LETTER
Enterobacter cloacae is an important nosocomial bacterium that is prevalent in hospital intensive care units (ICUs) (1–4). The presence of carbapenem-hydrolyzing beta-lactamases, such as Klebsiella pneumoniae carbapenemase (KPC) encoded by the blaKPC gene, underlies multidrug resistance in E. cloacae and other pathogens (5–7).
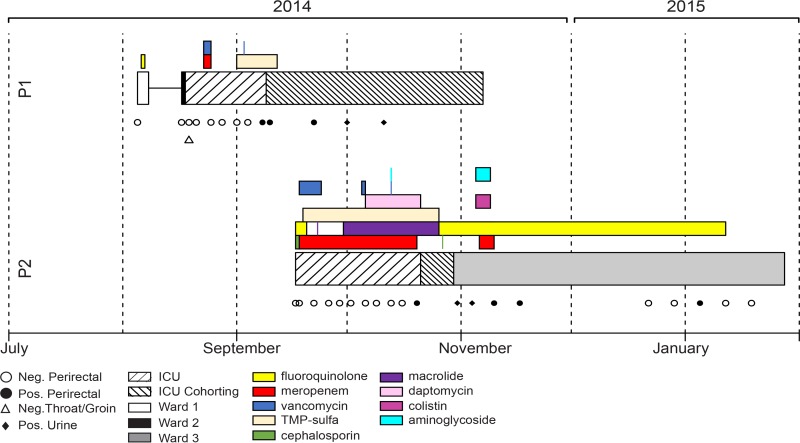
Active microbial surveillance at the NIH Clinical Center, which tests patients on admission and at regular intervals thereafter, detected multidrug-resistant E. cloacae colonization in two patients with overlapping stays in the ICU (Fig. 1). Both patients were undergoing treatment for a malignancy. Salient features of these two cases included multiple negative cultures at and after admission, administration of broad-spectrum antibiotics, and subsequent positive surveillance culture for blaKPC-positive E. cloacae leading to transfer to cohorted care (Fig. 1).
FIG 1.
Patient location, antibiotics received, and clinical culture results during the patients' hospital stay. The segment lengths indicate the duration of antibiotic receipt and stay in areas of the hospital or as an outpatient for each patient. Cultures are indicated as symbols below each patient trace (open symbols, negative; solid symbols, positive for blaKPC-positive E. cloacae). TMP-Sulfa, trimethoprim-sulfamethoxazole; P1, patient 1; P2, patient 2.
The timing and features of these two cases, along with shared carbapenem resistance, triggered an investigation. It was postulated that transmission of the E. cloacae strain from patient 1 to patient 2 might have occurred through a shared medical team. The patients did not share a room or nursing staff, and other potential sources of E. cloacae were negative, including: environmental samples (e.g., shared equipment, sinks, and surfaces) and perirectal swab cultures for all patients housed in the ICU in the weeks prior.
The isolates from patient 1 and 2 were both resistant to doripenem and ertapenem, but patient 2's isolate had increased resistance to imipenem and meropenem (see Table S1 in the supplemental material). Genomic sequencing, standard in our institute for all blaKPC-positive organisms (8), demonstrated that both isolates shared the blaKPC gene flanked by an IS26 element and the ISKpn6 tnpA gene, a previously observed genetic context (9). However, major differences between the two patient E. cloacae isolates were identified, ruling out direct transmission (Fig. 2). Patient 1's isolate is a sequence type 191 (ST191) E. cloacae strain with a 4.8-Mbp chromosome, a 50-kb ST6 IncN blaKPC-2 plasmid related to pKPC-47e, and additional 80- and 51-kb plasmids (lengths estimated from reference-based scaffolding) (10). The isolate from patient 1 is similar to E. cloacae ECNIH4, which was cultured in 2012 from a sink drain in our institution; however, sequence differences (including 35 nucleotide differences distributed along the chromosomes and insertion/deletions within the plasmids) help distinguish between these two isolates. Patient 2's isolate is an ST53 E. cloacae strain with a 5.1-Mbp chromosome, a 21-kb plasmid (pKPC-98f) carrying blaKPC-2, and additional plasmids of 79 and 144 kb.
FIG 2.
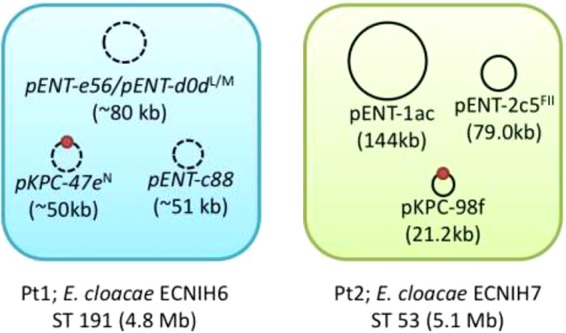
ECNIH6 and ECNIH7 carry three nonoverlapping plasmids and belong to different sequence types. The ECNIH6 genome is a shotgun assembly scaffolded on the closest reference (ECNIH4). Plasmid names and sizes are inferred from the reference; ordered and oriented contigs are represented by a dashed plasmid backbone. The blaKPC gene is marked as a red circle for both strains. Plasmid incompatibility groups (PlasmidFinder version 1.3) are indicated by superscripted letters.
In summary, genomic sequencing does not support nosocomial transmission of the blaKPC-positive E. cloacae strain from patient 1 to patient 2. Our epidemiologic and genomic results, including the absence of any matching isolates in a setting of extensive surveillance among a highly immunocompromised patient population, led us to postulate that these apparent acquisitions may rather represent rare transmission not detected by environmental surveillance or low-level gastrointestinal colonization on admission not detected by surveillance cultures. Administration of broad-spectrum antibiotics may have selected for the colonizing antimicrobial-resistant organisms, yielding a positive culture. These findings emphasize the importance of genomics for clarification of cases of suspected nosocomial transmission.
Accession number(s).
The whole-genome sequencing data can be retrieved at NCBI BioProject no. PRJNA279652 and PRJNA279659.
Availability of data.
Isolates can be obtained from K.M.F.; a material transfer agreement is necessary.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the National Human Genome Research Institute and NIH Clinical Center Intramural Research Programs.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00915-17.
REFERENCES
- 1.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 2.Jarvis WR, Martone WJ. 1992. Predominant pathogens in hospital infections. J Antimicrob Chemother 29(Suppl A):19–24. [DOI] [PubMed] [Google Scholar]
- 3.Vincent J-L, Rello J, Marshall J, Silva E, Anzueto A, Martin CD, Moreno R, Lipman J, Gomersall C, Sakr Y, Reinhart K, EPIC II Group of Investigators. 2009. International study of the prevalence and outcomes of infection in intensive care units. JAMA 302:2323–2329. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
- 4.Tabah A, Koulenti D, Laupland K, Misset B, Valles J, Bruzzi de Carvalho F, Paiva JA, Cakar N, Ma X, Eggimann P, Antonelli M, Bonten MJM, Csomos A, Krueger WA, Mikstacki A, Lipman J, Depuydt P, Vesin A, Garrouste-Orgeas M, Zahar J-R, Blot S, Carlet J, Brun-Buisson C, Martin C, Rello J, Dimopoulos G, Timsit J-F. 2012. Characteristics and determinants of outcome of hospital-acquired bloodstream infections in intensive care units: the EUROBACT International Cohort Study. Intensive Care Med 38:1930–1945. doi: 10.1007/s00134-012-2695-9. [DOI] [PubMed] [Google Scholar]
- 5.Hossain A, Ferraro MJ, Pino RM, Dew RB, Moland ES, Lockhart TJ, Thomson KS, Goering RV, Hanson ND. 2004. Plasmid-mediated carbapenem-hydrolyzing enzyme KPC-2 in an Enterobacter sp. Antimicrob Agents Chemother 48:4438–4440. doi: 10.1128/AAC.48.11.4438-4440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nordmann P, Poirel L. 2002. Emerging carbapenemases in Gram-negative aerobes. Clin Microbiol Infect 8:321–331. doi: 10.1046/j.1469-0691.2002.00401.x. [DOI] [PubMed] [Google Scholar]
- 7.Walsh TR, Toleman MA, Poirel L, Nordmann P. 2005. Metallo-beta-lactamases: the quiet before the storm? Clin Microbiol Rev 18:306–325. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dotson GA, NISC Comparative Sequencing Program, Dekker JP, Palmore TN, Segre JA, Conlan S. 2016. Draft genome sequence of a Klebsiella pneumoniae carbapenemase-positive sequence type 111 Pseudomonas aeruginosa strain. Genome Announc 4:e01663-15. doi: 10.1128/genomeA.01663-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conlan S, Thomas PJ, Deming C, Park M, Lau AF, Dekker JP, Snitkin ES, Clark TA, Luong K, Song Y, Tsai Y-C, Boitano M, Dayal J, Brooks SY, Schmidt B, Young AC, Thomas JW, Bouffard GG, Blakesley RW, NISC Comparative Sequencing Program, Mullikin JC, Korlach J, Henderson DK, Frank KM, Palmore TN, Segre JA. 2014. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci Transl Med 6:254ra126. doi: 10.1126/scitranslmed.3009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Assefa S, Keane T, Otto T, Newbold C, Berriman M. 2009. ABACAS: algorithm-based automatic contiguation of assembled sequences. Bioinformatics 25:1968–1969. doi: 10.1093/bioinformatics/btp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.