Abstract
Actin is a profoundly influential protein; it impacts, among other processes, membrane morphology, cellular motility, and vesicle transport. Actin can polymerize into long filaments that push on membranes and provide support for intracellular transport. Actin filaments have polar ends: the fast-growing (barbed) end and the slow-growing (pointed) end. Depolymerization from the pointed end supplies monomers for further polymerization at the barbed end. Tropomodulins (Tmods) cap pointed ends by binding onto actin and tropomyosins (Tpms). Tmods and Tpms have been shown to regulate many cellular processes; however, very few studies have investigated their joint role in the nervous system. Recent data directly indicate that they can modulate neuronal morphology. Additional studies suggest that Tmod and Tpm impact molecular processes influential in synaptic signaling. To facilitate future research regarding their joint role in actin regulation in the nervous system, we will comprehensively discuss Tpm and Tmod and their known functions within molecular systems that influence neuronal development.
Introduction - Actin Regulation in Neuritogenesis
Morphological development of neurons is complex and involves the entirety of the cytoskeleton. Many signaling pathways converge on and cross talk through the cytoskeleton to drive changes in cell shape (for reviews see (da Silva and Dotti, 2002; Gomez and Letourneau, 2014; Hur et al., 2012; Vitriol and Zheng, 2012)). Growing neurites extend from the cell body; the tip of a growing neurite has a structure called a growth cone. Growth cones are composed of a lamellipodium-like veil and fine filopodia that extend from the leading edge of the growth cone and probe the local environment chemically and mechanically (Mattila and Lappalainen, 2008) to guide neurite outgrowth (Bentley and Toroian-Raymond, 1986). Early neurites specify into either dendrites or an axon. Small protrusions, so-called dendritic spines, form along the dendritic shafts and associate with axonal termini to form connections called synapses. Neurotransmitters are released from the presynaptic axon termini and diffuse through the synaptic cleft to the postsynaptic dendritic spine.
There are six known actin isoforms, αskeletal, αcardiac, αsmooth, γsmooth, βcyto and γcyto actins, named after either the type of muscle they are found in (cardiac, skeletal, or smooth) or after their subcellular localization (cytoplasmic) (Perrin and Ervasti, 2010). Neurons are only known to express the cytoplasmic β and γ actin isoforms (Chiba et al., 1990; McHugh et al., 1991). Monomeric, globular actin (G-actin) polymerizes to produce actin filaments (F-actin). The two ends of F-actin have different critical concentrations for incorporation and loss of monomers; actin filaments have a fast-growing (barbed) and a slow-growing (pointed) ends. Regulation of the balance between G-actin and F-actin in distinct subcellular compartments within neurons is instrumental in driving neuronal development and function for review see (Pak et al., 2008)). The pointed end of actin filaments has largely been ignored in neuronal studies; they have been considered only as a source of actin monomers as the filament depolymerizes.
Of several actin populations inside growth cones three are most important for neurite growth: bundled actin in filopodia, and branched actin in lamellipodia and actin arcs in the transition zone between the lamellipodia and the central, microtubule-rich region (for review see (Lowery and Van Vactor, 2009)). In the leading edge of growth cones, distally oriented barbed ends of actin filaments alter membrane morphology by extending and pushing on the cellular membrane. Simultaneously, actin monomers depolymerize from the proximally oriented pointed ends to provide a constant supply of monomers for repolymerization. There are hosts of proteins and signaling pathways that converge on actin dynamics at the leading edge (for review see (Ridley, 2011)). Spatial separation in cues gives rise to multiple sub-populations of actin filaments with distinct properties and functions. For example, in the lamellipodia of growth cones, actin is highly branched by ARP2/3 (Rotty et al., 2013), while bundled actin filaments in filopodia give them their narrow shape (Vignjevic et al., 2006).
In addition to its roles in neurite development, actin has extensive functions in the formation and behavior of both the pre- and post-synapse (for reviews see (Lei et al., 2016; Rust and Maritzen, 2015)). Actin provides structures for the localization and transport of synaptic vesicles to and from the readily releasable pool of vesicles in axon termini. The dynamic behavior of actin is necessary for fusion of vesicles with the membrane and may also play a role in endo- and exocytosis at the pre-synaptic membrane (for review see (Cingolani and Goda, 2008)). The presynaptic neuron releases signals released into the synaptic cleft. The postsynaptic neuron receives the signal through dendritic spines and then translate, process and further propagate the message.
Actin’s versatility arises from the incredible number of actin-associated proteins that alter actin dynamics or use actin as a structural support for transport. Modes of actin regulation include sequestering or activating actin monomers, stabilizing or destabilizing actin filaments, capping or uncapping actin filaments, and increasing growth rates of actin filaments, reviewed in (dos Remedios et al., 2003). Molecular motors utilize actin filaments to alter cell shape and transport vesicles (for reviews see (Arnold and Gallo, 2014; Hirokawa et al., 2010)).
There is still considerable work to do in order to fully understand the role of actin dynamics at the pointed ends in the nervous system. In the literature actin dynamics at the pointed ends is considered critical to control the supply of actin monomers to be polymerized at the barbed end. Pointed ends can be capped by tropomodulins (Tmods), which bind directly onto the pointed end of actin filaments and onto tropomyosins (Tpms), which form a co-polymer with the actin filament (Figure 1B) These proteins have been scantily studied in the nervous system even though they influence many aspects of cells, including cell shape and motility (summarized in Table 1); some evidence suggests that they are connected with disease pathology, such as Alzheimer’s disease, Down syndrome and epilepsy (summarized in Table 2). Tpms are so influential that they have been dubbed “the “master regulator of actin filaments” (Gunning et al., 2015b).
Figure 1.
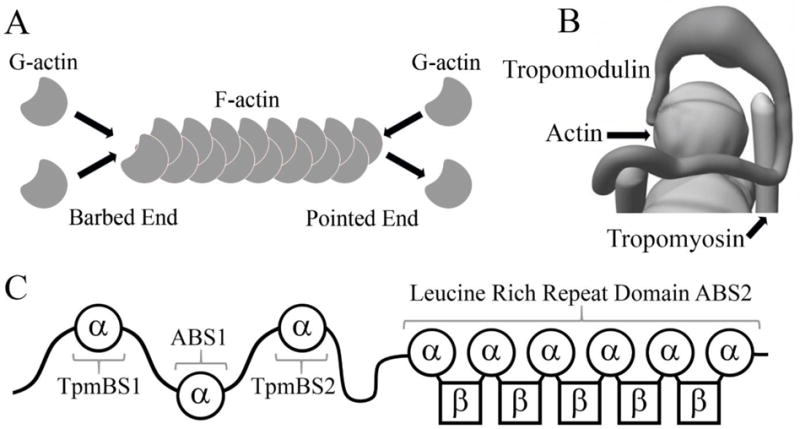
(A) Schematic of an actin filament. (B) Schematic of actin pointed-end capping by Tmod, modified from (Colpan et al., 2013) which was adapted from (Kostyukova et al., 2006). (C) Tmod structural domains and functional sites.
Table 1.
Expression and regulatory function of Tmods and Tpms in neurons
| Tmod isoform | Expression / Function | mRNA/Protein | System | Reference |
|---|---|---|---|---|
|
| ||||
| Tmod1 | Localizes to neuronal growth cones with developmental shift from proximal to a more uniform localization | Protein | Rat primary hippocampal neurons | (Fath et al., 2011) |
| Knockdown of Tmod1 leads to increased neurite extension | Protein | Mouse N2a neuroblastoma cell | (Fath et al., 2011) | |
| Increase in dendritic complexity in response to Tmod1 overexpression | Protein | Rat primary hippocampal neurons | (Gray et al., 2016) | |
| Increase in the number of filopodia/thin spines and total number of spines in response to Tmod1 overexpression | Protein | Rat primary hippocampal neurons | (Gray et al., 2016) | |
|
| ||||
| Tmod2 | Knockdown of Tmod2 leads to decreased neurite extension | Protein | Rat primary hippocampal neurons | (Fath et al., 2011) |
| Increase in dendritic complexity in response to Tmod2 overexpression | Protein | Rat primary hippocampal neurons | (Gray et al., 2016) | |
| Increase in the number of mushroom/stubby spines and total number of spines in response to Tmod2 overexpression | Protein | Rat primary hippocampal neurons | (Gray et al., 2016) | |
| Depletion leads reduced sensorimotor gating, hyperactivity and to enhanced LTP with deficits in learning and memory | mRNA/Protein | Knockout mice | (Cox et al., 2003) | |
| Compensatory upregulation of Tmod1 | Protein | Mouse hippocampal slices and cultured mouse hippocampal neurons | (Cox et al., 2003) | |
| Increased expression in response to chemically induced LTD by down regulation of miRNA-191 | Protein | (Hu et al., 2014) | ||
|
| ||||
| Tmod3 | Expressed in the brain | Protein | Protein | (Cox and Zoghbi, 2000) |
|
| ||||
| Overexpression did not impact complexity of dendritic arbor or dendritic spines | Protein | Rat primary hippocampal neurons | (Gray et al., 2016) | |
|
| ||||
| Tpm1.8/9 | Localization to growth cones in early developing neurons | Protein | Mouse primary cortical neurons | (Schevzov et al., 2005) |
| (Schevzov et al., 1997) | ||||
|
| ||||
| Tpm1.10 | Promotes reduction in growth cone size | Protein | B35 rat neuroblastoma cells | (Curthoys et al., 2014) |
|
| ||||
| Tpm1.11 | Present throughout the cell soma of the developing neuron | mRNA | Embryonal day 13 rat midbrain Mouse primary cortical neurons | (Hannan et al., 1995) |
| Promotes increase in neurite extension | Protein | Embryonal day 13 rat midbrain | (Hannan et al., 1996) | |
| Protein | B35 rat neuroblastoma cells | (Curthoys et al., 2014) | ||
|
| ||||
| Tpm1.12 | Localization in cell bodies and neurites | Protein | Rat primary mesencephalic and cortical neurons | (Stamm et al., 1993) |
| Promotes increase in growth cone size Promotes increase in filopodia formation Promotes increase in neurite branching |
Protein | B35 rat neuroblastoma cells | (Curthoys et al., 2014) | |
|
| ||||
| Tpm1.10/1.12 | Localization to the presynaptic compartment | Protein | Mouse primary hippocampal neurons | (Guven et al., 2011) |
| Rat cerebellar tissue | (Had et al., 1994) | |||
|
| ||||
| Tpm3.1–3.11 | Localization to the postsynaptic compartment | Protein | Mouse primary hippocampal neurons | (Guven et al., 2011) |
|
| ||||
| Tpm3.1 | Regulation of axonal and dendritic outgrowth and growth cone size | Protein | Tpm3.1-transgenic mice /mouse primary cortical neurons | (Schevzov et al., 2005) (Fath et al., 2010) |
| Regulation of the actin polymer in growth cones | Tpm3.1-transgenic mice/mouse primary hippocampal neurons | (Schevzov et al., 2008) | ||
|
| ||||
| Tpm3.1/2 | Localization to cell body in immature neurons, localization to the axon hillock in differentiating and recession to the cell body in mature neurons | mRNA | Mouse primary cortical neurons Adult rat cerebellum | (Hannan et al., 1995) |
| Localization to the developing axon, absent from the neuronal soma | Protein | Embryonic day 13 rat dorsal root ganglia | (Hannan et al., 1998) | |
| Regulation of axonal and dendritic outgrowth and growth cone size | Protein | Tpm3.1/2-knockout mice Mouse primary cortical neurons | (Fath et al., 2010) | |
| Distributed throughout cell body, not present in neurites | mRNA | Differentiated PC12 cells | (Hannan et al., 1995) | |
| Enriched at the periphery of cell body, extending into all neurites | Protein | Differentiated PC12 cells | (Hannan et al., 1995) | |
|
| ||||
| Tpm3.3/3.5 | Pyramidal cell bodies | Protein | Postnatal day 5 mouse cerebellum | (Vrhovski et al., 2003) |
| Compensatory upregulation in the brain in response to Tpm 3.4/3.7 knockout | Protein | Mouse brain | (Vrhovski et al., 2004) | |
|
| ||||
| Tpm3.4/3.7 | Enriched in the molecular layer, the Purkinje cells and the synaptic glomeruli with the granule cell layer | Protein | Adult rat cerebellum | (Dufour et al., 1998) |
| Pyramidal cell bodies | Protein | Postnatal day 5 mouse cerebellum | (Vrhovski et al., 2003) | |
| Compensatory upregulation in the brain in response to Tpm 3.1/2 knockout | Protein | Mouse brain | (Fath et al., 2010) | |
|
| ||||
| Tpm4.2 | Localization to the axonal pole of neurons, entering the proximal region of the axon | mRNA | Embryonal day 13 rat hindbrain | (Hannan et al., 1998) |
| Localization in the neuronal somata and axons | Protein | Embryonal day 13 rat midbrain | (Hannan et al., 1998) | |
| Localization to the postsynaptic compartment | Protein | Mouse primary hippocampal neurons | (Guven et al., 2011) | |
| Protein | Rat cerebellar tissue | (Had et al., 1994) | ||
| Promotes increase in growth cone size Promotes increase in filopodia formation Promotes increase in neurite branching | Protein | B35 rat neuroblastoma cells | (Curthoys et al., 2014) | |
It should be noted that the antibody (WSα/9d) used in this study may show some cross-reactivity with Tpm3.1/2
Table 2.
Dysregulation of Tmods and Tpms in the nervous system after injury and in disease
| Tmod isoform | Injury / Disease | Reference |
|---|---|---|
|
| ||
| Tmod 2 | Protein expression reduced in epilepsy | (Yang et al., 2006) |
| mRNA is increased 30% in ischemic stroke | (Chen et al., 2007) | |
| Protein expression found increased by 25% after methamphetamine exposure in rats | (Iwazaki et al., 2006) | |
| Protein expression tripled in Down syndrome | (Sun et al., 2011) | |
|
| ||
| Tpm isoform | Injury / Disease | Reference |
|
| ||
| Tpms [no discrimination between individual isoforms] | Abundant in Hirano body inclusions in Alzheimer’s disease | (Galloway et al., 1987) |
| Present in neurofibrillary tangles of Alzheimer’s disease, supranuclear palsy and Pick’s disease | (Galloway et al., 1990; Galloway and Perry, 1991) | |
| Found as component of Lewy bodies in diffuse Lewy body disease | (Galloway and Perry, 1991) | |
|
| ||
| Tpm3 isoforms [no discrimination between individual isoforms] | 2-fold increase in brains of Alzheimer’s disease cases after pull-down with Concanavalin A | (Owen et al., 2009) |
| Downregulation of Tpm3 isoform expression in Schizophrenia | (Martins-de-Souza et al., 2009) | |
|
| ||
| Tpm1 and Tpm3 isoforms [no discrimination between isoforms] | Increased expression in the white matter of brains from Alzheimer’s disease cases | (Castano et al., 2013) |
Tropomodulins and Tropomyosins – Discovery, Structure, and Regulation
Tmod Discovery and Isoforms
The first Tmod isoform (Tmod1) was identified in 1987 as a 43 kDa, Tpm-binding protein from erythrocyte membranes (Fowler, 1987). An additional three isoforms (Tmod2, Tmod3, and Tmod4) have since been identified (Almenar-Queralt et al., 1999; Cox and Zoghbi, 2000; Watakabe et al., 1996). Tmod1 and Tmod3 are expressed in many tissues including the brain; Tmod2 is expressed only in the brain (Almenar-Queralt et al., 1999; Cox and Zoghbi, 2000; Fowler, 1987; Watakabe et al., 1996) (see Table 1 for details on Tmod isoforms in the brain and their functional characterization). Tmod Structure. Tmods have four characterized binding sites, two actin binding sites and two Tpm binding sites (Fowler et al., 2003; Greenfield and Fowler, 2002; Greenfield et al., 2005; Kostyukova et al., 2006; Kostyukova et al., 2005). Tmods can be divided into two structurally and functionally distinct halves (Figure 1C). The N-terminal domain is largely disordered until binding to the pointed end (Kostyukova et al., 2000; Kostyukova et al., 2001). This domain contains two Tpm-binding sites (Tpm binding site 1 and 2) which flank the first actin-binding site (actin- binding site 1) (Greenfield and Fowler, 2002; Greenfield et al., 2005; Kostyukova et al., 2006; Kostyukova et al., 2005). Tmodl’s actin-binding site 1 binds actin across an extended surface which contains residues 64–77 of Tmod1 (Kostyukova et al., 2005) (Rao et al., 2014). In contrast to the N-terminal domain, the C-terminal domain is well-structured, and contains leucine-rich-repeat (LRR) domain (Krieger et al., 2002). The LRR domain is the other actin-binding site (actin-binding site 2) (Fowler et al., 2003; Rao et al., 2014). The LRR domain forms a repeated pattern of α-helices and β-strands, and the C-terminal α-helix blocks solvent from the domain’s hydrophobic core (Krieger et al., 2002), this motif is common among LRR containing proteins (for review see (Bella et al., 2008)). The βα-loops have residues with large side chains that create an extended ridge (Krieger et al., 2002). The αβ-loops have a series of asparagine residues that form hydrogen bonds in a motif called an asparagine ladder. Tmod2’s LRR domain has a structure similar to Tmod1, but was less resistant to denaturation by urea and more susceptible to protease degradation (Guillaud et al., 2014). The difference in stability of Tmod1’s and Tmod2’s LRR domains may be explained by difference in the orientation of the C-terminal α-helix, which is crucial for the LRR domain’s ability to bind actin (Colpan et al., 2016; Kostyukova and Hitchcock-DeGregori, 2004). These differences may contribute to the difference in affinity for actin monomers.
Tmod Expression Regulation
Little is known about the transcriptomic and genomic regulation of Tmods. Here we summarize knockout experiments and experiments that connect miRNAs and wnt/NF-кB pathways to Tmod expression levels. Tmod1 and Tmod3 knockout result in embryonic lethality due to disrupted heart formation and erythroid differentiation, respectively (Fritz-Six et al., 2003; Ono et al., 2005; Sui et al., 2014). In contrast, Tmod2 knockout is not lethal but results in impaired neurological development (Cox et al., 2003). Interestingly, in both Tmod1 and Tmod2 knockout, cells compensate for the lack of the specific isoform by altering expression and organization of the remaining isoforms (Cox et al., 2003; Moyer et al., 2010a);however, it is not enough to recover the lost function in either case.
Two miRNA’s have been shown to regulate Tmod expression; miRNA-23b-3p and miRNA-191 regulate Tmod1 and Tmod2 respectively. Erythroleukemia cells down-regulate miRNA-23b-3p in response to shear stress (Mu et al., 2015). Mu et al. speculate that Tmod1 is upregulated to further stabilized F-actin to increase resistance to shear stress. Tmod2 expression is increased in chemically induced long-term depression (LTD) by down-regulation of miRNA-191 (Hu et al., 2014).
Tmod1 expression has been connected to noncanonical wnt signaling pathways (Yun et al., 2007). Wnt signaling modulates many cellular functions including tissue development, cellular polarity and motility, reviewed in (Gomez-Orte et al., 2013; MacDonald et al., 2009; Rao and Kuhl, 2010). In neurons, wnt is a powerful modulator of development of nervous system affecting development of axons, dendrites and dendritic spines; components of the wnt signaling pathways are disrupted in Alzheimer’s Disease (for a review, see (Rosso and Inestrosa, 2013)). Tmod1 was up-regulated by non-canonical wnt signaling in L929 cells (Yun et al., 2007). This pathway regulated motility and altered cell shape, which are known to be altered by changes in Tmod expression (Fath et al., 2011; Fischer et al., 2003; Guillaud et al., 2014; Moroz et al., 2013a). We have shown that overexpression of Tmod1 increases the complexity of the dendritic arbor (Gray et al., 2016). These two observations suggest that upregulation of Tmod1 via wnt signaling could be a mechanism by which the brain may signal to increase dendritic complexity. Tmod regulation by post-translational modifications. Several post-translational modifications of Tmod2 were observed in a proteomic study, including fucosylation of serine 125, hydroxylation on asparagine 142, gamma-carboxylation on glutamic acids 276 and 280 and aspartic acid 282, and hyposin modification on lysine 316 (Sun et al., 2011). However, the study did not investigate the consequences of these modifications.
Tmods can be phosphorylated; for example, Tmod1 was found to be phosphorylated in cardiomyocytes (Bliss et al., 2014). TRPM7 phosphorylates Tmod1 (Dorovkov et al., 2008) and Tmod2 (Kostyukova, Dorovkov, unpublished) in vitro. Overexpression of a phospho-mimic Tmod1 in cardiomyocytes had weakened association with pointed ends and did not decrease thin filament length like either of the wild-type or phospho-null mutant (Bliss et al., 2014). TRPM7 is involved in actin regulation and cell death in neurons (for review see (Asrar and Aarts, 2013)). TRPM7 is known to inactivate LIMK1 which in turn is a suppressor of cofilin (Arber et al., 1998; Meng et al., 2003; Meng et al., 2002). Recently, inhibition and knockdown of TRPM7 was shown to increase the rate of progression through neuronal development stages and increase the length of axons (Turlova et al., 2016). Some of TPRM7’s effects could be through phosphorylation of Tmods, which are present in axonal shafts (Fath et al., 2011).
Tmod1 has been shown to be phosphorylated by Protein Kinase C (PKC) α (Wagner et al., 2002). PKCs are a family of calcium-dependent kinases that modulate the actin cytoskeleton (Larsson, 2006). In most cell types, activation of PKCs results in a loss of stress fibers and increases membrane ruffles and cellular migration.
Recently, Tmod3 was found to be phosphorylated by akt2 in actin-binding site 1 in insulin-treated adipocytes (Lim et al., 2015; Lim and Han, 2014). Phosphorylation of Tmod3 increased fusion of vesicles carrying the glucose transporter GLUT4 in the cellular membrane. Glucose-related signaling is known to affect hippocampal neural plasticity (reviewed in (Mainardi et al., 2015)).
Tpm Discovery and Isoforms
Bailey et al. first purified Tpm from skeletal and cardiac muscle in 1946, and described it as “an unusual protein of fibrous character” (Bailey, 1946). It was not until the 1970’s when non-muscle Tpms were found in the brain and other tissues (Fine et al., 1973). Early studies began to document the diversity of Tpms, noting variations in length, amino acid composition, affinity for actin, and began to speculate on the diversity of their functions. The Tpm family has expanded to include four genes (TPM1, TPM2, TPM3, and TPM4, previously α-Tm, β-Tm, γ-Tm, δ-Tm respectively) which give rise to pre-mRNA that can be alternatively spliced into 40 known isoforms (reviewed in (Gunning et al., 2008)). In this review, we will use a recently offered systematic nomenclature for Tpms (Geeves et al., 2015). When discussing closely numbered Tpms such as Tpm3.1 and Tpm3.2, we will refer to them as Tpm3.1/2. Neuronal cells express products of the TPM1, TPM3, and TPM4 genes. There are three brain-specific Tpm isoforms (Tpm1.10, Tpm1.11, Tpm1.12, previously TmBr1, TmBr2, and TmBr3) (Lees-Miller et al., 1990) (see Table 1 for details on Tpm isoforms in the brain and their functional characterization). Tpms are divided into two broad categories based on size: high molecular weight (HMW) or low molecular weight (LMW). HMW isoforms contain the 1a and 2a/b exons. LMW isoforms contain the 1b exon in place of the 1a exon and lack the 2a/b exons.
Tpm Structure
The structure and function of Tpms have been extensively reviewed recently (Gunning et al., 2015a; Gunning et al., 2015b). Tpm is an α-helical protein which dimerizes in a head-to-tail fashion to form a left-handed coiled-coil structure. Tpm binds to actin filaments through Tpm’s negative surface residues which interact with the positive residues in F-actin’s major groove (von der Ecken et al., 2015).
Tpm Regulation by post-translational modification
The most studied post-translational modification of Tpms is acetylation of its N-terminus. Acetylation supports the formation of the coiled-coil structure, local to the N-terminus (Brown et al., 2001). Disruption of Tpm’s coiled coil near the N-terminus by either mutagenesis or removing the acetyl group from the 1a exon of Tpms reduces actin binding (Moraczewska et al., 2000) and Tmod binding (Greenfield and Fowler, 2002). This is especially important for HMW Tpms; however, Tmods can effectively cap actin filaments decorated with LMW non-acetylated Tpms (Colpan et al., 2016; Kostyukova and Hitchcock-DeGregori, 2004; Kostyukova et al., 2007). Inhibition of the N-terminal acetyltransferase B complex (NatB), which acetylates Tpms, causes loss of actin stress fibers (Van Damme et al., 2012). Studies in yeast furthermore suggest a role for Tpm acetylation by NatB (Singer and Shaw, 2003) in regulating the motility of specific myosins (Coulton et al., 2010). Acetylation of yeast tropomyosin (Cdc8) influences the motility of class II myosins but does not impact on the motility of class I and V myosins (Coulton et al., 2010). Although there is evidence for the functional regulation of Tpms by acetylation in neuronal growth cones (discussed below), there is currently no evidence for acetylation of Tpms at synaptic connections in the brain.
Tpms containing the 9a exon can be specifically phosphorylated at S283 near the C-terminus (Mak et al., 1978) by PKCζ or DAP Kinase (Houle et al., 2007; Wu and Solaro, 2007). Phosphorylation of Tpm does not cause measurable changes in direct interactions with actin but alters the recruitment of myosins onto thin filaments (Rao et al., 2009). Structural studies indicate that this phosphorylation causes an increase in stiffness of the Tpm-Tpm binding junction (Lehman et al., 2015). Hippocampal neurons express PKCζ (Wu et al., 2012). There is a neural-specific splice variant of PKCζ (PKMζ) which has been implicated in learning and memory (Hernandez et al., 2003). PKCζ phosphorylation of Tpms may be a mechanism by which PKCζ can alter actin regulation in neurons and alter synaptic strength and plasticity. It is interesting that both Tmod1 and Tpms have been shown to be phosphorylated by different PKC isoforms; there may be a larger theme of pointed end regulation connected through calcium-activated PKCs. DAP kinase is associated with dendrite and axon retraction in ceramide-induced neuronal apoptosis (Pelled et al., 2002; Venkataraman and Futerman, 2000). Tpms are a logical target for DAP kinase because Tpms stabilize F-actin, which is lost during apoptosis.
Tropomodulins and Tropomyosins Regulate Actin Independently and Synergistically
Tmods and Tpms each have specific roles in modulating actin filaments and dynamics, they also synergistically form a cap on the pointed ends of filaments. Here we will discuss their respective and emergent roles in regulating actin. Tmods binding to G-actin can alter actin dynamics in two different modes: (1) by sequestering actin (Fischer et al., 2006) and (2) by nucleating actin filaments (Yamashiro et al., 2010). When Tmod binds one actin monomer it acts as an actin sequestering protein. When Tmod binds two actin monomers, it acts as a nucleator. Tmod2 has the strongest actin nucleating ability while Tmod1 has the weakest (Yamashiro et al., 2010).
Tpms bestow actin filaments with defined physical and mechanical properties. The vast diversity of Tpms generates a remarkable variety of actin filament sub-populations. Tpms alter the mechanical and physical properties of actin filaments by two broad mechanisms: (1) affecting stability and structure and (2) by modifying the recruitment of other actin-associated proteins.
As Tmods, Tpms also have isoform-specific impacts on actin polymerization rates (Janco et al., 2016; Kis-Bicskei et al., 2013; Robaszkiewicz et al., 2016) and either increase or decrease the rate. Tpms regulate actin branching by ARP2/3 (Blanchoin et al., 2001) in an isoform-specific manner (Brayford et al., 2016; Kis-Bicskei et al., 2013). They also have differential effects on the activity of the actin filament severing proteins, such as ADF/cofilin (Bryce et al., 2003; Curthoys et al., 2014; Gateva et al., 2017; Robaszkiewicz et al., 2016). The mechanical properties of actin filaments are influenced by the ability of Tpms to allow or block the access of the actin motor protein myosin II (Bryce et al., 2003). Tpms can uncap and anneal actin fragments that have been severed and capped by gelsolin (Ishikawa et al., 1989a; Ishikawa et al., 1989b). A single Tpm isoform can have a broad range of impacts on a filament, for example, Tpm3.1 can inhibit ARP2/3 recruitment (Brayford et al., 2016), reduce cofilin-1 binding to F-actin (Robaszkiewicz et al., 2016) and recruit myosin IIB (Bryce et al., 2003). Importantly, recent data highlight the complexity by which Tpm isoforms regulate actin dynamics in cells as compared to in vitro reconstituted systems (Gateva et al., 2017; Ostrowska et al., 2017). Cells overexpression of Tpm3.1 (Bryce et al., 2003) and Tpm4.2 (Curthoys et al., 2014) show increased inactivation of cofilin as indicated by an increase in phosphorylated cofilin over total cofilin levels. In contrast, Tpm3.1, Tpm3.2 (showing close homology to Tpm3.1) and Tpm4.2 did not inhibit severing of in vitro assembled actin filaments by cofilin (Gateva et al., 2017; Ostrowska et al., 2017). This apparent difference in Tpm activity in vitro and in vivo may be due to Tpm regulating upstream kinases and/or phosphatase of cofilin in a cellular environment which are absent in the reconstituted system in vitro. The study by Gateva and colleagues further suggests that structural differences of Tpms are contributing to their differential regulation of activity and access of other actin-associated proteins to actin filaments. While HMW Tpm1.6 and 1.7 protected actin filaments from cofilin-mediated disassembly and did not activate non-muscle myosin IIa, LMW Tpm3.1, 3.2 and 4.2 showed the opposite effect (Gateva et al., 2017; Ostrowska et al., 2017). Further experiments will be needed to establish the structure/function relationship of different Tpms in regulating the stability and dynamic properties of actin filaments.
Tmods are largely regarded as an actin pointed end capping protein (Gregorio et al., 1995; Weber et al., 1994). Only a single Tmod is necessary to cap the pointed end once bound (Kostyukova et al., 2006). Tmod caps by binding the pointed end through concerted action of its actin-binding sites and two Tpm binding sites (Fowler et al., 2003; Kostyukova et al., 2007). The interaction between Tmod and Tpm enhances the capping many fold (Weber et al., 1994). The affinities of Tmods’ binding sites vary between isoforms (Colpan et al., 2016; Uversky et al., 2011; Yamashiro et al., 2010). This gives rise to isoform specific variances in binding to different Tpm-decorated actin filaments (Colpan et al., 2016; Yamashiro et al., 2014). For instance, Tmod3 binds to the pointed ends of actin filaments decorated with Tpm3.1 more tightly than either Tmod1 or Tmod2 (Colpan et al., 2016). This variation in binding affinities give rise to additional degrees of complexity in actin regulation as Tmods can non-exclusively bind onto many types of filaments in a cellular subdomain.
Tropomodulins and Tropomyosins Regulate Neuronal Morphology
While Tpms and Tmods are intimately related at the pointed ends of actin filaments, only one paper has studied the importance of their interaction in neurons (Gray et al., 2016). Here we will explore the known roles of Tpms and Tmods on the formation of growth cones, dendrites, dendritic spines, and axons.
Influence of Tmod and Tpm on nervous system
Tmod2 expression was found throughout the brain, especially in the cortex, hippocampus, olfactory bulbs, brainstem and cerebellum (Cox et al., 2003). The knockout of Tmod2 in mouse line did not cause any gross anatomical differences; however, it did cause reduced sensorimotor gating, hyperactivity, and deficits in learning and memory (Cox et al., 2003). Studies of selective knockout of Tpm isoforms do not report any gross anatomical or behavioral changes (Fath et al., 2010; Vrhovski et al., 2004).
Growth Cones
Tmod1 and Tmod2 have different localization patterns in hippocampal neurons in cultures (Fath et al., 2011). Tmod2 has a diffuse localization and is enriched in the center of growth cones. Tmod1 was also found to be diffusely distributed but localized to lamellipodia and radial actin. Tmodl’s localization seems to be developmentally regulated, shifting from a distal to a more uniform localization during maturation. These differential localizations suggest that Tmod1 and Tmod2 may impact different actin sub-populations in neurons.
Several Tpm isoforms from the TPM1, TPM3 and TPM4 genes have been found in the growth cones of newly formed neurites, including the Tpm1.8/9, Tpm3.1/2 and Tpm4.2. The pool of Tpms in the growth cone is instrumental in regulating filamentous actin (Schevzov et al., 2008). Disruption of the expression of Tpm3.1/2 caused a modest decrease in the area of growth cones, a small increase in the growth cone protrusion rate, and increased the number of dendritic and axonal branches (Fath et al., 2010). Mice lacking Tpm3.1/2 had a compensatory upregulation of Tpms containing the 9c exon from the TPM3 gene which may result in partial functional compensation.
Although a detailed study on the subcellular interaction between Tmods and Tpms in the growth cones is still lacking, recent findings suggest a close interaction between Tpms and Tmods in growth cones (Gray et al., 2016). Proximity ligation assays (Figure 2) indicated proximity between exogenously expressed Tmod1 and Tmod2 with the endogenous Tpm3.1/Tpm3.2 in growth cones of developing hippocampal neurons (Gray et al., 2016).
Figure 2. Proximity ligation assay (PLA) demonstrating close proximity between Tmod2 and Tpm3.1/2.
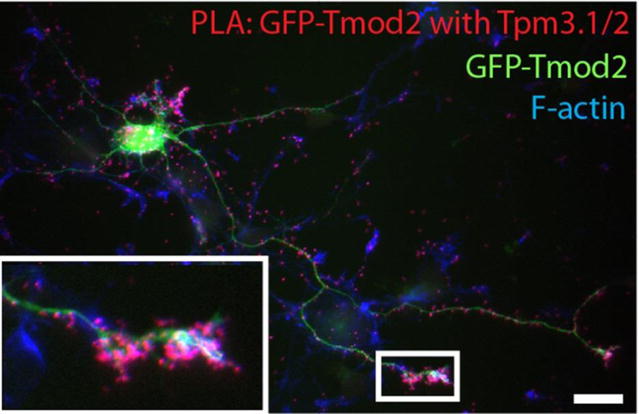
GFP-tagged Tmod2, expressed in mouse hippocampal neurons, cultured for 3 days shows close proximity to endogenous Tpm3.1/2. The red PLA-positive puncta, which indicate close proximity between Tmod and Tpm, are located in F-actin rich structures of the extending neurites. Note the growth cone enlarged in the zoomed insert (bottom left corner). Scale Bar = 20μm.
Dendrites
Recent data suggest isoform-specific roles of Tmod1, Tmod2, and Tmod3 in the development of the dendritic arbor in primary hippocampal neurons (Gray et al., 2016). Both Tmod1 and Tmod2 overexpression increased dendritic complexity. Tmod1 increased branching more distally from the cell body than Tmod2. The difference in dendritic branching mirrored the relative localization observed previously (Gray et al., 2016). Interestingly, Tmod3 had no effect on dendrite formation.
The most extensively studied Tpm in the context of neuronal morphogenesis is Tpm3.1. Overexpression of Tpm3.1 in cortical mouse neurons leads to an increase in axonal and dendritic arborization (Schevzov et al., 2005). Knockout of Tpm3.1/2 in mice leads to an increase in the branching of axons and dendrites in primary mouse neurons (Fath et al., 2010). These two pieces of information suggest that maintaining a precise pool of expressed Tpm3.1/2 is critical for the regulation of neurite arborization. Additionally, exogenous expression of Tpm1.7 increases dendritic complexity (Schevzov et al., 2005).
Dendritic Spines and the Synapse
In vivo and in vitro studies have connected brain-specific Tmod2 to synapse development (Cox et al., 2003; Gray et al., 2016; Hu et al., 2014). Knockout of Tmod2 increased long-term potentiation (LTP) in brain slices (Cox et al., 2003). This result was the first evidence implicating Tmod2 and synaptic signaling. Later, Tmod2 was found to be upregulated through altered expression of miRNA-191 after chemically induced long-term depression (LTD) (Hu et al., 2014) by treatment of neurons with glutamate, which overactivates NMDA receptors and weakens synaptic strength (Luscher and Malenka, 2012). Recently, Tmod2 overexpression was shown to cause an increase in the number of mature dendritic spines in hippocampal neurons (Gray et al., 2016).
There is some inconsistency between the observations that Tmod2 knockout causes an increase in LTP, and its overexpression causes spine maturation which is expected to enhance LTP. This inconsistency could be reconciled by (1) alterations to signaling pathways that act through Tmod2 or (2) altered expression of other proteins caused by the knockout. Hu et al. suggest that Tmod2 is a central point in a pathway that regulates spine dynamics (Hu et al., 2014). There are several post-translational modifications of Tmod2 (Sun et al., 2011) which may affect its function but are entirely unexplored in the literature. Perhaps these unstudied post-translational modifications regulate Tmod2 function in dendritic spines. An alternative explanation builds upon the observation that Tmod2 knockout causes an eight-fold upregulation of Tmod1 (Cox et al., 2003). Tmod1 overexpression causes an increase in the number of less mature filopodia and thin spines (Gray et al., 2016), which complicates the direct reasoning that reducing Tmod2 increases LTP.
Interestingly, Tmod1 and Tmod2 seem to differentially require their Tpm-binding abilities to impact neural morphology; Tmod1 requires its Tpm binding ability while Tmod2 does not (Gray et al., 2016). This is a clear indicator that Tmod-Tpm interactions are important in spinogenesis and depend on Tmod isoform. Previous work has shown the presence of products from both the TPM3 and the TPM4 gene in the postsynaptic compartment (Guven et al., 2011; Had et al., 1994). The presence of a diverse set of Tpm isoforms in this compartment and their differential interaction with Tmod1 and Tmod2 provides a potential mechanism of fine-tuning the regulation of the synaptic actin filament system. More information is needed to understand the full impact of the interactions between Tmods and Tpms in the synaptic compartment.
Axons
All we know about Tmods in axons is that both Tmod1, and Tmod2 have been found in axonal shafts seven days after plating (Fath et al., 2011). There is more information about the localization of Tpms with axonal development. Tpm1.8/9 mRNA and protein are spatially regulated in neurons with development; they initially localize to the base of a forming neurite, then along the axon and then is restricted into the soma at later stages of differentiation (Hannan et al., 1995; Weinberger et al., 1996). Expression of exogenous Tpm1.7 and Tpm3.1 alter neural morphology in an isoform-specific manner (Schevzov et al., 2005). Tpm3.1 overexpression increased both dendritic and axonal complexity while Tpm1.7 decreased dendritic complexity. Tpm3.1 overexpressing neurons also showed larger growth cones, increased recruitment of myosins and Tpm1.8 into growth cones. These effects are not seen in Tpm1.7 overexpressing neurons.
Tmod and Tpm in Neurological Disease
Despite increasing evidence for an important role of Tmods and Tpms in regulating morphogenesis and function of the nervous system, their roles in diseases of the nervous system are largely unknown, and no neurological disease-causing mutations have been found in these proteins to date. Aberrant expression of Tmods in the brain has been observed in several neurological conditions. Tmod2 mRNA increases after kainic acid induced seizures without an associated increase in protein amount (Sussman et al., 1994). Tmod2 and several other cytoskeletal proteins have reduced expression in epilepsy (Yang et al., 2006). Tmod2 mRNA is increased 30% after stroke (Chen et al., 2007). Tmod2 expression was found increased 25% after methamphetamine exposure in rats (Iwazaki et al., 2006) and tripled in Down syndrome (Sun et al., 2011). Possible connections between Tpms and neurodegenerative diseases are listed in Table 2 and have been recently reviewed in more detail (Brettle et al., 2016).
Future Directions
There is still considerable work to be done to understand actin pointed end regulation in cells of the nervous system and roles of Tmods and Tpms. Here we will explore some possible future directions for the studies of Tmods, Tpms and their influence in the nervous system.
Tmod3 is expressed in the brain but has been largely unstudied in the nervous system. In the only study of its role in neuronal development through overexpression, no impact was observed on either dendritic complexity or spine morphology (Gray et al., 2016). We hypothesize that Tmod3’s function is reserved for other purposes; one of these purposes could be regulating vesicle transport and their fusion with the cellular membrane. Recently, Tmod3 was found to be phosphorylated by Akt2 in adipocytes in response to insulin treatment (Lim et al., 2015). Fusion of GLUT4 carrying vesicles with the cellular membrane was found to be dependent on Tmod3 phosphorylation. Further investigation revealed that Tpm3.1 decoration defines F-actin sub-population impacted in this process (Kee et al., 2015). Tmod3’s function in neurons might be to regulate the dynamics of actin involved in vesicle transport. If this is the case, then Tmod3 should affect incorporation of membrane proteins and, by extension, synaptic signaling.
The impacts of Tmods on the axonal compartment are entirely unknown. The actin-spectrin network has a unique periodic structure along the axon (He et al., 2016; Xu et al., 2013). Tmods are known to be important players in regulating the length of actin filaments in the actin-spectrin network of red blood cells (Moyer et al., 2010b). Tmods may similarly define the length of actin filaments in the axonal actin-spectrin network. Furthermore, Akt2 is a known regulator of axonal length (Diez et al., 2012). This suggests that Tmods may be regulated by Akt2 in axons as well.
Two studies have used PC12 cells as a model system to study Tmods in neurite development (Guillaud et al., 2014; Moroz et al., 2013a). Although the impact of altered Tmod expression is quite different in primary neurons, the use of Tmod mutants hint at interesting differences in the function of Tmod’s LRR domain in neural morphology (Guillaud et al., 2014). Note, both Tmod1 and Tmod2 overexpression in hippocampal neurons increased the complexity of the dendritic arbor (Gray et al., 2016). In contrast, Tmod1 overexpression in PC12 cells had no impact on the number of neurite-like processes, while Tmod2 overexpression decreased them (Moroz et al., 2013b). Overexpression of either Tmod1 or Tmod2’s N-terminal domain reduces the number and length of neurite-like processes. Interestingly, overexpression of Tmod1 or a chimeric Tmod with Tmod2’s N-terminal domain with Tmod1’s LRR domain resulted in a “normal” phenotype, similar to Tmod1 overexpression. In cardiomyocytes, the LRR domain of Tmod1 is necessary for localization to the pointed ends (Tsukada et al., 2011). The LRR domains of Tmods may be similarly essential for shuttling Tmods to their respective locations in neurons. Additionally, a set of Tmod1 mutations was developed that specifically alter affinity for Tpm isoforms (Moroz et al., 2013a; Uversky et al., 2011); these mutations will be useful tools for further elucidating the functional significance of Tmod-Tpm interactions in the brain.
Beyond the shuttling of Tmods in neurons other significant questions remain to be answered about regulation of Tmods’ transcription and translation. One such question is: what is the mechanism that causes the upregulation of Tmod1 after Tmod2 knockout? As previously noted, Tmod2 knockout in mice resulted in an eight-fold increase of Tmod1 in the brain (Cox et al., 2003). The mechanism(s) of cross talk between the expressions of Tmod isoforms is unknown.
Another question lies in the observation that increased Tmod1 expression resulted in up-regulation of matrix metallopeptidase (MMP) 13 through stabilization of β-catenin (Ito-Kureha et al., 2015). MMPs are influential in the reorganization of the extracellular matrix. MMP9 is known to be upregulated by increasing β-catenin in neuronal stem cells (Ingraham et al., 2011) increasing spine length and reducing spine width (Michaluk et al., 2011). Tmod1 overexpression in hippocampal neurons increases dendritic spine length (Gray et al., 2016). Tmod1 stabilization may alter spine morphology via altering actin both directly and indirectly by stabilizing β-catenin and increasing MMP reorganization of the extracellular matrix.
Considerable effort has been spent on studying the effect of Tpm expression on the morphology of another type of neuron-like model cell line, B35 neuroblastoma cells. Altered expression of Tpm1.10, Tpm1.11, Tpm1.12 and Tpm4.2 (previously, TmBr1, TmBr2, TmBr3 and TM4 respectively) have a myriad of effects on B35 cell morphology (Curthoys et al., 2014). Tpm3.1 overexpression and knockout impact neurite extension and branching. Overexpression of Tpm3.1 increases the number and length of focal adhesions in B35 cells (Bach et al., 2009). Overexpression of Tpm1.12, Tpm3.1, and Tpm4.2 each increase the elastic modulus of B35 membranes (Jalilian et al., 2015). These data indicate the broad impacts Tpms can have in vivo. There is considerable work to be done in confirming Tpms’ effect in primary neurons and understanding the impacts of altered Tpm expression in neurons.
Conclusion
Morphological development of neurons is staggeringly complex, and there are many unanswered questions about the underlying mechanisms that drive and regulate development. Actin is an exceptionally influential protein in morphological development. Actin impacts, among other processes, membrane morphology, cellular motility, and vesicle transport. An essential part of behavior of actin filaments is the dynamics of the slow-growing, pointed end. The pointed end can be capped by the concerted binding of Tmod and Tpm onto the pointed end. Direct evidence and inferences from studies using other cell types indicate that these proteins impact the brain and neuronal morphology, and regulate synaptic plasticity and signaling. Although the present evidence highlights the importance of these proteins and their interactions, there is still work to be done. We outlined here some of the most pressing questions about the roles of Tmods and Tpms in the nervous system. Future work in this field will significantly expand on the regulation of the cytoskeleton and how it formulates the brain.
Highlights.
Review on the role of tropomodulins and tropomyosins in neurons
Discussion of the interaction between tropomodulin and tropomyosin in neurite outgrowth
Dysregulation of the actin tropomodulins, tropomyosins and the actin cytoskeleton in the diseased or injured nervous system
Acknowledgments
This work was supported by the Anne and Russ Fuller Interdisciplinary Research and Creative Activities Fellowship to KTG which allowed KTG to work in the lab of TF and Project Grant APP1083209 from the Australian National Health and Medical Research Council (NHMRC) (TF). KTG was supported by a NIH/NIGMS-funded predoctoral fellowship (T32 GM008336). We would like to thank Ms Holly Stefen for contributing images shown in Figure 2 of this article.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almenar-Queralt A, Lee A, Conley CA, Ribas de Pouplana L, Fowler VM. Identification of a novel tropomodulin isoform, skeletal tropomodulin, that caps actin filament pointed ends in fast skeletal muscle. J Biol Chem. 1999;274:28466–28475. doi: 10.1074/jbc.274.40.28466. [DOI] [PubMed] [Google Scholar]
- Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, Caroni P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393:805–809. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- Arnold DB, Gallo G. Structure meets function: actin filaments and myosin motors in the axon. Journal of neurochemistry. 2014;129:213–220. doi: 10.1111/jnc.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asrar S, Aarts M. TRPM7, the cytoskeleton and neuronal death. Channels (Austin) 2013;7:6–16. doi: 10.4161/chan.22824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach CT, Creed S, Zhong J, Mahmassani M, Schevzov G, Stehn J, Cowell LN, Naumanen P, Lappalainen P, Gunning PW, O’Neill GM. Tropomyosin isoform expression regulates the transition of adhesions to determine cell speed and direction. Molecular and cellular biology. 2009;29:1506–1514. doi: 10.1128/MCB.00857-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey K. Tropomyosin: a new asymmetric protein component of muscle. Nature. 1946;157:368. doi: 10.1038/157368b0. [DOI] [PubMed] [Google Scholar]
- Bella J, Hindle KL, McEwan PA, Lovell SC. The leucine-rich repeat structure. Cell Mol Life Sci. 2008;65:2307–2333. doi: 10.1007/s00018-008-8019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D, Toroian-Raymond A. Disoriented pathfinding by pioneer neurone growth cones deprived of filopodia by cytochalasin treatment. Nature. 1986;323:712–715. doi: 10.1038/323712a0. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD, Hitchcock-DeGregori SE. Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Current biology: CB. 2001;11:1300–1304. doi: 10.1016/s0960-9822(01)00395-5. [DOI] [PubMed] [Google Scholar]
- Bliss KT, Tsukada T, Novak SM, Dorovkov MV, Shah SP, Nworu C, Kostyukova AS, Gregorio CC. Phosphorylation of tropomodulin1 contributes to the regulation of actin filament architecture in cardiac muscle. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2014;28:3987–3995. doi: 10.1096/fj.13-246009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brayford S, Bryce NS, Schevzov G, Haynes EM, Bear JE, Hardeman EC, Gunning PW. Tropomyosin Promotes Lamellipodial Persistence by Collaborating with Arp2/3 at the Leading Edge. Current biology : CB. 2016;26:1312–1318. doi: 10.1016/j.cub.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettle M, Patel S, Fath T. Tropomyosins in the healthy and diseased nervous system. Brain research bulletin. 2016 doi: 10.1016/j.brainresbull.2016.06.004. [DOI] [PubMed] [Google Scholar]
- Brown JH, Kim KH, Jun G, Greenfield NJ, Dominguez R, Volkmann N, Hitchcock-DeGregori SE, Cohen C. Deciphering the design of the tropomyosin molecule. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8496–8501. doi: 10.1073/pnas.131219198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryce NS, Schevzov G, Ferguson V, Percival JM, Lin JJ, Matsumura F, Bamburg JR, Jeffrey PL, Hardeman EC, Gunning P, Weinberger RP. Specification of actin filament function and molecular composition by tropomyosin isoforms. Molecular Biology of the Cell. 2003;14:1002–1016. doi: 10.1091/mbc.E02-04-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castano EM, Maarouf CL, Wu T, Leal MC, Whiteside CM, Lue LF, Kokjohn TA, Sabbagh MN, Beach TG, Roher AE. Alzheimer disease periventricular white matter lesions exhibit specific proteomic profile alterations. Neurochemistry international. 2013;62:145–156. doi: 10.1016/j.neuint.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Liao WP, Lu Q, Wong WS, Wong PT. Upregulation of dihydropyrimidinase-related protein 2, spectrin alpha II chain, heat shock cognate protein 70 pseudogene 1 and tropomodulin 2 after focal cerebral ischemia in rats— a proteomics approach. Neurochemistry international. 2007;50:1078–1086. doi: 10.1016/j.neuint.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Chiba T, Nakamura Y, Sakiyama S. Developmental changes of expression of non-muscle (beta and gamma) actin mRNAs in the central nervous system studied by in situ hybridization. Neuroscience letters. 1990;112:31–36. doi: 10.1016/0304-3940(90)90317-3. [DOI] [PubMed] [Google Scholar]
- Cingolani LA, Goda Y. Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy.[erratum appears in Nat Rev Neurosci. 2008 Jun;9(6):494] Nature Reviews Neuroscience. 2008;9:344–356. doi: 10.1038/nrn2373. [DOI] [PubMed] [Google Scholar]
- Colpan M, Moroz NA, Gray KT, Cooper DA, Diaz CA, Kostyukova AS. Tropomyosin-binding properties modulate competition between tropomodulin isoforms. Archives of biochemistry and biophysics. 2016;600:23–32. doi: 10.1016/j.abb.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colpan M, Moroz NA, Kostyukova AS. Tropomodulins and tropomyosins: working as a team. Journal of muscle research and cell motility. 2013;34:247–260. doi: 10.1007/s10974-013-9349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulton AT, East DA, Galinska-Rakoczy A, Lehman W, Mulvihill DP. The recruitment of acetylated and unacetylated tropomyosin to distinct actin polymers permits the discrete regulation of specific myosins in fission yeast. J Cell Sci. 2010;123:3235–3243. doi: 10.1242/jcs.069971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox PR, Fowler V, Xu B, Sweatt JD, Paylor R, Zoghbi HY. Mice lacking tropomodulin-2 show enhanced long-term potentiation, hyperactivity, and deficits in learning and memory. Molecular and Cellular Neuroscience. 2003;23:1–12. doi: 10.1016/s1044-7431(03)00025-3. [DOI] [PubMed] [Google Scholar]
- Cox PR, Zoghbi HY. Sequencing, expression analysis, and mapping of three unique human tropomodulin genes and their mouse orthologs. Genomics. 2000;63:97–107. doi: 10.1006/geno.1999.6061. [DOI] [PubMed] [Google Scholar]
- Curthoys NM, Freittag H, Connor A, Desouza M, Brettle M, Poljak A, Hall A, Hardeman E, Schevzov G, Gunning PW, Fath T. Tropomyosins induce neuritogenesis and determine neurite branching patterns in B35 neuroblastoma cells. Mol Cell Neurosci. 2014;58C:11–21. doi: 10.1016/j.mcn.2013.10.011. [DOI] [PubMed] [Google Scholar]
- da Silva JS, Dotti CG. Breaking the neuronal sphere: regulation of the actin cytoskeleton in neuritogenesis. Nat Rev Neurosci. 2002;3:694–704. doi: 10.1038/nrn918. [DOI] [PubMed] [Google Scholar]
- Diez H, Garrido JJ, Wandosell F. Specific roles of Akt iso forms in apoptosis and axon growth regulation in neurons. PloS one. 2012;7:e32715. doi: 10.1371/journal.pone.0032715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorovkov MV, Beznosov SN, Shah S, Kotlianskaia L, Kostiukova AS. [Effect of mutations imitating the phosphorylation by TRPM7 kinase on the function of the N-terminal domain of tropomodulin] Biofizika. 2008;53:943–949. [PubMed] [Google Scholar]
- dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, Berry DA, Nosworthy NJ. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiological reviews. 2003;83:433–473. doi: 10.1152/physrev.00026.2002. [DOI] [PubMed] [Google Scholar]
- Dufour C, Weinberger RP, Gunning P. Tropomyosin isoform diversity and neuronal morphogenesis. Immunology and cell biology. 1998;76:424–429. doi: 10.1046/j.1440-1711.1998.00765.x. [DOI] [PubMed] [Google Scholar]
- Fath T, Chan YK Agnes, Vrhovski B, Clarke H, Curthoys N, Hook J, Lemckert F, Schevzov G, Tam P, Watson CM, Khoo PL, Gunning P. New aspects of tropomyosin-regulated neuritogenesis revealed by the deletion of Tm5NM1 and 2. Eur J Cell Biol. 2010;89:489–498. doi: 10.1016/j.ejcb.2009.11.028. [DOI] [PubMed] [Google Scholar]
- Fath T, Fischer RS, Dehmelt L, Halpain S, Fowler VM. Tropomodulins are negative regulators of neurite outgrowth. Eur J Cell Biol. 2011;90:291–300. doi: 10.1016/j.ejcb.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine RE, Blitz AL, Hitchcock SE, Kaminer B. Tropomyosin in brain and growing neurones. Nat New Biol. 1973;245:182–186. doi: 10.1038/newbio245182a0. [DOI] [PubMed] [Google Scholar]
- Fischer RS, Fritz-Six KL, Fowler VM. Pointed-end capping by tropomodulin3 negatively regulates endothelial cell motility. The Journal of cell biology. 2003;161:371–380. doi: 10.1083/jcb.200209057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer RS, Yarmola EG, Weber KL, Speicher KD, Speicher DW, Bubb MR, Fowler VM. Tropomodulin 3 binds to actin monomers. J Biol Chem. 2006;281:36454–36465. doi: 10.1074/jbc.M606315200. [DOI] [PubMed] [Google Scholar]
- Fowler VM. Identification and purification of a novel Mr 43,000 tropomyosin-binding protein from human erythrocyte membranes. The Journal of biological chemistry. 1987;262:12792–12800. [PubMed] [Google Scholar]
- Fowler VM, Greenfield NJ, Moyer J. Tropomodulin contains two actin filament pointed end-capping domains. J Biol Chem. 2003;278:40000–40009. doi: 10.1074/jbc.M306895200. [DOI] [PubMed] [Google Scholar]
- Fritz-Six KL, Cox PR, Fischer RS, Xu B, Gregorio CC, Zoghbi HY, Fowler VM. Aberrant myofibril assembly in tropomodulin1 null mice leads to aborted heart development and embryonic lethality. The Journal of cell biology. 2003;163:1033–1044. doi: 10.1083/jcb.200308164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway PG, Mulvihill P, Siedlak S, Mijares M, Kawai M, Padget H, Kim R, Perry G. Immunochemical demonstration of tropomyosin in the neurofibrillary pathology of Alzheimer’s disease. The American journal of pathology. 1990;137:291–300. [PMC free article] [PubMed] [Google Scholar]
- Galloway PG, Perry G. Tropomyosin distinguishes Lewy bodies of Parkinson disease from other neurofibrillary pathology. Brain research. 1991;541:347–349. doi: 10.1016/0006-8993(91)91036-z. [DOI] [PubMed] [Google Scholar]
- Galloway PG, Perry G, Gambetti P. Hirano body filaments contain actin and actin-associated proteins. J Neuropathol Exp Neurol. 1987;46:185–199. doi: 10.1097/00005072-198703000-00006. [DOI] [PubMed] [Google Scholar]
- Gateva G, Kremneva E, Reindl T, Kotila T, Kogan K, Gressin L, Gunning PW, Manstein DJ, Michelot A, Lappalainen P. Tropomyosin Isoforms Specify Functionally Distinct Actin Filament Populations In Vitro. Curr Biol. 2017;27:705–713. doi: 10.1016/j.cub.2017.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeves MA, Hitchcock-DeGregori SE, Gunning PW. A systematic nomenclature for mammalian tropomyosin isoforms. Journal of muscle research and cell motility. 2015;36:147–153. doi: 10.1007/s10974-014-9389-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Orte E, Saenz-Narciso B, Moreno S, Cabello J. Multiple functions of the noncanonical Wnt pathway. Trends in genetics : TIG. 2013;29:545–553. doi: 10.1016/j.tig.2013.06.003. [DOI] [PubMed] [Google Scholar]
- Gomez TM, Letourneau PC. Actin dynamics in growth cone motility and navigation. Journal of neurochemistry. 2014;129:221–234. doi: 10.1111/jnc.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray KT, Suchowerska AK, Bland T, Colpan M, Wayman G, Fath T, Kostyukova AS. Tropomodulin isoforms utilize specific binding functions to modulate dendrite development. Cytoskeleton (Hoboken) 2016;73:316–328. doi: 10.1002/cm.21304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield NJ, V, Fowler M. Tropomyosin requires an intact N-terminal coiled coil to interact with tropomodulin. Biophysical journal. 2002;82:2580–2591. doi: 10.1016/S0006-3495(02)75600-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield NJ, Kostyukova AS, Hitchcock-DeGregori SE. Structure and tropomyosin binding properties of the N-terminal capping domain of tropomodulin 1. Biophysical journal. 2005;88:372–383. doi: 10.1529/biophysj.104.051128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorio CC, Weber A, Bondad M, Pennise CR, Fowler VM. Requirement of pointed-end capping by tropomodulin to maintain actin filament length in embryonic chick cardiac myocytes. Nature. 1995;377:83–86. doi: 10.1038/377083a0. [DOI] [PubMed] [Google Scholar]
- Guillaud L, Gray KT, Moroz N, Pantazis C, Pate E, Kostyukova AS. Role of tropomodulin’s leucine rich repeat domain in the formation of neurite-like processes. Biochemistry. 2014;53:2689–2700. doi: 10.1021/bi401431k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning P, O’Neill G, Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiological reviews. 2008;88:1–35. doi: 10.1152/physrev.00001.2007. [DOI] [PubMed] [Google Scholar]
- Gunning PW, Ghoshdastider U, Whitaker S, Popp D, Robinson RC. The evolution of compositionally and functionally distinct actin filaments. Journal of cell science. 2015a;128:2009–2019. doi: 10.1242/jcs.165563. [DOI] [PubMed] [Google Scholar]
- Gunning PW, Hardeman EC, Lappalainen P, Mulvihill DP. Tropomyosin -master regulator of actin filament function in the cytoskeleton. Journal of cell science. 2015b;128:2965–2974. doi: 10.1242/jcs.172502. [DOI] [PubMed] [Google Scholar]
- Guven K, Gunning P, Fath T. TPM3 and TPM4 gene products segregate to the postsynaptic region of central nervous system synapses. Bioarchitecture. 2011;1:284–289. doi: 10.4161/bioa.1.6.19336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Had L, Faivre-Sarrailh C, Legrand C, Mery J, Brugidou J, Rabie A. Tropomyosin isoforms in rat neurons: the different developmental profiles and distributions of TM-4 and TMBr-3 are consistent with different functions. Journal of cell science. 1994;107(Pt 10):2961–2973. doi: 10.1242/jcs.107.10.2961. [DOI] [PubMed] [Google Scholar]
- Hannan AJ, Gunning P, Jeffrey PL, Weinberger RP. Structural compartments within neurons: developmentally regulated organization of microfilament isoform mRNA and protein. Molecular and cellular neurosciences. 1998;11:289–304. doi: 10.1006/mcne.1998.0693. [DOI] [PubMed] [Google Scholar]
- Hannan AJ, Henke RC, Weinberger RP, Sentry JW, Jeffrey PL. Differential induction and intracellular localization of SCG10 messenger RNA is associated with neuronal differentiation. Neuroscience. 1996;72:889–900. doi: 10.1016/0306-4522(95)00593-5. [DOI] [PubMed] [Google Scholar]
- Hannan AJ, Schevzov G, Gunning P, Jeffrey PL, Weinberger RP. Intracellular localization of tropomyosin mRNA and protein is associated with development of neuronal polarity. Molecular and cellular neurosciences. 1995;6:397–412. doi: 10.1006/mcne.1995.1030. [DOI] [PubMed] [Google Scholar]
- He J, Zhou R, Wu Z, Carrasco MA, Kurshan PT, Farley JE, Simon DJ, Wang G, Han B, Hao J, Heller E, Freeman MR, Shen K, Maniatis T, Tessier-Lavigne M, Zhuang X. Prevalent presence of periodic actin-spectrin-based membrane skeleton in a broad range of neuronal cell types and animal species. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:6029–6034. doi: 10.1073/pnas.1605707113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez AI, Blace N, Crary JF, Serrano PA, Leitges M, Libien JM, Weinstein G, Tcherapanov A, Sacktor TC. Protein kinase M zeta synthesis from a brain mRNA encoding an independent protein kinase C zeta catalytic domain. Implications for the molecular mechanism of memory. The Journal of biological chemistry. 2003;278:40305–40316. doi: 10.1074/jbc.M307065200. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Niwa S, Tanaka Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 2010;68:610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- Houle F, Poirier A, Dumaresq J, Huot J. DAP kinase mediates the phosphorylation of tropomyosin-1 downstream of the ERK pathway, which regulates the formation of stress fibers in response to oxidative stress. J Cell Sci. 2007;120:3666–3677. doi: 10.1242/jcs.003251. [DOI] [PubMed] [Google Scholar]
- Hu Z, Yu D, Gu QH, Yang Y, Tu K, Zhu J, Li Z. miR-191 and miR-135 are required for long-lasting spine remodelling associated with synaptic long-term depression. Nat Commun. 2014;5:3263. doi: 10.1038/ncomms4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur EM, Saijilafu, Zhou FQ. Growing the growth cone: remodeling the cytoskeleton to promote axon regeneration. Trends in neurosciences. 2012;35:164–174. doi: 10.1016/j.tins.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingraham CA, Park GC, Makarenkova HP, Crossin KL. Matrix metalloproteinase (MMP)-9 induced by Wnt signaling increases the proliferation and migration of embryonic neural stem cells at low O2 levels. The Journal of biological chemistry. 2011;286:17649–17657. doi: 10.1074/jbc.M111.229427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R, Yamashiro S, Matsumura F. Annealing of gelsolin-severed actin fragments by tropomyosin in the presence of Ca2+. Potentiation of the annealing process by caldesmon. The Journal of biological chemistry. 1989a;264:16764–16770. [PubMed] [Google Scholar]
- Ishikawa R, Yamashiro S, Matsumura F. Differential modulation of actin-severing activity of gelsolin by multiple isoforms of cultured rat cell tropomyosin. Potentiation of protective ability of tropomyosins by 83-kDa nonmuscle caldesmon. The Journal of biological chemistry. 1989b;264:7490–7497. [PubMed] [Google Scholar]
- Ito-Kureha T, Koshikawa N, Yamamoto M, Semba K, Yamaguchi N, Yamamoto T, Seiki M, Inoue J. Tropomodulin 1 expression driven by NF-kappaB enhances breast cancer growth. Cancer research. 2015;75:62–72. doi: 10.1158/0008-5472.CAN-13-3455. [DOI] [PubMed] [Google Scholar]
- Iwazaki T, McGregor IS, Matsumoto I. Protein expression profile in the striatum of acute methamphetamine-treated rats. Brain research. 2006;1097:19–25. doi: 10.1016/j.brainres.2006.04.052. [DOI] [PubMed] [Google Scholar]
- Jalilian I, Heu C, Cheng H, Freittag H, Desouza M, Stehn JR, Bryce NS, Whan RM, Hardeman EC, Fath T, Schevzov G, Gunning PW. Cell elasticity is regulated by the tropomyosin isoform composition of the actin cytoskeleton. PLoS One. 2015;10:e0126214. doi: 10.1371/journal.pone.0126214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janco M, Bonello TT, Byun A, Costre AC, Lebhar H, Dedova I, Gunning PW, Bocking T. The impact of tropomyosins on actin filament assembly is isoform specific. Bioarchitecture. 2016:1–15. doi: 10.1080/19490992.2016.1201619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee AJ, Yang L, Lucas CA, Greenberg MJ, Martel N, Leong GM, Hughes WE, Cooney GJ, James DE, Ostap EM, Han W, Gunning PW, Hardeman EC. An actin filament population defined by the tropomyosin Tpm3.1 regulates glucose uptake. Traffic. 2015;16:691–711. doi: 10.1111/tra.12282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kis-Bicskei N, Vig A, Nyitrai M, Bugyi B, Talian GC. Purification of tropomyosin Br-3 and 5NM1 and characterization of their interactions with actin. Cytoskeleton (Hoboken) 2013;70:755–765. doi: 10.1002/cm.21143. [DOI] [PubMed] [Google Scholar]
- Kostyukova A, Maeda K, Yamauchi E, Krieger I, Maeda Y. Domain structure of tropomodulin - Distinct properties of the N-terminal and C-terminal halves. Eur J Biochem. 2000;267:6470–6475. doi: 10.1046/j.1432-1327.2000.01738.x. [DOI] [PubMed] [Google Scholar]
- Kostyukova AS, Choy A, Rapp BA. Tropomodulin binds two tropomyosins: a novel model for actin filament capping. Biochemistry. 2006;45:12068–12075. doi: 10.1021/bi060899i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyukova AS, Hitchcock-DeGregori SE. Effect of the structure of the N terminus of tropomyosin on tropomodulin function. Journal of Biological Chemistry. 2004;279:5066–5071. doi: 10.1074/jbc.M311186200. [DOI] [PubMed] [Google Scholar]
- Kostyukova AS, Hitchcock-Degregori SE, Greenfield NJ. Molecular basis of tropomyosin binding to tropomodulin, an actin-capping protein. Journal of molecular biology. 2007;372:608–618. doi: 10.1016/j.jmb.2007.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyukova AS, Rapp BA, Choy A, Greenfield NJ, Hitchcock-DeGregori SE. Structural requirements of tropomodulin for tropomyosin binding and actin filament capping. Biochemistry. 2005;44:4905–4910. doi: 10.1021/bi047468p. [DOI] [PubMed] [Google Scholar]
- Kostyukova AS, Tiktopulo EI, Maeda Y. Folding properties of functional domains of tropomodulin. Biophysical journal. 2001;81:345–351. doi: 10.1016/S0006-3495(01)75704-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger I, Kostyukova A, Yamashita A, Nitanai Y, Maeda Y. Crystal structure of the C-terminal half of tropomodulin and structural basis of actin filament pointed-end capping. Biophys J. 2002;83:2716–2725. doi: 10.1016/S0006-3495(02)75281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson C. Protein kinase C and the regulation of the actin cytoskeleton. Cellular signalling. 2006;18:276–284. doi: 10.1016/j.cellsig.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Lees-Miller JP, Goodwin LO, Helfman DM. Three novel brain tropomyosin isoforms are expressed from the rat alpha-tropomyosin gene through the use of alternative promoters and alternative RNA processing. Molecular and cellular biology. 1990;10:1729–1742. doi: 10.1128/mcb.10.4.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman W, Medlock G, Li XE, Suphamungmee W, Tu AY, Schmidtmann A, Ujfalusi Z, Fischer S, Moore JR, Geeves MA, Regnier M. Phosphorylation of Ser283 enhances the stiffness of the tropomyosin head-to-tail overlap domain. Archives of biochemistry and biophysics. 2015;571:10–15. doi: 10.1016/j.abb.2015.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei W, Omotade OF, Myers KR, Zheng JQ. Actin cytoskeleton in dendritic spine development and plasticity. Current opinion in neurobiology. 2016;39:86–92. doi: 10.1016/j.conb.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CY, Bi X, Wu D, Kim JB, Gunning PW, Hong W, Han W. Tropomodulin3 is a novel Akt2 effector regulating insulin-stimulated GLUT4 exocytosis through cortical actin remodeling. Nature communications. 2015;6:5951. doi: 10.1038/ncomms6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CY, Han W. Tropomodulin3 as the link between insulin-activated AKT2 and cortical actin remodeling in preparation of GLUT4 exocytosis. Bioarchitecture. 2014;4:210–214. doi: 10.1080/19490992.2015.1031949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery LA, Van Vactor D. The trip of the tip: understanding the growth cone machinery. Nat Rev Mol Cell Biol. 2009;10:332–343. doi: 10.1038/nrm2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher C, Malenka RC. NMDA receptor-dependent long-term potentiation and long-term depression (LTP/LTD) Cold Spring Harbor perspectives in biology. 2012;4 doi: 10.1101/cshperspect.a005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Developmental cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainardi M, Fusco S, Grassi C. Modulation of hippocampal neural plasticity by glucose-related signaling. Neural plasticity. 2015;2015:657928. doi: 10.1155/2015/657928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak A, Smillie LB, Barany M. Specific phosphorylation at serine-283 of alpha tropomyosin from frog skeletal and rabbit skeletal and cardiac muscle. Proceedings of the National Academy of Sciences of the United States of America. 1978;75:3588–3592. doi: 10.1073/pnas.75.8.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins-de-Souza D, Gattaz WF, Schmitt A, Novello JC, Marangoni S, Turck CW, Dias-Neto E. Proteome analysis of schizophrenia patients Wernicke’s area reveals an energy metabolism dysregulation. BMC psychiatry. 2009;9:17. doi: 10.1186/1471-244X-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nature reviews Molecular cell biology. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- McHugh KM, Crawford K, Lessard JL. A comprehensive analysis of the developmental and tissue-specific expression of the isoactin multigene family in the rat. Developmental biology. 1991;148:442–458. doi: 10.1016/0012-1606(91)90263-3. [DOI] [PubMed] [Google Scholar]
- Meng Y, Zhang Y, Tregoubov V, Falls DL, Jia Z. Regulation of spine morphology and synaptic function by LIMK and the actin cytoskeleton. Reviews in the neurosciences. 2003;14:233–240. doi: 10.1515/revneuro.2003.14.3.233. [DOI] [PubMed] [Google Scholar]
- Meng Y, Zhang Y, Tregoubov V, Janus C, Cruz L, Jackson M, Lu WY, MacDonald JF, Wang JY, Falls DL, Jia Z. Abnormal spine morphology and enhanced LTP in LFMK-1 knockout mice. Neuron. 2002;35:121–133. doi: 10.1016/s0896-6273(02)00758-4. [DOI] [PubMed] [Google Scholar]
- Michaluk P, Wawrzyniak M, Alot P, Szczot M, Wyrembek P, Mercik K, Medvedev N, Wilczek E, De Roo M, Zuschratter W, Muller D, Wilczynski GM, Mozrzymas JW, Stewart MG, Kaczmarek L, Wlodarczyk J. Influence of matrix metalloproteinase MMP-9 on dendritic spine morphology. Journal of cell science. 2011;124:3369–3380. doi: 10.1242/jcs.090852. [DOI] [PubMed] [Google Scholar]
- Moraczewska J, Greenfield NJ, Liu Y, Hitchcock-DeGregori SE. Alteration of tropomyosin function and folding by a nemaline myopathy-causing mutation. Biophysical journal. 2000;79:3217–3225. doi: 10.1016/S0006-3495(00)76554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz N, Guillaud L, Desai B, Kostyukova AS. Mutations changing tropomodulin affinity for tropomyosin alter neurite formation and extension. PeerJ. 2013a;1:e7. doi: 10.7717/peerj.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz NA, Novak SM, Azevedo R, Colpan M, Uversky VN, Gregorio CC, Kostyukova AS. Alteration of tropomyosin-binding properties of tropomodulin-1 affects its capping ability and localization in skeletal myocytes. J Biol Chem. 2013b;288:4899–4907. doi: 10.1074/jbc.M112.434522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer JD, Nowak RB, Kim NE, Larkin SK, Peters LL, Hartwig J, Kuypers FA, Fowler VM. Tropomodulin1-null have a mild spherocytic elliptocytosis with appearance of Tropomodulin3 in red blood cells and disruption of the membrane skeleton. Blood. 2010a doi: 10.1182/blood-2010-02-268458. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer JD, Nowak RB, Kim NE, Larkin SK, Peters LL, Hartwig J, Kuypers FA, Fowler VM. Tropomodulin 1-null mice have a mild spherocytic elliptocytosis with appearance of tropomodulin 3 in red blood cells and disruption of the membrane skeleton. Blood. 2010b;116:2590–2599. doi: 10.1182/blood-2010-02-268458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu W, Wang X, Zhang X, Zhu S, Sun D, Ka W, Sung LA, Yao W. Fluid Shear Stress Upregulates E-Tmod41 via miR-23b-3p and Contributes to F-Actin Cytoskeleton Remodeling during Erythropoiesis. PloS one. 2015;10:e0136607. doi: 10.1371/journal.pone.0136607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y, Schwach C, Antin PB, Gregorio CC. Disruption in the tropomodulin1 (Tmod1) gene compromises cardiomyocyte development in murine embryonic stem cells by arresting myofibril maturation. Developmental biology. 2005;282:336–348. doi: 10.1016/j.ydbio.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Ostrowska Z, Robaszkiewicz K, Moraczewska J. Regulation of actin filament turnover by cofilin-1 and cytoplasmic tropomyosin isoforms. Biochim Biophys Acta. 2017;1865:88–98. doi: 10.1016/j.bbapap.2016.09.019. [DOI] [PubMed] [Google Scholar]
- Owen JB, Di Domenico F, Sultana R, Perluigi M, Cini C, Pierce WM, Butterfield DA. Proteomics-determined differences in the concanavalin-A-fractionated proteome of hippocampus and inferior parietal lobule in subjects with Alzheimer’s disease and mild cognitive impairment: implications for progression of AD. Journal of proteome research. 2009;8:471–482. doi: 10.1021/pr800667a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak CW, Flynn KC, Bamburg JR. Actin-Binding Proteins Take the Reins in Growth Cones. Nature Reviews. 2008;9:136–147. doi: 10.1038/nrn2236. [DOI] [PubMed] [Google Scholar]
- Pelled D, Raveh T, Riebeling C, Fridkin M, Berissi H, Futerman AH, Kimchi A. Death-associated protein (DAP) kinase plays a central role in ceramide-induced apoptosis in cultured hippocampal neurons. The Journal of biological chemistry. 2002;277:1957–1961. doi: 10.1074/jbc.M104677200. [DOI] [PubMed] [Google Scholar]
- Perrin BJ, Ervasti JM. The actin gene family: function follows isoform. Cytoskeleton (Hoboken) 2010;67:630–634. doi: 10.1002/cm.20475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao JN, Madasu Y, Dominguez R. Mechanism of actin filament pointed-end capping by tropomodulin. Science. 2014;345:463–467. doi: 10.1126/science.1256159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao TP, Kuhl M. An updated overview on Wnt signaling pathways: a prelude for more. Circulation research. 2010;106:1798–1806. doi: 10.1161/CIRCRESAHA.110.219840. [DOI] [PubMed] [Google Scholar]
- Rao VS, Marongelli EN, Guilford WH. Phosphorylation of tropomyosin extends cooperative binding of myosin beyond a single regulatory unit. Cell Motil Cytoskeleton. 2009;66:10–23. doi: 10.1002/cm.20321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ. Life at the leading edge. Cell. 2011;145:1012–1022. doi: 10.1016/j.cell.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Robaszkiewicz K, Ostrowska Z, Marchlewicz K, Moraczewska J. Tropomyosin isoforms differentially modulate the regulation of actin filament polymerization and depolymerization by cofilins. FEBS J. 2016;283:723–737. doi: 10.1111/febs.13626. [DOI] [PubMed] [Google Scholar]
- Rosso SB, Inestrosa NC. WNT signaling in neuronal maturation and synaptogenesis. Front Cell Neurosci. 2013;7:103. doi: 10.3389/fncel.2013.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nature reviews Molecular cell biology. 2013;14:7–12. doi: 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- Rust MB, Maritzen T. Relevance of presynaptic actin dynamics for synapse function and mouse behavior. Experimental cell research. 2015;335:165–171. doi: 10.1016/j.yexcr.2014.12.020. [DOI] [PubMed] [Google Scholar]
- Schevzov G, Bryce NS, Almonte-Baldonado R, Joya J, Lin JJ, Hardeman E, Weinberger R, Gunning P. Specific features of neuronal size and shape are regulated by tropomyosin isoforms. Molecular biology of the cell. 2005;16:3425–3437. doi: 10.1091/mbc.E04-10-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schevzov G, Fath T, Vrhovski B, Vlahovich N, Rajan S, Hook J, Joya JE, Lemckert F, Puttur F, Lin JJ, Hardeman EC, Wieczorek DF, O’Neill GM, Gunning PW. Divergent regulation of the sarcomere and the cytoskeleton. The Journal of biological chemistry. 2008;283:275–283. doi: 10.1074/jbc.M704392200. [DOI] [PubMed] [Google Scholar]
- Schevzov G, Gunning P, Jeffrey PL, Temm-Grove C, Helfman DM, Lin JJ, Weinberger RP. Tropomyosin localization reveals distinct populations of microfilaments in neurites and growth cones. Molecular and cellular neurosciences. 1997;8:439–454. doi: 10.1006/mcne.1997.0599. [DOI] [PubMed] [Google Scholar]
- Singer JM, Shaw JM. Mdm20 protein functions with Nat3 protein to acetylate Tpm1 protein and regulate tropomyosin-actin interactions in budding yeast. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:7644–7649. doi: 10.1073/pnas.1232343100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm S, Casper D, Lees-Miller JP, Helfman DM. Brain-specific tropomyosins TMBr-1 and TMBr-3 have distinct patterns of expression during development and in adult brain. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:9857–9861. doi: 10.1073/pnas.90.21.9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui Z, Nowak RB, Bacconi A, Kim NE, Liu H, Li J, Wickrema A, An XL, Fowler VM. Tropomodulin3-null mice are embryonic lethal with anemia due to impaired erythroid terminal differentiation in the fetal liver. Blood. 2014;123:758–767. doi: 10.1182/blood-2013-03-492710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Dierssen M, Toran N, Pollak DD, Chen WQ, Lubec G. A gel-based proteomic method reveals several protein pathway abnormalities in fetal Down syndrome brain. Journal of proteomics. 2011;74:547–557. doi: 10.1016/j.jprot.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Sussman MA, Sakhi S, Tocco G, Najm I, Baudry M, Kedes L, Schreiber SS. Neural tropomodulin: developmental expression and effect of seizure activity. Brain Res Dev Brain Res. 1994;80:45–53. doi: 10.1016/0165-3806(94)90088-4. [DOI] [PubMed] [Google Scholar]
- Tsukada T, Kotlyanskaya L, Huynh R, Desai B, Novak SM, Kajava AV, Gregorio CC, Kostyukova AS. Identification of Residues within Tropomodulin-1 Responsible for Its Localization at the Pointed Ends of the Actin Filaments in Cardiac Myocytes. Journal of Biological Chemistry. 2011;286:2194–2204. doi: 10.1074/jbc.M110.186924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlova E, Bae CY, Deurloo M, Chen W, Barszczyk A, Horgen FD, Fleig A, Feng ZP, Sun HS. TRPM7 Regulates Axonal Outgrowth and Maturation of Primary Hippocampal Neurons. Molecular neurobiology. 2016;53:595–610. doi: 10.1007/s12035-014-9032-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Shah SP, Gritsyna Y, Hitchcock-DeGregori SE, Kostyukova AS. Systematic analysis of tropomodulin/tropomyosin interactions uncovers fine-tuned binding specificity of intrinsically disordered proteins. J Mol Recognit. 2011;24:647–655. doi: 10.1002/jmr.1093. [DOI] [PubMed] [Google Scholar]
- Van Damme P, Lasa M, Polevoda B, Gazquez C, Elosegui-Artola A, Kim DS, De Juan-Pardo E, Demeyer K, Hole K, Larrea E, Timmerman E, Prieto J, Arnesen T, Sherman F, Gevaert K, Aldabe R. N-terminal acetylome analyses and functional insights of the N-terminal acetyltransferase NatB. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:12449–12454. doi: 10.1073/pnas.1210303109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman K, Futerman AH. Ceramide as a second messenger: sticky solutions to sticky problems. Trends in cell biology. 2000;10:408–412. doi: 10.1016/s0962-8924(00)01830-4. [DOI] [PubMed] [Google Scholar]
- Vignjevic D, Kojima S, Aratyn Y, Danciu O, Svitkina T, Borisy GG. Role of fascin in filopodial protrusion. The Journal of cell biology. 2006;174:863–875. doi: 10.1083/jcb.200603013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitriol EA, Zheng JQ. Growth cone travel in space and time: the cellular ensemble of cytoskeleton, adhesion, and membrane. Neuron. 2012;73:1068–1081. doi: 10.1016/j.neuron.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Ecken J, Muller M, Lehman W, Manstein DJ, Penczek PA, Raunser S. Structure of the F-actin-tropomyosin complex. Nature. 2015;519:114–117. doi: 10.1038/nature14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrhovski B, Lemckert F, Gunning P. Modification of the tropomyosin isoform composition of actin filaments in the brain by deletion of an alternatively spliced exon. Neuropharmacology. 2004;47:684–693. doi: 10.1016/j.neuropharm.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Vrhovski B, Schevzov G, Dingle S, Lessard JL, Gunning P, Weinberger RP. Tropomyosin isoforms from the gamma gene differing at the C-terminus are spatially and developmentally regulated in the brain. Journal of neuroscience research. 2003;72:373–383. doi: 10.1002/jnr.10586. [DOI] [PubMed] [Google Scholar]
- Wagner LM, V, Fowler M, Takemoto DJ. The interaction and phosphorylation of tropomodulin by protein kinase Calpha in N/N 1003A lens epithelial cells. Molecular vision. 2002;8:394–406. [PubMed] [Google Scholar]
- Watakabe A, Kobayashi R, Helfman DM. N-tropomodulin: a novel isoform of tropomodulin identified as the major binding protein to brain tropomyosin. Journal of cell science. 1996;109(Pt 9):2299–2310. doi: 10.1242/jcs.109.9.2299. [DOI] [PubMed] [Google Scholar]
- Weber A, Pennise CR, Babcock GG, Fowler VM. Tropomodulin Caps the Pointed Ends of Actin-Filaments. Journal of Cell Biology. 1994;127:1627–1635. doi: 10.1083/jcb.127.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger R, Schevzov G, Jeffrey P, Gordon K, Hill M, Gunning P. The molecular composition of neuronal microfilaments is spatially and temporally regulated. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1996;16:238–252. doi: 10.1523/JNEUROSCI.16-01-00238.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DM, Lu J, Zheng YL, Zhang YQ, Hu B, Cheng W, Zhang ZF, Li MQ. Small interfering RNA-mediated knockdown of protein kinase C zeta attenuates domoic acid-induced cognitive deficits in mice. Toxicological sciences : an official journal of the Society of Toxicology. 2012;128:209–222. doi: 10.1093/toxsci/kfs124. [DOI] [PubMed] [Google Scholar]
- Wu SC, Solaro RJ. Protein kinase C zeta. A novel regulator of both phosphorylation and de-phosphorylation of cardiac sarcomeric proteins. The Journal of biological chemistry. 2007;282:30691–30698. doi: 10.1074/jbc.M703670200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Zhong G, Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 2013;339:452–456. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro S, Gokhin DS, Sui ZH, Bergeron SE, Rubenstein PA, Fowler VM. Differential Actin-regulatory Activities of Tropomodulin1 and Tropomodulin3 with Diverse Tropomyosin and Actin Isoforms*. Journal of Biological Chemistry. 2014;289:11616–11629. doi: 10.1074/jbc.M114.555128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro S, Speicher KD, Speicher DW, Fowler VM. Mammalian tropomodulins nucleate actin polymerization via their actin monomer binding and filament pointed end-capping activities. The Journal of biological chemistry. 2010;285:33265–33280. doi: 10.1074/jbc.M110.144873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JW, Czech T, Felizardo M, Baumgartner C, Lubec G. Aberrant expression of cytoskeleton proteins in hippocampus from patients with mesial temporal lobe epilepsy. Amino acids. 2006;30:477–493. doi: 10.1007/s00726-005-0281-y. [DOI] [PubMed] [Google Scholar]
- Yun S, Rim Y, Jho EH. Induced expression of the transcription of tropomodulin 1 by Wnt5a and characterization of the tropomodulin 1 promoter. Biochem Biophys Res Commun. 2007;363:727–732. doi: 10.1016/j.bbrc.2007.09.033. [DOI] [PubMed] [Google Scholar]


