ABSTRACT
The global intensification of antiretroviral therapy (ART) can lead to increased rates of HIV drug resistance (HIVDR) mutations in treated and also in ART-naive patients. ART-naive HIV-1-infected patients from Cameroon were subjected to a multimethod HIVDR analysis using amplification-refractory mutation system (ARMS)-PCR, Sanger sequencing, and longitudinal next-generation sequencing (NGS) to determine their profiles for the mutations K103N, Y181C, K65R, M184V, and T215F/Y. We processed 66 ART-naive HIV-1-positive patients with highly diverse subtypes that underlined the predominance of CRF02_AG and the increasing rate of F2 and other recombinant forms in Cameroon. We compared three resistance testing methods for 5 major mutation sites. Using Sanger sequencing, the overall prevalence of HIVDR mutations was 7.6% (5/66) and included all studied mutations except K65R. Comparing ARMS-PCR with Sanger sequencing as a reference, we obtained a sensitivity of 100% (5/5) and a specificity of 95% (58/61), caused by three false-positive calls with ARMS-PCR. For 32/66 samples, we obtained NGS data and we observed two additional mismatches made up of minority variants (7% and 18%) that might not be clinically relevant. Longitudinal NGS analyses revealed changes in HIVDR mutations in all five positive subjects that could not be attributed to treatment. In one of these cases, superinfection led to the temporary masking of a resistant virus. HIVDR mutations can be sensitively detected by ARMS-PCR and sequencing methods with comparable performances. Longitudinal changes in HIVDR mutations have to be considered even in the absence of treatment.
KEYWORDS: human immunodeficiency virus, HIV, drug resistance mutations, ARMS-PCR, Sanger sequencing, next-generation sequencing, transmitted drug resistance, drug-naive patients, superinfection, subtype diversity, longitudinal changes
INTRODUCTION
Antiretroviral therapy (ART) is the key element to achieve viral suppression, prevent viral transmission, and reduce HIV/AIDS-related deaths. The developing world has experienced a rapid scale-up of ART over the past few years. As of December 2015, 17 million people living with HIV were accessing ART, up from 400,000 recorded at the end of 2003 (>40-fold increase) (1–8). In Cameroon, more than 180,000 patients were receiving ART in 2015, representing less than 50% of ART-eligible patients, and yet this is a great increase, since in 2005 only 16,500 HIV-infected patients in Cameroon had access to ART. In 2010, the Cameroon National HIV/AIDS Control Committee planned to provide coverage to more than 80% of the people aged 15 and older who need ART by 2020. This coverage is likely to increase with the implementation of the “90-90-90” and “95-95-95” UNAIDS initiatives (3, 6, 9).
Cameroon, like many resource-constrained settings (RCS), uses the WHO public health approach for the treatment of HIV-infected patients. This approach recommends the standardized and simplified treatment protocol for first-line treatment consisting of two nucleoside reverse transcriptase inhibitors (NRTIs) (tenofovir disoproxil [TDF] or zidovudine [AZT] plus lamivudine [3TC] or emtricitabine [FTC]) plus one non-NRTI (NNRTI) (efavirenz [EFV] or nevirapine [NVP]) (9). The second-line treatment is based on 2 NRTIs (TDF or AZT plus 3TC or FTC) plus a ritonavir-boosted protease inhibitor (atazanavir-ritonavir or lopinavir-ritonavir). Of note, TDF and AZT are among the most-used antiretroviral drugs included in first- and second-line regimens in Cameroon; however, TDF was introduced in Cameroon only in 2010, i.e., 1 year prior to sample collection of the current study. The third-line treatment usually contains a regimen of a booster PI (darunavir-ritonavir) plus an integrase inhibitor (raltegravir or dolutegravir) (unpublished data).
Though ART has been quite successful, one obstacle that remains is the emergence of HIV drug resistance (HIVDR) mutations. The monitoring of HIVDR mutations is needed for the management of the increasing proportion of individuals presenting for HIV/AIDS care to ensure that they receive optimal ART (10–17). A recent analysis of 287 studies, of which 151 were conducted in sub-Saharan Africa and South or Southeast Asia, reveals that between 10% and 30% of patients receiving a standard first-line NRTI/NNRTI-containing regimen will develop viral failure at some point during their treatment (18–20). As the number of patients with acquired drug resistance has increased, so has the proportion of newly infected patients with transmitted drug resistance (20–24). Drug-resistant virus can be transmitted to previously uninfected individuals or, rarely, in case of superinfection, to previously infected individuals (25–31). Transmitted HIVDR mutations have the potential to persist for many years, even in the absence of drug pressure; however, they gradually revert to wild type if they reduce viral fitness (25, 32). Transmitted HIVDR mutations are of increasing public health concern; data are, however, limited in resource-constrained settings where HIVDR mutation testing is not routinely performed. In the recently launched “test and treat” era (3, 4), HIV drug-naive patient samples are becoming more scarce in RCS, which impedes studies on ART-naive subjects and transmitted HIVDR mutations.
In developed countries, Sanger sequencing-based drug resistance assays are used to select the appropriate regimens for initiating or switching ART (23, 33). Of note, standard sequencing detects only variants representing >20% of the viral quasispecies. Studies conducted with ART-naive patients have shown a 2- to 3-fold increase in the prevalence of transmitted HIVDR mutations when next-generation sequencing (NGS) technology was used to detect mutations (34). There are reports that low-frequency NNRTI mutations (≥1%) might be attributed to a dose-dependent treatment failure in a few individuals (35); however, general implications are still under debate. Minority variants (<5% of quasispecies) can occur during PCR amplification or due to polymerase incorporation errors during sequencing, and low-abundance variants (<20% of quasispecies) mostly seem to have limited impact on the clinical response (35, 36). In resource-constrained settings, the use of Sanger and NGS is restricted because of their high cost and intense laboratory infrastructure requirements (37, 38).
Point mutation assays are generally PCR-based assays with the capacity to detect specific genomic point mutations that confer drug resistance. They have been developed as an alternative for genotyping assays, among which amplification-refractory mutation system (ARMS)-PCR uses the difference in extension efficiency between primers with matched and mismatched 3′ bases to identify mutations based solely on the presence of PCR products via gel electrophoresis. ARMS-PCR was developed in 1989 and was further optimized by our group and others to identify HIVDR mutations with high performance compared to that of standard sequencing (39–44). So far, no study has compared point mutation assays with NGS. In this study, we compared the performance of ARMS-PCR to those of standard (Sanger) sequencing and NGS to detect HIVDR mutations in antiretroviral-naive patients from an area with high genetic diversity. Longitudinal NGS analyses enabled us to monitor reversion as well as acquisition of HIVDR mutations in the absence of apparent antiretroviral drug pressure.
RESULTS
HIV-1 genetic diversity in the study population.
The phylogenetic analysis of reverse transcriptase (rt) sequences revealed a wide range of HIV-1 subtypes, among which recombinant strains dominated, along with a smaller fraction of pure subtypes (Fig. 1). CRF02_AG was the most common clade identified in 71.2% of our study subjects, followed by F2 (9.1%), A1 (4.6%), CRF37_cpx (4.6%), CRF01_AE (1.5%), D (1.5%), G (1.5%), CRF11_cpx (1.5%), CRF18_cpx (1.5%), A1G (1.5%), and URF_F2U (1.5%). Our findings confirm the predominance of CRF02_AG and the increased emergence of subtype F2 (22, 45–50), which necessitates further monitoring of HIV-1 diversity in whole-genome sequences in Cameroon and a large sample size. For each patient, the rt sequences obtained using Sanger sequencing and longitudinal NGS, if available, clustered together, enabling a comparative HIVDR mutation analysis.
FIG 1.
HIV-1 genetic diversity of the study subjects. (A) Pie chart showing the HIV-1 subtype distribution of our study population according to the phylogenetic analysis of the rt sequences and according to HIV BLAST (https://www.hiv.lanl.gov); (B) phylogenetic tree of rt sequences (HIV region from positions 2723 to 3225 according to HXB2 numbering) of the study population generated with Sanger sequencing (red) and next-generation sequencing (NGS) (green), including longitudinal time points, together with reference sequences (black) from the Los Alamos sequence database (https://www.hiv.lanl.gov). For the NGS analysis, consensus sequences were generated for each longitudinal time point per study subject using DNASTAR's SeqMan Pro. Neighbor-joining phylogenetic trees were generated using MEGA and FigTree software. The number following the study subject's identifier represents the sample collection time point. The black bar indicates the genetic distance. Subject MDC192 (gray asterisk) is either CRF02_AG or CRF36_cpx.
HIVDR mutation profiles of the study subjects.
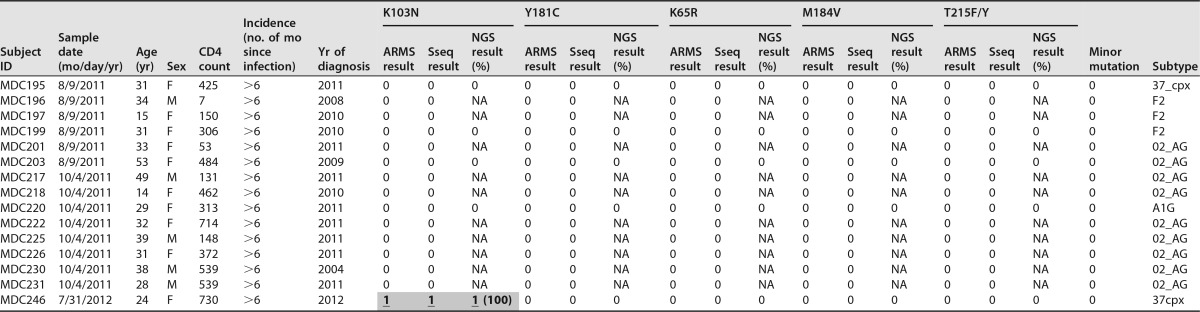
In this study, we focused on the five major HIVDR mutations in Cameroon according to their population-based prevalence, the on-site-applied antiretroviral drugs, and the mutation scoring (Table 1) (22, 47, 51, 52). For three NRTI mutations (K65R, M184V, and T215F/Y) and two NNRTI mutations (K103N and Y181C), we have developed and optimized the ARMS-PCR procedure (39) (Fig. 2; Table 2). We performed, in addition to ARMS-PCR, Sanger sequencing and NGS. Using double-stranded Sanger sequencing as our gold standard, we observed an overall prevalence of major HIVDR mutations in 7.6% (5/66) of patients. The application of ARMS-PCR and Sanger sequencing for 66 patients and 5 mutation sites provided a total of 330 patient/mutation data sets that were used for comparative analyses (Table 3). Using Sanger sequencing, we detected a total of 8 HIVDR mutations out of 330 data sets; all the mutations studied were present except K65R. Two of the patients with major HIVDR mutations harbored additional minor mutations, as detected by sequencing. ARMS-PCR detected all HIVDR mutations observed with Sanger sequencing (8/8), yielding 100% sensitivity. Three false-positive calls decreased the ARMS-PCR specificity to 95% (63/66). NGS was obtained for 32 patients, and we observed variants with two additional significant HIVDR mutations, T215F and K103N, representing 7% and 18% of viral quasispecies, respectively (Tables 3 and 4).
TABLE 1.
Selected drug resistance mutations for comparative ARMS-PCR, Sanger sequencing, and NGS analyses
| ARV class | Mutation(s)a | Prevalence (%) | Drug(s) affected | Mutation score |
|---|---|---|---|---|
| NRTI | K65R | TDF | 60 (high-level resistance) | |
| M184V | 39–77.2 | 3TC, FTC | 60 (high-level resistance) | |
| T215Y/F | 11–24 | AZT | 45 (low-level resistance) | |
| NNRTI | K103N | 14–44 | NVP, EFV | 60 (high-level resistance) |
| Y181C | 18–19.8 | NVP | 60 (high-level resistance) |
The selection of drug resistance mutations for ARMS-PCR, Sanger sequencing, and NGS was based on (i) the prevalence of the mutations in Cameroon (22, 47, 51, 52), (ii) the drugs deployed in Cameroon and affected by the mutation, and (iii) the mutation score (http://hivdb.stanford.edu/DR/asi/releaseNotes/index.html).
FIG 2.
ARMS-PCR results, exemplarily shown for the M184V and K65R mutations. An ethidium bromide-stained 2% agarose gel was loaded with ARMS-PCR samples of 8 study subjects and the negative-control subject (Neg Ctrl) with M184V (A) and one study subject (MDC007), the positive control (Pos Ctrl), and a negative-control subject with K65R (B). For each sample, ARMS-PCR was run with a wild-type (Wt) and mutant-type (Mut) reverse primer. The expected product sizes are 328 bp for M184V and 185 bp for K65R. The presence of 2 bands (Mut and Wt) is indicative of viruses with the respective mutation, while the presence of only one band (Wt) is indicative of viruses without mutation. Our ARMS-PCR results suggest that subjects MDC007, MDC058, and MDC091 harbored virus with the M184V mutation (yellow arrows) and that patients MDC043, MDC044, MDC068, MDC080, and MDC086 did not harbor virus with M184V. MDC007 did not harbor virus with the K65R mutation. M, Gene Ruler low-range marker.
TABLE 2.
Primers used in ARMS-PCR to detect the K65R, M184V, T215Y/F, K103N, and Y181C mutationsa
| ARV class | Mutation(s) | Nature of mutation or primer name | Primer sequence | HXB2 location | Product length (bp) |
|---|---|---|---|---|---|
| NRTI | K65R | WT | 5′-CTAATTTTCTCCATTTAGTACTATCTTTAT-3′ | 2772–2743 | 185c |
| Mut | 5′-CTAATTTTCTCCATTTAGTACTATCTTGTC-3′ | 2772–2743 | |||
| M184V | WT | 5′-TAAATCAGATCCTACATACAAATCATACAT-3′ | 3128–3099 | 328b | |
| Mut | 5′-TAAGTCAGATCCTACATATAAATCATCCTC-3′ | 3128–3099 | |||
| T215Y/F | WT | 5′-TTCTTTCTGATGTTTCTTATCTGGTGTGGT-3′ | 3221–3192 | 421b | |
| Mut | 5′-TTCTTTCTGATGTTTCTTATCTGGTGGGWA-3′ | 3221–3192 | |||
| Common forward 1 | 5′-CTCAAGACTTCTGGGAGGTCT-3′ | 2800–2820 | |||
| NNRTI | K103N | WT | 5′-TCTCCCACATCCAGTACTGTTACTGATGTT-3′ | 2887–2858 | 300c |
| Mut | 5′-TCTCCCACATCCAGTACTGTTACTGAGTTR-3′ | 2887–2858 | |||
| Y181C | WT | 5′-ATCCTACATATAAATCATCCACATATTGRT-3′ | 3120–3091 | 533c | |
| Mut | 5′-ATCCTACATATAAATCATCCACATATGGRC-3′ | 3120–3091 | |||
| Common forward 2 | 5′-AGCCAGGAATGGATGGCCCAA-3′ | 2587–2607 |
WT, wild-type reverse primer; Mut, mutant reverse primer; HXB2 location, primer location according to the HIV-1 subtype B reference sequence.
The product length when combined with that of Common forward1.
The product length when combined with that of Common forward2.
TABLE 3.
Characteristics of study subjects and comparative HIVDR mutation profiles for 5 mutations using 3 testing methodsa
Detected drug resistance mutations are shaded in gray. “0” indicates the absence of the respective drug resistance mutation (<5% of quasispecies; not considered to be significant), and “1” indicates the presence of the respective drug resistance mutation (≥5% of quasispecies). ARMS, ARMS-PCR; Sseq, Sanger sequencing; NGS, next-generation sequencing; F, female; M, male; NA, not applicable. Shaded underlined boldface numbers indicate calls positive by all 3 testing methods, and shaded italic numbers indicate false-positive calls by ARMS-PCR and NGS for minorities. Minor HIVDR mutations are indicated if present. The HIV subtypes were determined with HIV BLAST (LANL database) and phylogenetic analysis of the reverse transcriptase sequences generated by standard sequencing and NGS (Fig. 1).
b MDC192 is either subtype CRF02_AG or subtype CRF36_cpx.
TABLE 4.
Longitudinal drug resistance mutation analysis using next-generation sequencing, Sanger sequencing, and ARMS-PCRa
| Subject ID | Time point | Sample date (mo/day/yr) | K103N result(s) | Y181C result(s) | K65R result(s) | M184V result(s) | T215F/Y result(s) |
|---|---|---|---|---|---|---|---|
| MDC008 | 1 | 2/8/2011 | 0 | 0 | 0 | 0 | 0 |
| MDC008 | 2 | 8/11/2011 | 0 | 0 | 0 | 0 | 0 |
| MDC008 | 3 | 2/4/2012 | 0 | 0 | 0 | 0 | 0 |
| MDC044 | 1 | 4/26/2011 | 0 | 0 | 0 | 0 | 0 |
| MDC044 | 3 | 6/19/2012 | 0 | 0 | 0 | 0 | 0 |
| MDC044 | 7 | 1/12/2015 | 0 | 0 | 0 | 0 | 0 |
| MDC058 | 1 | 4/26/2011 | 0 | 1 (99.4) | 0 | 1 (100) | 1 (96.2) |
| MDC058 | 3 | 10/13/2014 | 0 | 0 | 0 | 0 | 0 |
| MDC068 | 1 | 4/26/2011 | 0 | 0 | 0 | 0 | 0 |
| MDC068 | 3 | 6/19/2012 | 0 | 0 | 0 | 0 | 0 |
| MDC068 | 4 | 5/12/2014 | 0 | 0 | 0 | 0 | 0 |
| MDC086 | 1 | 5/31/2011 | 0 | 0 | 0 | 0 | 0 |
| MDC086 | 3 | 10/16/2012 | 0 | 0 | 0 | 0 | 0 |
| MDC086 | 4 | 2/17/2014 | 0 | 0 | 0 | 0 | 0 |
| MDC091 | 1 | 5/31/2011 | 1 (100), 1S, 1A | 0 | 0 | 1 (100), 1S, 1A | 1 (7), 0S, 0A |
| MDC091 | 2 | 11/8/2011 | 1 (100), 1S, 1A | 0 | 0 | 1 (100), 1S, 1A | 0, 0S, 0A |
| MDC093 | 1 | 5/31/2011 | 0 | 0 (1.6) | 0 | 0 | 0 |
| MDC093 | 3 | 7/31/2012 | 0 | 0 | 0 | 0 | 0 |
| MDC093 | 5 | 2/17/2014 | 0 | 0 | 0 | 0 | 0 |
| MDC098 | 1 | 5/31/2011 | 0 | 0, 0S, 0A | 0 | 0 | 0 |
| MDC098 | 3 | 6/19/2012 | 0 | 0 | 0 | 0 | 0 |
| MDC098 | 5 | 11/17/2014 | 0 | 1 (89.5), 1S, 1A | 0 | 0 | 0 |
| MDC100 | 1 | 5/31/2011 | 0 | 0 | 0 | 0 | 0 |
| MDC100 | 3 | 6/19/2012 | 0 | 0 | 0 | 0 | 0 |
| MDC100 | 5 | 11/17/2014 | 0 | 0 | 0 | 0 | 0 |
| MDC128 | 1 | 6/28/2011 | 0 | 0 | 0 | 0 | 0 |
| MDC128 | 3 | 6/19/2012 | 0 | 0 | 0 | 0 | 0 |
| MDC128 | 6 | 10/13/2014 | 0 | 0 | 0 | 0 | 0 |
| MDC131 | 1 | 6/28/2011 | 0 | 0 | 0 | 0 | 0 |
| MDC131 | 3 | 6/19/2012 | 0 | 0 | 0 | 0 | 0 |
| MDC131 | 6 | 8/25/2014 | 0 | 0 | 0 | 0 | 0 |
| MDC146 | 1 | 6/28/2011 | 0 | 0 | 0 | 0 | 0 |
| MDC146 | 3 | 7/31/2012 | 0 | 0 | 0 | 0 | 0 |
| MDC146 | 6 | 4/13/2015 | 0 | 0 | 0 | 0 | 0 |
| MDC166 | 1 | 8/9/2011 | 1 (100) | 0 | 0 | 0 | 0 |
| MDC166 | 3 | 10/16/2012 | 1 (100) | 0 | 0 | 1 (100) | 0 |
| MDC179 | 1 | 8/9/2011 | 0 | 0 | 0 | 0 | 0 |
| MDC179 | 3 | 6/19/2012 | 0 | 0 | 0 | 0 | 0 |
| MDC179 | 5 | 10/8/2013 | 0 | 0 | 0 | 0 | 0 |
| MDC189 | 1 | 8/9/2011 | 0 | 0 | 0 | 0 | 0 |
| MDC189 | 2 | 2/14/2012 | 0 | 0 | 0 | 0 | 0 |
| MDC189 | 3 | 4/14/2014 | 0 | 0 | 0 | 0 | 0 |
| MDC192 | 1 | 8/9/2011 | 0 | 0 | 0 | 0 | 0 |
| MDC192 | 2 | 2/14/2012 | 0 | 0 | 0 | 0 | 0 |
| MDC192 | 3 | 6/18/2013 | 0 | 0 | 0 | 0 | 0 |
| MDC194 | 1 | 8/9/2011 | 0 | 0 | 0 | 0 | 0 |
| MDC194 | 2 | 6/18/2013 | 0 | 0 | 0 | 0 | 0 |
| MDC195 | 1 | 8/9/2011 | 0 | 0 | 0 | 0 | 0 |
| MDC195 | 2 | 2/14/2012 | 0 | 0 | 0 | 0 | 0 |
| MDC203 | 1 | 8/9/2011 | 0 | 0 | 0 | 0 | 0 |
| MDC203 | 2 | 3/26/2013 | 0 | 0 | 0 | 0 | 0 |
| MDC220 | 1 | 10/4/2011 | 0 | 0 | 0 | 0 | 0 |
| MDC220 | 3 | 10/16/2012 | 0 | 0 | 0 | 0 | 0 |
| MDC220 | 4 | 10/8/2013 | 0 | 0 | 0 | 0 | 0 |
| MDC246 | 1 | 7/31/2012 | 1 (100), 1S, 1A | 0 | 0 | 0 | 0 |
| MDC246 | 2 | 10/16/2012 | 0, 0S, 0A | 0 | 0 | 0 | 0 |
| MDC246 | 6 | 4/13/2015 | 1 (18), 0S, 0A | 0 | 0 | 0 | 0 |
All study subjects with multiple time points were analyzed for changes in drug resistance mutations using NGS. The study subjects MDC058, MDC091, MDC098, MDC166, and MDC246 showed an HIV drug resistance mutation change in the longitudinal analysis (highlighted in gray). “0” indicates the absence of the respective drug resistance mutation, and “1” indicates the presence of the respective drug resistance mutation, with a threshold of 5% positive calls of total sequences. The numbers in parentheses indicate the percentages of NGS quasispecies harboring the respective mutation. Three study subjects with longitudinal changes in DRM were also longitudinally screened with Sanger sequencing (S) and ARMS-PCR (A).
ARMS-PCR's performance compared to that of sequencing.
Sanger sequencing and NGS completely matched each other in the detection of a majority of the 32 analyzed variants and differed only in two cases, i.e., with minority variants (7% and 18% of quasispecies), detected exclusively by NGS (Tables 3 to 5). Taking Sanger sequencing as the standard, 63 out of 66 patients (95.4%) studied had similar results for ARMS-PCR for the five mutations tested. Three patients presented discordant results for M184V or T215F/Y, representing false-positive calls by ARMS-PCR, while no false negatives were observed. The sensitivity, the specificity, the negative predictive value (NPV), and the positive predictive value (PPV) were calculated for each mutation, for the 66 patients in total, and for the entirety of the 330 data sets (Table 5). ARMS-PCR provided 100% sensitivity, thereby giving a 100% NPV for all the mutations tested. The highest specificity (100%) was obtained for mutations K103N and Y181C, while the lowest specificity (92.9%) was obtained for the detection of the M184V mutation. The highest PPV was 100% for the K103N and Y181C mutations; the lowest (50%) was obtained for the T215F/Y mutation.
TABLE 5.
Performance of ARMS-PCR compared to standard (Sanger) sequencing in the detection of HIV drug resistance mutationsa
| Mutation(s) | No. of patients or data sets with indicated test result |
Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| ARMS + | Sanger sequencing + | ARMS true + | ARMS false + | ARMS true − | ARMS false − | |||||
| K65R | 0 | 0 | 0 | 0 | 66 | 0 | NA | NA | NA | NA |
| M184V | 5 | 3 | 3 | 2 | 63 | 0 | 100 | 97 | 60 | 100 |
| T215Y/F | 2 | 1 | 1 | 1 | 65 | 0 | 100 | 98 | 50 | 100 |
| K103N | 3 | 3 | 3 | 0 | 63 | 0 | 100 | 100 | 100 | 100 |
| Y181C | 1 | 1 | 1 | 0 | 65 | 0 | 100 | 100 | 100 | 100 |
| All (66 patients) | 8 | 5 | 5 | 3 | 58 | 0 | 100 | 95 | 63 | 100 |
| All (330 data sets) | 11 | 8 | 8 | 3 | 319 | 0 | 100 | 99 | 73 | 100 |
+, positive test result; −, negative test result; PPV: positive predictive value; NPV, negative predictive value; NA, not applicable. The sensitivity, specificity, PPV, and NPV of ARMS-PCR were calculated using standard sequencing as the gold standard (https://www.medcalc.org/calc/diagnostic_test.php). The 330 data sets included the comparative data of the first sample time points (Table 3) and excluded the selective longitudinal comparisons (Table 4).
Prevalence of major NRTI-associated mutations.
Among the 66 studied individuals, 4.5% harbored one or more of the major NRTI mutations (M184V and/or T215F/Y) (Table 6). The K65R mutation, which causes intermediate-/high-level resistance to TDF, didanosine (ddI), abacavir (ABC), and stavudine (d4T) and low-/intermediate-level resistance to 3TC and FTC, was completely absent. The M184V mutation is the most common NRTI resistance mutation and is known to cause high-level in vitro resistance to 3TC and FTC and low-level resistance to ddI and ABC. 3TC, FTC, TDF, and AZT constitute the NRTIs used in first-line treatment in Cameroon. Accordingly, M184V was the most prevalent HIVDR mutation in our study, detected in 3 patients (4.5%) by sequencing and in 5 patients (7.6%) by ARMS-PCR. The T215F/Y mutation is a thymidine analogue mutation (TAM) which causes intermediate-/high-level resistance to AZT and d4T, low-level resistance to ddI, and potentially low-level resistance to ABC and TDF. Two out of 66 patients (3.0%) harbored viruses with the T215F mutation, as determined by ARMS-PCR, while only 1/66 patients (1.5%) harbored virus with the T215F mutation by Sanger sequencing.
TABLE 6.
NRTI and NNRTI drug resistance mutations among study subjects, determined by standard (Sanger) sequencing and ARMS-PCRa
| ARV class | Mutation(s) | No. (%) of mutations by: |
|
|---|---|---|---|
| Standard sequencing | ARMS-PCR | ||
| NRTI | M184V or T215Y/F or K65R | 3 (4.5) | 6 (9.1) |
| K65R | 0 (0) | 0 (0) | |
| M184V | 3 (4.5) | 5 (7.6) | |
| T215Y/F | 1 (1.5) | 2 (3.0) | |
| M184V + T215Y/F | 1 (1.5) | 1 (1.5) | |
| M184V + T215Y/F + K65R | 0 (0) | 0 (0) | |
| NNRTI | K103N or Y181C | 4 (6.1) | 4 (6.1) |
| K103N | 3 (4.5) | 3 (4.5) | |
| Y181C | 1 (1.5) | 1 (1.5) | |
| K103N + Y181C | 0 (0) | 0 (0) | |
Both absolute and relative numbers (percentages) are shown. Percentages were calculated by dividing the absolute number of the respective drug resistance mutations by the total number of study subjects (66) and multiplying that number by 100 (as presented in Table 3).
Of note, 1 patient (1.5%) was found to concomitantly harbor viruses with 2 NRTI-associated mutations (M184V and T215F) by Sanger sequencing and ARMS-PCR, and 2 patients were found to harbor these mutations by NGS.
Prevalence of major NNRTI-associated mutations.
Using Sanger sequencing as a standard, 6.1% of the 66 study subjects carried a major NNRTI mutation (K103N or Y181C) (Table 6). K103N is a nonpolymorphic mutation that causes high-level resistance to NVP and EFV, i.e., the two NNRTIs used for first-line treatment in Cameroon. The two assays (ARMS-PCR and Sanger sequencing) detected 3 patients out of 66 (4.5%) harboring viruses with the K103N mutation. Y181C is another nonpolymorphic mutation selected in patients receiving NVP, etravirine (ETR), and rilpivirine (RPV). It reduces susceptibility to NVP, ETR, RPV, and EFV by >50-fold, 5-fold, 3-fold, and 2-fold, respectively. Only 1 patient out of 66 (1.5%) harbored a virus with the Y181C mutation detected by all three genotyping methods.
Of note, no patient concomitantly harbored viruses with 2 NNRTI-associated mutations (K103N and Y181C), but 2/66 (3.0%) concomitantly harbored viruses with NRTI- and NNRTI-associated mutations.
Longitudinal changes in transmitted HIVDR mutations.
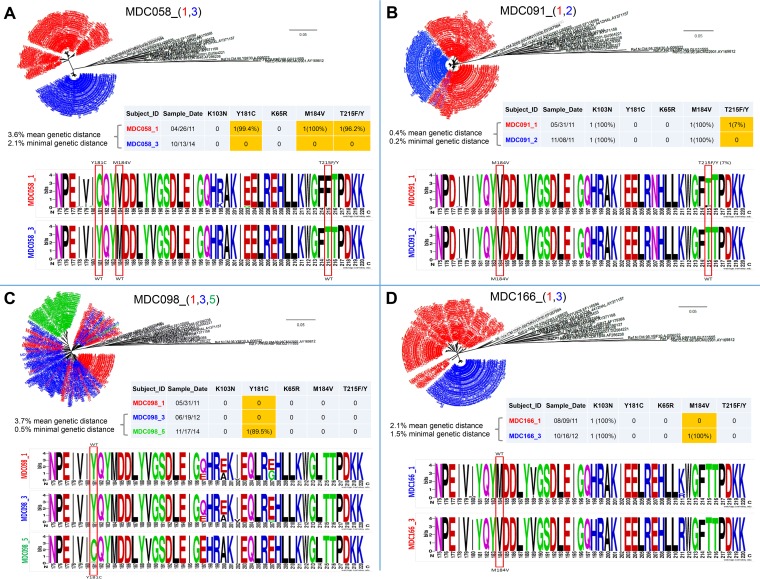
For 21 subjects, we were able to collect longitudinal ART-naive plasma samples spanning up to 4 years. The multiple time points were used to study the dynamics of HIVDR mutations in the absence of antiretroviral drug pressure by NGS and for 3 study subjects also by Sanger sequencing and ARMS-PCR (Table 4). Five individuals exhibited changes in HIVDR mutation profiles that included the loss (subjects MDC058 and MDC091), gain (MDC098 and MDC166), and sequential loss and gain (MDC246) of HIVDR mutations. To investigate the occurrence of superinfection as a possible cause for the change in mutation profile, we performed a detailed phylogenetic/genetic distance analysis and studied the sequence compositions longitudinally with sequence logos (Fig. 3 and Fig. 4). Within subject MDC058, all three HIVDR mutations (Y181C, M184V, and T215F) completely reverted to wild type within 42 months. Subject MDC091 exhibited a reversion in that 7% of minority variants carrying the T215F mutation evolved back to wild type within 6 months. In contrast, both subjects MDC098 and MDC166 adopted HIVDR mutations. MDC098 acquired Y181C within 17 months; MDC166 gained M184V in 14 months. For these 4 individuals (Fig. 3), the occurrence of superinfection could be excluded due to a gradual phylogenetic evolution (minimal genetic distance of ≤1% per year), which was obvious from the phylogenetic trees and sequence logos (adjacent regions of HIVDR sites). In contrast, MDC246 changed her 100% K103N positive profile within 3 months to become completely sensitive. The genetic distance of ≥2.7% in pol and 16% in env (≥1% per year) is strongly indicative of a superinfection with complete replacement of the drug-resistant variants by sensitive superinfecting variants (Fig. 4). At 3.5 years and despite the absence of apparent drug pressure, the pol mutant variants reemerged as a minority population (18% of quasispecies) detectable by NGS but not by Sanger sequencing or ARMS-PCR.
FIG 3.
Phylogenetic and sequence analysis of study subjects with longitudinal changes in drug resistance mutations in the absence of superinfection. Longitudinal changes in HIV drug resistance (HIVDR) mutations were observed for the four study subjects MDC058 (A), MDC091 (B), MDC098 (C), and MDC166 (D). Using the NGS pol sequences, a multi-time-point phylogenetic tree (top), a drug resistance pattern and genetic distance analysis (middle), and a sequence logo analysis (bottom) were performed for each study subject. (Top) Phylogenetic trees were generated using the MEGA (neighbor-joining method) and FigTree softwares. Sequences in black are references downloaded from the HIV sequence database (https://www.hiv.lanl.gov). Numbers in parentheses after the patient IDs and a low dash indicate the sampling time points, and the corresponding sequences are shown with the same colors in the phylogenetic tree. The bar indicates the genetic distance. (Middle) Table with drug resistance patterns for each time point. Discordant longitudinal HIVDR mutations are highlighted in yellow. Mean genetic distances between time points and minimal genetic distances between two NGS sequences from different time points (in percentages) were calculated in MEGA. (Bottom) Sequence logo analysis was performed with the WebLogo online tool (weblogo.berkeley.edu). The red rectangles indicate the presence of HIVDR mutations with/without longitudinal change. The analysis was performed from amino acid positions 175 to 220 of the product of the rt gene and includes all mutations of interest. (A) MDC058 changed from mutant types C, V, and Fs to wild-type Y, M, and T at positions 181, 184, and 215, respectively. The changes in the rt sequences occurred between time points 1 and 3 within 42 months. (B) MDC091 changed from mutant type F (7% prevalence) to wild-type T at position 215. The changes in the rt sequences occurred between time points 1 and 2 within 6 months. (C) MDC098 changed from wild-type Y to mutant type C at position 181. The changes in rt sequences occurred between time points 3 and 5 within 17 months. (D) MDC166 changed from wild-type M to mutant type V at position 184. The changes in the rt sequences occurred between time points 1 and 3 within 14 months.
FIG 4.
Phylogenetic and sequence analysis of a superinfected study subject with longitudinal changes in drug resistance mutations. Longitudinal changes in HIVDR mutations were observed for the superinfected study subject MDC246. Using the NGS pol sequences, a multi-time-point phylogenetic tree (top), a drug resistance pattern and genetic distance analysis (middle), and a sequence logo analysis (bottom) were performed. (Top) Phylogenetic trees were generated using the MEGA (neighbor-joining method) and FigTree softwares. Sequences in black are references downloaded from the HIV sequence database (https://www.hiv.lanl.gov). Numbers in parentheses following the patient ID and a low dash indicate the sampling time points, and the corresponding sequences are shown with the same colors in the phylogenetic tree. The bar indicates the genetic distance. (Middle) Table with drug resistance patterns for each time point. Discordant longitudinal HIVDR mutations are highlighted in yellow. Mean genetic distances between time points and minimal genetic distances between two NGS sequences from different time points (in percentages) were calculated in MEGA for pol and additionally for env using Sanger sequences. (Bottom) Sequence logo analysis was performed with the WebLogo online tool (weblogo.berkeley.edu). The red rectangle indicates the presence of HIVDR mutations with longitudinal change. The analysis was performed from amino acid positions 80 to 125 of the rt gene product and includes the site of interest, K103. MDC246 changed from mutant type K103N to wild-type K after the occurrence of superinfection in a window of 2.5 months. At the last time point, 6, both primary and superinfecting variants are present with a mixture of the K103N mutant and K103 wild-type signatures.
DISCUSSION
The current study describes the HIVDR mutation profile of 66 ART-naive individuals, longitudinal changes of HIVDR mutations in the absence of reported drug pressure, and the comparative analysis of three genotypic drug resistance assays. As demonstrated in this and other studies, there is a considerable plasticity at the mutation sites, with reversions and even new acquisitions occurring in the absence of reported drug history. Our longitudinal NGS analysis highlighted the potential of both NRTI and NNRTI mutations to revert to drug sensitivity, induced either by superinfection or intrinsically. Correspondingly, other studies reported a varying persistence of transmitted HIVDR mutations dependent on their fitness costs (25, 32, 53, 54). While NNRTI mutations are known to gradually revert after a mean time of 3 years, NRTIs exhibit greater differences. M184V and T215F/Y pay high viral fitness burdens, which drive prompt reversals, potentially within 1 year. This is exemplified in our study subject MDC058, in whom the three HIVDR mutations M184V, T215F, and Y181C completely reverted within 3.5 years. The 3.6%/2.1% mean/minimal genetic distances of NGS pol sequences after 3.5 years underline the pronounced genetic changes, without providing indications for superinfection (≤1% genetic distance/year). We surprisingly observed, besides reversion, two cases of HIVDR mutation acquisition in the absence of reported ART (Y181C in MDC098, M184V in MDC166). Since almost the complete swarms of analyzed quasispecies changed their mutation pattern at the respective sites, spontaneous changes seem unlikely. Although MDC098 and MDC166 self-reported as “drug naive,” hidden drug pressures through occasional drug intake, e.g., through available drugs from HIV-positive family members or friends, appear more plausible. MDC246 exhibited both loss and acquisition of detectable drug-resistant variants under the influence of superinfection. Cases of HIV superinfection and masking of preexisting resistance in the absence of drug pressure have been reported (55, 56). Most likely, the initial drug-resistant variants harboring the K103N mutation were replaced/masked in the plasma from MDC246's second time point by superinfecting variants with significant genetic distance (≥2.7%/3 months) (55). Of interest, after 3.5 years, the mutant type reappeared at a low percentage (18%) despite the absence of reported drug pressure, which is indicative of the occurrence of low-level reactivation from reservoirs.
In addition to performing the longitudinal HIVDR mutation analysis, we determined the cross-sectional drug resistance profiles for all samples at time point 1. We observed major NRTI and NNRTI resistance mutations. Based on Sanger sequencing, the M184V and K103N mutations were the most prevalent (4.5% each), followed by Y181C and T215F/Y (1.5% each), while K65R was absent. The rates of HIVDR mutations obtained in this study (0 to 4.5%) are slightly lower than in our previous study (0 to 15%) (39) but are in the range of those from other studies (1 to 12%) conducted on ART-naive patients in Cameroon between 2008 and 2014 (11, 22, 47, 52, 57). The predominance of the M184V and K103N mutations is in line with several recent studies and is the likely consequence of the first-line NRTI and NNRTI regimens with 3TC or FTC and NVP or EFV, which exercise drug pressure on the respective sites (9, 37). The absence of the K65R mutation can be ascribed to the recent introduction of tenofovir in our study population. Our samples were collected only 1 year after TDF was introduced in 2010 (58), which, combined with a high genetic barrier, massively reduced the risk of the development and transmission of TDF-associated mutations like K65R. Other studies, conducted in Cameroon between 2008 and 2013, verified the very low prevalence of the K65R mutation (<2% of quasispecies) in ART-naive patients (11, 22, 47, 57).
The current analysis of HIVDR mutations was further applied for the first comparison of ARMS-PCR with NGS and Sanger sequencing. As expected, NGS and Sanger sequencing showed a good match in their performances, with only two deviations caused by minority variants (<20% of quasispecies), which is under the detection limit of Sanger sequencing and putatively of minor clinical importance (34, 59). In direct comparison to Sanger sequencing, ARMS-PCR performed very well, with 100% sensitivity based on no false-negative determinations for all studied mutations. Specificity also reached 100% for K103N and Y181C; however, for M184V and T215F/Y, specificity dropped to 93% and 97%, respectively, as evidenced by three repeated false-positive calls. Due to our first side-by-side analysis with NGS, we could exclude minority variants or sequence variances at the primer binding sites as the cause for our false-positive calls via ARMS-PCR. In turn, it is possible that mispriming was assisted/sustained by sequence regions surrounding the HIVDR mutation positions and/or secondary/tertiary structural features of the template DNA (12). A limitation of the current resistance assay analysis is the inherent low number of drug-resistant mutations in drug-naive samples.
However, the apparently good performance of ARMS-PCR with drug-naive samples is in line with the results of our previous study from 2015 on 75 ART-naive and -experienced patients with a total of 79 HIVDR mutations (39). The ARMS-PCR assay was able to detect M184V, T215F/Y, K103N, and Y181C with sensitivities of 97%, 86%, 91%, and 70% and specificities of 91%, 95%, 100%, and 97%, respectively. In Thailand, a duplex ARMS-PCR assay was successfully applied to detect K103N/Y181C and Q151M/T215Y mutations in 45 naive and treated HIV-1 CRF01_AE-infected patients that achieved good performance ratings (96% sensitivity and 98% specificity) similar to those of Sanger sequencing (60). The suitability of ARMS-PCR was also demonstrated in two studies carried out in India that screened 60 individuals for mutations at codons 70, 184, and 215 (43) and 25 children for mutations at codons 103 and 215 (44). The good performance of ARMS-PCR comes with a fast turnaround time and low cost but has the limitation of providing information on only select mutations. Since every single codon requires its own specific PCR, the investigation of multisite resistance patterns in a large number of patients is laboratory intensive. The efficient application of point mutation assays needs active surveillance to identify active trends in clinically significantly transmitted and acquired HIVDR mutations (19). Nonetheless, the ability of ARMS-PCR to sensitively detect key mutations makes it a very pragmatic tool for determining major HIVDR mutations in resource-constrained settings.
MATERIALS AND METHODS
Ethical considerations.
This study was approved by the Institutional Ethical Review Board of New York University School of Medicine, New York, New York, USA, and by the Institutional Review Board of Cameroon's Ministry of Public Health. Written informed consent was obtained from all the study subjects.
Study samples and subjects.
Our study included 66 HIV-positive patients, all of whom were ART naive according to the study questionnaire and patient information and recruited between February 2011 and March 2012 at the Medical Diagnostic Center (MDC) in Yaoundé, Cameroon. A detailed questionnaire including demographic information was administered to each patient. Seventy-five percent of the patients were women, the median age was 34 years, and the median CD4 cell count was 330 cells/mm3. The diagnosis of HIV infection had occurred for most patients between 2008 and 2012. Incidence testing of the samples from the first study time point revealed that among the 66 HIV-positive patients, 3 had recently become infected (<6 months since infection), while the majority (n = 63) had already been at a more chronic stage of infection (>6 months). A one-by-one description for each subject, including clinical, personal, and drug resistance parameters, is provided in Table 3.
RNA extraction and RT-PCR.
Virus in 500 μl of plasma was concentrated by centrifugation at 14,000 × g for 1 h at 4°C prior to RNA extraction. After removal of 360 μl of supernatant, the virus pellet was resuspended in the remaining 140 μl of supernatant by being vortexed, and viral RNA was extracted using the QIAamp viral RNA minikit according to the manufacturer's instructions (Qiagen, Inc., Valencia, CA, USA) (61). A total of 2.5 μl of RNA was used for the amplification of 1,750 bp of the pol region, as previously described (39).
ARMS-PCR.
Amplification-refractory mutation system-PCR (ARMS-PCR) was used to detect wild-type and mutant sequences at different codons (K65R, M184V, T215Y/F, K103N, and Y181C) as previously described (39); the sequences and HXB2 locations of the primers are found in Table 2. HIVDR mutations were selected based on their prevalence, their effect on the first-line regimen recommended by the WHO for resource-constrained settings, and the Stanford HIV drug resistance database mutation scoring system (http://hivdb.stanford.edu/DR/asi/releaseNotes/index.html). ARMS-PCR results were compared to Sanger sequencing and next-generation sequencing (NGS) data. Double-stranded Sanger sequencing (62) was used as a gold standard for level of sequence identification, and the sensitivity, specificity, and positive and negative predictive values of ARMS-PCR were calculated as previously described (39).
Sensitivity, specificity, NPV, and PPV have the common property of being based on counts of mutations, which refers to a binomial variable. In order to estimate the adequacy of our sample size for estimating the four parameters, we determined the precision of the probability (P) of mutation. The standard error of estimating P with a sample size of 66 is ≤0.007575758, so the half-length of the 95% confidence interval is ≤0.01484848. Thus, our sample size of 66 estimates a P with a ≤2.97% error.
Sanger (standard) sequencing.
Two microliters of the RT-PCR product was used for nested pol PCRs and consecutive traditional Sanger sequencing (Macrogen) with RTPOLF2 and RTPOLR2 primers or Pol2 forward and Pol2 reverse as previously described (HXB2 positions 2723 to 3225) (39, 61). Sanger env sequences (HXB2 positions 6684 to 7784) were generated by nested PCRs with second-round EnvB and Gp120 IN primers as described in reference 61.
NGS using MiSeq.
NGS was performed on a region of the pol gene (HXB2 positions 2723 to 3225). Briefly, viral RNA was reverse transcribed, amplified, and sequenced using a MiSeq next-generation sequencing platform (Illumina, Inc., San Diego, CA, USA) with the Nextera index primer sets and analyzed with MEGA5.2 (63), FigTree1.4.3 (64), and Lasergene12 (DNASTAR, Inc., Madison, WI, USA) as described in reference 61. The protocol was modified from a previous ∼500-bp-long read from the 454 NGS-based protocol (65–67) to the paired-end 2× 300-bp read protocol of MiSeq. The resulting MiSeq reads were analyzed and segregated into unique amplicons. Similar amplicons were combined into a single consensus sequence. Consensus sequences that contained >0.02% of the total number of amplicons for that sample were used for all subsequent analyses (64, 66). A representative sequence from each phylogenetically distinct population for every sample on a given MiSeq run was aligned and examined on a neighbor-joining tree for the presence of cross-contamination. Any minor variant that colocalized with a major viral population from another unrelated sample in the run was removed (details of the NGS protocol are described in reference 61. The calibrated population resistance tool (http://cpr.stanford.edu/cpr.cgi) was used to analyze the NGS sequences for drug resistance mutations. Only mutations present in ≥5% consensus sequences were considered significant. For phylogenetic analysis and sequence logos, all individual consensus sequences per longitudinal NGS time point and study subject were averaged to one major consensus sequence using DNASTAR's SeqMan Pro.
Phylogenetic analysis.
Reverse transcriptase (rt) sequences (HIV region from positions 2723 to 3225 according to HXB2 numbering) were aligned with reference sequences of HIV-1 group M subtypes and circulating recombinant forms (CRFs) from the Los Alamos HIV sequence database (https://www.hiv.lanl.gov). Neighbor-joining phylogenetic trees were created using MEGA5.2 software (Kimura 2-parameter model, 200 bootstrap replications) and FigTree1.4.3 (63, 64). Subtyping was based on phylogenetic and HIV BLAST (https://www.hiv.lanl.gov) analyses of the rt sequences, as an approximate estimate of the whole-genome subtypes. To calculate the mean genetic distance between different time points of each patient, sequences were grouped according to time point in MEGA and analyzed with the compute mean distance analysis. To calculate the minimal genetic distance between two time points, a pairwise distance analysis was performed for all individual NGS sequences between two different time points, followed by a screening for the lowest pairwise distance (59). Genetic distances between Sanger env sequences were calculated accordingly for the HIV region from positions 6684 to 7784.
Drug resistance genotyping.
The pol DNA sequences were analyzed for drug resistance mutations using the Stanford University HIV database genotypic-resistance interpretation algorithm (http://hivdb.stanford.edu/index.html). Mutations in the study sequences were defined as differences from the consensus B reference sequence and were further characterized as NRTI or NNRTI resistance mutations.
Sequence logos.
To compare the amino acid compositions of the longitudinal NGS consensus sequences, a sequence logo analysis was performed with the WebLogo online tool (weblogo.berkeley.edu) from amino acid positions 175 to 220 of the rt gene product (including the mutations of major interest).
Accession number(s).
The Sanger and NGS consensus pol sequences are available from GenBank with the accession numbers KT758206, KT758208, KT758243, KT758259, KT758186, KT758199, KT758249, KT758262, KT758270, KY475637 to KY475720, KY931691 to KY931732, and MF278283 to MF278286. The whole set of NGS pol sequences are available upon request. The Sanger env sequences are available from GenBank with the accession numbers MF278287 to MF278289.
ACKNOWLEDGMENTS
We thank the individuals who donated their blood samples for this study and the Cameroon Ministry of Public Health for support. We also thank Caroline Kakam, Bladine Asaah, Michael Tuen, and Flavia Camacho for their assistance in sample collection, manuscript preparation, and methodological guidance. We also thank Daniel Bruno and Craig Martens for assistance in NGS data generation and initial analysis.
This study was supported by National Institutes of Health grants AI083142 (A.J.N., L.A.A., M.K.G., P.N.N., R.D.) and TW009604 (A.J.N., G.N., E.A., J.S.B., A.N.B., L.A.A., J.M., M.K.G., P.N.N., R.D.) and in part by the Division of Intramural Research, NIAID, NIH (A.D.R., A.R.K., S.F.P., T.C.Q.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. We have no conflicts of interest to disclose.
REFERENCES
- 1.Bigna JJ, Fonkoue L, Tchatcho MF, Dongmo CN, Soh DM, Um JL, Sime PS, Affana LA, Woum AR, Noumegni SR, Tabekou A, Wanke AM, Taffe HR, Tchoukouan ML, Anyope KO, Ella SB, Mouaha BV, Kenne EY, Mbessoh UI, Tchapmi AY, Tene DF, Voufouo SS, Zogo SM, Nouebissi LP, Satcho KF, Tchoumo WJ, Basso MF, Tcheutchoua BD, Agbor AA. 2014. Association of academic performance of premedical students to satisfaction and engagement in a short training program: a cross sectional study presenting gender differences. BMC Res Notes 7:105. doi: 10.1186/1756-0500-7-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.National AIDS Control Committee. 2010. The impact of HIV and AIDS in Cameroon through 2020. National AIDS Control Committee, Yaoundé, Cameroon: http://hivhealthclearinghouse.unesco.org/library/documents/impact-hiv-and-aids-cameroon-through-2020. [Google Scholar]
- 3.UNAIDS. 2014. UNAIDS fast track 95-95-95 target. UNAIDS, Washington, DC: http://www.unaids.org/sites/default/files/media_asset/JC2686_WAD2014report_en.pdf. [Google Scholar]
- 4.UNAIDS. 2014. 90-90-90. An ambitious treatment target to help end the aids epidemic. UNAIDS, Washington, DC: http://www.unaids.org/sites/default/files/media_asset/90-90-90_en.pdf. [Google Scholar]
- 5.UNAIDS. 2015. UNAIDS fact sheet 2015. UNAIDS, Washington, DC: http://www.unaids.org/en/resources/fact-sheet. [Google Scholar]
- 6.WHO/UNAIDS. 2003. Global initiative to provide antiretroviral therapy to 3 million people with HIV/AIDS in developing countries by the end of 2005. WHO, Geneva, Switzerland: http://www.who.int/3by5/publications/documents/en. [Google Scholar]
- 7.Gunthard HF, Saag MS, Benson CA, del Rio C, Eron JJ, Gallant JE, Hoy JF, Mugavero MJ, Sax PE, Thompson MA, Gandhi RT, Landovitz RJ, Smith DM, Jacobsen DM, Volberding PA. 2016. Antiretroviral drugs for treatment and prevention of HIV infection in adults: 2016 recommendations of the International Antiviral Society—USA panel. JAMA 316:191–210. doi: 10.1001/jama.2016.8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuznik A, Iliyasu G, Habib AG, Musa BM, Kambugu A, Lamorde M. 2016. Initiation of antiretroviral therapy based on the 2015 WHO guidelines. AIDS 30:2865–2873. doi: 10.1097/QAD.0000000000001251. [DOI] [PubMed] [Google Scholar]
- 9.Gilks CF, Crowley S, Ekpini R, Gove S, Perriens J, Souteyrand Y, Sutherland D, Vitoria M, Guerma T, De Cock K. 2006. The WHO public-health approach to antiretroviral treatment against HIV in resource-limited settings. Lancet 368:505–510. doi: 10.1016/S0140-6736(06)69158-7. [DOI] [PubMed] [Google Scholar]
- 10.Gill VS, Lima VD, Zhang W, Wynhoven B, Yip B, Hogg RS, Montaner JS, Harrigan PR. 2010. Improved virological outcomes in British Columbia concomitant with decreasing incidence of HIV type 1 drug resistance detection. Clin Infect Dis 50:98–105. doi: 10.1086/648729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aghokeng AF, Monleau M, Eymard-Duvernay S, Dagnra A, Kania D, Ngo-Giang-Huong N, Toni TD, Toure-Kane C, Truong LX, Delaporte E, Chaix ML, Peeters M, Ayouba A. 2014. Extraordinary heterogeneity of virological outcomes in patients receiving highly antiretroviral therapy and monitored with the World Health Organization public health approach in sub-Saharan Africa and Southeast Asia. Clin Infect Dis 58:99–109. doi: 10.1093/cid/cit627. [DOI] [PubMed] [Google Scholar]
- 12.Clutter DS, Jordan MR, Bertagnolio S, Shafer RW. 2016. HIV-1 drug resistance and resistance testing. Infect Genet Evol 46:292–307. doi: 10.1016/j.meegid.2016.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villabona-Arenas CJ, Vidal N, Guichet E, Serrano L, Delaporte E, Gascuel O, Peeters M. 2016. In-depth analysis of HIV-1 drug resistance mutations in HIV-infected individuals failing first-line regimens in West and Central Africa. AIDS 30:2577–2589. doi: 10.1097/QAD.0000000000001233. [DOI] [PubMed] [Google Scholar]
- 14.Van Laethem K, Theys K, Vandamme AM. 2015. HIV-1 genotypic drug resistance testing: digging deep, reaching wide? Curr Opin Virol 14:16–23. doi: 10.1016/j.coviro.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Mali SN, Sapkal PM. 2015. HIV drug resistance: an overview. Int J Res Methodol 1(1): 72–82. http://ijrm.humanjournals.com/wp-content/uploads/2015/10/4.Suraj-Narayan-Mali-Prasanna-Mahendra-Sapkal.pdf. [Google Scholar]
- 16.Onywera H, Maman D, Inzaule S, Auma E, Were K, Fredrick H, Owiti P, Opollo V, Etard JF, Mukui I, Kim AA, Zeh C. 2017. Surveillance of HIV-1 pol transmitted drug resistance in acutely and recently infected antiretroviral drug-naive persons in rural western Kenya. PLoS One 12:e0171124. doi: 10.1371/journal.pone.0171124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips AN, Lampe FC, Smith CJ, Geretti AM, Rodger A, Lodwick RK, Cambiano V, Tsintas R, Johnson MA. 2010. Ongoing changes in HIV RNA levels during untreated HIV infection: implications for CD4 cell count depletion. AIDS 24:1561–1567. doi: 10.1097/QAD.0b013e32833a6056. [DOI] [PubMed] [Google Scholar]
- 18.Barth RE, van der Loeff MF, Schuurman R, Hoepelman AI, Wensing AM. 2010. Virological follow-up of adult patients in antiretroviral treatment programmes in sub-Saharan Africa: a systematic review. Lancet Infect Dis 10:155–166. doi: 10.1016/S1473-3099(09)70328-7. [DOI] [PubMed] [Google Scholar]
- 19.Rhee SY, Blanco JL, Jordan MR, Taylor J, Lemey P, Varghese V, Hamers RL, Bertagnolio S, Rinke de Wit TF, Aghokeng AF, Albert J, Avi R, Avila-Rios S, Bessong PO, Brooks JI, Boucher CA, Brumme ZL, Busch MP, Bussmann H, Chaix ML, Chin BS, D'Aquin TT, De Gascun CF, Derache A, Descamps D, Deshpande AK, Djoko CF, Eshleman SH, Fleury H, Frange P, Fujisaki S, Harrigan PR, Hattori J, Holguin A, Hunt GM, Ichimura H, Kaleebu P, Katzenstein D, Kiertiburanakul S, Kim JH, Kim SS, Li Y, Lutsar I, Morris L, Ndembi N, Ng KP, Paranjape RS, Peeters M, Poljak M, Price MA, Ragonnet-Cronin ML, Reyes-Teran G, Rolland M, Sirivichayakul S, Smith DM, Soares MA, Soriano VV, Ssemwanga D, Stanojevic M, Stefani MA, Sugiura W, Sungkanuparph S, Tanuri A, Tee KK, Truong HH, van de Vijver DA, Vidal N, Yang C, Yang R, Yebra G, Ioannidis JP, Vandamme AM, Shafer RW. 2015. Geographic and temporal trends in the molecular epidemiology and genetic mechanisms of transmitted HIV-1 drug resistance: an individual-patient- and sequence-level meta-analysis. PLoS Med 12:e1001810. doi: 10.1371/journal.pmed.1001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frentz D, Boucher CA, van de Vijver DA. 2012. Temporal changes in the epidemiology of transmission of drug-resistant HIV-1 across the world. AIDS Rev 14:17–27. [PubMed] [Google Scholar]
- 21.WHO. 2012. HIV drug resistance report. WHO, Geneva, Switzerland. [Google Scholar]
- 22.Ceccarelli L, Salpini R, Moudourou S, Cento V, Santoro MM, Fokam J, Takou D, Nanfack A, Dori L, Torimiro J, Sarmati L, Andreoni M, Perno CF, Colizzi V, Cappelli G. 2012. Characterization of drug resistance mutations in naive and ART-treated patients infected with HIV-1 in Yaounde, Cameroon. J Med Virol 84:721–727. doi: 10.1002/jmv.23244. [DOI] [PubMed] [Google Scholar]
- 23.Hirsch MS, Gunthard HF, Schapiro JM, Brun-Vezinet F, Clotet B, Hammer SM, Johnson VA, Kuritzkes DR, Mellors JW, Pillay D, Yeni PG, Jacobsen DM, Richman DD. 2008. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society—USA panel. Clin Infect Dis 47:266–285. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar]
- 24.Steegen K, Carmona S, Bronze M, Papathanasopoulos MA, van Zyl G, Goedhals D, MacLeod W, Sanne I, Stevens WS. 2016. Moderate levels of pre-treatment HIV-1 antiretroviral drug resistance detected in the first South African National Survey. PLoS One 11:e0166305. doi: 10.1371/journal.pone.0166305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castro H, Pillay D, Cane P, Asboe D, Cambiano V, Phillips A, Dunn DT, UK Collaborative Group on HIV Drug Resistance. 2013. Persistence of HIV-1 transmitted drug resistance mutations. J Infect Dis 208:1459–1463. doi: 10.1093/infdis/jit345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castro E, Zhao H, Cavassini M, Mullins JI, Pantaleo G, Bart PA. 2014. HIV-1 superinfection with a triple-class drug-resistant strain in a patient successfully controlled with antiretroviral treatment. AIDS 28:1840–1844. doi: 10.1097/QAD.0000000000000342. [DOI] [PubMed] [Google Scholar]
- 27.Martin F, Lee J, Thomson E, Tarrant N, Hale A, Lacey CJ. 2016. Two cases of possible transmitted drug-resistant HIV: likely HIV superinfection and unmasking of pre-existing resistance. Int J STD AIDS 27:66–69. doi: 10.1177/0956462415571671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pernas M, Casado C, Fuentes R, Perez-Elias MJ, Lopez-Galindez C. 2006. A dual superinfection and recombination within HIV-1 subtype B 12 years after primoinfection. J Acquir Immune Defic Syndr 42:12–18. doi: 10.1097/01.qai.0000214810.65292.73. [DOI] [PubMed] [Google Scholar]
- 29.Pingen M, Nijhuis M, de Bruijn JA, Boucher CA, Wensing AM. 2011. Evolutionary pathways of transmitted drug-resistant HIV-1. J Antimicrob Chemother 66:1467–1480. doi: 10.1093/jac/dkr157. [DOI] [PubMed] [Google Scholar]
- 30.Pingen M, Nouwen JL, Dinant S, Albert J, Mild M, Brodin J, Simen BB, Walsh S, Kayser M, van der Ende ME, Schutten M, Boucher CA. 2012. Therapy failure resulting from superinfection by a drug-resistant HIV variant. Antivir Ther 17:1621–1625. doi: 10.3851/IMP2267. [DOI] [PubMed] [Google Scholar]
- 31.Smith DM, Wong JK, Hightower GK, Ignacio CC, Koelsch KK, Petropoulos CJ, Richman DD, Little SJ. 2005. HIV drug resistance acquired through superinfection. AIDS 19:1251–1256. doi: 10.1097/01.aids.0000180095.12276.ac. [DOI] [PubMed] [Google Scholar]
- 32.Mourad R, Chevennet F, Dunn DT, Fearnhill E, Delpech V, Asboe D, Gascuel O, Hue S, UK HIV Drug Resistance Database & the Collaborative HIV, Anti-HIV Drug Resistance Network. 2015. A phylotype-based analysis highlights the role of drug-naive HIV-positive individuals in the transmission of antiretroviral resistance in the UK. AIDS 29:1917–1925. doi: 10.1097/QAD.0000000000000768. [DOI] [PubMed] [Google Scholar]
- 33.Little SJ, Frost SD, Wong JK, Smith DM, Pond SL, Ignacio CC, Parkin NT, Petropoulos CJ, Richman DD. 2008. Persistence of transmitted drug resistance among subjects with primary human immunodeficiency virus infection. J Virol 82:5510–5518. doi: 10.1128/JVI.02579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charpentier C, Lee GQ, Rodriguez C, Visseaux B, Storto A, Fagard C, Molina JM, Katlama C, Yazdanpanah Y, Harrigan PR, Descamps D. 2015. Highly frequent HIV-1 minority resistant variants at baseline of the ANRS 139 TRIO trial had a limited impact on virological response. J Antimicrob Chemother 70:2090–2096. doi: 10.1093/jac/dkv048. [DOI] [PubMed] [Google Scholar]
- 35.Li JZ, Paredes R, Ribaudo HJ, Svarovskaia ES, Metzner KJ, Kozal MJ, Hullsiek KH, Balduin M, Jakobsen MR, Geretti AM, Thiebaut R, Ostergaard L, Masquelier B, Johnson JA, Miller MD, Kuritzkes DR. 2011. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA 305:1327–1335. doi: 10.1001/jama.2011.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Charpentier C, Gody JC, Mbitikon O, Moussa S, Matta M, Pere H, Fournier J, Longo Jde D, Belec L. 2012. Virological response and resistance profiles after 18 to 30 months of first- or second-/third-line antiretroviral treatment: a cross-sectional evaluation in HIV type 1-infected children living in the Central African Republic. AIDS Res Hum Retroviruses 28:87–94. doi: 10.1089/AID.2011.0035. [DOI] [PubMed] [Google Scholar]
- 37.Fokam J, Salpini R, Santoro MM, Cento V, D'Arrigo R, Gori C, Perno CF, Colizzi V, Nanfack A, Gwom LC, Cappelli G, Takou D. 2011. Performance evaluation of an in-house human immunodeficiency virus type-1 protease-reverse transcriptase genotyping assay in Cameroon. Arch Virol 156:1235–1243. doi: 10.1007/s00705-011-0982-3. [DOI] [PubMed] [Google Scholar]
- 38.Saravanan S, Vidya M, Balakrishnan P, Kumarasamy N, Solomon SS, Solomon S, Kantor R, Katzenstein D, Ramratnam B, Mayer KH. 2009. Evaluation of two human immunodeficiency virus-1 genotyping systems: ViroSeq 2.0 and an in-house method. J Virol Methods 159:211–216. doi: 10.1016/j.jviromet.2009.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nanfack AJ, Agyingi L, Noubiap JJ, Ngai JN, Colizzi V, Nyambi PN. 2015. Use of amplification refractory mutation system PCR assay as a simple and effective tool to detect HIV-1 drug resistance mutations. J Clin Microbiol 53:1662–1671. doi: 10.1128/JCM.00114-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, Smith JC, Markham AF. 1989. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res 17:2503–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Laethem K, Van Vaerenbergh K, Schmit JC, Sprecher S, Hermans P, De Vroey V, Schuurman R, Harrer T, Witvrouw M, Van Wijngaerden E, Stuyver L, Van Ranst M, Desmyter J, De Clercq E, Vandamme AM. 1999. Phenotypic assays and sequencing are less sensitive than point mutation assays for detection of resistance in mixed HIV-1 genotypic populations. J Acquir Immune Defic Syndr 22:107–118. doi: 10.1097/00126334-199910010-00001. [DOI] [PubMed] [Google Scholar]
- 42.Ketloy C, Sirivichayakul S, Ruxrungtham K. 2009. A low cost duplex polymerase chain reaction to detect common HIV-1 CRF 01AE reverse transcriptase inhibitors resistance-associated mutations: 103N/181C and 151M/215Y. Asian Biomed 3:11. [Google Scholar]
- 43.Sachdeva N, Sehgal S, Arora SK. 2005. Frequency of drug-resistant variants of HIV-1 coexistent with wild-type in treatment-naive patients of India. J Int AIDS Soc 7:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sehgal S, Pasricha N, Singh S. 2008. High rate of mutation K103N causing resistance to nevirapine in Indian children with acquired immunodeficiency syndrome. Indian J Med Microbiol 26:372–374. doi: 10.4103/0255-0857.43583. [DOI] [PubMed] [Google Scholar]
- 45.Aghokeng AF, Vergne L, Mpoudi-Ngole E, Mbangue M, Deoudje N, Mokondji E, Nambei WS, Peyou-Ndi MM, Moka JJ, Delaporte E, Peeters M. 2009. Evaluation of transmitted HIV drug resistance among recently-infected antenatal clinic attendees in four Central African countries. Antivir Ther 14:401–411. [DOI] [PubMed] [Google Scholar]
- 46.Agyingi L, Mayr LM, Kinge T, Orock GE, Ngai J, Asaah B, Mpoame M, Hewlett I, Nyambi P. 2014. The evolution of HIV-1 group M genetic variability in southern Cameroon is characterized by several emerging recombinant forms of CRF02_AG and viruses with drug resistance mutations. J Med Virol 86:385–393. doi: 10.1002/jmv.23846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burda ST, Viswanath R, Zhao J, Kinge T, Anyangwe C, Tinyami ET, Haldar B, Powell RL, Jarido V, Hewlett IK, Nyambi PN. 2010. HIV-1 reverse transcriptase drug-resistance mutations in chronically infected individuals receiving or naive to HAART in Cameroon. J Med Virol 82:187–196. doi: 10.1002/jmv.21677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Courtney CR, Agyingi L, Fokou A, Christie S, Asaah B, Meli J, Ngai J, Hewlett I, Nyambi PN. 2016. Monitoring HIV-1 group M subtypes in Yaounde, Cameroon reveals broad genetic diversity and a novel CRF02_AG/F2 infection. AIDS Res Hum Retroviruses 32:381–385. doi: 10.1089/aid.2015.0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koizumi Y, Ndembi N, Miyashita M, Lwembe R, Kageyama S, Mbanya D, Kaptue L, Numazaki K, Fujiyama Y, Ichimura H. 2006. Emergence of antiretroviral therapy resistance-associated primary mutations among drug-naive HIV-1-infected individuals in rural western Cameroon. J Acquir Immune Defic Syndr 43:15–22. doi: 10.1097/01.qai.0000226793.16216.55. [DOI] [PubMed] [Google Scholar]
- 50.Ndongmo CB, Pieniazek D, Holberg-Petersen M, Holm-Hansen C, Zekeng L, Jeansson SL, Kaptue L, Kalish ML. 2006. HIV genetic diversity in Cameroon: possible public health importance. AIDS Res Hum Retroviruses 22:812–816. doi: 10.1089/aid.2006.22.812. [DOI] [PubMed] [Google Scholar]
- 51.Fokam J, Salpini R, Santoro MM, Cento V, Perno CF, Colizzi V, Ndumbe PM, Fokunang Ntungen C, Ndiang Tetang SM, Nanfack AJ, Takou Komego DA, Cappelli G. 2011. Drug resistance among drug-naive and first-line antiretroviral treatment-failing children in Cameroon. Pediatr Infect Dis J 30:1062–1068. doi: 10.1097/INF.0b013e31822db54c. [DOI] [PubMed] [Google Scholar]
- 52.Ndembi N, Abraha A, Pilch H, Ichimura H, Mbanya D, Kaptue L, Salata R, Arts EJ. 2008. Molecular characterization of human immunodeficiency virus type 1 (HIV-1) and HIV-2 in Yaounde, Cameroon: evidence of major drug resistance mutations in newly diagnosed patients infected with subtypes other than subtype B. J Clin Microbiol 46:177–184. doi: 10.1128/JCM.00428-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jain V, Sucupira MC, Bacchetti P, Hartogensis W, Diaz RS, Kallas EG, Janini LM, Liegler T, Pilcher CD, Grant RM, Cortes R, Deeks SG, Hecht FM. 2011. Differential persistence of transmitted HIV-1 drug resistance mutation classes. J Infect Dis 203:1174–1181. doi: 10.1093/infdis/jiq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang WL, Kouyos RD, Boni J, Yerly S, Klimkait T, Aubert V, Scherrer AU, Shilaih M, Hinkley T, Petropoulos C, Bonhoeffer S, Gunthard HF, Swiss HIV Cohort Study. 2015. Persistence of transmitted HIV-1 drug resistance mutations associated with fitness costs and viral genetic backgrounds. PLoS Pathog 11:e1004722. doi: 10.1371/journal.ppat.1004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koelsch KK, Smith DM, Little SJ, Ignacio CC, Macaranas TR, Brown AJ, Petropoulos CJ, Richman DD, Wong JK. 2003. Clade B HIV-1 superinfection with wild-type virus after primary infection with drug-resistant clade B virus. AIDS 17:F11–F16. doi: 10.1097/00002030-200305020-00001. [DOI] [PubMed] [Google Scholar]
- 56.Koning FA, Badhan A, Shaw S, Fisher M, Mbisa JL, Cane PA. 2013. Dynamics of HIV type 1 recombination following superinfection. AIDS Res Hum Retroviruses 29:963–970. doi: 10.1089/aid.2013.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aghokeng AF, Kouanfack C, Eymard-Duvernay S, Butel C, Edoul GE, Laurent C, Koulla-Shiro S, Delaporte E, Mpoudi-Ngole E, Peeters M. 2013. Virological outcome and patterns of HIV-1 drug resistance in patients with 36 months' antiretroviral therapy experience in Cameroon. J Int AIDS Soc 16:18004. doi: 10.7448/IAS.16.1.18004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Republic of Cameroon, Ministry of Public Health. 2010. Directives nationales de prevention et de prise en charge du VIH au Cameroun. Ministry of Public Health, Yaoundé, Republic of Cameroon: http://www.emtct-iatt.org/wp-content/uploads/2016/01/Cameroon20141432236210.pdf. [Google Scholar]
- 59.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 60.Ketloy C, Sirivichayakul S, Ruxrungtham K. 2009. A low cost duplex polymerase chain reaction to detect common HIV-1 CRF 01AE reverse transcriptase inhibitors resistance-associated mutations: 103N/181C and 151M/215Y. Asian Biomed 3:611–621. [Google Scholar]
- 61.Courtney CR, Mayr L, Nanfack AJ, Banin AN, Tuen M, Pan R, Jiang X, Kong XP, Kirkpatrick AR, Bruno D, Martens CA, Sykora L, Porcella SF, Redd AD, Quinn TC, Nyambi PN, Durr R. 2017. Contrasting antibody responses to intrasubtype superinfection with CRF02_AG. PLoS One 12:e0173705. doi: 10.1371/journal.pone.0173705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanger F, Nicklen S, Coulson AR. 1977. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rambaut A. 2012. FigTree, v.1.4.3. http://tree.bio.ed.ac.uk/software/figtree.
- 65.Lehman DA, Wamalwa DC, McCoy CO, Matsen FA, Langat A, Chohan BH, Benki-Nugent S, Custers-Allen R, Bushman FD, John-Stewart GC, Overbaugh J. 2012. Low-frequency nevirapine resistance at multiple sites may predict treatment failure in infants on nevirapine-based treatment. J Acquir Immune Defic Syndr 60:225–233. doi: 10.1097/QAI.0b013e3182515730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Redd AD, Collinson-Streng A, Martens C, Ricklefs S, Mullis CE, Manucci J, Tobian AA, Selig EJ, Laeyendecker O, Sewankambo N, Gray RH, Serwadda D, Wawer MJ, Porcella SF, Quinn TC, Rakai Health Sciences Program. 2011. Identification of HIV superinfection in seroconcordant couples in Rakai, Uganda, by use of next-generation deep sequencing. J Clin Microbiol 49:2859–2867. doi: 10.1128/JCM.00804-11. [DOI] [PMC free article] [PubMed] [Google Scholar]