ABSTRACT
Colistin and polymyxin B remain part of the last line of antibiotics for multidrug-resistant Gram-negative bacteria, such as carbapenem-resistant Enterobacteriaceae. Current joint EUCAST-CLSI recommendations are for broth microdilution (BMD) to be performed for MIC testing of colistin. Commercial susceptibility testing methods were evaluated and compared against the reference BMD, using a susceptibility breakpoint of ≤2 mg/liter for both colistin and polymyxin B. Seventy-six Enterobacteriaceae were included, of which 21 were mcr-1 positive (18 Escherichia coli isolates, 2 Klebsiella pneumoniae isolates, and 1 Enterobacter aerogenes isolate). Rates of essential agreement (EA) of colistin test results between BMD and Vitek 2, Sensititre, and Etest were 93.4%, 89.5%, and 75.0%, respectively. Rates of EA of polymyxin B test results between BMD and Vitek 2, Sensititre, and Etest were 96.1%, 96.1%, and 48.7%, respectively. A positive MIC correlation with a categorical agreement of >90% was achieved for Sensititre (colistin Spearman's ρ = 0.863, and polymyxin B Spearman's ρ = 0.877) and Vitek 2 (polymyxin B [only] Spearman's ρ = 0.8917). Although a positive MIC correlation (Spearman's ρ = 0.873) with the reference method was achieved for colistin testing with Vitek 2, categorical agreement was <90%, with very major error rates of 36%. Correlation with the Etest MIC was lower, with very major error rates of 12% (colistin) and 26.1% (polymyxin B). MicroScan (colistin) categorical agreement was 88.2%, with a very major error rate of 4%. Colistin MICs for 15 of the 21 mcr-1-positive isolates were >2 mg/liter, and polymyxin MICs for 17 of them were >2 mg/liter by broth microdilution. The use of a lower breakpoint of ≤1 mg/liter further improves detection of mcr-1 for all testing methods. However, further data on the correlation between MICs and clinical outcome are required to determine the most suitable breakpoint to guide clinical management.
KEYWORDS: colistin, polymyxin B, susceptibility testing, mcr-1
INTRODUCTION
Multidrug-resistant organisms (MDROs) represent a progressive burden on health care systems globally. Carbapenem-resistant Enterobacteriaceae (CRE) are now reported worldwide. CRE infections are difficult to treat, and there are limited options available. The polymyxins (colistin and polymyxin B) are antibiotics which are used for treating infections with CRE. Polymyxins initially fell out of favor among clinicians due to concerns over high rates of nephrotoxicity and neurotoxicity (1). In recent years, the use of polymyxins has been on the rise, with increasing rates of CRE infection. Combination therapy is often used for additive or synergistic antimicrobial activity (2, 3).
Chromosomal and plasmid-mediated resistance to polymyxins has been described. Mutations that result in alteration of the target site of action (lipopolysaccharide) result in elevated MICs of these drugs (4). Plasmid-mediated polymyxin resistance (mcr-1 and mcr-2) is also a reality and, as with plasmid-mediated carbapenemase resistance genes, is now disseminated globally (5, 6). The presence of the mcr-1 or mcr-2 plasmid increases the MICs of colistin and polymyxin B for CRE and may contribute to treatment failure. Susceptibility testing plays a paramount role in guiding antimicrobial treatment, as treatment benefits need to be balanced against potential toxicities associated with this class of antibiotics.
The European Committee on Antimicrobial Susceptibility Testing (EUCAST) has published breakpoints for colistin for Enterobacteriaceae, Pseudomonas aeruginosa, and Acinetobacter spp. with a susceptible breakpoint of ≤2 mg/liter and a resistant breakpoint of >2 mg/liter (7). The Clinical and Laboratory Standards Institute (CLSI) has published breakpoints for colistin and polymyxin B for Pseudomonas aeruginosa and Acinetobacter spp. (8). The CLSI currently does not have colistin or polymyxin B breakpoints for Enterobacteriaceae but has published colistin epidemiological cutoff values for Escherichia coli, Klebsiella pneumoniae, Raoultella ornithinolytica, Enterobacter aerogenes, and Enterobacter cloacae. The recommendation is to not provide susceptible, intermediate, or resistant interpretations but instead to define tested isolates as either wild type or non-wild type (8).
A joint recommendation by the CLSI and EUCAST released in 2016 recommended the ISO-20776 standard broth microdilution (BMD) method for MIC testing of colistin (9). Other testing methods, such as agar dilution, disk diffusion, and gradient diffusion, are not currently recommended. The use of reference broth microdilution methods for susceptibility testing may not be practical in diagnostic microbiology laboratories, depending on individual laboratory workloads. A reliable and reproducible susceptibility testing method is required for diagnostics. We investigated commercially available broth microdilution methods (Sensititre [Thermo Fisher Scientific, MA, USA], MicroScan [Beckman Coulter, CA, USA], and the Vitek 2 system [bioMérieux, Marcy l'Etoile, France]) on carbapenemase-resistant Enterobacteriaceae and mcr-1-positive isolates. Colistin and polymyxin B Etests (bioMérieux, Marcy l'Etoile, France) were also performed. All commercial tests were compared against the reference broth microdilution method.
RESULTS
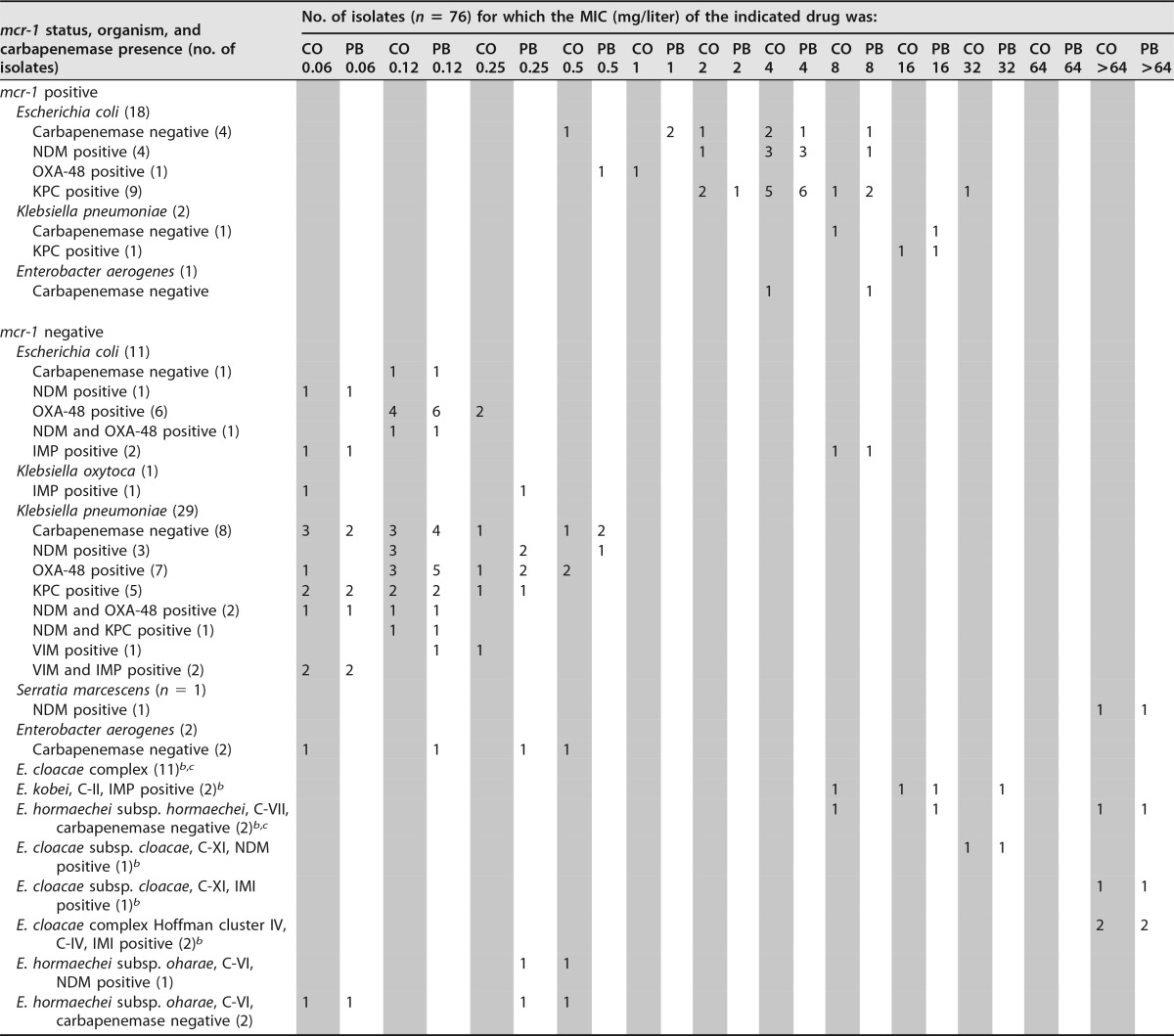
MICs for E. coli ATCC 25922 were between 0.25 and 0.5 mg/liter by all testing methods, which is within the expected range for all tests (0.25 to 2 mg/liter for colistin and polymyxin B) (8). Proteus mirabilis ATCC 12453 demonstrated resistance by all testing methods. The distributions of colistin and polymyxin B MICs determined by BMD for all isolates are presented in Table 1. Overall, 51 (67.1%) and 49 (64.5%) isolates were susceptible to colistin and polymyxin B, respectively, by BMD. Twenty-one of the 76 isolates were mcr-1 positive. For the majority of mcr-1-positive isolates, MICs of colistin and polymyxin B were >2 mg/liter. For four isolates, MICs of colistin only were ≤2 mg/liter, for two isolates, MICs of polymyxin B only were ≤2 mg/liter, and for two isolates, MICs of both colistin and polymyxin B were ≤2 mg/liter (ranging from 0.5 to 2 mg/liter for colistin and polymyxin B). The other drug-method combinations and their sensitivities for demonstrating phenotypic resistance in mcr-1-positive isolates are shown in Table 2. Both Sensititre and MicroScan had 100% sensitivity when colistin was tested; Sensititre and Vitek 2 had equal sensitivities of 95.2% when polymyxin B was tested. Sensitivities of other methods for detecting mcr-1 were <95%.
TABLE 1.
Broth microdilution colistin and polymyxin B MIC distributions for all test isolatesa
CO, colistin; PB, polymyxin B.
b E. cloacae complex isolates belonging to hsp60 genetic clusters associated with colistin heteroresistance.
c One isolate from cluster VII resulted in a VME by Sensititre, with heteroresistance demonstrated upon repeat testing.
TABLE 2.
Sensitivities of colistin and polymyxin B susceptibility testing methods for the detection of mcr-1-positive isolatesa
| Drug | Method | % of isolates susceptible at a breakpoint of ≤2 mg/liter | % of isolates susceptible at a breakpoint of ≤1 mg/liter |
|---|---|---|---|
| Colistin | BMD | 71.4 | 90.5 |
| Sensititre | 100 | 100.0 | |
| Vitek 2 | 42.9 | 95.2 | |
| Etest | 76.2 | 95.2 | |
| MicroScan | 100 | NA | |
| Polymyxin B | BMD | 81.0 | 85.7 |
| Sensititre | 95.2 | 100.0 | |
| Vitek 2 | 95.2 | 100.0 | |
| Etest | 66.7 | 95.2 |
We tested 21 mcr-1-positive isolates. NA, not applicable, as the lowest MIC interpretation possible for MicroScan (colistin) is 2 mg/liter.
Both colistin and polymyxin B had the same MIC50 and MIC90. The overall MIC50 and MIC90 were 0.25 mg/liter and 16 mg/liter, respectively. For mcr-1-positive isolates, the MIC50 was 4 mg/liter and the MIC90 was 8 mg/liter. The highest MICs were found for Enterobacter cloacae complex isolates that were mcr-1 negative and the intrinsically resistant Serratia marcescens isolate.
The overall performance characteristics of the different methods compared to those of BMD are presented in Table 3. Spearman correlation between Vitek 2 or Sensititre MICs and BMD MICs was higher than between Etest MICs and BMD MICs. There was also >90% categorical agreement (CA) between Sensititre (colistin and polymyxin B), Vitek 2 (polymyxin B), or Etest (colistin) and BMD. Although Vitek 2 (colistin) and Etests (colistin and polymyxin B) demonstrated close to 90% CA, the overall very major error (VME) rate was high (12% to 36%). This is despite positive correlation with MICs from Vitek 2 (colistin) (Spearman's ρ = 0.873) and likely reflects that the MICs were elevated but remained close to the breakpoint. Essential agreement was particularly poor for Etests and could be related to the poor diffusion of polymyxin molecules, resulting in a narrow zone of inhibition close to the MIC.
TABLE 3.
Performance characteristics of colistin and polymyxin B susceptibility testing methods in comparison to broth microdilutiona
| Drug | Method | No. of isolates that were susceptible | No. of isolates that were resistant | No. (%) of isolates exhibiting EA | No. (%) of isolates exhibiting CA | No. of isolates exhibiting VMEs (%) | No. of isolates exhibiting MEs (%) | Spearman's coefficient |
|---|---|---|---|---|---|---|---|---|
| Colistin | Vitek 2 | 60 | 16 | 71 (93.4)b | 67 (88.2) | 9 (36.0) | 0 (0) | 0.873c |
| Sensititre | 46 | 30 | 68 (89.5) | 69 (90.1)b | 1 (4) | 6 (11.8) | 0.863c | |
| Etest | 51 | 25 | 57 (75.0) | 70 (92.1)b | 3 (12.0) | 3 (5.9) | 0.600c | |
| MicroScan | 44 | 32 | NAd | 67 (88.2) | 1 (4) | 8 (15.8) | NAd | |
| Polymyxin B | BMD | 49 | 27 | NA | NA | NA | NA | NA |
| Vitek 2 | 47 | 29 | 73 (96.1)b | 72 (94.7)b | 1 (3.7) | 3 (6.1) | 0.917c | |
| Sensititre | 47 | 29 | 73 (96.1)b | 72 (94.7)b | 1 (3.7) | 3 (6.1) | 0.877c | |
| Etest | 53 | 23 | 38 (48.7) | 68 (89.5) | 6 (26.1) | 1 (1.9) | 0.534c |
BMD, broth microdilution; EA, essential agreement; CA, categorical agreement; VME, very major error; ME, major error; NA, not applicable. Spearman's coefficient indicates concordance of MICs with those of BMD.
The testing method-drug combination meets CLSI M52 recommendations for acceptable EA or CA performance.
P value < 0.001.
EA and Spearman's correlation coefficient were not determined for MicroScan due to a narrow MIC range.
A single VME was demonstrated for each of the following method-drug combinations: MicroScan (colistin), Sensititre (colistin and polymyxin B), and Vitek 2 (polymyxin B). Higher VME rates were demonstrated for Vitek 2 testing with colistin (36.0%) and for Etests (colistin [12.0%] and polymyxin B [26.1%]). MicroScan had eight major errors (MEs) for colistin, Vitek 2 had zero MEs for colistin and three MEs for polymyxin B, Sensititre had six MEs for colistin and three MEs for polymyxin B, and Etest had three MEs for colistin and one ME for polymyxin B. All isolates for which we found MEs were mcr-1-positive isolates without phenotypic resistance as determined by BMD based on the current breakpoints, with the exception of two isolates tested with MicroScan (colistin).
A total of 10 mcr-1-negative isolates were resistant to both colistin and polymyxin B by BMD. Of these, one was Escherichia coli and eight were Enterobacter cloacae complex strains. Serratia marcescens also tested resistant, as expected. This isolate was included, as it was part of the collection of carbapenem-resistant Enterobacteriaceae. Eight of 11 mcr-1-negative E. cloacae complex isolates were resistant to colistin and polymyxin on the basis of BMD MICs (Table 1). Resistance to colistin was not detected by Vitek 2, but resistance to polymyxin B was detected by Vitek 2 for two of the resistant E. cloacae complex isolates; the same bacterial suspension was used for both Vitek 2 cards. For another isolate, the MICs of both colistin and polymyxin B were low (≤0.25 mg/liter) when it was tested by Sensititre; this isolate accounted for the only VMEs for Sensititre testing. Repeat testing of this isolate with Sensititre resulted in visible growth in the colistin (4 mg/liter) and polymyxin B (2 mg/liter) wells, with no growth in other wells. Subculture confirmed pure growth of E. cloacae complex in these wells. This result was considered uninterpretable given the presence of more than one skip well. Heteroresistance to colistin and polymyxin B was suspected, and phylogenetic clustering was performed using hsp60 sequencing of all E. cloacae complex isolates (10, 11) (Table 1). The isolate with heteroresistance by Sensititre, together with all other E. cloacae complex isolates, was found to belong to clusters that had been previously characterized and associated with heteroresistance (Table 1).
None of the commercial testing methods for both colistin and polymyxin B met the CLSI-recommended performance standards for commercial antimicrobial susceptibility testing systems (essential agreement [EA] ≥ 90%, CA ≥ 90%, VME ≤ 1.5%, ME ≤ 3.0%) (12).
DISCUSSION
Other studies have reviewed different susceptibility testing methods; the reference methods used vary and include agar dilution and broth microdilution with polysorbate-80 supplementation. These methods are not recommended, and broth microdilution without polysorbate-80 supplementation is currently recommended as the reference method (9). Caution is required in interpreting studies in which other reference methods were used. Colistin Etests have had various levels of error rates reported, and the Etest is currently not recommended as a testing method (9, 13–17). The highest rate of very major errors (VMEs) reported for colistin after an Etest is 41.5% (13). Disk diffusion testing has also been found to be unreliable (15–17). Vitek 2 was previously reported as a good testing method for colistin (13, 14, 16); this was not reproduced in our study. A VME rate of 36% for colistin testing by Vitek 2 was demonstrated. Conversely, polymyxin B results from Vitek 2 had better concordance with BMD results. Published studies on MicroScan susceptibility testing for colistin and polymyxin B are currently limited; categorical agreement (CA) of 87.3% for MicroScan (colistin) was reported by Lee et al. when it was compared to agar dilution as the reference method (14). Hindler and Humphries compared Sensititre with colistin to broth microdilution supplemented with polysorbate-80 (18). No VMEs were reported, and 2 major errors (MEs) were reported for P. aeruginosa isolates (18). There are otherwise no studies evaluating Sensititre for colistin and polymyxin B susceptibility testing. Further studies are required to confirm our findings.
The commercial testing methods in this study did not meet the recommendations published by the CLSI in terms of VMEs and MEs compared to results of broth microdilution as the reference method. Vitek 2 (colistin) and Etests (colistin and polymyxin B) have VMEs well in excess of the ≤1.5% rate recommended by the CLSI (12). A single VME each was demonstrated for Sensititre (colistin and polymyxin B), MicroScan (colistin), and Vitek 2 (polymyxin B). Although the percentage VME exceeds CLSI recommendations, this may be due to the limited number of resistant isolates (denominator) by broth microdilution in this study (25 were colistin resistant and 27 were polymyxin B resistant). The MEs for all methods (except Vitek 2 colistin testing and Etest polymyxin B testing) also exceeded the CLSI recommendation of ≤3.0% (12). However, except with two isolates tested by MicroScan (colistin), all MEs occurred with mcr-1-positive isolates.
Plasmid-mediated colistin and polymyxin B resistance (mcr-1) has disseminated worldwide and was present long before it was first described in 2015 (5). Although acquisition of the mcr-1 gene results in elevated MICs of the polymyxins, some remain within a 2-fold dilution of the susceptibility breakpoint. However, the clinical significance of MICs for mcr-1-positive isolates that are still within the susceptible range is unclear. To gain insight into why some isolates had colistin and polymyxin B MICs of ≤2 mg/liter, we examined all the E. coli isolates for the mcr-1 coding sequence (data not shown) and the approximately 200-bp region upstream which has been shown to carry the −35 and −10 promoter boxes of mcr-1 (19). We did not find indels, which suggests that mcr-1 would have been expressed per normal. Studies have shown that even though horizontal transfer of mcr-1 typically results in elevated MICs of colistin and polymyxin B, the MICs for some transformed isolates can remain within the susceptible range (2 mg/liter) (5). Using a susceptible breakpoint of ≤2 mg/liter, Sensititre and MicroScan had the highest sensitivity for detecting the potential presence of the mcr-1 gene. The use of a susceptible breakpoint of ≤1 mg/liter for both colistin and polymyxin B may be able to identify isolates which are more likely to harbor the mcr-1 resistance gene (Table 2). One additional mcr-1-negative isolate was reported as resistant to colistin by the Etest, but none were reported by other testing methods.
A susceptible breakpoint of ≤2 mg/liter was used in this study for both colistin (based on EUCAST breakpoints) and polymyxin B. Although the breakpoint of 2 mg/liter was also applied to polymyxin B in our study, this is an arbitrary breakpoint, as the pharmacokinetics of both agents are dissimilar despite their being from the same class of antibiotics. The MICs of colistin and polymyxin B are also not interchangeable and cannot be inferred from one another for Enterobacteriaceae (20). Also, the correlation of colistin and polymyxin B MICs and clinical outcomes is limited. Current breakpoints from EUCAST result in dichotomous interpretations of susceptible or resistant to colistin without an intermediate category. The introduction of an intermediate category was simulated for two scenarios (susceptible, ≤2 mg/liter; intermediate, 4 mg/liter; resistant, ≥8 mg/liter; and susceptible, ≤1 mg/liter; intermediate, 2 mg/liter; resistant, ≥4 mg/liter), and results are presented in Table 4. In both scenarios, the numbers of VMEs were reduced for Vitek 2 (colistin) and Etest (colistin and polymyxin B). In addition, the numbers of MEs decreased for all testing methods. Many of these VMEs and MEs were reclassified as minor errors (mEs), reflecting that the MICs were largely within one dilution difference but resulted in different interpretations. The introduction of an intermediate category at 4 mg/liter resulted in 11 colistin-resistant mcr-1-positive and 10 polymyxin B-resistant mcr-1-positive isolates to be reclassified into the intermediate category. The use of 2 mg/liter as an intermediate classification resulted in four of six colistin-susceptible (66.7%) and one of four polymyxin B-susceptible (25%) mcr-1-positive isolates to be reclassified from susceptible to intermediate. This reflects the fact that the majority of MICs for mcr-1-positive isolates remained close to or, for some isolates, below the breakpoints (Table 1). There was also an overall reduction in VMEs and MEs across the commercial susceptibility testing methods, which may reflect the current status of uncertainty in susceptibility testing for the polymyxins. Other considerations include the limited data available on the correlation between MICs, the presence of mcr-1 genes, and outcomes. An intermediate interpretation may be useful in the interim until these issues are addressed.
TABLE 4.
Performance characteristics of tests based on two simulated breakpoints in comparison to those of broth microdilutiona
| Simulated breakpoints | Drug | Method | No. of isolates that were: |
No. (%) of isolates exhibiting: |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Susceptible | Resistant | Intermediate | CA | VMEs | MEs | mEs | |||
| Susceptible at ≤2 mg/liter; intermediate at 4 mg/liter; resistant at ≥8 mg/liter | Colistin | Vitek 2 | 60 | 12 | 4 | 62 (81.6) | 2 (14.3) | 0 (0.0) | 11 (14.5) |
| Sensititre | 46 | 29 | 1 | 58 (76.3) | 1 (7.1) | 5 (9.8) | 12 (15.8) | ||
| Etest | 51 | 9 | 16 | 63 (82.9) | 1 (7.1) | 1 (2.0) | 9 (11.8) | ||
| MicroScan | 44 | 29 | 3 | 56 (73.7) | 1 (7.1) | 5 (9.8) | 14 (18.4) | ||
| Polymyxin B | Vitek 2 | 47 | 26 | 3 | 64 (84.2) | 1 (5.9) | 2 (4.1) | 9 (11.8) | |
| Sensititre | 47 | 22 | 7 | 66 (86.8) | 1 (5.9) | 0 (0.0) | 9 (18.4) | ||
| Etest | 53 | 12 | 11 | 62 (81.6) | 1 (5.9) | 0 (0.0) | 13 (17.1) | ||
| Susceptible at ≤1 mg/liter; intermediate at 2 mg/liter; resistant at ≥4 mg/liter | Colistin | Vitek 2 | 49 | 16 | 11 | 66 (86.8) | 2 (8.0) | 0 (0.0) | 6 (7.9) |
| Sensititre | 46 | 30 | 0 | 69 (90.8) | 1 (4.0) | 2 (4.3%) | 4 (5.3) | ||
| Etest | 46 | 25 | 5 | 69 (90.8) | 1 (4.0) | 1 (2.1) | 5 (6.6) | ||
| MicroScan | NA | NA | NA | NA | NA | NA | NA | ||
| Polymyxin B | Vitek 2 | 46 | 29 | 1 | 71 (93.4) | 1 (3.7) | 1 (2.1) | 3 (3.9) | |
| Sensititre | 46 | 29 | 1 | 71 (93.4) | 1 (3.7) | 2 (4.2) | 2 (2.6) | ||
| Etest | 47 | 22 | 7 | 68 (89.5) | 1 (3.7) | 1 (2.1) | 6 (7.9) | ||
CA, categorical agreement; VMEs, very major errors; MEs, major errors; mEs, minor errors (minor errors are errors which do not constitute a VME or ME and are related to an intermediate interpretation for either the reference method or the test method); NA, not applicable because the lowest MIC interpretation possible for MicroScan (colistin) is 2 mg/liter.
Confounding the interpretation of colistin and polymyxin B susceptibility testing is heteroresistance to polymyxins among Gram negatives (21–23). Poor reliability and repeatability of colistin susceptibility testing for Enterobacter cloacae and Enterobacter aerogenes were first reported in 2013 (24). For some isolates, a higher number of heteroresistant colonies was detectable when the isolate was cultured in relatively higher polymyxin B concentrations. A wide range of MICs may need to be tested in order to improve detection of heteroresistance. Performing a population analysis may not be practical for many laboratories. Heteroresistance to colistin among strains of the Enterobacter cloacae complex is hsp60 genetic cluster dependent (11). There are 12 clusters (C-I to C-XII) of which the species name could be assigned confidently to specific clusters. Strains from clusters I, II, IV, VII, IX, X, XI, and XII are usually heteroresistant, whereas those from clusters III, V, VI, VIII, and XIII are typically susceptible. All isolates with resistant phenotypes in our study correlated with genetic clusters associated with heteroresistance. Determination of E. cloacae nomenspecies by hsp60 sequencing may be considered an adjunct to phenotypic susceptibility testing. However, the direct impact of heteroresistance on treatment outcomes has not been completely elucidated, especially as colistin and polymyxin B are often given in combination with other antimicrobials, as they are often employed as a last line of defense against MDROs.
There is currently a paucity of data in multiple areas in relation to the use of polymyxins in clinical treatment. The issues related to susceptibility testing methods remain unresolved. Further studies on susceptibility testing with a focus on the correlation between MICs and clinical outcomes are required to guide treatment. The clinical significance of acquisition of mcr-1 genes and the impact of heteroresistance on phenotypic susceptibility testing also require further study. Only when there is further clarity on these issues can suitable breakpoints be implemented confidently.
MATERIALS AND METHODS
Seventy-six Enterobacteriaceae were included in this study. Carbapenem-resistant Enterobacteriaceae and known mcr-1-positive isolates from human screening and clinical samples were used. All isolates were characterized for mcr and carbapenemase carriage as previously described (5, 25).
All susceptibility tests for colistin and polymyxin B were performed in parallel with commercial methods and broth microdilution. Sulfate salts of colistin and polymyxin B (Sigma, St. Louis, MO, USA) were dissolved in cation-adjusted BBL Mueller-Hinton II broth (CAMHB) (BD, Franklin Lakes, NJ, USA), and broth microdilution was performed in accordance with the CLSI reference method (26). Dilutions to the MIC range of 0.06 mg/liter to 64 mg/liter were performed in untreated 96-well polystyrene microplates (Greiner, Frickenhausen, Germany). Supplementation with polysorbate-80 was not performed, as per current recommendations (9).
The Sensititre (GNX3F plates) and Vitek 2 (N315 and N275 cards) systems and Etests (bioMérieux) were used to test colistin and polymyxin B susceptibility. MicroScan (NM44) was used to test colistin susceptibility only because polymyxin B was not available on any MicroScan drug panel. Susceptibility testing was performed as per the manufacturers' instructions. E. coli ATCC 25922 (American Type Culture Collection) was used as the drug-susceptible control strain (27), while intrinsically resistant Proteus mirabilis ATCC 12453 served as the polymyxin-resistant control strain. The possible range of MIC readings for each method and drug combination was as follows: for BMD (colistin and polymyxin B), ≤0.06 to >64 mg/liter; for Sensititre (colistin and polymyxin B), ≤0.25 to >4 mg/liter; for MicroScan (colistin only), ≤2 to >4 mg/liter; for Vitek 2 (colistin and polymyxin B), ≤0.5 to ≥16 mg/liter; for Etest (colistin), ≤0.016 to >256 mg/liter; and for Etest (polymyxin B), ≤0.064 to >1,024 mg/liter.
EUCAST breakpoints were used for interpretation of colistin MIC results (susceptible, ≤2 mg/liter; resistant, >2 mg/liter). In the absence of official breakpoints for polymyxin B for Enterobacteriaceae, a breakpoint of ≤2 mg/liter for susceptible and >2 mg/liter for resistant isolates was used. Comparison of commercial methods was made against broth microdilution as the reference method. Rates of essential agreement (EA), categorical agreement (CA), very major errors (VMEs), and major errors (MEs) were determined. EA was defined as a MIC result within a 2-fold dilution of the BMD result. This was adjusted for differences in the ranges of MICs that could be determined by the respective systems. EA was not determined for MicroScan, as the MIC ranges were limited to ≤2 mg/liter, 4 mg/liter, and >4 mg/liter. Categorical agreement was defined as agreement in the interpretation of the MICs of the commercial kit and BMD. VMEs occurred where the tested method MIC interpretation was susceptible and the BMD MIC interpretation was resistant. MEs occurred where the tested method MIC interpretation was resistant and the BMD MIC interpretation was susceptible. The VME rates were calculated using the number of isolates resistant by BMD as the denominator, while the ME rates were calculated using the number of isolates susceptible by BMD as the denominator.
ACKNOWLEDGMENTS
We thank Thermo Fisher Scientific for providing MicroScan (NM44) plates.
This research was funded by a Singapore Infectious Diseases Initiative short-term research grant provided to Jeanette Teo (NRYLL068OM).
Footnotes
For a commentary on this article, see https://doi.org/10.1128/JCM.00888-17.
REFERENCES
- 1.Landman D, Georgescu C, Martin DA, Quale J. 2008. Polymyxins revisited. Clin Microbiol Rev 21:449–465. doi: 10.1128/CMR.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falagas ME, Lourida P, Poulikakos P, Rafailidis PI, Tansarli GS. 2014. Antibiotic treatment of infections due to carbapenem-resistant enterobacteriaceae: systematic evaluation of the available evidence. Antimicrob Agents Chemother 58:654–663. doi: 10.1128/AAC.01222-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nabarro LEB, Veeraraghavan B. 2015. Combination therapy for carbapenem-resistant Enterobacteriaceae: increasing evidence, unanswered questions, potential solutions. Eur J Clin Microbiol Infect Dis 34:2307–2311. doi: 10.1007/s10096-015-2486-7. [DOI] [PubMed] [Google Scholar]
- 4.Olaitan AO, Morand S, Rolain JM. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 6.Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, Malhotra-Kumar S. 2016. Identification of a novel plasmid-mediated colistin resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 21(27):30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 7.EUCAST. 2017. European Committee on Antimicrobial Susceptibility Testing breakpoint tables for interpretation of MICs and zone diameters, version 7.1. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_7.1_Breakpoint_Tables.pdf.
- 8.CLSI. 2017. Performance standards for antimicrobial susceptibility testing, 27th ed CLSI document M100-S27 CLSI, Wayne, PA. [Google Scholar]
- 9.EUCAST. 2016. Recommendations for MIC determination of colistin (polymyxin E) as recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group. EUCAST http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_documents/Recommendations_for_MIC_determination_of_colistin_March_2016.pdf.
- 10.Hoffmann H, Roggenkamp A. 2003. Population genetics of the nomenspecies Enterobacter cloacae. Appl Environ Microbiol 69:5306–5318. doi: 10.1128/AEM.69.9.5306-5318.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guérin F, Isnard C, Sinel C, Morand P, Dhalluin A, Cattoir V, Giard JC. 2016. Cluster-dependent colistin hetero-resistance in Enterobacter cloacae complex. J Antimicrob Chemother 71:3058–3061. doi: 10.1093/jac/dkw260. [DOI] [PubMed] [Google Scholar]
- 12.CLSI. 2017. Verification of commercial microbial identification and antimicrobial susceptibility testing systems, 1st ed CLSI document M52-Ed1 Clinical and Laboratory Standards Institute, Wayne PA. [Google Scholar]
- 13.Dafopoulou K, Zarkotou O, Dimitroulia E, Hadjichristodoulou C, Gennimata V, Pournaras S, Tsakris A. 2015. Comparative evaluation of colistin susceptibility testing methods among carbapenem-nonsusceptible Klebsiella pneumoniae and Acinetobacter baumannii clinical isolates. Antimicrob Agents Chemother 59:4625–4630. doi: 10.1128/AAC.00868-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee SY, Shin JH, Lee K, Joo MY, Park KH, Shin MG, Suh SP, Ryang DW, Kim SH. 2013. Comparison of the Vitek 2, MicroScan, and Etest methods with the agar dilution method in assessing colistin susceptibility of bloodstream isolates of Acinetobacter species from a Korean University Hospital. J Clin Microbiol 51:1924–1926. doi: 10.1128/JCM.00427-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moskowitz SM, Garber E, Chen Y, Clock SA, Tabibi S, Miller AK, Doctor M, Saiman L. 2010. Colistin susceptibility testing: evaluation of reliability for cystic fibrosis isolates of Pseudomonas aeruginosa and Stenotrophomonas maltophilia. J Antimicrob Chemother 65:1416–1423. doi: 10.1093/jac/dkq131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lo-Ten-Foe JR, de Smet AM, Diederen BM, Kluytmans JA, van Keulen PH. 2007. Comparative evaluation of the Vitek 2, disk diffusion, Etest, broth microdilution, and agar dilution susceptibility testing methods for colistin in clinical isolates, including heteroresistant Enterobacter cloacae and Acinetobacter baumannii strains. Antimicrob Agents Chemother 51:3726–3730. doi: 10.1128/AAC.01406-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maalej SM, Meziou MR, Rhimi FM, Hammami A. 2011. Comparison of disc diffusion, Etest and agar dilution for susceptibility testing of colistin against Enterobacteriaceae. Lett Appl Microbiol 53:546–551. doi: 10.1111/j.1472-765X.2011.03145.x. [DOI] [PubMed] [Google Scholar]
- 18.Hindler JA, Humphries RM. 2013. Colistin MIC variability by method for contemporary clinical isolates of multidrug-resistant Gram-negative bacilli. J Clin Microbiol 51:1678–1684. doi: 10.1128/JCM.03385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poirel L, Kieffer N, Brink A, Coetze J, Jayol A, Nordmann P. 2016. Genetic features of MCR-1-producing colistin-resistant Escherichia coli isolates in South Africa. Antimicrob Agents Chemother 60:4394–4397. doi: 10.1128/AAC.00444-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sader HS, Rhomberg PR, Farrell DJ, Jones RN. 2015. Differences in potency and categorical agreement between colistin and polymyxin B when testing 15,377 clinical strains collected worldwide. Diagn Microbiol Infect Dis 83:379–381. doi: 10.1016/j.diagmicrobio.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 21.Napier BA, Band V, Burd EM, Weiss DS. 2014. Colistin heteroresistance in Enterobacter cloacae is associated with cross-resistance to the host antimicrobial lysozyme. Antimicrob Agents Chemother 58:5594–5597. doi: 10.1128/AAC.02432-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jayol A, Nordmann P, Brink A, Poirel L. 2015. Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob Agents Chemother 59:2780–2784. doi: 10.1128/AAC.05055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hawley JS, Murray CK, Jorgensen JH. 2008. Colistin heteroresistance in Acinetobacter and its association with previous colistin therapy. Antimicrob Agents Chemother 52:351–352. doi: 10.1128/AAC.00766-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landman D, Salamera J, Quale J. 2013. Irreproducible and uninterpretable polymyxin B MICs for Enterobacter cloacae and Enterobacter aerogenes. J Clin Microbiol 51:4106–4111. doi: 10.1128/JCM.02129-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teo J, Ngan G, Balm M, Jureen R, Krishnan P, Lin R. 2012. Molecular characterization of NDM-1 producing Enterobacteriaceae isolates in Singapore hospitals. Western Pac Surveill Response J 3:19–24. doi: 10.5365/wpsar.2011.2.4.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.CLSI. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard—10th ed. CLSI document M07-A10 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 27.Jones RN, Anderegg TR, Swenson JM. 2005. Quality control guidelines for testing Gram-negative control strains with polymyxin B and colistin (polymyxin E) by standardized methods. J Clin Microbiol 43:925–927. doi: 10.1128/JCM.43.2.925-927.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]