 downregulation, ALDH1 expression was probed in HEC1A/RIBO cells. Similar to the effects observed above using the shRNA lentiviral vector, the EMP2 ribozyme construct also significantly reduced total ALDH1 expression, further strengthening the direct regulation of ALDH1 by EMP2 (Figure 2F, left).
Our in vitro results suggested that EMP2 levels directly alter ALDH1 expression, but it is known that ALDH activity can be regulated by a number of factors including immune cells and cytokines within the microenvironment in vivo33, 34. To investigate the regulation of ALDH1 by EMP2 in vivo, cells were analyzed from tumors created from cells that overexpressed EMP2 as well as HEC1A/RIBO. We have previously shown that HEC1A/EMP2 grow rapidly while the HEC1A/RIBO cells produce small, poorly vascularized tumors in Balb/c nude animals16. Similar to the in vitro data, EMP2 expression positively regulated ALDH1 expression (Figure 2F, right).
To confirm the results above, we extended our analysis to another endometrial cancer cell line Ishikawa. Cells were generated with increased or reduced expression of EMP2, and cells were placed under a hypoxic condition. Similar to HEC1A, Ishikawa cells showed significant modulation of HIF-1α and CD44 expression by western blot analysis relative to EMP2 levels, and this effect was specific as no change in EpCAM expression was observed (Figure 3A). However, unlike HEC1A cells, Ishikawa cells express significant levels of CD133, but similar to EpCAM, its expression was not significantly altered by changes in EMP2 expression.
Figure 3. EMP2 promotes ALDH1 expression in Ishikawa cells.
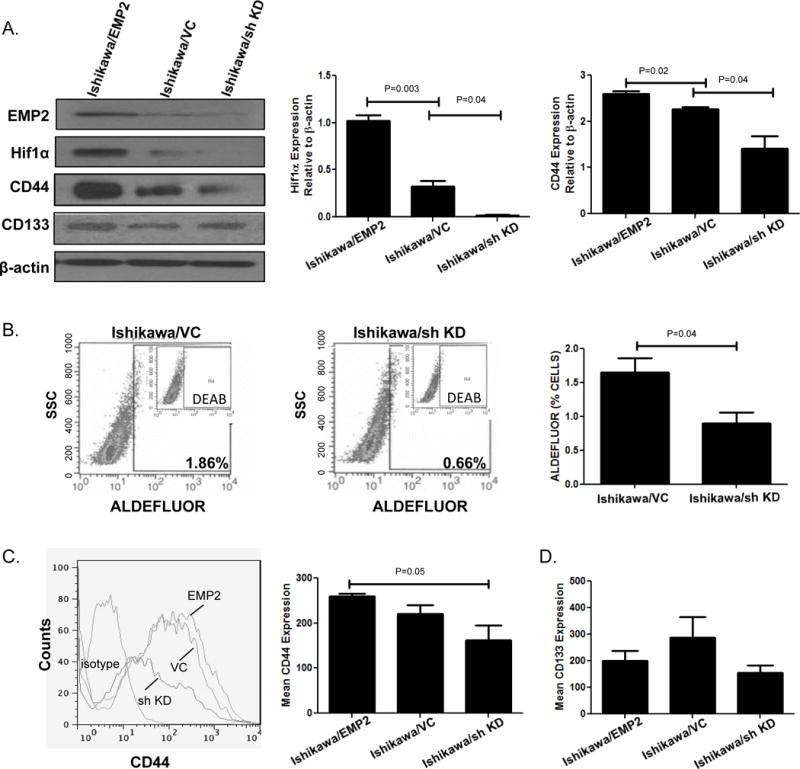
A. Ishikawa cells that overexpress EMP2, express a vector control, or have a reduction in EMP2 levels using a sh KD vector were probed for EMP2, HIF-1α, CD44, and CD133 relative to β-Actin. Right, quantitation of HIF-1α and CD44 expression from three independent experiments. Data is expressed as the average expression relative to β-Actin levels ± SEM. B. Representative staining of ALDH activity measured in Ishikawa/VC and Ishikawa/sh KD cells. Right, Average ± SEM of ALDH activity measured from three independent experiments. C. CD44 expression was measured using flow cytometry in cells with modulated EMP2 expression. Right, mean of CD44 expression from three independent experiments ± SEM. D. Average mean fluorescent intensity (MFI) ± SEM from three independent experiments of CD133 expression in Ishikawa cells with modified EMP2 levels.
As ALDH1 expression was below detection in Ishikawa cells by western blot analysis, we next determined if ALDH activity was altered in these cells using the ALDEFLUOR assay. Ishikawa cells with shRNA targeted reduction of EMP2 significantly decreased the percentage of cells showing ALDH activity compared to control cells (0.9±0.2 vs. 1.6±0.2, respectively; Figure 3B). Concordant with the western results above, a concomitant change in the surface expression between CD44 and EMP2was observed using flow cytometry (Figure 3C). Similarly, CD133 surface expression was not altered by changes in EMP2 levels (Figure 3D).
EMP2 regulates tumor growth in Ishikawa cells
In HEC1A cells, EMP2 levels have been shown to regulate tumor growth17. To determine if this response was specific to HEC1A or more robust, Ishikawa cells were injected into BALB/c nude mice. Tumor growth was monitored for 46 days, and significantly, EMP2 increased the rate of growth of these cells. In contrast, reduction in EMP2 levels reduced tumor growth or in some cases prevented tumor initiation (Figure 4A). To determine if the microenvironment could also alter ALDH1 expression, tumors were fixed and stained for ALDH1. Upregulation of EMP2 significantly increased the number of ALDH1 positive cells compared to the Ishikawa/VC. In the Ishikawa/sh KD, few if any ALDH1+ cells were observed throughout the entire tumor (Figure 4B).
Figure 4. EMP2 promotes tumor growth in Ishikawa cells.
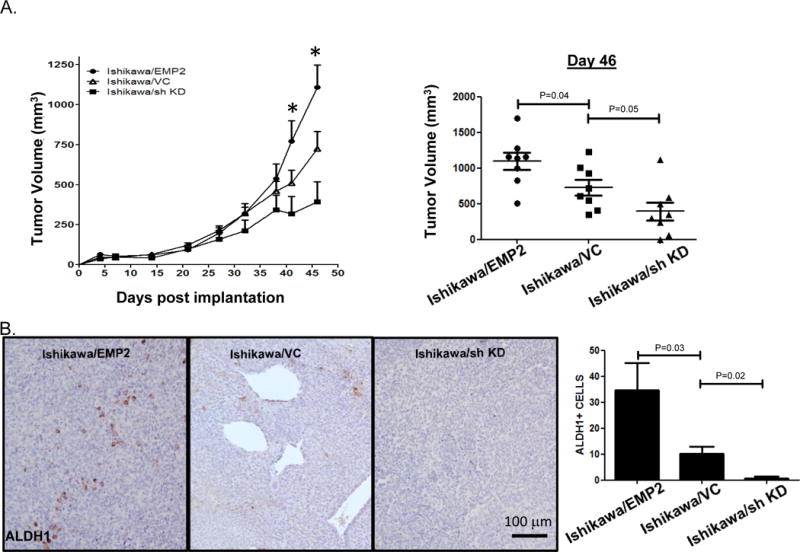
A. 1×106 Ishikawa cells with modified EMP2 levels (Ishikawa/EMP2, Ishikawa/VC, and Ishikawa/sh KD) were injected into injected s.c. into BALB/c Fox Chase SCID mice, and tumor growth monitored for 46 days. Two way ANOVA, p<0.05. Tumor volume on day 46 is shown to the right with groups compared using Student’s t test. B. Tumors were then harvested and ALDH1 expression measured using IHC. Magnification, 200X. ALDH+ cells were enumerated in 4 independent fields from at least 3 tumors with results depicted as the average ± SEM shown to the far right.
EMP2 and ALDH1 are co-expressed in endometrial cancer
Our results thus far suggested that EMP2 promotes ALDH1 expression and activity. While much is known about the role of ALDH activity and expression in breast cancer35, less is known about its role in endometrial cancer. In breast cancer, ALDH1 correlates poor prognosis and properties associated with cancer stem cells also called tumor initiating cells. To determine if ALDH activity translates into differences in endometrial tumor initiation, ALDEFLUOR positive and negative HEC1A and HEC1B cells were sorted and injected into animals (Figure 5A). In HEC1A cells, while the 50,000 cell injection site showed small differences in tumor size relative to ALDH activity, only ALDH positive cells formed tumors at the 5000 cell injection site, supporting the idea that these cells have a more potent tumor initiation capacity than ALDH negative cells (Figure 5B). To validate these cells, HEC1B cells were sorted and injected at 50,000 cells. By day 62, significant differences in tumor volume and weight were observed which suggested that ALDH+ cells had a greater capacity to initiate tumor growth than ALDH- cells.
Figure 5. ALDH1+ cells show enhanced tumor initiation.
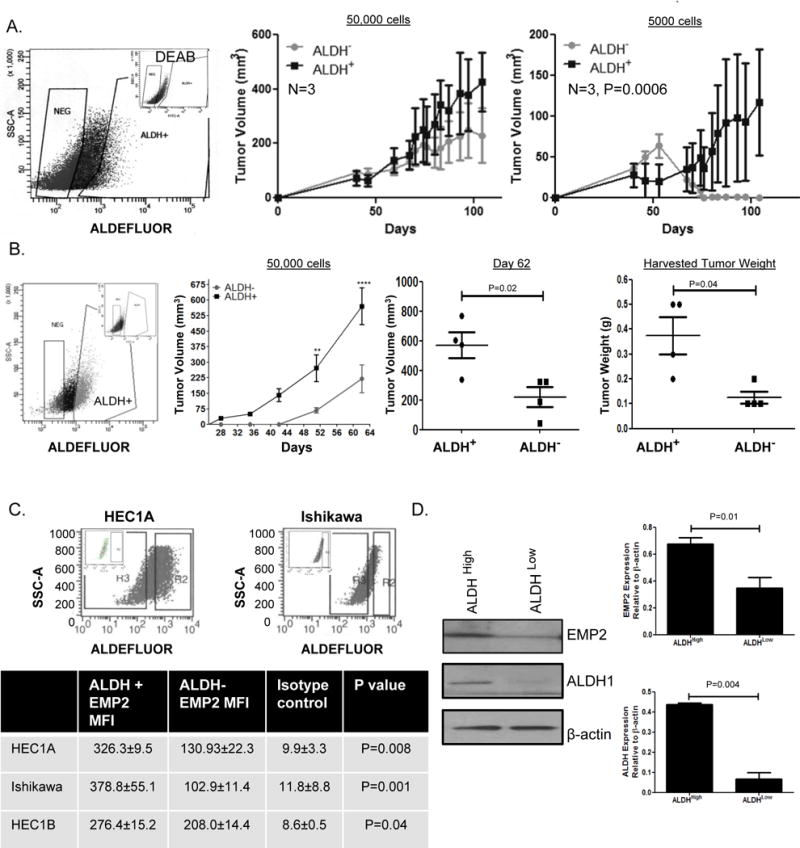
A. ALDH+ and ALDH- HEC1A cells were flow sorted and immediately injected into BALB/c nude mice at 50,000 or 5000 cells. Experiment was repeated, with a representative experiment shown. N=3. B. ALDH+ and ALDH- HEC1B cells were flow sorted and immediately injected into BALB/c Fox Chase SCID mice at 50,000 cells. Tumors were monitored for 62 days. Two way ANOVA, p= 0.005. Significance was further determined at Day 62 using the final tumor volume (right) and final tumor weight (far right) using Student’s t test. N=4. C. EMP2 expression was measured in HEC1A, Ishikawa, and HEC1B cells with high or low ALDH activity. The average mean fluorescent intensity (MFI) of EMP2 ± SEM is presented in the table below. D. To confirm the results, HEC1A were sorted into ALDH+ and ALDH- populations. Western blot analysis was used to confirm EMP2 and ALDH1 expression in the two populations with average ± SEM presented to the right.
As ALDH1 expression is heterogeneous within cell lines and within patient tumors35–37, we hypothesized that EMP2 expression may vary in a concordant manner. HEC1A, HEC1B, and Ishikawa cells were stained for EMP2 and ALDH activity. While both ALDH positive and negative populations express EMP2, its total expression was three-fold higher in both HEC1A and Ishikawa cells with high ALDH activity compared to cells with low/no activity (Figure 5C). In HEC1B cells, a similar trend was observed. ALDH+ cells showed a 1.5 fold increase in EMP2 expression compared to the ALDH- cells.
To confirm these results using a different method, HEC1A cells were sorted by ALDEFLUOR activity as described above, and EMP2 expression quantitated by western blot analysis. HEC1A cells with higher levels of ALDH activity correlated with a significant increase in EMP2 levels compared to those with no minimal activity (Figure 5D).
Although ALDH1 expression has been observed in up to 45% of patients with endometrial carcinomas, its expression is heterogeneous with often less than 10% of the cells staining positive38, 39. To determine the relationship between EMP2 and ALDH isoforms in human endometrial tumors, the provisional TCGA uterine corpus endometrial cancer database was queried to examine EMP2 mRNA levels in 248 tumors with available RNA sequencing data (RNA Seq V2 RSEM). Several ALDH isoforms correlated with EMP2 expression, with both ALDH1A1 and ALDH1A3 showing a Pearson correlation coefficient of 0.62 and 0.91, respectively (Table 1). To confirm the co-expression, a small panel of endometrial tumors were double stained for both EMP2 and ALDH1 proteins. No ALDH1 expression was seen in 1/15 (6.7%) patients with an additional 5/15 (33.3%) showing less than 10% of the cells showing any positivity. While ALDH1 expression was observed within the tumor parenchyma, some samples showed marked staining within the stroma as well. In contrast, EMP2 expression was present on 13/15 patients with tumor cells showing both cytoplasmic and membrane expression (Figure 6B). While the majority of cells within the tumor express EMP2, some variation in expression intensity was observed in the fluorescent images. Two representative samples are shown with enlarged insets provided (Figure 6). Select patients with ALDH1 expression within the tumor also show co-expression of EMP2 in those cells, and this co-staining pattern is highlighted by the asterisks.
Table 1. Correlation of EMP2 and ALDH isoforms in endometrial cancer.
Using The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov), EMP2 expression was queried in 248 sequenced endometrial carcinoma patients. Genes with the highest correlation to EMP2 were identified and all ALDH isoforms identified by the screen are listed below.
| Correlated Gene | Pearson’s Coefficient |
|---|---|
|
| |
| ALDH1B1 | 0.39 |
| ALDH6A1 | 0.47 |
| ALDH7A1 | 0.55 |
| ALDH3B1 | 0.55 |
| ALDH1A1 | 0.62 |
| ALDH18A1 | 0.71 |
| ALDH2 | 0.82 |
| ALDH1A3 | 0.91 |
| ALDH3A1 | 0.94 |
Figure 6. EMP2 and ALDH1 are co-expressed in primary endometrial cancer tumors.
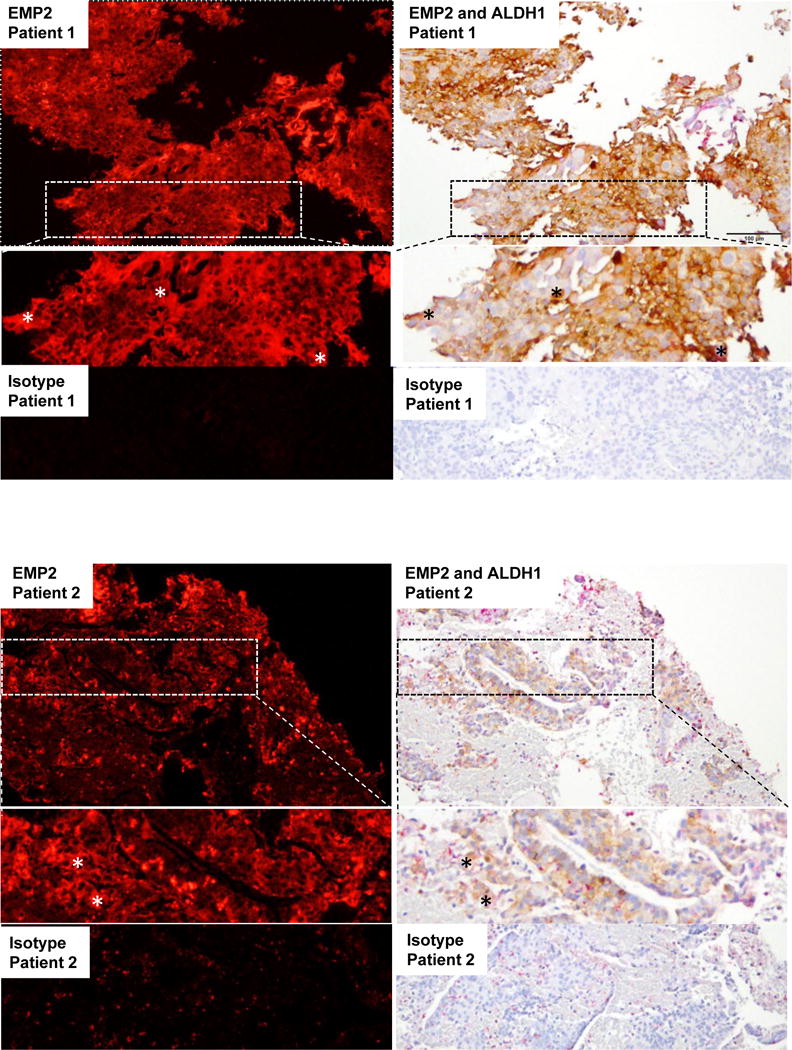
15 endometrial cancer tumors were double stained for EMP2 and ALDH1. EMP2 expression was visualized using the fluorescent agent VectorRED while ALDH1 staining was detected using DAB. Two representative patients are shown compared to the isotype control at a 200X magnification. *, An enlarged inset is also provided, highlighting areas of EMP2 and ALDH1 co-positivity.
In order to confirm the staining and ensure that the co-expression was not due to an artifact of the double stain, sequential sections were stained for ALDH1 or EMP2 individuals. Similar to the double staining results found above, in 14 out of 15 tumors, ALDH1 and EMP2 showed co-expression within the parenchyma of the tumor resulting in Spearman’s rank correlation coefficient of R = 0.62, p = 0.01 between EMP2 and ALDH expression (Supplementary Figure S1).
Anti-EMP2 IgG1 targets ALDH+ cancer stem cells
The above data suggested that downregulation of EMP2 was sufficient to reduce markers and activities associated with CSCs. To determine if antibody targeting could produce a similar effect, HEC1A/EMP2 or HEC1A/VC cells were incubated with control IgG1 or the anti-EMP2 IgG1 overnight in a hypoxic chamber. Anti-EMP2 IgG1 significantly reduced the expression of ALDH1 and CD44 expression in both cell lines by western blot analysis (Figure 7A). To confirm and expand these results, a panel of endometrial cancer cells were treated with a control human IgG antibodies or with the anti-EMP2 IgG1. A dose escalation in RL95-2 cells showed that ALDH activity began to decrease with 10μg/ml with a >50% reduction at 100 μg/ml (Figure 7B; Supplementary Figure S2). To expand on this effect, HEC1A, Ishikawa, and HEC1B were treated with 50 μg/ml of the anti-EMP2 antibody or an isotype control. In all three cell lines, greater than a 50% reduction in ALDH activity was observed (Figure 7C).
Figure 7. Anti-EMP2 IgG1 reduces ALDH expression in vitro.
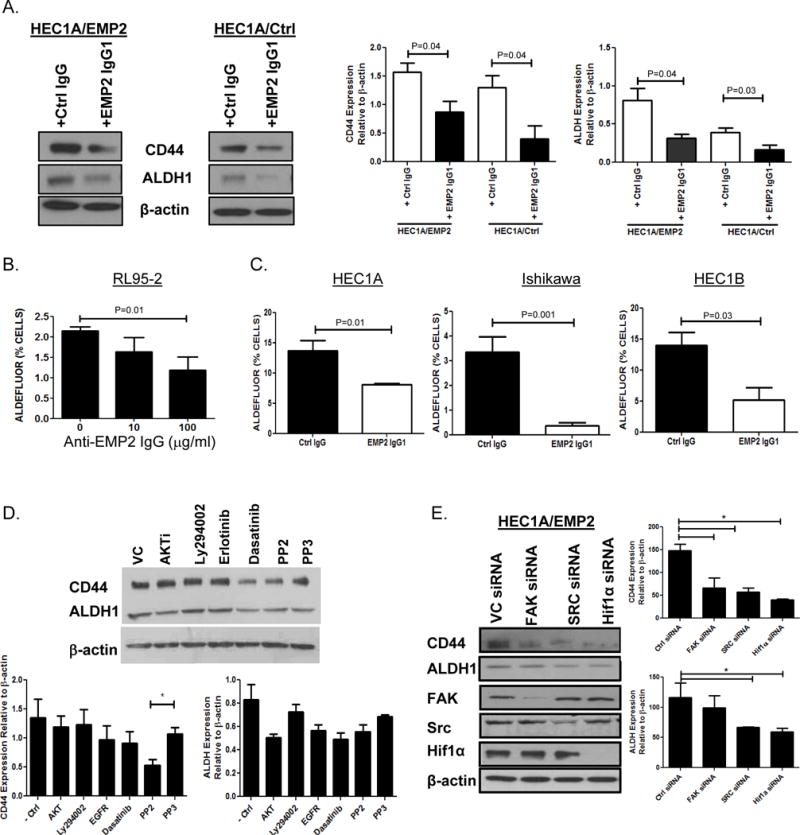
A. HEC1A/VC or HEC1A/EMP2 cells were treated with anti-EMP2 IgG1 or human IgG at 50μg/ml for 24 hrs. Left, representative western blots showing ALDH1 and CD44 expression relative to β-Actin. Right, relative expression of ALDH1 and CD44 expression from three independent experiments normalized to β-Actin. B. RL95-2 cells were treated with 10 or 100μg/ml of anti-EMP2 IgG1 for 24 hours. ALDH activity was measured using the ALDEFLUOR assay. C. HEC1A (left), Ishikawa cells (middle), and HEC1B (right) were treated with control or anti-EMP2 IgG1 antibody with 50μg/ml. ALDEFLUOR activity was measured after 24 hours with the average activity from three independent experiments ± SEM. D. HEC1A/EMP2 cells were treated with common inhibitors targeting the AKT, PI3K, EGFR, and FAK/Src, pathways under hypoxic conditions for 24 hours. PP3 as well as a vehicle control (saline) were included as controls. Total CD44 and ALDH1 levels were measured using western blot analysis and a representative graph is included. Right, Levels of CD44 and ALDH1 expression were quantitated to relative to β-Actin from three independent experiments. Results are presented as the mean ±SEM. *, p<0.05. E. HEC1A/EMP2 cells were treated with FAK, Src, and HIF-1α siRNA under hypoxic conditions. CD44, ALDH1, FAK, Src, HIF-1α, and β-Actin levels were measured using western blot analysis with a representative graph included on the left. Right, Levels of CD44 and ALDH1 expression were quantitated to relative to β-Actin from three independent experiments. Results are presented as the mean ±SEM.
To characterize the mechanism by which EMP2 regulates CD44 and ALDH expression, HEC1A/EMP2 cells were treated with common inhibitors to several characterized pathways to determine their effects on CD44 and ALDH1 expression under hypoxic conditions for 24 hours. Under the treated conditions, no cell death was detectable using trypan blue exclusion (data not shown). As shown in Figure 7D, in contrast to the anti-EMP2 IgG1, although the greatest effect appeared to be with an AKTi and dasatinib, none of the drugs tested significantly reduced ALDH1 expression, and only PP2, a small molecule inhibitor of the Src family of kinases, significantly reduced CD44 expression. In order to dissociate unexpected effects of these inhibitors from their predicted targets, FAK, Src, and HIF-1α siRNA constructs were tested as these pathways are activated by EMP2 and have been linked to CSC formation19, 40–42. Using the HEC1A/EMP2 cell line under hypoxic conditions, knockdown of FAK, Src, and HIF-1α all significantly reduced expression of CD44 (Figure 7E). In contrast, with regard to ALDH1, only cells transiently transfected with Src or HIF-1α siRNA showed a significant reduction in its expression.
EMP2 IgG1 improves endometrial cancer survival through reduction in CSCs
Several studies have shown that ALDH cells exhibit a higher proliferative index as well as increased metastatic capacity36, 39, 43. Initially, the ability of the anti-EMP2 IgG1 to reduce CSCs was determined using ALDH1 expression as a surrogate. Ishikawa cells were s.c. injected into BALB/c nude mice. When tumors approached 100mm3, they were treated with either saline or the anti-EMP2 IgG1. Significantly, twice a week treatment reduced tumor load which persisted throughout the experimental timeframe. Furthermore, this change in tumor volume correlated with reduced tumor weight (Figure 8A). To understand if this reduction corresponded to a change in the number of tumor initiating cells, ALDH1 expression was evaluated as a surrogate. Pockets of ALDH1 cells were observed in the vehicle control xenografts. However, few cells expressed this marker after anti-EMP2 treatment (Figure 8B).
Figure 8. Anti-EMP2 IgG1 reduces CSCs in vivo.
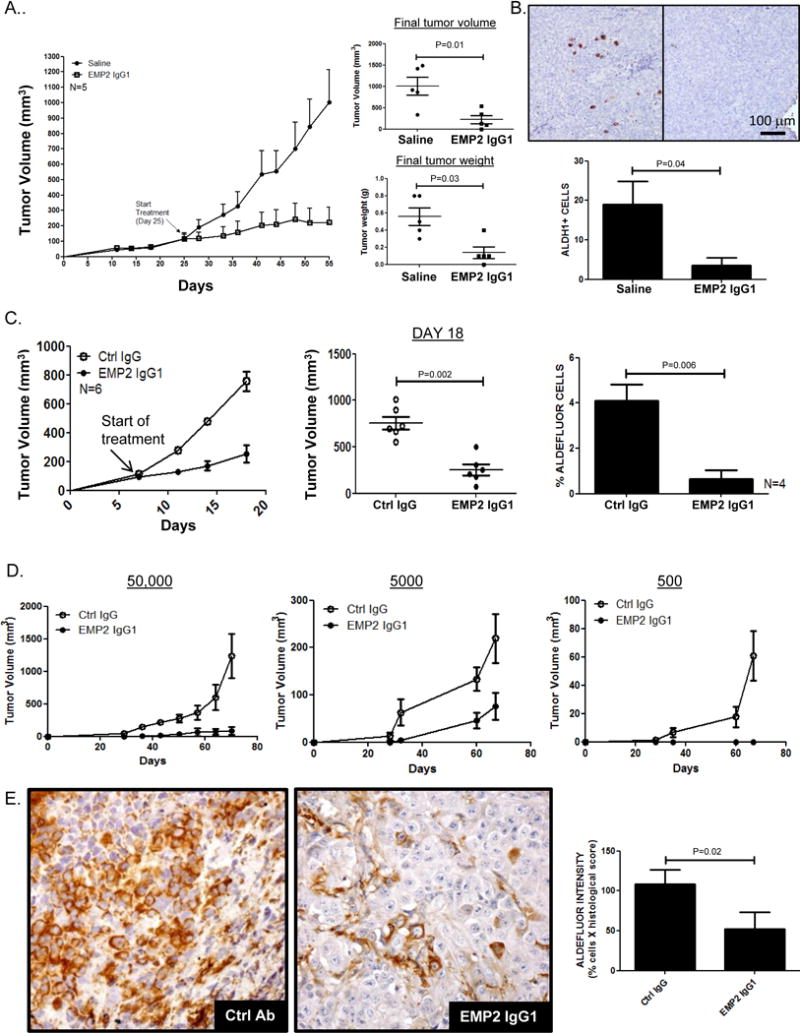
A. One million Ishikawa cells were implanted s.c. into BALB/c nude mice. When tumors reached ~100mm3, they were treated with anti-EMP2 IgG1 at 10mg/kg or with saline twice a week. N=5. Tumors were harvested after 30 days of treatment and weighed. Individual tumor volumes and weights are shown and compared using Student’s t test. B. ALDH1 expression was determined by immunohistochemical staining of tumors. Below, ALDH1+ cells were enumerated from 4 animals per group, and results expressed as the average ± SEM. B. HEC1A tumors implanted into the right shoulder of BALB/c nude mice. When tumors reached ~150mm3, they were treated with anti-EMP2 IgG1 or control IgG antibody at 10mg/kg twice a week. N=6. C. Following three treatments, tumors were harvested and disassociated. ALDEFLUOR activity was measured using flow cytometry as previously described. N=4. D. Cells dissociated from the anti-EMP2 IgG1 or control IgG treated tumors were reinjected into secondary animals without any further treatment at 50,000, 5000, or 500 cells/animal, and tumor growth measured. N=6. E. Immunohistochemical quantitation of ALDH1 expression in secondary tumors formed from the 50,000 cell reinjection site. Right, ALDH1 expression was quantitated from 5 animals per group, and results expressed as the average ± SEM.
In order to translate the effect of anti-EMP2 antibodies on tumor initiating cells, the ability of the antibody to reduce secondary tumor formation was evaluated. Serial transplantation is a common method to investigate long-term tumorigenic potential of cancer cells, and it has been shown that cells with high ALDH activity show an enhanced repopulating and metastatic capacity44–46. To test the idea that anti-EMP2 antibodies alter the ability of the CSCs within the tumor, subcutaneous xenografts using HEC1A wild-type cells were created. Similar to the results obtained in Ishikawa cells, i.p. injections of anti-EMP2 IgG1 significantly reduced tumor load compared to control human antibodies (Figure 8C, p=0.0002). Moreover, consistent with the in vitro data, isolation of cells within the tumor post anti-EMP2 treatment showed a significant reduction in the number of ALDEFLUOR positive cells compared to the control treated tumors (Figure 8C).
To next determine if anti-EMP2 treatment altered the tumor initiating capacity of the remaining cells, tumor cells were re-injected using serial dilutions into athymic nude mice without any further treatment. At all cell re-injection sites, tumors from previously control IgG treated tumors enhanced the repopulating capacity in new recipients compared to those treated with the anti-EMP2 IgG1(Figure 8C). The combined data was analyzed using the Extreme Limiting Dilution Analysis, a method recognized to compute the capacity for subpopulations to reinitiate tumors47, 48. In the case of anti-EMP2 therapy, all reinjected tumor cells treated with control IgG formed tumors, and this was reinforced by the 1:1 tumor initiation capacity of these cells (Table 2). In contrast, those treated with EMP2 IgG1 showed a significant reduction in their capacity to form tumors, particularly at the lowest cell numbers, and this translated to CSC frequency of 1 in 3500 (Table 2). To understand if the reduction in ALDH expression was durable even in these secondary animals, tumors were stained by immunohistochemistry for ALDH1 expression. Secondary tumors from the anti-EMP2 treated group continued to maintain an approximate four-fold reduction in ALDH1 expression (Figure 8D). These results suggest that anti-EMP2 IgG1 treatment reduces tumor initiating capacity in part via its regulation of ALDH1 expression.
Table 2. Anti-EMP2 IgG1 inhibits secondary tumor formation.
The take rates of secondary tumors created from HEC1A tumors treated previously with control or anti-EMP2 IgG1 were tabulated. The extreme limiting dilution analysis (ELDA) was used to compare the frequency of cancer stem cells between the control and anti-EMP2 IgG1 treated animals48.
| Treatment | ||
|---|---|---|
| # Cells Injected | Ctrl IgG | EMP2 IgG1 |
| 50,000 | 6/6 | 6/6 |
| 5000 | 6/6 | 5/6 |
| 500 | 6/6 | 0/6 |
| CSC Frequency* (95% CI) |
1:1 (1–535) |
1:3528 (1421–8758) |
p=1.2×10−5; chi-square=19.2
Discussion
EMP2 is a tetraspan protein implicated in endometrial cancer progression and survival20, 21. However, the mechanism by which it contributes to tumorigenicity remains ambiguous. Using a global approach, we analyzed the reciprocal regulation of genes altered by EMP2 modulation. EMP2 caused pleiotropic changes within the cell as 109 genes were reciprocally regulated using a 2 fold criteria. Of particular note, using Ingenuity Pathways Analysis software, we found that EMP2 modulation enriches for biological processes involved with cellular movement, development and survival (Supplementary table 2). Within the groups, several genes associated with cancer stem cells were identified and validated, with a focus on understanding the regulation of ALDH1 by EMP2.
In endometrial cancer, identification of cancer CSCs remains a challenge. Several markers have been implicated in tumor initiation including CD133+ and ALDH1+ 14, 15, 24, 49. However, there is no conclusive evidence suggesting that either is a universal marker for the endometrial cancer stem cell population. CD133/prominin-1 is a 120-kDa pentaspan membrane protein expressed on normal differentiated epithelia from many tissues. Although tumor cells with CD133 expression are thought to have a higher tumor initiation potential, cells without this antigen have shown to successfully form tumors24, 50. In endometrioid adenocarcinomas, purified CD133+ cells were found to be resistant to cisplatin-induced and paclitaxel-induced cytotoxicity and expressed a gene signature consisting of another cancer stem cell associated gene CD4424. Other cancer stem cell markers include the family of aldehyde dehydrogenases (ALDH), with a focus on ALDH1. Although they have been better characterized in breast cancer, limited data does exist for ALDH1 in endometrial cancer35, 36, 43, and in both malignancies ALDH1 positivity serves as an independent prognostic indicator for poor survival35, 36, 39. Several groups have shown that functionally, CSCs can most reliably defined by tumor initiation51. In results presented in this paper, ALDH activity was sufficient to serve as a surrogate for cancer stem cells as ALDH positive cells from HEC1A and HEC1B cells showed enhanced tumor initiating potential compared to ALDH negative populations.
We have previously shown that EMP2 expression promoted tumor initiation and growth in HEC1A cells16, 17, and in this paper, we extend this phenotype to Ishikawa cells. In both cell lines, high levels of EMP2 within the tumor showed increased tumor burden which could be reduced via inhibition of EMP2 expression. While EMP2 levels do not significantly alter HEC1A endometrial cancer cell growth in vitro17, studies have shown that EMP2 expression alters the tumor microenvironment16. While it is hard to know if EMP2 differentially affects CSCs versus the bulk tumor population in vivo, higher levels of EMP2 resulted in higher ALDH1 expression with a reciprocal reduction observed with knockdown of EMP2. To determine if this association could be visualized in primary tumors, the co-expression of EMP2 and ALDH genes was initially analyzed using the Cancer Genome Atlas (http://cancergenome.nih.gov)/. Several isoforms of ALDH correlated with EMP2, with both ALDH1A1 and ALDH1A3 notably ranking highly based on the Pearson’s correlation coefficient. While both of these isoforms have been implicated as functional CSC markers36, 52, ALDH1A1 was also found to correlate with EMP2 levels in our microarray screen. To translate this association, endometrial tumor tissue was co-stained for both ALDH1 and EMP2. Whereas EMP2 was detectable in both ALDH positive and negative cells within this small sampling of women with endometrial cancer, ALDH1 expression was only found in EMP2 positive parenchyma.
We have previously shown that anti-EMP2 IgG1 appears to reduce tumor load and as a monotherapy can improve survival16. In this study, we suggest that the mechanism of action of the antibody may be through inhibiting both CD44 and ALDH expression, two prominent cancer stem cell markers, ultimately producing a reduction in tumor load. This brings into question how does EMP2 regulate CSC formation? Previous studies demonstrated that ALDH1 in breast cancer can be regulated by a number of factors including hypoxia, HER2, and ERK activation53–55, and in this study, we show that ALDH1 can be regulated transcriptionally by changes in EMP2 expression. While hypoxia and EMP2 expression can act synergistically to increase the expression of ALDH1 in endometrial cancer cells, in the absence of EMP2, hypoxia is not sufficient to turn on its expression.
Mechanistically, EMP2 is known to increase FAK and Src activation leading to an increase in cellular migration, and studies have reported that FAK and Src inhibitors can suppress ALDH expression56, 57. In endometrial cancer, similar results were obtained. In HEC1A/EMP2 cells, Src and HIF-1α siRNA both significantly reduced ALDH1 expression suggesting that targeting this pathway may be beneficial therapeutically. However, inhibition of these pathways using the Src family kinase inhibitors PP2 or dasatinib were not effective in reducing ALDH1 expression suggesting that either the treatment condition or off-target effects limited ability of these proteins to inhibit Src and/or HIF-1α activation in endometrial cancer cell lines.
Although anti-EMP2 IgG1 reduced the numbers of endometrial cancer stem cells, several questions remain. First, as EMP2 is expressed on both CSC and non-CSC populations, it is not known if anti-EMP2 IgG1 preferentially affects the CSC population. Although we predict that it induces apoptosis of these cells, additional experiments will be needed to confirm if it eliminates these cells through apoptosis or via differentiation. Second, mechanistically, it is unclear how EMP2 regulates ALDH1 expression. While EMP2 expression appears to be upregulated early in endometrial cancer pathogenesis21, ALDH1 expression displays a more discrete expression profile and is more easily detectable in later stages14. Although it is possible that EMP2 upregulation in cancer may indirectly regulate CSC gene expression through a non-specific increase in genomic instability, this appears unlikely given that anti-EMP2 IgG1 as well as EMP2 sh KD constructs are able to specifically reduce ALDH and CD44 expression. Alternatively, while high ALDH1 mRNA is seen with high levels of EMP2, translational regulation may reside in part with microRNA expression. In support of this idea, several microRNAs have been implicated in the regulation of both CD44 and ALDH58, 59, but additional experiments will be needed to further determine factors that influence the translation of these proteins.
In conclusion, this work describes the novel regulation of cancer stem cell genes through EMP2. EMP2 promotes ALDH expression and activity, providing further evidence for the supposition that EMP2 is an oncoprotein. Moreover, the ability of anti-EMP2 IgG1 to reduce the expression of these markers as well as prevent secondary tumor formation suggests it may be a first in class treatment for endometrial cancer stem cells.
Materials and Methods
Cell lines
The human endometrial adenocarcinoma cell line HEC1A, RL95-2, HEC1B and Ishikawa were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in McCoys or DMEM media supplemented with 10% fetal bovine serum, 1% L-glutamine, 1% sodium pyruvate, and 1% penicillin-streptomycin at 37°C in a humidified 5% CO2 according to the supplier’s recommendations. Cell lines were used within 3 months after resuscitation of frozen aliquots and were authenticated based on viability, recovery, growth, morphology, and isoenzymology by the supplier and were tested monthly for mycoplasma (Lonza). Stably transfected cells with a human EMP2-GFP fusion protein have been previously described60, 61. Stably infected endometrial cancer lines containing a non-targeting shRNA control or a shRNA knockdown of EMP2 (sh KD; TRCN0000322911) in pLKO.1-puro were generated as per manufacturer’s instructions (Sigma-Aldrich). All experiments were done with sub-confluent cells.
In some experiments, FAK, Src, and Hif1α levels were decreased by transiently transfecting HEC1A/EMP2 and HEC1A/VC cells with 120 picomoles siRNA duplexes (FAK-SMARTpool L-0031640-00, Src-SMARTpool L-003175-00, Hif1α-SMARTpool-004018-00; ThermoScientific) and a lipophilic transfection reagent (Lipofectamine 2000; Invitrogen) for 6 hours and then placed in a 0.5% hypoxic chamber for 24 hours. As a negative control, cells were transfected with 120 picomoles scrambled control siRNA (D-001206-13-05; ThermoScientific). Cells were harvested for Western blot analysis 24 hours post-transfection as detailed below.
RNA isolation, Affymetrix analysis, and qPCR
Total RNA was purified from cells by using RNeasy Plus kit (Qiagen). RNA quantity and purity were determined by using a NanoDrop ND-1000, and RNA integrity was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Gene expression analyses was performed on an Affymetrix U133 Plus 2.0 human oligonucleotide microarrays containing over 47,000 transcripts and variants, including 38,500 well-characterized human genes by the Jonsson Comprehensive Cancer Center Genomics Shared Resource / UCLA Clinical Microarray Core. Preparation of cRNA, hybridizations, washes, and detection were done as recommended by the supplier. Data were analyzed with Partek genomics Suite 6.6, RMA normalized and then assessed using a parametric test assuming unequal variances. Gene expression between HEC1A/EMP2, HEC1A/VC, and HEC1A/sh KD were simultaneously compared with genes that produced a greater than 2 fold difference with p <0.05 considered significant. The data presented in this publication has been deposited into the NCBI Gene Expression Omnibus and is accessible through GEO Series accession number GSE92886.
To validate cancer stem cell gene transcription, PCR primers were designed by Life Technologies. Reverse transcription-PCR (RT-PCR) was carried out by using SuperScript One-Step RT-PCR with Platinum Taq (Invitrogen) by using 200 ng of RNA in a Bio-Rad MyCycler. PCR samples were analyzed on 1.5% agarose gels and imaged on a Gel Doc XR (Bio-Rad). First strand cDNA for quantitative PCR (qPCR) was synthesized by using the RT2 First Strand cDNA Kit (SABiosciences). Gene expression was determined using the RT2 Profiler PCR Array qPCR kit and detected with the RT2 SYBR Green qPCR Master Mix (SABioscience) according to the manufacturer’s protocol and run on ABI 7900HT with standard 96 block (Applied Biosystems). Expression analysis was conducted by using the manufacturer's online analysis tool by the core facility and gene expression was normalized to housekeeping genes. Differential expression was measured as the fold expression relative to the control cell line.
Preparation of Xenografts
Ethical Treatment of Animals Statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Animal Research Committee at the University of California, Los Angeles. All efforts were made to minimize animal suffering.
BALB/c nude and Fox Chase SCID female mice (6–8 weeks) were obtained from Charles River Laboratories (Wilmington, MA) and maintained at the University of California, Los Angeles. 4 × 106 HEC-1A cells or 1 × 106 Ishikawa cells were suspended in 1:1 saline with matrigel (BD Biosciences) solution and injected s.c. into the right flank of female athymic mice. In some experiments, Ishikawa cells or engineered Ishikawa cells were mixed with basement membrane extract type III (Trevigen). When tumors reached at least 100 mm3, animals were randomly sorted into test groups with the aim of keeping the average between groups the same. Systemic treatments were administered using 10 mg/kg dose of anti-EMP2 IgG1, control IgG (Sigma), an anti-VEGF IgG1 antibody (Bevacizumab; Genentech), or saline as indicated in the figure legends twice a week by intraperitoneal injection. Tumors were measured using calipers, and the volume calculated with the formula: length x width2/2. Mice were euthanized once tumors approached 1.5 cm in diameter. Tumors were excised, fixed in formalin, and then processed for hematoxylin and eosin staining by the Tissue Procurement Laboratory at UCLA.
Production of the anti-EMP2 IgG1
The anti-EMP2 IgG1 has been previously described62. In order to minimize batch variation, the antibody is currently produced in bulk by Lake Pharma on a contractual basis according to their standard practices. Each production run includes a certificate of analysis testing both the size of both native (160kD) and reduced forms of the antibody by electropherogram. In addition, the affinity of the antibody is verified against a specific and scrambled EMP2 peptide using ForteBio Octet with an affinity between 5–8 nM considered acceptable.
Preparation of Single Cell Suspensions of Tumor Cells
In some experiments, single cell suspensions of primary tumors were prepared from the xenografts treated with anti-EMP2 IgG1 or control IgG1. Single cell suspensions were created as previously described63. Briefly, xenograft tumors were cut in half and then minced by using sterile blades. To obtain single cell suspensions, the tumor pieces were mixed with ultra-pure collagenase III and hyaluronidase in DMEM and allowed to incubate at 37°C for 1 h. After incubation, cells were filtered through a 40 μm nylon mesh and washed with saline. Cells were counted and resuspended in a saline and Matrigel mix (1:1) and injected into the mammary fat pad at 50,000, 5,000, or 500 cells. Tumor size was monitored, and mice were euthanized once tumors reached 1.5 cm in diameter. Tumors were isolated, fixed and processed for hematoxylin and eosin staining as previously described20.
Flow cytometry
Samples were removed from the plate using a HBSS-EDTA buffer. Cells were immediately suspended in a HBSS buffer containing 0.05% sodium azide, 1% FBS, and 0.5% BSA. All incubations and washes were performed in this buffer. Primary antibodies used were: EpCAM- Cy5, CD44-APC (dilution 1:100), or an anti-EMP2 IgG1 (10 μg/ml). Incubation was performed for 30 min. on ice, followed by washing. For detection of EMP2, the secondary antibody used was goat anti-human Fc-PE (Invitrogen; 1:100). After incubation, cells were washed once with HBSS, and analysis was performed using a FACSCalibur (Becton Dickinson, Palo Alto, CA, USA) flow cytometer.
ALDEFLUOR assay
ALDH activity was assessed in various endometrial cancer cell lines using the ALDEFLUOR kit (StemCell Technologies). Briefly, cells were incubated in ALDEFLUOR assay buffer containing ALDH substrate (1μmol/L per 1 × 106 cells). For each condition, a sample of cells was stained under identical conditions with 50 mmol/L of diethylaminobenzaldehyde, an ALDH inhibitor, as a negative control. In some experiments, ALDH positive or ALDH negative cells were sorted using the FACSAriaII (BD Biosciences).
Immunohistochemistry
For staining from xenograft models, tumors were excised, fixed in formalin, and then processed for hematoxylin and eosin staining by the Tissue Procurement Laboratory (TPCL) at UCLA. For staining of patient samples, sections were obtained from the TPCL on de-identified material with appropriate permission from the UCLA Institutional Review Board.
Single Staining Immunohistochemistry
Single staining immunohistochemistry was performed using the DAKO Envision+ HRP Mouse (DAB+, K400711-2) and Rabbit (DAB+, K401111-2) kits. Briefly, 4 μm sections of paraffin embedded tissue were deparaffinized in xylene and rehydrated using graded alcohols. For heat-induced epitope retrieval, the slides were heated in a 95°C water bath with 0.01 M sodium citrate buffer (pH 6.0) for 25 minutes and cooled to room temperature for approximately 30 minutes. In order to remove the endogenous peroxidase activity, the HIER treated slides were blocked in DAKO Peroxidase Blocking Reagent for 10 minutes. Subsequently, the tissues were blocked with DAKO Serum-Free Protein Block (X0909) for 30 minutes, and incubated in primary antibody overnight at 4°C. The primary antibodies were diluted in DAKO Antibody Diluent (S0809) using the specifications previously mentioned. The following day the tissues were treated with DAKO Labeled Polymer-HRP anti-Mouse or anti-Rabbit for 30 minutes, visualized using diaminobenzidine, dehydrated with graded alcohol, and counterstained using diluted Harris Modified Method Hematoxylin (Fisherbrand, SH25-500D).
In order to quantitate EMP2 or ALDH1 expression within the tumor, sequential sections were stained and then scored by a pathologist (V.F.) using the following formula (H score): [3(%a) + 2(%b) + 1(%c)]/100, where a, b, and c is the percentage of cells staining at intensity 3, 2, and 1, respectively.
Double Staining Immunohistochemistry
Our double staining immunohistochemistry required both the DAKO Envision+ HRP Mouse kit (DAB+, K400711-2), Vector Vectastain ABC-Alkaline Phosphatase Kit (AK-5001), Vector Red Alkaline Phosphatase Substrate kit (SK-5100). Following the same procedure as the single staining immunohistochemistry, the sections were deparaffinized, re-hydrated, HIER treated, and blocked with DAKO Peroxidase Blocking Reagent and DAKO Serum-free Protein Block. Both the anti-EMP2 and anti-ALDH antibodies were mixed together in the DAKO Antibody Diluent and incubated overnight at 4°C. The subsequent day, sections were treated with DAKO Labeled Polymer-HRP anti-Mouse for 30 minutes, and ALDH antigenic sites were visualized using diaminobenzidine. EMP2 staining was then detected by incubating the sample with biotinylated anti-rabbit followed by the Streptavidin reagent from the Vector ABC-Alkaline Phosphatase Kit for 30 minutes. Staining was visualized with Vector Red. The slides were then dehydrated in graded alcohol and counterstained with hematoxylin. Samples were analyzed using an Olympus BX51 light microscope using a 20 X objective connected to a DP72 digital camera.
TCGA Data
The co-expression of EMP2 and ALDH mRNA was obtained through TCGA’s online data portal site (http://cancergenome.nih.gov). 248 sequenced cases of endometrial cancer (study?id=ucec_tcga) were evaluated for EMP2 expression (RNA Seq V2 RSEM)) and then queried for transcripts with the highest expression correlation. The co-expression of EMP2 and ALDH transcripts were ranked based on Pearson's correlation coefficient.
SDS-PAGE/Western Blot Analysis
Cells were resuspended in Laemmli sample buffer (62.5 mM Tris-Cl, pH 6.8, 10% glycerol, 2% SDS, 0.01% bromophenol blue, 2% β-mercaptoethanol). As EMP2 contains multiple glycosylation sites64, N-linked glycans were cleaved using peptide N-glycanase (PNGase; New England Biolabs, Beverly, MA). Eluates were treated as per manufacturer’s instructions at 37°C for 2 h. Proteins were separated by SDS-PAGE, transferred to a nitrocellulose membrane (Amersham Biosciences), and stained with Ponceau S (Sigma-Aldrich, St. Louis, MO) to determine transfer efficiency. Membranes were blocked with 10% low fat milk in PBS containing 0.1% Tween 20 and probed with EMP2 antisera (1:1000), anti-ALDH (BD Biosciences), anti-HIF-1α (BD Biosciences), anti-CD44 (R&D Systems) or β-actin (US Biologicals). Protein bands were visualized using a horseradish peroxidase-labeled secondary antibody (BD Biosciences; Southern Biotechnology Associates, Birmingham, AL) followed by chemiluminescence (ECL; Millipore). Band intensities were quantified using the NIH program Image J as above. To account for loading variability β-actin was used to normalize each sample. At least three independent experiments were performed and the results were evaluated for statistical significance using an unpaired Student’s t-test. A level of P<0.05 was considered to be statistically significant.
In some experiments, cells were treated with antibodies or inhibitors to determine the contribution to specific pathways to cancer stem cell marker expression of CD44 and ALDH1. HEC1A/EMP2 cells were treated with 100μg/ml control or EMP2 IgG1, 10μm of the FAK-Src small molecule inhibitor PP2 or the small molecule control PP365, 5μm of AKT inhibitor VIII66, the EGFR inhibitor Erlotinib67; 10μM, Genentech) or the Src family tyrosine kinase inhibitor Dasatinib68; 10nM, Bristol-Myers Squibb) under hypoxic conditions. Efficacy of inhibitors was tested at the manufacturer’s recommended dosage, and potential toxicity was measured using trypan-blue exclusion. Samples were harvested and probed by SDS-PAGE/Western blot analysis as above.
Statistical analysis
The number of mice used per group is indicated within the figure legend. For treatment studies, the minimum sample size was calculated at N=5 using G power with the α level set to 0.05 assuming a 50% reduction in tumor volume with anti-EMP2 IgG1 treatment69. Differences in in vitro phenotypic changes or in vivo tumor growth were evaluated using Student’s unpaired t-test at a 95% confidence level (GraphPad Prism version 5.0; GraphPad Software, La Jolla, CA). Differences in the rate of growth over time in ALDH+ and ALDH- tumors were determined through two-way ANOVA (GraphPad Prism version 5.0; GraphPad Software, La Jolla, CA). Extreme limiting dilution analysis (ELDA) was performed in accordance with Hu and Smyth48. The frequency of secondary tumor formation was calculated using ELDA software at: http://bioinf.wehi.edu.au/software/elda. In all cases, P values <0.05 were considered significant.
Supplementary Material
Acknowledgments
This work was generously supported by the Early Detection Research Network NCI CA-86366 (L. Goodglick), CA016042 (University of California at Los Angeles Jonsson Comprehensive Cancer Center Flow Cytometry Core), Charles Drew University/UCLA NIH U54-CA-143931 (J.L. and M.W.), the NIH/National Center for Advancing Translational Sciences (NCATS) UL1TR000124, and NCI R01 CA163971 (M. W.).
Footnotes
Conflict of interest
M.W., L.K.G, and J.B. are inventors on the University of California patents related to the anti- EMP2 IgG1 antibody presented in this work. They are also founders of Paganini Biopharma. MSW has financial holdings and is a scientific advisor for OncoMed Pharmaceuticals, Verastem, Paganini and MedImmune and receives research support from Dompe Pharmaceuticals and MedImmune. No other authors have competing interests.
Supplementary Information
Supplementary information accompanies the paper on the Oncogene website (http//www.nature.com/onc).
References
- 1.Dossus L, Allen N, Kaaks R, Bakken K, Lund E, Tjonneland A, et al. Reproductive risk factors and endometrial cancer: the European Prospective Investigation into Cancer and Nutrition. Int J Cancer. 127:442–451. doi: 10.1002/ijc.25050. [DOI] [PubMed] [Google Scholar]
- 2.Whitcomb BP. Gynecologic malignancies. Surg Clin North Am. 2008;88:301–317. vi. doi: 10.1016/j.suc.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 4.Salvesen HB, Haldorsen IS, Trovik J. Markers for individualised therapy in endometrial carcinoma. The Lancet Oncology. 2012;13:e353–e361. doi: 10.1016/S1470-2045(12)70213-9. [DOI] [PubMed] [Google Scholar]
- 5.Boman BM, Wicha MS. Cancer stem cells: a step toward the cure. J Clin Oncol. 2008;26:2795–2799. doi: 10.1200/JCO.2008.17.7436. [DOI] [PubMed] [Google Scholar]
- 6.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 7.Glinsky GV. Stem cell origin of death-from-cancer phenotypes of human prostate and breast cancers. Stem Cell Rev. 2007;3:79–93. doi: 10.1007/s12015-007-0011-9. [DOI] [PubMed] [Google Scholar]
- 8.Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 9.Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea–a paradigm shift. Cancer Res. 2006;66:1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. discussion 1895–1886. [DOI] [PubMed] [Google Scholar]
- 10.Soltysova A, Altanerova V, Altaner C. Cancer stem cells. Neoplasma. 2005;52:435–440. [PubMed] [Google Scholar]
- 11.Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eyler CE, Rich JN. Survival of the fittest: cancer stem cells in therapeutic resistance and angiogenesis. J Clin Oncol. 2008;26:2839–2845. doi: 10.1200/JCO.2007.15.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levina V, Marrangoni AM, DeMarco R, Gorelik E, Lokshin AE. Drug-selected human lung cancer stem cells: cytokine network, tumorigenic and metastatic properties. PLoS One. 2008;3:e3077. doi: 10.1371/journal.pone.0003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hubbard SA, Friel AM, Kumar B, Zhang L, Rueda BR, Gargett CE. Evidence for Cancer Stem Cells in Human Endometrial Carcinoma. Cancer Research. 2009;69:8241–8248. doi: 10.1158/0008-5472.CAN-08-4808. [DOI] [PubMed] [Google Scholar]
- 15.Kyo S. Endometrial Cancer Stem Cells: Are They a Possible Therapeutic Target? Current Obstetrics and Gynecology Reports. 2013;2:1–10. [Google Scholar]
- 16.Gordon LK, Kiyohara M, Fu M, Braun J, Dhawan P, Chan A, et al. EMP2 regulates angiogenesis in endometrial cancer cells through induction of VEGF. Oncogene. 2013;32:5369–5376. doi: 10.1038/onc.2012.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu M, Rao R, Sudhakar D, Hogue CP, Rutta Z, Morales S, et al. Epithelial membrane protein-2 promotes endometrial tumor formation through activation of FAK and Src. PLoS One. 2011;6:e19945. doi: 10.1371/journal.pone.0019945. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Liang D, Ma Y, Liu J, Trope C, Holm R, Nesland J, et al. The hypoxic microenvironment upgrades stem-like properties of ovarian cancer cells. BMC Cancer. 2012;12:201. doi: 10.1186/1471-2407-12-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wicha MS, Liu S, Dontu G. Cancer Stem Cells: An Old Idea—A Paradigm Shift. Cancer Research. 2006;66:1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. [DOI] [PubMed] [Google Scholar]
- 20.Wadehra M, Natarajan S, Seligson DB, Williams CJ, Hummer AJ, Hedvat C, et al. Expression of epithelial membrane protein-2 is associated with endometrial adenocarcinoma of unfavorable outcome. Cancer. 2006;107:90–98. doi: 10.1002/cncr.21957. [DOI] [PubMed] [Google Scholar]
- 21.Habeeb O, Goodglick L, Soslow RA, Rao RG, Gordon LK, Schirripa O, et al. Epithelial membrane protein-2 expression is an early predictor of endometrial cancer development. Cancer. 2010;116:4718–4726. doi: 10.1002/cncr.25259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mamat S, Ikeda J-i, Tian T, Wang Y, Luo W, Aozasa K, et al. Transcriptional Regulation of Aldehyde Dehydrogenase 1A1 Gene by Alternative Spliced Forms of Nuclear Factor Y in Tumorigenic Population of Endometrial Adenocarcinoma. Genes & Cancer. 2011;2:979–984. doi: 10.1177/1947601911436009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ginestier C, Wicinski J, Cervera N, Monville F, Finetti P, Bertucci F, et al. Retinoid signaling regulates breast cancer stem cell differentiation. Cell cycle (Georgetown, Tex) 2009;8:3297. doi: 10.4161/cc.8.20.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rutella S, Bonanno G, Procoli A, Mariotti A, Corallo M, Prisco MG, et al. Cells with Characteristics of Cancer Stem/Progenitor Cells Express the CD133 Antigen in Human Endometrial Tumors. Clinical Cancer Research. 2009;15:4299–4311. doi: 10.1158/1078-0432.CCR-08-1883. [DOI] [PubMed] [Google Scholar]
- 25.Györffy B, Surowiak P, Kiesslich O, Denkert C, Schäfer R, Dietel M, et al. Gene expression profiling of 30 cancer cell lines predicts resistance towards 11 anticancer drugs at clinically achieved concentrations. International Journal of Cancer. 2006;118:1699–1712. doi: 10.1002/ijc.21570. [DOI] [PubMed] [Google Scholar]
- 26.Dedes KJ, Wetterskog D, Ashworth A, Kaye SB, Reis-Filho JS. Emerging therapeutic targets in endometrial cancer. Nat Rev Clin Oncol. 2011;8:261–271. doi: 10.1038/nrclinonc.2010.216. [DOI] [PubMed] [Google Scholar]
- 27.Leblanc M, Poncelet C, Soriano D, Walker-Combrouze F, Madelenat P, Scoazec J, et al. Alteration of CD44 and cadherins expression: possible association with augmented aggressiveness and invasiveness of endometrial carcinoma. Virchows Archiv. 2001;438:78–85. doi: 10.1007/s004280000269. [DOI] [PubMed] [Google Scholar]
- 28.Ayhan A, Tok EC, Bildirici I, Ayhan A. Overexpression of CD44 variant 6 in human endometrial cancer and its prognostic significance. Gynecologic Oncology. 2001;80:355–358. doi: 10.1006/gyno.2000.6014. [DOI] [PubMed] [Google Scholar]
- 29.Xing F, Okuda H, Watabe M, Kobayashi A, Pai SK, Liu W, et al. Hypoxia-induced Jagged2 promotes breast cancer metastasis and self-renewal of cancer stem-like cells. Oncogene. 2011;30:4075–4086. doi: 10.1038/onc.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samanta D, Gilkes DM, Chaturvedi P, Xiang L, Semenza GL. Hypoxia-inducible factors are required for chemotherapy resistance of breast cancer stem cells. Proceedings of the National Academy of Sciences. 2014 doi: 10.1073/pnas.1421438111. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Kim R-J, Park J-R, Roh K-J, Choi AR, Kim S-R, Kim P-H, et al. High aldehyde dehydrogenase activity enhances stem cell features in breast cancer cells by activating hypoxia-inducible factor-2α. Cancer Letters. 2013;333:18–31. doi: 10.1016/j.canlet.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 32.Wadehra M, Dayal M, Mainigi M, Ord T, Iyer R, Braun J, et al. Knockdown of the tetraspan protein epithelial membrane protein-2 inhibits implantation in the mouse. DevBiol. 2006;292:430–441. doi: 10.1016/j.ydbio.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 33.Ye J, Wu D, Wu P, Chen Z, Huang J. The cancer stem cell niche: cross talk between cancer stem cells and their microenvironment. Tumor Biology. 2014;35:3945–3951. doi: 10.1007/s13277-013-1561-x. [DOI] [PubMed] [Google Scholar]
- 34.Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 2015;16:225–238. doi: 10.1016/j.stem.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ginestier C, Hur M, Charafe-Jauffret E, Monville F, Dutcher J, Brown M. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Charafe-Jauffret E, Ginestier C, Iovino F, Tarpin C, Diebel M, Esterni B, et al. Aldehyde Dehydrogenase 1–Positive Cancer Stem Cells Mediate Metastasis and Poor Clinical Outcome in Inflammatory Breast Cancer. Clinical Cancer Research. 2010;16:45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanei T, Morimoto K, Shimazu K, Kim SJ, Tanji Y, Taguchi T, et al. Association of Breast Cancer Stem Cells Identified by Aldehyde Dehydrogenase 1 Expression with Resistance to Sequential Paclitaxel and Epirubicin-Based Chemotherapy for Breast Cancers. Clinical Cancer Research. 2009;15:4234–4241. doi: 10.1158/1078-0432.CCR-08-1479. [DOI] [PubMed] [Google Scholar]
- 38.Daoud SA, Hosni HN, Hareedy AA. Immunohistochemical Expressions of ER, PR and ALDH1 in Endometrial Hyperplasia and Carcinoma. Journal of American Science. 2013;9 [Google Scholar]
- 39.Rahadiani N, Ikeda J-i, Mamat S, Matsuzaki S, Ueda Y, Umehara R, et al. Expression of aldehyde dehydrogenase 1 (ALDH1) in endometrioid adenocarcinoma and its clinical implications. Cancer Science. 2011;102:903–908. doi: 10.1111/j.1349-7006.2011.01864.x. [DOI] [PubMed] [Google Scholar]
- 40.Luo M, Fan H, Nagy T, Wei H, Wang C, Liu S, et al. Mammary Epithelial-Specific Ablation of the Focal Adhesion Kinase Suppresses Mammary Tumorigenesis by Affecting Mammary Cancer Stem/Progenitor Cells. Cancer Research. 2009;69:466–474. doi: 10.1158/0008-5472.CAN-08-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franovic A, Holterman CE, Payette J, Lee S. Human cancers converge at the HIF-2α oncogenic axis. Proceedings of the National Academy of Sciences. 2009;106:21306–21311. doi: 10.1073/pnas.0906432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mendez O, Zavadil J, Esencay M, Lukyanov Y, Santovasi D, Wang S-C, et al. Knock down of HIF-1alpha in glioma cells reduces migration in vitro and invasion in vivo and impairs their ability to form tumor spheres. Molecular Cancer. 2010;9:133. doi: 10.1186/1476-4598-9-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Croker A, Allan A. Inhibition of aldehyde dehydrogenase (ALDH) activity reduces chemotherapy and radiation resistance of stem-like ALDHhiCD44+ human breast cancer cells. Breast Cancer Res Treat. 2012;133:75–87. doi: 10.1007/s10549-011-1692-y. [DOI] [PubMed] [Google Scholar]
- 44.Charafe-Jauffret E, Ginestier C, Birnbaum D. Breast cancer stem cells: tools and models to rely on. BMC Cancer. 2009;9:202. doi: 10.1186/1471-2407-9-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu H, Clauser KR, Tam WL, Fröse J, Ye X, Eaton EN, et al. A breast cancer stem cell niche supported by juxtacrine signalling from monocytes and macrophages. Nat Cell Biol. 2014;16:1105–1117. doi: 10.1038/ncb3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct Populations of Cancer Stem Cells Determine Tumor Growth and Metastatic Activity in Human Pancreatic Cancer. Cell stem cell. 1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 47.O’Brien CA, Kreso A, Jamieson CHM. Cancer Stem Cells and Self-renewal. Clinical Cancer Research. 2010;16:3113–3120. doi: 10.1158/1078-0432.CCR-09-2824. [DOI] [PubMed] [Google Scholar]
- 48.Hu Y, Smyth GK. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. Journal of Immunological Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 49.Mirantes C, Espinosa I, Ferrer I, Dolcet X, Prat J, Matias-Guiu X. Epithelial-to-mesenchymal transition and stem cells in endometrial cancer. Human Pathology. 2013;44:1973–1981. doi: 10.1016/j.humpath.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 50.Wu Y, Wu PY. CD133 as a marker for cancer stem cells: progresses and concerns. Stem cells and development. 2009;18:1127–1134. doi: 10.1089/scd.2008.0338. [DOI] [PubMed] [Google Scholar]
- 51.Kreso A, Dick John E. Evolution of the Cancer Stem Cell Model. Cell stem cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 52.Marcato P, Dean CA, Pan D, Araslanova R, Gillis M, Joshi M, et al. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem cells (Dayton, Ohio) 2011;29:32–45. doi: 10.1002/stem.563. [DOI] [PubMed] [Google Scholar]
- 53.Korkaya H, Paulson A, Iovino F, Wicha MS. HER2 regulates the mammary stem/progenitor cell population driving tumorigenesis and invasion. Oncogene. 2008;27:6120–6130. doi: 10.1038/onc.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Conley SJ, Gheordunescu E, Kakarala P, Newman B, Korkaya H, Heath AN, et al. Antiangiogenic agents increase breast cancer stem cells via the generation of tumor hypoxia. Proceedings of the National Academy of Sciences. 2012 doi: 10.1073/pnas.1018866109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alam M, Ahmad R, Rajabi H, Kharbanda A, Kufe D. MUC1-C Oncoprotein Activates ERK→C/EBPβ Signaling and Induction of Aldehyde Dehydrogenase 1A1 in Breast Cancer Cells. Journal of Biological Chemistry. 2013;288:30892–30903. doi: 10.1074/jbc.M113.477158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kurebayashi J, Kanomata N, Moriya T, Kozuka Y, Watanabe M, Sonoo H. Preferential antitumor effect of the Src inhibitor dasatinib associated with a decreased proportion of aldehyde dehydrogenase 1-positive cells in breast cancer cells of the basal B subtype. BMC Cancer. 2010;10:568–568. doi: 10.1186/1471-2407-10-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shapiro IM, Kolev VN, Vidal CM, Kadariya Y, Ring JE, Wright Q, et al. Merlin deficiency predicts for FAK inhibitor sensitivity: A synthetic lethal relationship. Science translational medicine. 2014;6:237ra268–237ra268. doi: 10.1126/scitranslmed.3008639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alonso-Alconada L, Muinelo-Romay L, Madissoo K, Diaz-Lopez A, Krakstad C, Trovik J, et al. Molecular profiling of circulating tumor cells links plasticity to the metastatic process in endometrial cancer. Molecular cancer. 2014;13:223. doi: 10.1186/1476-4598-13-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nandy SB, Subramani R, Rajamanickam V, Lopez-Valdez R, Arumugam A, Boopalan T, et al. microRNA alterations in ALDH positive mammary epithelial cells: a crucial contributing factor towards breast cancer risk reduction in case of early pregnancy. BMC Cancer. 2014;14:644. doi: 10.1186/1471-2407-14-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wadehra M, Dayal M, Mainigi M, Ord T, Iyer R, Braun J, et al. Knockdown of the tetraspan protein epithelial membrane protein-2 inhibits implantation in the mouse. Dev Biol. 2006;292:430–441. doi: 10.1016/j.ydbio.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 61.Wadehra M, Forbes A, Pushkarna N, Goodglick L, Gordon LK, Williams CJ, et al. Epithelial membrane protein-2 regulates surface expression of alphavbeta3 integrin in the endometrium. Dev Biol. 2005;287:336–345. doi: 10.1016/j.ydbio.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 62.Fu M, Maresh EL, Helguera GF, Kiyohara M, Qin Y, Ashki N, et al. Rationale and preclinical efficacy of a novel anti-EMP2 antibody for the treatment of invasive breast cancer. Mol Cancer Ther. 2014;13:902–915. doi: 10.1158/1535-7163.MCT-13-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tirino V, Desiderio V, Paino F, Papaccio G, De Rosa M. Methods for Cancer Stem Cell Detection and Isolation. In: Singh SR, editor. Somatic Stem Cells. Vol. 879. Humana Press; 2012. pp. 513–529. [DOI] [PubMed] [Google Scholar]
- 64.Wadehra M, Forbes A, Pushkarna N, Goodglick L, Gordon LK, Williams CJ, et al. Epithelial membrane protein-2 regulates surface expression of alphavbeta3 integrin in the endometrium. DevBiol. 2005;287:336–345. doi: 10.1016/j.ydbio.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 65.Morales SA, Mareninov S, Prasad P, Wadehra M, Braun J, Gordon LK. Collagen gel contraction by ARPE-19 cells is mediated by a FAK-Src dependent pathway. Exp Eye Res. 2007;85:790–798. doi: 10.1016/j.exer.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 66.Barnett SF, Defeo-Jones D, Fu S, Hancock PJ, Haskell KM, Jones RE, et al. Identification and characterization of pleckstrin-homology-domain-dependent and isoenzyme-specific Akt inhibitors. Biochem J. 2005;385:399–408. doi: 10.1042/BJ20041140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang MY, Lu KV, Zhu S, Dia EQ, Vivanco I, Shackleford GM, et al. Mammalian target of rapamycin inhibition promotes response to epidermal growth factor receptor kinase inhibitors in PTEN-deficient and PTEN-intact glioblastoma cells. Cancer Res. 2006;66:7864–7869. doi: 10.1158/0008-5472.CAN-04-4392. [DOI] [PubMed] [Google Scholar]
- 68.Kao J, Salari K, Bocanegra M, Choi Y-L, Girard L, Gandhi J, et al. Molecular Profiling of Breast Cancer Cell Lines Defines Relevant Tumor Models and Provides a Resource for Cancer Gene Discovery. PLoS One. 2009;4:e6146. doi: 10.1371/journal.pone.0006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Faul F, Erdfelder E, Buchner A, Lang A-G. Statistical power analyses using G*Power 3.1: Tests for correlation and regression analyses. Behavior Research Methods. 2009;41:1149–1160. doi: 10.3758/BRM.41.4.1149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


