Abstract
Wilson disease is an autosomal-recessive disorder of hepatocellular copper deposition caused by pathogenic variants in the copper-transporting gene, ATP7B. Early detection and treatment are critical to prevent lifelong neuropsychiatric, hepatic, and systemic disabilities. Due to the marked heterogeneity in age of onset and clinical presentation, the diagnosis of Wilson disease remains challenging to physicians today. Direct sequencing of the ATP7B gene is the most sensitive and widely used confirmatory testing method, and concurrent biochemical testing improves diagnostic accuracy. More than 600 pathogenic variants in ATP7B have been identified, with single-nucleotide missense and nonsense mutations being the most common, followed by insertions/deletions, and, rarely, splice site mutations. The prevalence of Wilson disease varies by geographic region, with higher frequency of certain mutations occurring in specific ethnic groups. Wilson disease has poor genotype–phenotype correlation, although a few possible modifiers have been proposed. Improving molecular genetic studies continue to advance our understanding of the pathogenesis, diagnosis, and screening for Wilson disease.
INTRODUCTION
In this chapter, we will discuss the inheritance, gene frequency, variants, genotype–phenotype correlation, and modifiers of the ATP7B gene, and the clinical molecular diagnosis and population screening for Wilson disease.
INHERITANCE
Wilson disease is a monogenic autosomal-recessive condition and carriers do not manifest any symptoms. Autosomal-recessive conditions are not usually present in consecutive generations, but may occur in populations with particularly high carrier frequency of Wilson disease (F. Wu et al., 2015). Our group and others have reported the presence of Wilson disease in two or more successive generations within the same family, reflecting a “pseudo-dominant” inheritance (Dziezyc et al., 2011, 2014; Bennett et al., 2013; H. Park et al., 2015). Therefore, the diagnosis of Wilson disease should not be excluded simply due to a misleading family history consistent with an autosomal-dominant inheritance pattern. Furthermore, recent studies have also identified Wilson disease due to atypical forms of inheritance, such as the presence of three concurrent mutations in a single patient or segmental uniparental disomy (Coffey et al., 2013). Uniparental disomy occurs when both homologs of a chromosome originate from a single parent. These findings have implications for clinical practice and genetic counseling, as clinicians may need to consider genotyping asymptomatic parents or obtaining full sequencing of ATP7B to confirm that pathogenic variants occur in trans.
ATP7B GENE ANDATPASE
Wilson disease is caused by homozygous or compound heterozygous mutations in the ATP7B gene (OMIM# 606882), which encodes a transmembrane copper-transporting P-type ATPase of the same name. Currently, ATP7B is the only identified gene known to cause Wilson disease (Bull et al., 1993; Petrukhin et al., 1993; Tanzi et al., 1993). Mutations in the ATP7B gene have been reported in almost all exons. Previous studies have reported individuals with both biochemical and clinical diagnosis of Wilson disease in the absence of two ATP7B mutations, raising the possibility of a second causative gene (Lovicu et al., 2006; Kenney and Cox, 2007; S. Park et al., 2007; Mak and Lam, 2008; Nicastro et al., 2010; Coffey et al., 2013). Nonetheless, ATP7B remains the only known gene responsible for Wilson disease.
Human dietary intake of copper is about 1.5–2.5 mg/day, which is absorbed in the stomach and duodenum, bound to circulating albumin, and transported to the liver for regulation and excretion (Culotta and Scott, 2016). The uptake of copper occurs on the basolateral side of hepatocytes via copper transporter 1 (CTR1), as illustrated in Figure 3.1. A specific copper chaperone, antioxidant protein 1 (ATOX1), delivers copper to the Wilson disease protein, ATP7B, by copper-dependent protein–protein interactions (Walker et al., 2004). Within hepatocytes, ATP7B performs two important functions in either the trans-Golgi network (TGN) or in cytoplasmic vesicles. In the TGN, ATP7B activates ceruloplasmin by packaging six copper molecules into apoceruloplasmin, which is then secreted into the plasma. In the cytoplasm, ATP7B sequesters excess copper into vesicles and excretes it via exocytosis across the apical canalicular membrane into bile (Bull et al., 1993; Tanzi et al., 1993; Yamaguchi et al., 1999; Cater et al., 2007). Due to the binary role of the ATP7B transporter in both the synthesis and excretion of copper, defects in its function lead to copper accumulation and the progressive features of Wilson disease (Fig. 3.1).
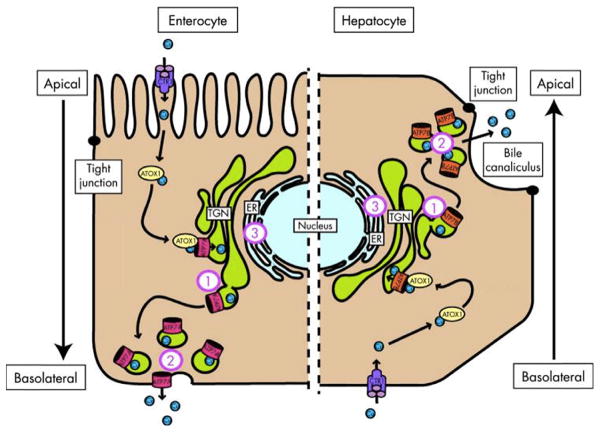
Fig. 3.1.
Schematic representation of copper-induced relocalization of ATP7A and ATP7B. The left side of the diagram represents an enterocyte and the right side represents a hepatocyte. On both sides, copper enters the cell through copper transporter 1 (CTR1) and is escorted by copper chaperone antioxidant protein 1 (ATOX1) to ATP7A or ATP7B in the trans-Golgi network (TGN). When copper levels rise above a certain threshold, ATP7A and ATP7B excrete copper into the plasma on the basolateral side of the enterocyte and into the bile on the apical side of the hepatocyte. Defects in localization of ATP7B may lead to copper accumulation at the (1) TGN due to unresponsiveness, (2) cell periphery, and (3) endoplasmic reticulum (ER) due to misfolding. (Reproduced from de Bie et al., 2007.)
MOLECULAR STRUCTURE OFATP7B
ATP7B is located on 13q14.3 and contains 20 introns and 21 exons, for a total genomic length of 80 kb (Bull et al., 1993; Petrukhin et al., 1993; Tanzi et al., 1993). The gene is synthesized in the endoplasmic reticulum, then relocated to the TGN within hepatocytes. ATP7B is most highly expressed in the liver, but is also found in the kidney, placenta, mammary glands, brain, and lung.
ATP7B (P-TYPE ATPASE) PROTEIN STRUCTURE AND FUNCTION
ATP7B belongs to class 1B (PIB) of the highly conserved P-type ATPase superfamily, which is responsible for the transport of copper and other heavy metals across cellular membranes (Gourdon et al., 2011). The protein contains 1465 amino acids, a phosphatase domain (A-domain), phosphorylation domain (P-domain, amino acid residues 971–1035), nucleotide-binding domain (N-domain, amino acid residues 1240–1291), and M-domain, which is comprised of eight transmembrane ion channels (Fig. 3.2) (Cater et al., 2004, 2007; Lenartowicz and Krzeptowski, 2010).
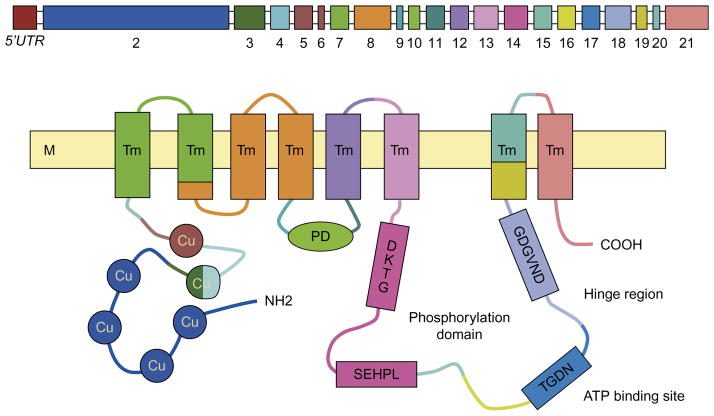
Fig. 3.2.
Schematic representation of ATP7B gene and corresponding human ATP7B protein. Top diagram shows 5’UTR promoter region and exons separated by introns. Bottom diagram shows the domain organization of human copper ATPase. Conserved amino acid motifs are present at the core structure of each functional domain, i.e., TGDN and GDGVND at the A-domain, DKTG at the P-domain, and SEHPL in the N-domain. M, phospholipidic bilayer of the membrane; Cu, the metal-binding domains of the trasmembrane cation channel; Tm, transmembrane domains; PD, phosphatase domain. (Reproduced from Fanni et al., 2005.)
Unique amino acid motifs are present at the core structure of each domain, such as TGEA at the A-domain, DKTGT at the P-domain, and SEHPL in the N-domain. Specifically, the N-terminal metal-binding domain (MBD) is composed of six copper-binding sites, each with the conserved sequence motif GMXCXXC (Fatemi and Sarkar, 2002; Sazinsky et al., 2006). These MBDs play a central role in accepting copper from copper chaperone ATOX1 through protein–protein interactions. Previous studies have demonstrated unequal impact of MBDs on ATP7B activity, with MBD 5 and 6 having stronger effects on the catalytic activation of ATP7B than MBDs 1–4 (Lutsenko et al., 1997).
The active transport of copper across membranes is a complex process that begins with ATP7B binding copper at the N-terminal domain and transporting it across cellular membranes, using ATP as an energy source (Fig. 3.2). Next, free copper binds intracellularly to GG motifs in the MBDs, followed by transport on to the Cys-Pro-Cys (CPC) sequence motifs in MBD 6. Finally, dephosphorylation of acyl-phosphate at the A-domain discharges copper across the cellular membrane. Mutations causing copper accumulation may occur at any of these steps (Huster et al., 2006; Schushan et al., 2012).
Although the mechanism by which the histadine-containing SEHPL motif affects copper transport remains to be elucidated, it is clear that histidine-to-glutamate substitution at amino acid 1069 (p.H1069Q) in this motif is the most common cause of Wilson disease in northern Europeans. In the hepatocytes of patients homozygous for p.H1069Q, ATP7B was found in the endoplasmic reticulum instead of its usual TGN location, suggesting abnormal protein trafficking (Huster et al., 2003). Insect models with the p.H1069Q mutation in SF9 cells showed decreased ATP-mediated catalytic phosphorylation but no major protein misfolding, suggesting a role for p.H1069Q in the orientation of the ATP7B catalytic site for ATP binding prior to hydrolysis (Tsivkovskii et al., 2003).
VARIANTS IN THE ATP7B GENE
More than 600 pathogenic variants in ATP7B have been identified, with single-nucleotide missense and nonsense mutations being the most common, followed by insertions/deletions and splice site mutations (Human Gene Mutation Database, accessed 29 April 2016; Stenson et al. 2014). Other rare genetic mechanisms that have been reported in the literature include whole-exon deletions, promoter region mutations, three concurrent pathogenic variants, and monogenic disomy (Coffey et al., 2013; Bandmann et al., 2015). Mutation “hotspots” in ATP7B have also been reported to vary by geographic region (see regional gene frequency section, below). The majority of pathogenic mutations are located in the M- and N-domains in presymptomatic patients or in those with hepatic symptoms (S. Park et al., 2007). The common mutations in ATP7B seen in various populations are listed in Figure 3.3.
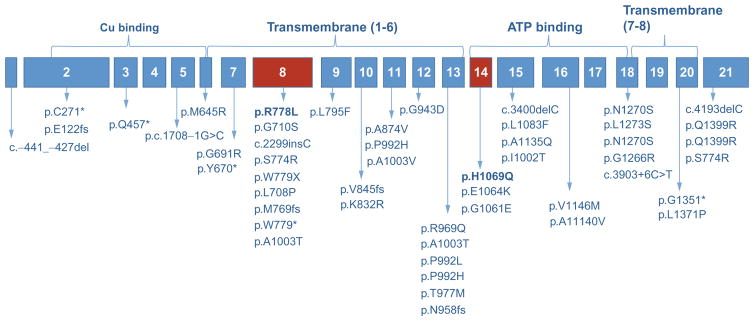
Fig. 3.3.
Schematic of the ATP7B gene with common mutation sites, including p.H1069Q (rs76151636), p.R778L (rs28942074), p.E1064K (rs376910645), c.3400delC, and p.Ala1135fs (rs137853281). Please refer to Table 3.1 for more details.
The p.H1069Q mutation is one of the most common mutations, with a population allelic frequency of 10–40% (30–70% among Caucasians). Most patients are compound heterozygotes, carrying different mutations on each copy of the chromosome (Usta et al., 2014). The p.H1069Q mutation occurs when histidine of the conserved SEHPL motif in the N-domain of ATP7B is replaced by glutamic acid, resulting in N-domain protein misfolding, abnormal phosphorylation in the P-domain, and decreased ATP binding affinity (Rodriguez-Granillo et al., 2008). This mutation also leads to decreased heat stability and abnormal localization of the protein to the TGN (Ralle et al., 2010).
Other common mutations in ATP7B include p. E1064A, p.R778L, p.G943S, and p.M769V. Mutations in p.E1064A, also found in the SEHPL motif, completely disable ATP binding affinity but do not result in protein misfolding, transport abnormalities, or thermal instability. The p.R778L mutation affects transmembrane transport of copper (Dmitriev et al., 2011). The p.G943S and p.M769V mutations result in defective copper metabolism but preserved ceruloplasmin levels (Okada et al., 2010).
A substantial proportion of Wilson disease-associated missense mutations, including p.H1069Q and p.R778L, result in markedly decreased level of the protein caused by enhanced degradation (Payne et al., 1998; de Bie et al., 2007; van den Berghe et al., 2009). Other prevalent mutations, such as protein-truncating nonsense mutations (~13% of known point mutations) (Merle et al., 2010) and frameshift mutations (Vrabelova et al., 2005), are predicted to cause decay of mRNA (Mendell et al., 2004; Chang et al., 2007) or a severely truncated protein, resulting in absent or diminished levels of protein. It is therefore expected that most patients with Wilson disease have absent or significantly reduced levels of ATP7B.
REGIONAL GENE FREQUENCY
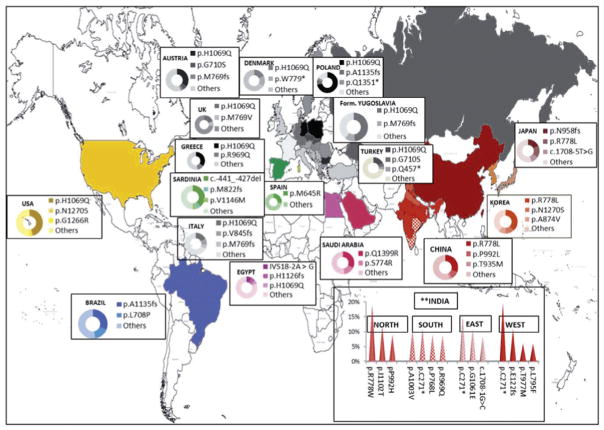
The prevalence of Wilson disease varies by geographic region, with higher prevalence of specific mutations reported in certain populations (Ferenci, 2006) (see Chapter 2 for more details). A list of the common regional variants of ATP7B mutations and geographic clustering of mutations are shown in Table 3.1 and Figure 3.4, respectively.
Table 3.1.
Regional distribution of common Wilson disease mutations by geographic location
| Prevalent mutations
|
|||||||
|---|---|---|---|---|---|---|---|
| Region | AF (%) | Protein | Nucleotide | RS | Exon | Type | Domain |
| Europe | |||||||
| Austria (Ferenci, 2006) | 34.1 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| 6.4 | p.Gly710Ser | c.2128G>A | 8 | Missense | TM2 | ||
| 3.6 | p.Met769fs | c.2298_2299insC | rs137853287 | 8 | Premature stop | TM4 | |
| Benelux (Ferenci, 2006) | 53 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| Bulgaria (Todorov et al., 2005) | 58.8 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| Canary Islands (García Villarreal et al., 2000) | 64 | p.Leu708Pro | c.2123 T>C | 8 | Missense | TM2 | |
| Czech Republic (Vrabelova et al., 2005) | 57 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| Denmark (Møller et al., 2011) | 18 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| 16 | p.Trp779* | c.2336G>A | rs137853283 | 8 | Nonsense | TM4 | |
| France (Bost et al., 2012) | 15 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| Germany (Ferenci, 2006) | 47.9 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| Germany (East, former) (Caca et al., 2000) | 63 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| Greece (Panagiotakaki et al., 2004; | 35 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| Dedoussis et al., 2005; Gomes and Dedoussis, 2016) | |||||||
| 12 | p.Arg969Gln | c.2906G>A | rs774028495 | 13 | Missense | TM6 | |
| Hungary (Firneisz et al., 2002; Folhoffer et al., 2007) | 42.9 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| Iceland (Thomas et al., 1995a; Hofer et al., 2012) | 100 | p.Tyr670* | c.2007_2013del | 7 | Nonsense | TM1 | |
| Italy (Loudianos et al., 1999) | 17.5 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| 9 | p.Val845fs | c.2530delA | rs755709270 | 10 | Premature stop | Td | |
| 6 | p.Met769fs | c.2298_2299insC | rs137853287 | 8 | Premature stop | TM4 | |
| Netherlands (Stapelbroek et al., 2004) | 33 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| Poland (Gromadzka et al., 2005) | 72 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| 7.3 | p.Ala1135fs | c.3400delC | rs137853281 | 15 | Premature stop | ATP loop | |
| 3.7 | p.Gln1351* | c.4051C>T | 20 | Nonsense | |||
| Romania (Iacob et al., 2012) | 38.1 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| Russia (Ivanova-Smolenskaya et al., 1997) | 49 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| Sardinia (Figus et al., 1995) | 60.5 | c.-441_-427del | 5prime | Unknown | Promoter | ||
| 8.5 | p.Met822fs | c.2463delC | 10 | Deletion | TM4/Td | ||
| 7.9 | p.Val1146Met | c.3436G>A | 16 | Missense | ATP loop | ||
| Serbia | 38.4 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| (Tomić et al., 2013) | |||||||
| 11.6 | p.Met769fs | c.2304dupC | 8 | Missense | TM4 | ||
| 9.3 | p.Ala1003Thr | c.3007G>A | rs1801247 | 13 | Missense | TM6/Ph | |
| Spain (Margarit et al., 2005) | 27 | p.Met645Arg | c.1934 T>G | rs121907998 | 6 | Missense | Cu6/TM1 |
| Sweden (Shah et al., 1997) | 38 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| Turkey (Ferenci, 2006; Simsek Papur et al., 2013) | 17.4 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| 5.3 | p.Gly710Ser | c.2128G>A | rs772595172 | 8 | Missense | TM2 | |
| 4.53 | p.Gln457* | c.1369C>T | 3 | Nonsense | Cu4/Cu5 | ||
| UK (Coffey et al., 2013) | 19 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| 8 | p.Met769Val | c.2305A>G | 8 | Missense | TM4 | ||
| Yugoslavia (former) (Loudianos et al., 1999) | 48.9 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| 11.4 | p.Met769fs | c.2298_2299insC | rs137853287 | 8 | Premature stop | TM4 | |
| Asia | |||||||
| China (Gu et al., 2003; Z.-Y. Wu et al., 2003; Wang et al., 2011; Wei et al., 2014) | 31 | p.Arg778Leu | c.2332C>T | rs28942074 | 8 | Missense | TM4 |
| 10 | p.Pro992Leu | c.2975C>T | rs201038679 | 13 | Missense | TM6/Ph | |
| 9.6 | p.Ile1148Thr | c.3443 T>C | rs60431989 | 16 | Missense | ATP loop | |
| 3.3 | p.Thr935Met | c.2804C>T | 12 | Missense | TM5 | ||
| 19 | p.Arg778Leu | c.2332C>T | rs28942074 | 8 | Missense | TM4 | |
| North India (S. Kumar et al., 2006; Gupta et al., 2007) | |||||||
| 12 | p.Ile1102Thr | c.3305 T>C | rs560952220 | 15 | Missense | ATP loop | |
| 9 | p.Pro992His | c.2975C>A | 13 | Missense | TM6/Ph | ||
| South India (Santhosh et al., 2006; S. S. Kumar et al., 2012) | 11 | p.Ala1003Val | c.3008C>T | 13 | Missense | TM6/Ph | |
| 11 | p.Cys271* | c.813C>A | rs572147914 | 2 | Nonsense | Cu3 | |
| 9 | p.Pro768Leu | c.2303C>T | 8 | Missense | TM4 | ||
| 9 | p.Arg969Gln | c.2906G>A | rs121907996 | 13 | Missense | TM6 | |
| East India (Gupta et al., 2005) | 16 | p.Cys271* | c.813C>A | rs572147914 | 2 | Nonsense | Cu3 |
| 11 | p.Gly1061Glu | c.3182G>A | 14 | Missense | ATP loop | ||
| 8.5 | c.1708-1G>C | rs137853280 | 5 | Splice | Cu6 | ||
| West India (Aggarwal and Bhatt, 2013; Aggarwal et al., 2013) | 20 | p.Cys271* | c.813C>A | rs572147914 | 2 | Nonsense | Cu3 |
| 11 | p.Glu122fs | c.365_366delins | 2 | Ins/Del | Cu1 | ||
| TTCGAAGC | |||||||
| 6 | p.Thr977Met | c.2930C>T | rs72552255 | 13 | Missense | TM6 | |
| 6 | p.Leu795Phe | c.2383C>T | 9 | Missense | TM4/Td | ||
| Japan (Okada et al., 2000; Tatsumi et al., 2010) | 17.95 | p.Asn958fs | c.2871delC | 13 | Premature stop | TM5/TM6 | |
| 16.7 | p.Arg778Leu | c.2332C>T | rs28942074 | 8 | Missense | TM4 | |
| 10.5 | c.1708-5 T>G | 5 | Splice | Cu6 | |||
| Korea (E. K. Kim et al., 1998; Yoo, 2002; G.-H. Kim et al., 2008; Song et al., 2012) | 37.9 | p.Arg778Leu | c.2332C>T | rs28942074 | 8 | Missense | TM4 |
| 12.1 | p.Asn1270Ser | c.3809A>G | rs121907990 | 18 | Missense | ATP hinge | |
| 9.4 | p.Ala874Val | c.2621C>T | rs376355660 | 11 | Missense | TM5 | |
| 8 | p.Leu1083Phe | c.3247C>T | 15 | Missense | ATP loop | ||
| Lebanon (Usta et al., 2014) | 44.7 | p. Ala1003Thr | c.2299insC | rs137853287 | 8 | Missense | TM4 |
| Saudi Arabia (Al Jumah et al., 2004; Majumdar et al., 2004) | 32 | p.Gln1399Arg | c.4196A>G | 21 | Missense | After TM8 | |
| 16 | p.Ser774Arg | c.2230 T>C | rs535217574 | 21 | Missense | TM3 | |
| Taiwan (Lee et al., 2000; Wan et al., 2006) | 29.6 | p.Arg778Leu | c.2332C>T | rs28942074 | 8 | Missense | TM4 |
| 8.9 | p.Pro992Leu | c.2975C>T | rs201038679 | 13 | Missense | TM6 | |
| 4.8 | p.Gly943Asp | c.2828G>A | 12 | Missense | TM5 | ||
| Thailand (Panichareon et al., 2011) | 10.52 | p.Arg778Leu | c.2332C>T | rs28942074 | 8 | Missense | TM4 |
| 7.89 | p.Leu1371Pro | c.4112 T>C | 20 | Missense | TM8 | ||
| Iran (Zali et al., 2011) | 19 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| Africa | |||||||
| Egypt (Abdelghaffar et al., 2008; Abdel Ghaffar et al., 2011) | 42.2 | IVS18+6 T>C | c.3903+6C>T | rs2282057 | 18 | Splice | |
| 40.6 | p.Ala11140Val | c.3419C>T | 16 | Missense | ATP loop | ||
| 26.5 | p.Lys832Arg | c.2495A>G | rs1061472 | 10 | Missense | TM4/Td | |
| Americas | |||||||
| USA (Kuppala et al., 2009) | 40.3 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| 1.9 | p.Asn1270Ser | c.3809A>G | rs121907990 | 18 | Missense | ATP hinge | |
| 1.9 | p.Gly1266Arg | c.3796G>A | rs121907992 | 18 | Missense | ATP hinge | |
| Brazil (Deguti et al., 2004; Machado et al., 2008; Bem et al., 2013) | 37.1 | p.His1069Gln | c.3207C>A | rs76151636 | 14 | Missense | ATP loop |
| 31.25 | p.Ala1135fs | c.3400delC | rs137853281 | 15 | Premature stop | ATP loop | |
| 11.4 | p.Ala1135GlnfsX13 | c.3402delC | rs137853281 | 15 | Premature stop | ATP loop | |
| p.Leu708Pro | c.2123 T>C | 8 | Missense | TM2 | |||
| Costa Rica (Shah et al., 1997) | 61 | p.Asn1270Ser | c.3809A>G | rs121907990 | 18 | Missense | ATP hinge |
| Venezuela (Paradisi et al., 2015) | 26.9 | p.Ala1135GlnfsX13 | c.3402delC | rs137853281 | 15 | Premature stop | ATP loop |
| 9.6 | p.Gly691Arg | c.2071G>A | 7 | Missense | TM2 | ||
AF, RS,
Fig. 3.4.
Prevalence of ATP7B mutation by geographic region; the darker the gradient, the higher the allelic frequency. (Reproduced from Gomes and Dedoussis, 2016, with permission from Taylor and Francis.)
GENOTYPE–PHENOTYPE CORRELATION
Direct genotype–phenotype relationships in Wilson disease have been difficult to establish, despite several studies examining correlation (Panagiotakaki et al., 2004; Vrabelova et al., 2005; Nicastro et al., 2010; Cocoş et al., 2014; Usta et al., 2014). The numerous low-frequency and compound heterozygous nature of Wilson disease obfuscate the process of characterizing its numerous genetic variants and their clinical consequences. Descriptions of phenotypes are limited to age of onset and presenting symptoms, both of which may be affected by inaccurate diagnostic criteria, delayed diagnosis, and practitioner selection bias. Therefore, the marked variability in phenotype of Wilson disease is likely attributable to an amalgamation of genetic, metabolic, and environmental factors (Leggio et al., 2006).
The most consistent genotype–phenotype correlation in Wilson disease is that the most severe, early-onset disease with predominantly hepatic presentation is associated with mutations causing absent ATPase activity. Convincing studies have demonstrated fulminant hepatic disease in mouse models such as the toxic milk (tx) mouse and the Jackson tx mouse (txj), which harbor point mutations causing loss of ATP7B function, but not affecting ATP7B synthesis (Theophilos et al., 1996; Coronado et al., 2001; La Fontaine et al., 2001; Huster et al., 2006).
Genetic polymorphisms in ATP7B, other genes, and epigenetic factors have been shown to impact disease phenotype by affecting ATP7B protein structure and function. Of the over 600 mutations associated with Wilson disease, the majority are missense mutations that completely inactivate the copper-transporting function of ATP7B (Lutsenko, 2014). In general, individuals with protein-truncating mutations have earlier onset of disease by decreasing protein stability and quantity (Merle et al., 2010). However, other studies have demonstrated partial preservation of copper-transporting function, perhaps explaining the milder phenotypes associated with certain mutations (Rodriguez-Granillo et al., 2008; Dmitriev et al., 2011; Huster et al., 2012). Individuals with the R778L mutation have been shown to have an earlier onset of disease and predominantly hepatic presentation (Z. Y. Wu et al., 2003). In contrast, individuals with the H1069Q substitution have been shown to have mean onset of symptoms of 20–22 years old and predominantly neurologic presentations (Stapelbroek et al., 2004; Kalita et al., 2010). There is also some evidence that Kayser–Fleischer rings are more common in H1069Q homozygous patients in Hungary at time of diagnosis than in compound heterozygous individuals (Folhoffer et al., 2007).
Moreover, pathogenic variants may affect ATP7B targeting from the TGN to cytosolic vesicles. For instance, the p.Met875Val mutation results in a less stable protein and causes reversible ATP7B localization defects. Under a low-copper environment, the p.Gly875Arg variant is sequestered in the endoplasmic reticulum. However, addition of exogenous copper to the cellular growth medium stabilizes the protein, allowing it to complete its intended journey to the TGN and overcoming its disease-causing phenotype. Theoretically, patients with this specific variant may be more sensitive to dietary copper deficiency (Gupta et al., 2011).
The timing and location of copper buildup can also preferentially alter the hepatic transcriptome, based on homozygous ATP7B−/− mouse models. Proteomic analyses of mRNA profiles at each of these disease stages reflect unique patterns (Huster et al., 2006; Ralle et al., 2010). In the initial stage, mRNA for proteins responsible for cell cycle regulation, splicing, and cholesterol synthesis is present (Burkhead et al., 2011). This leads to early accumulation of copper bound to metallothioneins in the cytosol and free copper in the nuclei. In the progressive stage, mRNA changes throughout the cell are present, including the endoplasmic reticulum, mitochondria, and endocytic pathways causing copper to pathologically accumulate within hepatocytes. In the later stages, mRNA for lysosomal and endosomal proteins is upregulated. In these final stages, copper concentrations decrease in the cytosol and nuclei, and accumulate in the membranous cellular compartment, causing bile duct proliferation and hepatic neoplastic changes. Therefore, the location of copper accumulation may convey more specific prognostic information about disease progression rather than total copper levels.
Other studies have compared homozygotes to compound heterozygotes of the same mutation to establish genotype–phenotype correlations. A study of 76 members of a large, consanguineous Lebanese family showed an association between c.2299insC and hepatic disease and between the p.Ala1003Thr mutation and neurologic disease (Usta et al., 2014).
Other candidate polymorphisms that are thought to modify the clinical phenotype of Wilson disease include MTHFR (Gromadzka et al., 2005), COMMD1 (Weiss, 2006), ATOX1 (Simon, 2008), XIAP (Weiss et al., 2010), PNPLA3 and hepatic steatosis (Stättermayer et al., 2012), and DMT1 (Przybyłkowski et al., 2014), although none of these genes has been demonstrated to have significant diagnostic or predictive value.
Significant phenotypic variation of Wilson disease exists between individuals with the same mutation, individuals within the same family, and even between monozygotic twins (Członkowska et al., 2009; Kegley et al., 2010). While some studies have documented high intra-familial concordance of clinical symptoms and biochemical results (Hofer et al., 2012; Chabik et al., 2014; Ferenci et al., 2015), others have reported a wide range in age of onset and presenting symptoms amongst siblings (Ala et al., 2007; Taly et al., 2007) and families carrying the same mutation (Takeshita et al., 2002). Indeed, disparate clinical presentations in monozygotic twins raise the suspicion for epigenetic modifiers in Wilson disease. See Chapter 4 for more details about the genetic and environmental modifiers of Wilson disease.
CLINICAL MOLECULAR DIAGNOSIS
The current gold standard for Wilson disease diagnosis is direct Sanger sequencing of ATP7B gene or molecular testing for familial mutations that were previously identified. Historically, most pathogenic variants in ATP7B were identified using a combination of polymerase chain reaction (PCR)/restriction fragment length polymorphism (RFLP), single-strand conformation polymorphism (SSCP), denaturing gradient gel electrophoresis (DGGE), temporal temperature gradient electrophoresis (TTGE), denaturing high-performance liquid chromatography (DHPLC), and Sanger sequencing (Loudianos et al., 1999; Shimizu et al., 1999; Margarit et al., 2005; Vrabelova et al., 2005; G. H. Kim et al., 2008). The critical demerits of this complex tiered approach are that the detection rate is not high enough to find most mutations and the turnaround time is often extended. Although regional clusters of specific mutations have been well described, a customized screening approach taking into account these regional variants may be complicated by ethnically diverse populations and inaccurate information provided with samples. Biochemical results are often imprecise, as elevations in urinary copper excretion tend to occur late in the disease process and fewer than 40% of presymptomatic patients excrete copper less than 100 μg/day (Sternlieb and Scheinberg, 1968; Nakayama et al., 2008). For these reasons, direct sequencing of the ATP7B gene has become the preferred standard and provided the greatest yield in clinical molecular diagnosis. Please refer to Chapter 14 for details about the diagnosis of Wilson disease.
Starting the diagnostic process with molecular testing may significantly reduce the need for invasive liver biopsy. Liver copper content alone was found to be insufficient to exclude Wilson disease, as levels may not be elevated in some affected patients. In several studies on patients with possible Wilson disease based on biochemical and clinical tests, disease-causing mutations in both alleles were identified in about 80% of them. Currently available screening tests may not definitely rule out the disease and no single test could permit de novo diagnosis. Of note, many patients may not possess the characteristic findings and may present when their clinical disease is relatively mild. Inappropriate treatment for false-positive cases has the potential of inducing copper deficiency, which can result in hematologic and neurologic sequelae (N. Kumar et al., 2003). These findings reinforce the need for reliable clinical diagnostic criteria and underscore the benefits of DNA testing prior to invasive procedures (Ferenci, 2005).
Multiplex PCR is used to amplify all 21 exons and splice sites of ATP7B, including promoter regions. Although the large deletions or duplications cannot be detected with this conventional Sanger sequencing method, the chance of these being present in Wilson disease appears low (Stenson et al., 2012). If clinical suspicion is still high with only one pathogenic variant present, then multiplex ligation-dependent probe amplification (MLPA) test should be considered. Microarray-based comparative genomic hybridization is another option to evaluate partial or full gene deletions or duplications with higher sensitivity. Cases with only one copy of mutation present should be carefully reviewed in the context of other biochemical and clinical findings. Molecular genetic testing using direct mutation analysis is very effective in identifying affected patients and presymptomatic siblings of probands (Manolaki et al., 2009).
Wilson disease is an autosomal-recessive disorder, which means that there is a 25% chance that a sibling of the index case also has Wilson disease. Once homozygous or compound heterozygous mutations in ATP7B have been established in the index patient, mutation detection becomes valuable in family screening. The same genotype in asymptomatic family members confirms diagnosis of the disease, thus allowing for early treatment before the onset of complications. In family members in whom clinical and biochemical features are uncertain, the demonstration of either heterozygous (carrier) or wild-type gene sequence prevents unnecessary treatment (Chang et al., 2007).
If the proband has secured a diagnosis of Wilson disease on the basis of clinical and biochemical evidence, but testing for ATP7B mutations is not available, family screening can be done by haplotype analysis of polymorphic markers flanking the disease gene (Thomas et al., 1995b; Gupta et al., 2005; Przybyłkowski et al., 2014). In this instance, the rare possibility of recombination events (typically 0.5–5% of cases) needs to be considered. The rate of recombination is dependent on which flanking markers are studied. Microsatellite or single-nucleotide polymorphisms in the ATP7B lateral wing are used for haplotyping, which is useful for screening relatives of patients with previously identified familial mutations. False-positive results may occur if haplotyping is used on patients with low-probability gene recombinations.
Genetic testing for ATP7B mutations can be valuable to confirm a diagnosis of Wilson disease, especially when presentation is unusual (Caprai et al., 2006). Attention has been drawn to this situation by the molecular confirmation of early-onset hepatic disease in a 3-year-old child (Wilson et al., 2000). Mutation analysis has also confirmed late-onset disease, including the case of two siblings in their 70s – the oldest reported patients so far at time of diagnosis (Nanji et al., 1997; Gupta et al., 2005; Perri et al., 2005; Weitzman et al., 2014).
ATP7B mutation analysis makes an important contribution to clinical practice. Unfortunately, systematic genetic testing for Wilson disease is still difficult and fairly expensive due to the plethora of different mutations, the occurrence of regulatory mutations in non-coding sequence, the large size of the gene, and the limitations of currently available methods. However, technical advances allowing high-throughput screening could be applied to the disease (Bost et al., 2012; Lepori et al., 2012). This new apparatus can sequence six million basepairs of DNA per hour with accuracy greater than 99%. Such advances might permit specialized laboratories to sequence the entire genomic Wilson disease gene from patients, including not only the translated exons, but also the important noncoding sequences that are not normally investigated, to detect all variants.
Interpretation of variants of uncertain significance has become a major challenge for accurate interpretation, genetic counseling, and prevention. Screening family members may help with the interpretation of variants of uncertain significance, but not all variants can be resolved with this approach. Functional analysis is often necessary; however, no clinical functional analysis is yet available. A computational approach to predict significance of mutations is often helpful but a further concrete model is required to demonstrate the efficacy in aiding the clinical decision.
POPULATION SCREENING
The purpose of newborn screening is to identify treatable congenital conditions that can affect a child’s long-term health and development. Recent tandem mass spectrometry (MS/MS) applications have markedly expanded the ability to screen for >50 metabolic diseases from a single dried blood spot. In addition to the original Wilson–Jungner classic screening criteria (Wilson and Jungner, 1968), the American College of Medical Genetics convened the Newborn Screening Expert Group to develop a uniform screening panel in 2006. Of the primary tenants, Wilson disease is an ideal target for screening, given its relatively high prevalence and availability of effective treatment (Hahn et al., 2002; Roberts et al., 2008). Unfortunately, despite extensive discussion on the need for population screening, no cost-effective biomarkers or methods for early screening have been developed for Wilson disease yet. Several small pilot studies have been conducted using ceruloplasmin as a marker, with limited outcomes (Yamaguchi et al., 1999; Hahn et al., 2002; Owada et al., 2002; Schilsky and Shneider, 2002; Kroll et al., 2006). Ceruloplasmin alone is not sufficient to screen for Wilson disease in newborns, as a substantial number of newborns present with physiologically low ceruloplasmin. Ceruloplasmin assay around 3 years of age may be the most appropriate population-screening method, but mandatory health checkups at this age are not universally available in the USA and worldwide.
Many treatable congenital disorders are caused by mutations that result in absent or diminished levels of proteins; thus, protein biomarkers have enormous potential in the diagnosis/screening of congenital disorders. Liquid chromatography mass spectrometry with multiple reaction monitoring (LC-MRM-MS) has emerged as a robust technology that enables highly precise, specific, multiplex quantification of signature proteotypic peptides as stoichiometric surrogates of biomarker proteins.
Our lab is currently exploring the use of peptide immunoaffinity enrichment (Whiteaker et al. 2010, 2011) to quantify ATP7B in DBS. These promising proof-of-concept data open up the possibility of screening for Wilson disease in newborns. Further clinical validation on a large-scale pilot study will be required to determine the efficacy of the assay.
CONCLUSION
Wilson disease is an autosomal-recessive disease due to pathogenic mutations in ATP7B. ATP7B is the only identified gene known to cause Wilson disease, and encodes a transmembrane copper-transporting ATPase of the same name. While biochemical testing and clinical criteria may assist in the early diagnosis and treatment, the current gold standard for Wilson disease diagnosis is direct Sanger sequencing of ATP7B or molecular testing for known familial mutations. Genotype–phenotype correlations have been studied extensively but direct causations remain nebulous. Modifier genes may affect the penetrance and phenotypes but a large-scale study for clinical validation is warranted. The overall worldwide prevalence of Wilson disease is 1 in 30 000 individuals, with significant geographic variation. The most common mutation in Northern America and Europe is the missense mutation p.H1069Q and the most common mutation in East Asian populations is the missense p.R778L. Ceruloplasmin alone is insufficient to screen for Wilson disease in newborns. While peptide immunoaffinity assays show promise for newborn screening, further large-scale clinical studies are required to determine efficacy of these population-based screening methods for Wilson’s disease.
References
- Abdel Ghaffar TY, Elsayed SM, Elnaghy S, et al. Phenotypic and genetic characterization of a cohort of pediatric Wilson disease patients. BMC Pediatr. 2011;11(1):56. doi: 10.1186/1471-2431-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelghaffar TY, Elsayed SM, Elsobky E, et al. Mutational analysis of ATP7B gene in Egyptian children with Wilson disease: 12 novel mutations. J Hum Genet. 2008;53(8):681–687. doi: 10.1007/s10038-008-0298-7. [DOI] [PubMed] [Google Scholar]
- Aggarwal A, Bhatt M. Update on Wilson disease. Int Rev Neurobiol. 2013;110:313–348. doi: 10.1016/B978-0-12-410502-7.00014-4. [DOI] [PubMed] [Google Scholar]
- Aggarwal A, Chandhok G, Todorov T, et al. Wilson disease mutation pattern with genotype–phenotype correlations from Western India: confirmation of p.C271* as a common Indian mutation and identification of 14 novel mutations. Ann Hum Genet. 2013;77(4):299–307. doi: 10.1111/ahg.12024. [DOI] [PubMed] [Google Scholar]
- Al Jumah M, Majumdar R, Al Rajeh S, et al. A clinical and genetic study of 56 Saudi Wilson disease patients: identification of Saudi-specific mutations. Eur J Neurol. 2004;11(2):121–124. doi: 10.1046/j.1351-5101.2003.00729.x. [DOI] [PubMed] [Google Scholar]
- Ala A, Walker AP, Ashkan K, et al. Wilson’s disease. Lancet (London, England) 2007;369(9559):397–408. doi: 10.1016/S0140-6736(07)60196-2. [DOI] [PubMed] [Google Scholar]
- American College of Medical Genetics Newborn Screening Expert Group. Newborn screening: toward a uniform screening panel and system – executive summary. Pediatrics. 2006;117(5 Pt 2):S296–S307. doi: 10.1542/peds.2005-2633I. [DOI] [PubMed] [Google Scholar]
- Bandmann O, Weiss KH, Kaler SG. Wilson’s disease and other neurological copper disorders. Lancet Neurol. 2015;14(1):103–113. doi: 10.1016/S1474-4422(14)70190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bem RS, Raskin S, de Muzzillo DA, et al. Wilson’s disease in Southern Brazil: genotype–phenotype correlation and description of two novel mutations in ATP7B gene. Arq Neuropsiquiatr. 2013;71(8):503–507. doi: 10.1590/0004-282X20130078. [DOI] [PubMed] [Google Scholar]
- Bennett JT, Schwarz KB, Swanson PD, et al. An exceptional family with three consecutive generations affected by Wilson disease. JIMD Reports. 2013;10(Chapter 206):1–4. doi: 10.1007/8904_2012_206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bost M, Piguet-Lacroix G, Parant F, et al. Molecular analysis of Wilson patients: direct sequencing and MLPA analysis in the ATP7B gene and Atox1 and COMMD1 gene analysis. J Trace Elem Med Biol. 2012;26(2–3):97–101. doi: 10.1016/j.jtemb.2012.04.024. [DOI] [PubMed] [Google Scholar]
- Bull PC, Thomas GR, Rommens JM, et al. The Wilson disease gene is a putative copper transporting P-type ATPase similar to the Menkes gene. Nat Genet. 1993;5(4):327–337. doi: 10.1038/ng1293-327. [DOI] [PubMed] [Google Scholar]
- Burkhead JL, Ralle M, Wilmarth P, et al. Elevated copper remodels hepatic RNA processing machinery in the mouse model of Wilson’s disease. J Mol Biol. 2011;406(1):44–58. doi: 10.1016/j.jmb.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caca K, Ferenci P, Kuhn HJ, et al. High prevalence of the ATP7B-H1069Q mutation in Wilson disease patients from Eastern Germany. J Hepatol. 2000;32:134. doi: 10.1016/s0168-8278(01)00219-7. [DOI] [PubMed] [Google Scholar]
- Caprai S, Loudianos G, Massei F, et al. Direct diagnosis of Wilson disease by molecular genetics. J Pediatr. 2006;148(1):138–140. doi: 10.1016/j.jpeds.2005.07.036. [DOI] [PubMed] [Google Scholar]
- Cater MA, Forbes J, La Fontaine S, et al. Intracellular trafficking of the human Wilson protein: the role of the six N-terminal metal-binding sites. Biochem J. 2004;380(3):805–813. doi: 10.1042/BJ20031804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cater MA, La Fontaine S, Mercer JFB. Copper binding to the N-terminal metal-binding sites or the CPC motif is not essential for copper-induced trafficking of the human Wilson protein (ATP7B) Biochem J. 2007;401(1):143–153. doi: 10.1042/BJ20061055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabik G, Litwin T, Członkowska A. Concordance rates of Wilson’s disease phenotype among siblings. J Inherit Metab Dis. 2014;37(1):131–135. doi: 10.1007/s10545-013-9625-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y-F, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76(1):51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Cocoş R, Sendroiu A, Schipor S, et al. Genotype–phenotype correlations in a mountain population community with high prevalence of Wilson’s disease: genetic and clinical homogeneity. PLoS One. 2014;9(6):e98520. doi: 10.1371/journal.pone.0098520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey AJ, Durkie M, Hague S, et al. A genetic study of Wilson’s disease in the United Kingdom. Brain. 2013;136(Pt 5):1476–1487. doi: 10.1093/brain/awt035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronado V, Nanji M, Cox DW. The Jackson toxic milk mouse as a model for copper loading. Mamm Genome. 2001;12:793–795. doi: 10.1007/s00335-001-3021-y. [DOI] [PubMed] [Google Scholar]
- Culotta V, Scott RA. Metals in Cells. John Wiley; Chichester: 2016. [Google Scholar]
- Członkowska A, Gromadzka G, Chabik G. Monozygotic female twins discordant for phenotype of Wilson’s disease. Mov Disord. 2009;24(7):1066–1069. doi: 10.1002/mds.22474. [DOI] [PubMed] [Google Scholar]
- de Bie P, van de Sluis B, Burstein E, et al. Distinct Wilson’s disease mutations in ATP7B are associated with enhanced binding to COMMD1 and reduced stability of ATP7B. Gastroenterology. 2007;133(4):1316–1326. doi: 10.1053/j.gastro.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedoussis GVZ, Genschel J, Sialvera TE, et al. Wilson disease: high prevalence in a mountainous area of Crete. Ann Hum Genet. 2005;69(Pt 3):268–274. doi: 10.1046/j.1529-8817.2005.00171.x. [DOI] [PubMed] [Google Scholar]
- Deguti MM, Genschel J, Cancado EL, et al. Wilson disease: novel mutations in the ATP7B gene and clinical correlation in Brazilian patients. Hum Mutat. 2004;23(4):398. doi: 10.1002/humu.9227. [DOI] [PubMed] [Google Scholar]
- Dmitriev OY, Bhattacharjee A, Nokhrin S, et al. Difference in stability of the N-domain underlies distinct intracellular properties of the E1064A and H1069Q mutants of copper-transporting ATPase ATP7B. J Biol Chem. 2011;286(18):16355–16362. doi: 10.1074/jbc.M110.198101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziezyc K, Groamadzka G, Członkowska A. Wilson’s disease in consecutive generations of one family. Park Rel Dis. 2011;17:577–578. doi: 10.1016/j.parkreldis.2011.04.013. [DOI] [PubMed] [Google Scholar]
- Dziezyc K, Litwin T, Chabik G, et al. Families with Wilson’s disease in subsequenr generations: clinical and genetic analysis. Mov Dis. 2014;29:1828–1832. doi: 10.1002/mds.26057. [DOI] [PubMed] [Google Scholar]
- Fanni D, Pilloni L, Orru S, et al. Expression of ATP7B in normal human liver. Eur J Histochem. 2005;49(4):371–378. doi: 10.4081/965. [DOI] [PubMed] [Google Scholar]
- Fatemi N, Sarkar B. Molecular mechanism of copper transport in Wilson disease. Environ Health Perspect. 2002;110(Suppl 5):695–698. doi: 10.1289/ehp.02110s5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenci P. Wilson’s disease. Clin Gastroenterol Hepatol. 2005;3(8):726–733. doi: 10.1016/s1542-3565(05)00484-2. [DOI] [PubMed] [Google Scholar]
- Ferenci P. Regional distribution of mutations of the ATP7B gene in patients with Wilson disease: impact on genetic testing. Hum Genet. 2006;120(2):151–159. doi: 10.1007/s00439-006-0202-5. [DOI] [PubMed] [Google Scholar]
- Ferenci P, Litwin T, Seniow J, et al. Encephalopathy in Wilson disease: copper toxicity or liver failure? J Clin Exp Hepatol. 2015;5:S88–S95. doi: 10.1016/j.jceh.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figus A, Arigius A, Loudianos G, et al. Molecular pathology and haplotype analysis of Wilson disease in Mediterranean populations. Am J Hum Genet. 1995;57(6):1318–1324. [PMC free article] [PubMed] [Google Scholar]
- Firneisz G, Lakatos PL, Szalay F, et al. Common mutations of ATP7B in Wilson disease patients from Hungary. Am J Med Genet. 2002;108(1):23–28. doi: 10.1002/ajmg.10220. [DOI] [PubMed] [Google Scholar]
- Folhoffer A, Ferenci P, Csak T, et al. Novel mutations of the ATP7B gene among 109 Hungarian patients with Wilson’s disease. Eur J Gastroenterol Hepatol. 2007;19(2):105–111. doi: 10.1097/01.meg.0000223904.70492.0b. [DOI] [PubMed] [Google Scholar]
- García Villarreal L, Daniels S, Shaw SH, et al. High prevalence of the very rare Wilson disease gene mutation Leu708Pro in the island of Gran Canaria (Canary Islands, Spain): a genetic and clinical study. Hepatology (Baltimore, Md) 2000;32(6):1329–1336. doi: 10.1053/jhep.2000.20152. [DOI] [PubMed] [Google Scholar]
- Gomes A, Dedoussis GV. Geographic distribution of ATP7B mutations in Wilson disease. Ann Hum Biol. 2016;43(1):1–8. doi: 10.3109/03014460.2015.1051492. [DOI] [PubMed] [Google Scholar]
- Gourdon P, Liu XY, Skjorringe T, et al. Crystal structure of a copper-transporting PIB-type ATPase. Nature. 2011;475(7354):59–64. doi: 10.1038/nature10191. [DOI] [PubMed] [Google Scholar]
- Gromadzka G, Schmidt HH, Genschel J, et al. Frameshift and nonsense mutations in the gene for ATPase7B are associated with severe impairment of copper metabolism and with an early clinical manifestation of Wilson’s disease. Clin Genet. 2005;68(6):524–532. doi: 10.1111/j.1399-0004.2005.00528.x. [DOI] [PubMed] [Google Scholar]
- Gu Y-H, Kodama H, Du SL, et al. Mutation spectrum and polymorphisms in ATP7B identified on direct sequencing of all exons in Chinese Han and Hui ethnic patients with Wilson’s disease. Clin Genet. 2003;64(6):479–484. doi: 10.1046/j.1399-0004.2003.00179.x. [DOI] [PubMed] [Google Scholar]
- Gupta A, Aikath D, Neogi R, et al. Molecular pathogenesis of Wilson disease: haplotype analysis, detection of prevalent mutations and genotype–phenotype correlation in Indian patients. Hum Genet. 2005;118(1):49–57. doi: 10.1007/s00439-005-0007-y. [DOI] [PubMed] [Google Scholar]
- Gupta A, Chattopadhyay I, Dey S, et al. Molecular pathogenesis of Wilson disease among Indians: a perspective on mutation spectrum in ATP7B gene, prevalent defects, clinical heterogeneity and implication towards diagnosis. Cell Mol Neurobiol. 2007;27(8):1023–1033. doi: 10.1007/s10571-007-9192-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Bhattacharjee A, Dmitriev OY, et al. Cellular copper levels determine the phenotype of the Arg875 variant of ATP7B/Wilson disease protein. Proc Natl Acad Sci U S A. 2011;108(13):5390–5395. doi: 10.1073/pnas.1014959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn SH, Lee SY, Jang YJ, et al. Pilot study of mass screening for Wilson’s disease in Korea. Mol Genet Metab. 2002;76(2):133–136. doi: 10.1016/s1096-7192(02)00026-4. [DOI] [PubMed] [Google Scholar]
- Hofer H, Willheim-Polli C, Knoflauch P, et al. Identification of a novel Wilson disease gene mutation frequent in Upper Austria: a genetic and clinical study. J Hum Genet. 2012;57(9):564–567. doi: 10.1038/jhg.2012.65. [DOI] [PubMed] [Google Scholar]
- Huster D, Hoppert M, Lutsenko S, et al. Defective cellular localization of mutant ATP7B in Wilson’s disease patients and hepatoma cell lines. Gastroenterology. 2003;124(2):335–345. doi: 10.1053/gast.2003.50066. [DOI] [PubMed] [Google Scholar]
- Huster D, Finegold MJ, Morgan CT, et al. Consequences of copper accumulation in the livers of the Atp7b–/– (Wilson disease gene) knockout mice. Am J Pathol. 2006;168(2):423–434. doi: 10.2353/ajpath.2006.050312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huster D, Kuhne A, Bhattacharjee A, et al. Diverse functional properties of Wilson disease ATP7B variants. Gastroenterology. 2012;142(4):947–956. e5. doi: 10.1053/j.gastro.2011.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacob R, Iacob S, Nastase A, et al. The His1069Gln mutation in the ATP7B gene in Romanian patients with Wilson’s disease referred to a tertiary gastroenterology center. J Gastrointestin Liver Dis. 2012;21(2):181–185. [PubMed] [Google Scholar]
- Ivanova-Smolenskaya I, et al. 5-29-06 Molecular analysis in Russian families with Wilson’s disease. J Neurol Sci. 1997;150:S314. [Google Scholar]
- Kalita J, Somarajan BI, Misra UK, et al. R778L, H1069Q, and I1102T mutation study in neurologic Wilson disease. Neurol India. 2010;58(4):627–630. doi: 10.4103/0028-3886.68678. [DOI] [PubMed] [Google Scholar]
- Kegley KM, Sellers MA, Ferber MJ, et al. Fulminant Wilson’s disease requiring liver transplantation in one monozygotic twin despite identical genetic mutation. Am J Transplant. 2010;10(5):1325–1329. doi: 10.1111/j.1600-6143.2010.03071.x. [DOI] [PubMed] [Google Scholar]
- Kenney SM, Cox DW. Sequence variation database for the Wilson disease copper transporter, ATP7B. Hum Mutat. 2007;28(12):1171–1177. doi: 10.1002/humu.20586. [DOI] [PubMed] [Google Scholar]
- Kim EK, Yoo OJ, Song KY, et al. Identification of three novel mutations and a high frequency of the Arg778Leu mutation in Korean patients with Wilson disease. Hum Mutat. 1998;11(4):275–278. doi: 10.1002/(SICI)1098-1004(1998)11:4<275::AID-HUMU4>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Kim G-H, Jang JY, Park JY, et al. Estimation of Wilson’s disease incidence and carrier frequency in the Korean population by screening ATP7B major mutations in newborn filter papers using the SYBR green intercalator method based on the amplification refractory mutation system. Genet Test. 2008;12(3):395–399. doi: 10.1089/gte.2008.0016. [DOI] [PubMed] [Google Scholar]
- Kroll CA, Ferber MJ, Dawson BD, et al. Retrospective determination of ceruloplasmin in newborn screening blood spots of patients with Wilson disease. Mol Genet Metab. 2006;89(1–2):134–138. doi: 10.1016/j.ymgme.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Kumar N, Gross JB, Ahlskog JE. Myelopathy due to copper deficiency. Neurology. 2003;61(2):273–274. doi: 10.1212/01.wnl.0000073542.02761.5f. [DOI] [PubMed] [Google Scholar]
- Kumar S, Thapa B, Kaur G, et al. Analysis of most common mutations R778G, R778L, R778W, I1102T and H1069Q in Indian Wilson disease patients: correlation between genotype/phenotype/copper ATPase activity. Mol Cell Biochem. 2006;294(1–2):1–10. doi: 10.1007/s11010-005-9028-z. [DOI] [PubMed] [Google Scholar]
- Kumar SS, Kurian G, Eapen CE, et al. Genetics of Wilson’s disease: a clinical perspective. Indian J Gastroenterol. 2012;31(6):285–293. doi: 10.1007/s12664-012-0237-6. [DOI] [PubMed] [Google Scholar]
- Kuppala D, et al. Wilson disease mutations in the American population: identification of five novel mutations in ATP7B. Open Hepatol J. 2009;1(1):1–4. [Google Scholar]
- La Fontaine S, Theophilos MB, Firth SD, et al. Effect of the toxic milk mutation (tx) on the function and intracellular localization of Wnd, the murine homologue of the Wilson copper ATPase. Hum Mol Genet. 2001;10(4):361–370. doi: 10.1093/hmg/10.4.361. [DOI] [PubMed] [Google Scholar]
- Lee CC, Wu JY, Tsai FJ, et al. Molecular analysis of Wilson disease in Taiwan: identification of one novel mutation and evidence of haplotype-mutation association. J Hum Genet. 2000;45(5):275–279. doi: 10.1007/s100380070015. [DOI] [PubMed] [Google Scholar]
- Leggio L, Addolorato G, Loudianos G, et al. Genotype–phenotype correlation of the Wilson disease ATP7B gene. Am J Med Genet A. 2006;140A(8):933. doi: 10.1002/ajmg.a.31191. [DOI] [PubMed] [Google Scholar]
- Lenartowicz M, Krzeptowski W. Structure and function of ATP7A and ATP7B proteins – Cu-transporting ATPases. Postepy Biochem. 2010;56:317–327. [PubMed] [Google Scholar]
- Lepori M-B, Zappu A, Incollu S, et al. Mutation analysis of the ATP7B gene in a new group of Wilson’s disease patients: contribution to diagnosis. Mol Cell Probes. 2012;26(4):147–150. doi: 10.1016/j.mcp.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Loudianos G, Dessi V, Lovicu M, et al. Mutation analysis in patients of Mediterranean descent with Wilson disease: identification of 19 novel mutations. J Med Genet. 1999;36(11):833–836. [PMC free article] [PubMed] [Google Scholar]
- Lovicu M, Dessi V, Lepori MB, et al. The canine copper toxicosis gene MURR1 is not implicated in the pathogenesis of Wilson disease. J Gastroenterol. 2006;41(6):582–587. doi: 10.1007/s00535-006-1807-0. [DOI] [PubMed] [Google Scholar]
- Lutsenko S. Modifying factors and phenotypic diversity in Wilson’s disease. Ann N Y Acad Sci. 2014;1315(1):56–63. doi: 10.1111/nyas.12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutsenko S, Petrukhin K, Cooper MJ, et al. N-terminal domains of human copper-transporting adenosine triphosphatases (the Wilson’s and Menkes disease proteins) bind copper selectively in vivo and in vitro with stoichiometry of one copper per metal-binding repeat. J Biol Chem. 1997;272(30):18939–18944. doi: 10.1074/jbc.272.30.18939. [DOI] [PubMed] [Google Scholar]
- Machado AAC, Deguti MM, Genschel J, et al. Neurological manifestations and ATP7B mutations in Wilson’s disease. Parkinsonism Relat Disord. 2008;14(3):246–249. doi: 10.1016/j.parkreldis.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Majumdar R, Al-Jumah M, Zaidan R. A rare homozygous missense mutation in ATP7B exon 19 in a case of Wilson disease. Eur Neurol. 2004;51(1):52–54. doi: 10.1159/000075092. [DOI] [PubMed] [Google Scholar]
- Mak CM, Lam C-W. Diagnosis of Wilson’s disease: a comprehensive review. Crit Rev Clin Lab Sci. 2008;45(3):263–290. doi: 10.1080/10408360801991055. [DOI] [PubMed] [Google Scholar]
- Manolaki N, Nikolopoulou G, Daikos GL, et al. Wilson disease in children: analysis of 57 cases. J Pediatr Gastroenterol Nutr. 2009;48(1):72–77. doi: 10.1097/MPG.0b013e31817d80b8. [DOI] [PubMed] [Google Scholar]
- Margarit E, Bach V, Gomez D, et al. Mutation analysis of Wilson disease in the Spanish population - identification of a prevalent substitution and eight novel mutations in the ATP7B gene. Clin Genet. 2005;68(1):61–68. doi: 10.1111/j.1399-0004.2005.00439.x. [DOI] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, et al. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36(10):1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- Merle U, Weiss KH, Eisenbach C, et al. Truncating mutations in the Wilson disease gene ATP7B are associated with very low serum ceruloplasmin oxidase activity and an early onset of Wilson disease. BMC Gastroenterol. 2010;10(1):8. doi: 10.1186/1471-230X-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller LB, Horn N, Jeppesen TD, et al. Clinical presentation and mutations in Danish patients with Wilson disease. Eur J Hum Genet. 2011;19(9):935–941. doi: 10.1038/ejhg.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Kubota M, Katoh Y, et al. Early and pre-symptomatic detection of Wilson’s disease at the mandatory 3-year-old medical health care examination in Hokkaido Prefecture with the use of a novel automated urinary ceruloplasmin assay. Mol Genet Metab. 2008;94(3):363–367. doi: 10.1016/j.ymgme.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Nanji MS, Nguyen VT, Kawasoe JH, et al. Haplotype and mutation analysis in Japanese patients with Wilson disease. Am J Hum Genet. 1997;60(6):1423–1429. doi: 10.1086/515459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro E, Ranucci G, Vajro P, et al. Re-evaluation of the diagnostic criteria for Wilson disease in children with mild liver disease. Hepatology (Baltimore, Md) 2010;52(6):1948–1956. doi: 10.1002/hep.23910. [DOI] [PubMed] [Google Scholar]
- Okada T, Shiono Y, Hayashi H, et al. Mutational analysis of ATP7B and genotype–phenotype correlation in Japanese with Wilson’s disease. Hum Mutat. 2000;15(5):454–462. doi: 10.1002/(SICI)1098-1004(200005)15:5<454::AID-HUMU7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Okada T, Shinono Y, Kaneko Y, et al. High prevalence of fulminant hepatic failure among patients with mutant alleles for truncation of ATP7B in Wilson’s disease. Scand J Gastroenterol. 2010;45(10):1232–1237. doi: 10.3109/00365521.2010.492527. [DOI] [PubMed] [Google Scholar]
- Owada M, Suzuki K, Fukushi M, et al. Mass screening for Wilson’s disease by measuring urinary holoceruloplasmin. J Pediatr. 2002;140(5):614–616. doi: 10.1067/mpd.2002.122731. [DOI] [PubMed] [Google Scholar]
- Panagiotakaki E, Tzetis M, Manolaki N, et al. Genotype–phenotype correlations for a wide spectrum of mutations in the Wilson disease gene (ATP7B) Am J Med Genet A. 2004;131A(2):168–173. doi: 10.1002/ajmg.a.30345. [DOI] [PubMed] [Google Scholar]
- Panichareon B, Taweechue K, Thongnoppakhun W, et al. Six novel ATP7B mutations in Thai patients with Wilson disease. Eur J Med Genet. 2011;54(2):103–107. doi: 10.1016/j.ejmg.2010.10.008. [DOI] [PubMed] [Google Scholar]
- Paradisi I, De Freitas L, Arias S. Most frequent mutation c.3402delC (p.Ala1135GlnfsX13) among Wilson disease patients in Venezuela has a wide distribution and two old origins. Eur J Med Genet. 2015;58(2):59–65. doi: 10.1016/j.ejmg.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Park S, Park JY, Kim GH, et al. Identification of novel ATP7Bgene mutations and their functional roles in Korean patients with Wilson disease. Hum Mutat. 2007;28(11):1108–1113. doi: 10.1002/humu.20574. [DOI] [PubMed] [Google Scholar]
- Park H, Park DK, Kim MS, et al. Pseudo-dominant inheritance in Wilson’s disease. Neurol Sci. 2015;37(1):153–155. doi: 10.1007/s10072-015-2394-8. [DOI] [PubMed] [Google Scholar]
- Payne AS, Kelly EJ, Gitlin JD. Functional expression of the Wilson disease protein reveals mislocalization and impaired copper-dependent trafficking of the common H1069Q mutation. Proc Natl Acad Sci U S A. 1998;95(18):10854–10859. doi: 10.1073/pnas.95.18.10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perri RE, Hahn SH, Ferber MJ, et al. Wilson disease –keeping the bar for diagnosis raised. Hepatology (Baltimore, Md) 2005;42(4):974–974. doi: 10.1002/hep.20893. [DOI] [PubMed] [Google Scholar]
- Petrukhin K, Fischer SG, Pirastu M, et al. Mapping, cloning and genetic characterization of the region containing the Wilson disease gene. Nat Genet. 1993;5(4):338–343. doi: 10.1038/ng1293-338. [DOI] [PubMed] [Google Scholar]
- Przybyłkowski A, Gromadzka G, Członkowska A. Polymorphisms of metal transporter genes DMT1 and ATP7A in Wilson’s disease. J Trace Elem Med Biol. 2014;28(1):8–12. doi: 10.1016/j.jtemb.2013.08.002. [DOI] [PubMed] [Google Scholar]
- Ralle M, Huster D, Vogt S, et al. Wilson disease at a single cell level: intracellular copper trafficking activates compartment-specific responses in hepatocytes. J Biol Chem. 2010;285(40):30875–30883. doi: 10.1074/jbc.M110.114447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts EA, Schilsky ML American Association for Study of Liver Diseases (AASLD) Diagnosis and treatment of Wilson disease: an update. Hepatology (Baltimore, Md) 2008;47(6):2089–2111. doi: 10.1002/hep.22261. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Granillo A, Sedlak E, Wittung-Stafshede P. Stability and ATP binding of the nucleotide-binding domain of the Wilson disease protein: effect of the common H1069Q mutation. J Mol Biol. 2008;383(5):1097–1111. doi: 10.1016/j.jmb.2008.07.065. [DOI] [PubMed] [Google Scholar]
- Santhosh S, Shaji RV, Eapen CE, et al. ATP7B mutations in families in a predominantly Southern Indian cohort of Wilson’s disease patients. Indian J Gastroenterol. 2006;25(6):277–282. [PubMed] [Google Scholar]
- Sazinsky MH, Mandal AK, Arguello JM, et al. Structure of the ATP binding domain from the Archaeoglobus fulgidus Cu+-ATPase. J Biol Chem. 2006;281(16):11161–11166. doi: 10.1074/jbc.M510708200. [DOI] [PubMed] [Google Scholar]
- Schilsky ML, Shneider B. Population screening for Wilson’s disease. J Pediatr. 2002;140(5):499–501. doi: 10.1067/mpd.2002.124769. [DOI] [PubMed] [Google Scholar]
- Schushan M, Bhattacharjee A, Ben-Tai N, et al. A structural model of the copper ATPase ATP7B to facilitate analysis of Wilson disease-causing mutations and studies of the transport mechanism. Metallomics : Integrated Biometal Science. 2012;4(7):669–678. doi: 10.1039/c2mt20025b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah AB, Chernov I, Zhang HT, et al. Identification and analysis of mutations in the Wilson disease gene (ATP7B): population frequencies, genotype–phenotype correlation, and functional analyses. Am J Hum Genet. 1997;61(2):317–328. doi: 10.1086/514864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu N, Nakazono H, Takeshita Y, et al. Molecular analysis and diagnosis in Japanese patients with Wilson’s disease. Pediatr Int. 1999;41(4):409–413. doi: 10.1046/j.1442-200x.1999.01092.x. [DOI] [PubMed] [Google Scholar]
- Simon I. Analysis of the human Atox 1 homologue in Wilson patients. World J Gastroenterol. 2008;14(15):2383. doi: 10.3748/wjg.14.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek Papur O, Akman SA, Cakmur R, et al. Mutation analysis of ATP7B gene in Turkish Wilson disease patients: identification of five novel mutations. Eur J Med Genet. 2013;56(4):175–179. doi: 10.1016/j.ejmg.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Song M-J, Lee ST, Lee MK, et al. Estimation of carrier frequencies of six autosomal-recessive Mendelian disorders in the Korean population. J Hum Genet. 2012;57(2):139–144. doi: 10.1038/jhg.2011.144. [DOI] [PubMed] [Google Scholar]
- Stapelbroek JM, Bollen CW, van Amstel JK, et al. The H1069Q mutation in ATP7B is associated with late and neurologic presentation in Wilson disease: results of a meta-analysis. J Hepatol. 2004;41(5):758–763. doi: 10.1016/j.jhep.2004.07.017. [DOI] [PubMed] [Google Scholar]
- Stättermayer AF, Rutter K, Beinhardt S, et al. Genetic factors associated with histologic features of the liver and treatment outcome in chronic hepatitis C patients. Z Gastroenterol. 2012;50(05):P51. [Google Scholar]
- Stenson PD, Ball EV, Mort M, et al. The Human Gene Mutation Database (HGMD) and its exploitation in the fields of personalized genomics and molecular evolution. Curr Protoc Bioinformatics. 2012;Chapter 1(Unit 1.13) doi: 10.1002/0471250953.bi0113s39. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, et al. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133(1):1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternlieb I, Scheinberg IH. Prevention of Wilson’s disease in asymptomatic patients. New Engl J Med. 1968;278:352–359. doi: 10.1056/NEJM196802152780702. [DOI] [PubMed] [Google Scholar]
- Takeshita Y, Shimizu N, Yamaguchi Y, et al. Two families with Wilson disease in which siblings showed different phenotypes. J Hum Genet. 2002;47(10):0543–0547. doi: 10.1007/s100380200082. [DOI] [PubMed] [Google Scholar]
- Taly AB, Meenakshi-Sundaram S, Sinha S, et al. Wilson disease: description of 282 patients evaluated over 3 decades. Medicine. 2007;86(2):112–121. doi: 10.1097/MD.0b013e318045a00e. [DOI] [PubMed] [Google Scholar]
- Tanzi RE, Petrukhin K, Chernov I, et al. The Wilson disease gene is a copper transporting ATPase with homology to the Menkes disease gene. Nat Genet. 1993;5(4):344–350. doi: 10.1038/ng1293-344. [DOI] [PubMed] [Google Scholar]
- Tatsumi Y, Hattori A, Hayashi H, et al. Current state of Wilson disease patients in central Japan. Intern Med (Tokyo, Japan) 2010;49(9):809–815. doi: 10.2169/internalmedicine.49.2931. [DOI] [PubMed] [Google Scholar]
- Theophilos MB, Cox DW, Mercer JF. The toxic milk mouse is a murine model of Wilson disease. Hum Mol Genet. 1996;5(10):1619–1624. doi: 10.1093/hmg/5.10.1619. [DOI] [PubMed] [Google Scholar]
- Thomas GR, Jensson O, Gudmundsson G, et al. Wilson disease in Iceland: a clinical and genetic study. Am J Hum Genet. 1995a;56(5):1140–1146. [PMC free article] [PubMed] [Google Scholar]
- Thomas GR, Roberts EA, Walshe JM, et al. Haplotypes and mutations in Wilson disease. Am J Hum Genet. 1995b;56(6):1315–1319. [PMC free article] [PubMed] [Google Scholar]
- Todorov T, Savov A, Jelev H, et al. Spectrum of mutations in the Wilson disease gene (ATP7B) in the Bulgarian population. Clin Genet. 2005;68(5):474–476. doi: 10.1111/j.1399-0004.2005.00516.x. [DOI] [PubMed] [Google Scholar]
- Tomić A, Dobricic V, Novakovic I, et al. Mutational analysis of ATP7B gene and the genotype–phenotype correlation in patients with Wilson’s disease in Serbia. Vojnosanit Pregl. 2013;70(5):457–462. doi: 10.2298/vsp1305457t. [DOI] [PubMed] [Google Scholar]
- Tsivkovskii R, Efremov RG, Lutsenko S. The role of the invariant His-1069 in folding and function of the Wilson’s disease protein, the human copper-transporting ATPase ATP7B. J Biol Chem. 2003;278:13302–13308. doi: 10.1074/jbc.M300034200. [DOI] [PubMed] [Google Scholar]
- Usta J, Wehbeh A, Rida K, et al. Phenotype–genotype correlation in Wilson disease in a large Lebanese family: association of c.2299insC with hepatic and of p. Ala1003Thr with neurologic phenotype. PLoS One. 2014;9(11):e109727. doi: 10.1371/journal.pone.0109727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berghe PVE, Stapelbroek JM, Krieger E, et al. Reduced expression of ATP7B affected by Wilson disease-causing mutations is rescued by pharmacological folding chaperones 4-phenylbutyrate and curcumin. Hepatology (Baltimore, Md) 2009;50(6):1783–1795. doi: 10.1002/hep.23209. [DOI] [PubMed] [Google Scholar]
- Vrabelova S, Letocha O, Borsky M, et al. Mutation analysis of the ATP7B gene and genotype/phenotype correlation in 227 patients with Wilson disease. Mol Genet Metab. 2005;86(1–2):277–285. doi: 10.1016/j.ymgme.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Walker JM, Huster D, Ralle M, et al. The N-terminal metal-binding site 2 of the Wilson’s disease protein plays a key role in the transfer of copper from Atox1. J Biol Chem. 2004;279(15):15376–15384. doi: 10.1074/jbc.M400053200. [DOI] [PubMed] [Google Scholar]
- Wan L, Tsai CH, Tsai Y, et al. Mutation analysis of Taiwanese Wilson disease patients. Biochem Biophys Res Commun. 2006;345(2):734–738. doi: 10.1016/j.bbrc.2006.04.136. [DOI] [PubMed] [Google Scholar]
- Wang L-H, Huang YQ, Shang X, et al. Mutation analysis of 73 southern Chinese Wilson’s disease patients: identification of 10 novel mutations and its clinical correlation. J Hum Genet. 2011;56(9):660–665. doi: 10.1038/jhg.2011.76. [DOI] [PubMed] [Google Scholar]
- Wei Z, Huang Y, Liu A, et al. Mutational characterization of ATP7B gene in 103 Wilson’s disease patients from Southern China: identification of three novel mutations. Neuroreport. 2014;25(14):1075–1080. doi: 10.1097/WNR.0000000000000216. [DOI] [PubMed] [Google Scholar]
- Weiss KH. Copper toxicosis gene MURR1 is not changed in Wilson disease patients with normal blood ceruloplasmin levels. World J Gastroenterol. 2006;12(14):2239. doi: 10.3748/wjg.v12.i14.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss KH, Runz H, Noe B, et al. Genetic analysis of BIRC4/XIAP as a putative modifier gene of Wilson disease. J Inherit Metab Dis. 2010;33(3):233–240. doi: 10.1007/s10545-010-9123-5. [DOI] [PubMed] [Google Scholar]
- Weitzman E, Pappo O, Weiss P, et al. Late onset fulminant Wilson’s disease: a case report and review of the literature. World J Gastroenterol. 2014;20(46):17656–17660. doi: 10.3748/wjg.v20.i46.17656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteaker JR, Zhao L, Anderson L, et al. An automated and multiplexed method for high throughput peptide immunoaffinity enrichment and multiple reaction monitoring mass spectrometry-based quantification of protein biomarkers. Mol Cell Proteomics. 2010;9(1):184–196. doi: 10.1074/mcp.M900254-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteaker JR, Lin C, Kennedy J, et al. A targeted proteomics-based pipeline for verification of biomarkers in plasma. Nat Biotechnol. 2011;29(7):625–634. doi: 10.1038/nbt.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JMG, Jungner G. Public Health Paper no. 34. World Health Organization; Geneva: 1968. Principles and practice of screening for disease. [Google Scholar]
- Wilson DC, Phillips MJ, Cox DW, et al. Severe hepatic Wilson’s disease in preschool-aged children. J Pediatr. 2000;137(5):719–722. doi: 10.1067/mpd.2000.108569. [DOI] [PubMed] [Google Scholar]
- Wu Z-Y, Lin MT, Murong SX, et al. Molecular diagnosis and prophylactic therapy for presymptomatic Chinese patients with Wilson disease. Arch Neurol. 2003;60(5):737–741. doi: 10.1001/archneur.60.5.737. [DOI] [PubMed] [Google Scholar]
- Wu F, Wang J, Pu C, et al. Wilson’s disease: a comprehensive review of the molecular mechanisms. Int J Mol Sci. 2015;16(3):6419–6431. doi: 10.3390/ijms16036419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Aoki T, Arashima S, et al. Mass screening for Wilson’s disease: results and recommendations. Pediatr Int. 1999;41(4):405–408. doi: 10.1046/j.1442-200x.1999.01096.x. [DOI] [PubMed] [Google Scholar]
- Yoo HW. Identification of novel mutations and the three most common mutations in the human ATP7B gene of Korean patients with Wilson disease. Genet Med. 2002;4(6 Suppl):43S–48S. doi: 10.1097/00125817-200211001-00009. [DOI] [PubMed] [Google Scholar]
- Zali N, Mohebbi SR, Estaghamat S, et al. Prevalence of ATP7B gene mutations in Iranian patients with Wilson disease. Hepatitis Monthly. 2011;11(11):890–894. doi: 10.5812/kowsar.1735143X.762. [DOI] [PMC free article] [PubMed] [Google Scholar]