ABSTRACT
The metabolism of methane is an important part of the biogeochemical cycling of carbon. Methane is also a major contributor to climate change. A specialized group of microbes that consume methane, the methanotrophs, represent a natural filter preventing even faster accumulation of methane in the atmosphere. Methanotrophy can proceed via both anaerobic and aerobic modes. The anaerobic methanotrophs, represented by both archaea and bacteria, all appear to be engaged in syntrophic interdependencies with other species, to overcome the energetic barriers of methane metabolism in the absence of oxygen. In contrast, aerobic methanotrophy can be carried out by pure cultures of bacteria. However, a concept of communal function in aerobic methane oxidation has been gaining momentum, based on data regarding the natural cooccurrence of specific functional guilds and results from laboratory manipulations. The mechanistic details of how and why the methanotrophs share their carbon with other species, and whether and what they gain in return, are still sparse. In this minireview, we highlight recent studies that led to this new concept of community function in aerobic methane oxidation. We first describe stable isotope probing experiments employing heavy-carbon-labeled methane and tracing methane carbon consumption. We then present an analysis of data on microcosm community dynamics. We further discuss the role of a synthetic community approach in elucidating the principles of carbon flow and species cooperation in methane consumption. Finally, we touch on the role of lanthanides, which are rare Earth elements previously thought to be biologically inert, in bacterial metabolism of methane.
KEYWORDS: community function, methane metabolism, Methylococcaceae, Methylophilaceae, rare Earth element switch
INTRODUCTION
Methane is produced both abiogenically, through processes in the Earth's crust (1, 2), and biogenically, through microbial methanogenesis (3), and its metabolism is a significant part of the global carbon cycle. Therefore, microbes that consume methane, the methanotrophs, play a major role in methane emission mitigation (4). Methane oxidation takes place in both oxic and anoxic environments, the former carried out by aerobic methanotrophs that are bacteria (5, 6) and the latter carried out by anaerobic methanotrophs that are archaea (i.e., anaerobic methane-oxidizing [ANME] archaea) (7, 8) or bacteria (9). Aerobic methanotrophy was first discovered in the early twentieth century (10, 11), and the physiology and biochemistry of this mode of methanotrophy have been characterized in pure cultures of methanotrophs belonging to Alphaproteobacteria and Gammaproteobacteria and, more recently, Verrucomicrobia (6) (Fig. 1A). The unique biochemistry employed by these microbes involves enzymes for oxygen-dependent methane oxidation, i.e., particulate and/or soluble methane monooxygenases, converting methane into methanol (12). Methanol is further oxidized to formaldehyde, formate, and CO2 by the respective enzymes and pathways, while biomass is built from formaldehyde, formate, CO2, or a combination (6). Specific genes involved in each of these functions, in combination, represent genetic signatures of aerobic methanotrophs (13).
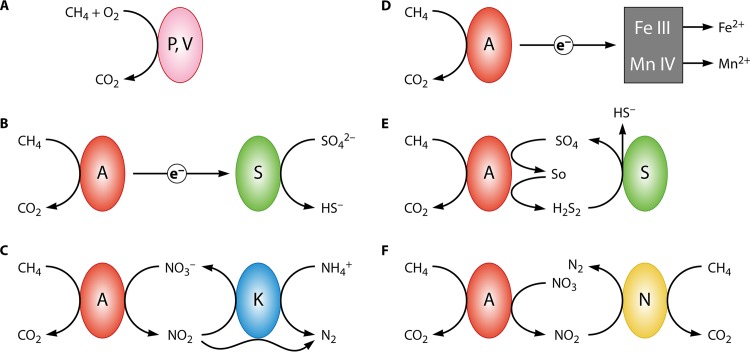
FIG 1.
Different modes of methanotrophy and different types of syntrophy in methanotrophy. (A) Aerobic methanotrophy. Aerobic methanotrophs require oxygen and can grow as pure cultures; in nature, however, they appear to form consortia with other bacteria. P, Proteobacteria; V, Verrucomicrobia. (B) AOM by ANME archaea (A) linked to sulfate reduction by sulfate-reducing bacteria (S), involving extracellular electron transfer. (C) AOM by ANME archaea linked to anaerobic ammonium oxidation by Kuenenia (K). (D) AOM by ANME archaea via direct electron transfer to insoluble metals. (E) AOM by ANME archaea directly coupled to sulfate reduction. (F) AOM by ANME archaea linked to denitrification, in which NC10 bacteria (N) assist with nitrite removal, linking its reduction to oxygen-dependent methane oxidation. The source of oxygen for the latter is unclear.
The biochemistry of anaerobic oxidation of methane (AOM) by archaea is very different and involves the same enzymes and cofactors that are involved in methanogenesis, acting in reverse (14, 15). Because the energetics of this process are severely constrained, the process can take place only through syntrophic cooperation, involving interspecies electron transfer or other interdependencies (16). Accordingly, ANME organisms have never been obtained in pure cultures (7, 8). Syntrophy in AOM may involve sulfate-reducing bacteria (SRB) as partners, based on the well-documented cooccurrence and tight physical association of ANME archaea and SRB (17–19) (Fig. 1B). AOM may also be linked to denitrification (20, 21). This metabolism was proposed to involve a different syntrophic partner, the anaerobic ammonium-oxidizing bacteria of the genus Kuenenia (Planctomycetes) (20) (Fig. 1C). Insoluble iron(III) and manganese(IV) were also proposed as electron acceptors for AOM, and these processes may take place via extracellular metal reduction (22) (Fig. 1D). Direct coupling of AOM to sulfate reduction by ANME archaea, not requiring SRB partners, was also proposed (23) (Fig. 1E).
Bacterial AOM linked to denitrification and carried out by species of the NC10 phylum has also been described; these species appear to be syntrophic, as they have never been obtained in pure cultures (9, 24). Indeed, NC10 bacteria are often enriched, along with ANME archaea, in denitrifying reactors (20, 24) (Fig. 1F). However, NC10 bacteria do not use the reverse methanogenesis pathway. Instead, they use the pathway in which methane is activated by oxygen and converted to CO2, which is a typical aerobic methanotrophy pathway (9). The source of oxygen for methane activation in such a metabolic scheme is unclear, but the oxygen was proposed to originate from a dismutation reaction (9); this proposition is still awaiting biochemical demonstration.
While syntrophy in anaerobic methanotrophy has been well documented and is supported by the existence of the energetic constraints or other interdependencies described above, a concept of syntrophic behavior by aerobic bacterial methanotrophs has also existed for a while, based on observations of the cooccurrence of methanotrophs and nonmethanotrophs in a variety of natural and experimental settings (25, 26). In this minireview, we describe a series of experiments that support community functions in aerobic methane oxidation, identify specific syntrophic partnerships, and present some mechanisms for carbon transfer. We outline the potential of synthetic communities as models for studying syntrophic interactions, and we discuss challenges in interpreting the behavior of such communities in the laboratory, with relevance to mechanisms and principles underlying metabolic synergy among different microbial guilds in nature. We also address the potential role of lanthanides in shaping methane-utilizing communities, both in the laboratory and in nature.
AEROBIC METHANOTROPHS APPEAR TO SHARE CARBON WITH OTHER SPECIES
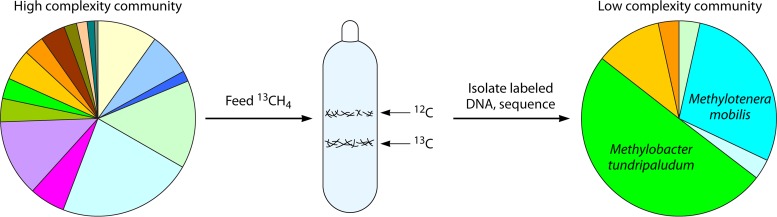
Aerobic methanotrophs have been studied as pure cultures for over 100 years, since they were first discovered (10, 11). However, it has been noted that some methanotrophs are difficult to isolate in pure cultures due to the persistent presence of contaminating organisms (25, 27), suggesting that methanotrophs tend to release organic carbon and feed their satellites (26, 28). The carbon released has been proposed to be free-for-all “public goods” (26, 28). Experimental evidence for the nature of the satellite communities consuming methane-derived carbon has been gained through the stable isotope probing (SIP) approach, a method that follows the fate of carbon by analyzing fractions of DNA labeled with 13C (29). A combination of SIP with 16S rRNA and functional gene profiling revealed that, in addition to bona fide methanotrophs, microbial guilds known for methanol utilization (alphaproteobacterial and betaproteobacterial methylotrophs), as well as guilds not known for methylotrophy, were feeding on carbon from methane (30, 31). However, the biochemistry of this process and the specific metabolic pathways involved remained unknown. Finer details of communities consuming methane, including genomic contents, were obtained through high-resolution metagenomics (a combination of SIP and metagenomic sequencing) applied to a lake sediment community (32). Sequencing of the DNA fraction labeled with heavy methane revealed two major consumers of the labeled carbon, namely, gammaproteobacterial methanotrophs of the family Methylococcaceae (mainly Methylobacter) and betaproteobacterial nonmethanotrophic methylotrophs of the family Methylophilaceae (mainly Methylotenera) (Fig. 2). Whole-genome metabolic reconstruction revealed that Methylobacter was capable of methane oxidation, while Methylotenera lacked methane oxidation genes. Both organisms possessed genes for denitrification, suggesting one mechanism for adaptation to hypoxic niches, such as methane/oxygen countergradients existing in lake sediments (33, 34). A follow-up metagenomic study that investigated the same methane-consuming sediment communities under conditions of oxygen and nitrate additions confirmed the correlation between the abundances of the populations of Methylococcaceae and Methylophilaceae. These data suggested not only that Methylophilaceae can obtain carbon from methane but also that there may be selective pressure for these organisms to work in unison with the methanotrophs (35). Supporting this observation was a study that employed Arctic permafrost samples, in which Methylophilaceae were found to be colabeled with Methylobacter by heavy methane in DNA-SIP experiments (36). A separate study that analyzed in situ transcriptomes in melting permafrost samples found that Methylobacter and Methylotenera cooccurred in all samples and both species were transcriptionally active (37).
FIG 2.
Schematic of an experimental design combining stable isotope probing with whole-genome shotgun sequencing. While Methylotenera cannot oxidize methane, it accumulates significant amounts of label through cross-feeding (32).
In marine environments, partnerships in methanotrophy appear to include additional actors. A bloom in methanotroph populations was observed in response to the natural gas spill during the Deepwater Horizon disaster of 2010, dominated by Gammaproteobacteria with a significant proportion of Methylococcaceae, and their abundances were correlated positively with the concentrations of hydrocarbons and negatively with the dissolved oxygen concentrations (38, 39). The nonmethanotrophic methylotroph populations that were part of these blooms were represented by Methylophaga and Methylophilaceae species, while nonmethylotrophic actors were represented by Flavobacteriaceae (38–40). Similar communities are typically found in microbial mats surrounding methane slips, which have constant supplies of methane (41, 42).
METHANE-FED MICROCOSM DYNAMICS SUGGEST THAT SYNTROPHY IN AEROBIC METHANE OXIDATION MAY BE SPECIES SPECIFIC
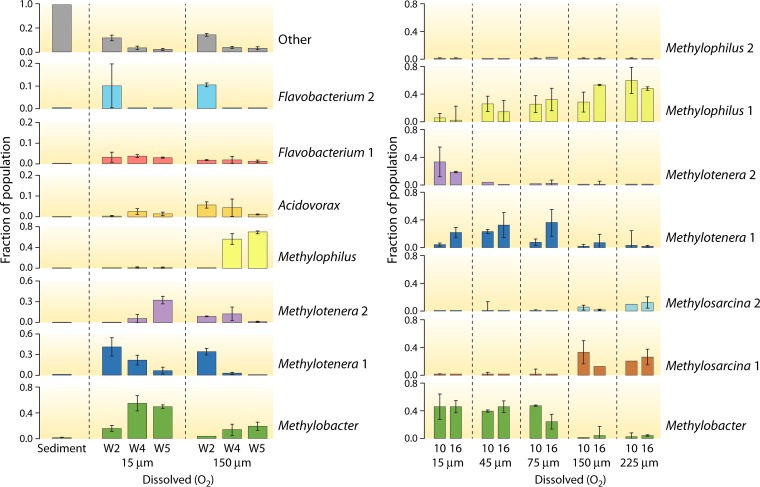
One way to support observations from the SIP experiments and the cooccurrence of species in natural environments, all hinting at a communal function in methane oxidation, is to test cooperative behavior in microcosm experiments. Such experiments were carried out with Lake Washington sediment samples, which were incubated under semi-in situ conditions under an atmosphere of methane, with weekly dilutions and transfers. This method allows competitive species to persist over time, while noncompetitive species are diluted out of the microcosm (43). Two oxygen-partial-pressure regimens were implemented, i.e., high and low, to mimic oxygen concentrations along the oxygen gradient in a natural lake sediment (33, 34). A very rapid decline in community complexity was observed after just several dilutions, with the two most competitive species being Methylococcaceae and Methylophilaceae in each microcosm (43). However, different community dynamics were observed between the low- and high-oxygen regimens, with the former selecting for Methylophilaceae of the Methylotenera type, including dynamics among different Methylotenera ecotypes (Fig. 3), and the latter selecting for Methylophilus types (43). In a separate series of experiments (44), a wider range of oxygen concentrations was used, with more frequent oxygen additions, and communities were profiled at 10 and 16 weeks, at which point they were assumed to be stable (Fig. 3). Among the Methylophilaceae, again Methylotenera types prevailed under low-oxygen conditions while Methylophilus types prevailed under high-oxygen conditions. In these experiments, however, an additional methanotroph actor, Methylosarcina, was observed and appeared to outcompete Methylobacter under high-oxygen conditions. In these microcosms, while Methylococcaceae and Methylophilaceae were the most abundant species, some low-abundance species also appeared nonrandom, as they were identified in each microcosm; these species were Bacteroidetes, mostly represented by Flavobacterium, and Burkholderiales, mostly represented by Acidovorax (43, 44). Independent time-series microcosm experiments were carried out more recently, followed by large-scale metagenomic sequencing, which uncovered very similar trends in community dynamics, with Methylobacter-Methylotenera partnerships persisting under low-oxygen conditions and Methylosarcina-Methylophilus partnerships persisting under high-oxygen conditions and Bacteroidetes and Burkholderiales species being present in each microcosm (M. E. Hernandez and L. Chistoserdova, unpublished data).
FIG 3.
Community dynamics in laboratory microcosms incubated under methane, with transfers and dilutions to identify species that are most competitive in communal methane consumption. The duration of experiments is shown at the bottom, in weeks (W). Sediment, sediment community at time zero. See references 43 (left) and 44 (right).
A similar experimental design was used to investigate methane-consuming communities in a landfill cover soil, at three different partial pressures of oxygen (45). In these experiments, communities also simplified rapidly to select for Methylocystis (alphaproteobacterial methanotrophs) and Methylophilaceae, with minor amounts of other bacteria, including Bacteroidetes. Overall, it was concluded that oxygen limitation promoted greater carbon transfer to the satellite community and resulted in greater community diversity (45). The results of these experiments strongly suggest that partner selection may be both species specific and environment specific. However, these findings disagree with the mechanism of public goods leaked by the methanotrophs, because, in the diverse multispecies communities present in natural samples, specific functional guilds (such as Methylophilaceae) appear to be most successful in consuming these goods.
WHAT ARE WE LEARNING FROM MANIPULATING SYNTHETIC COMMUNITIES?
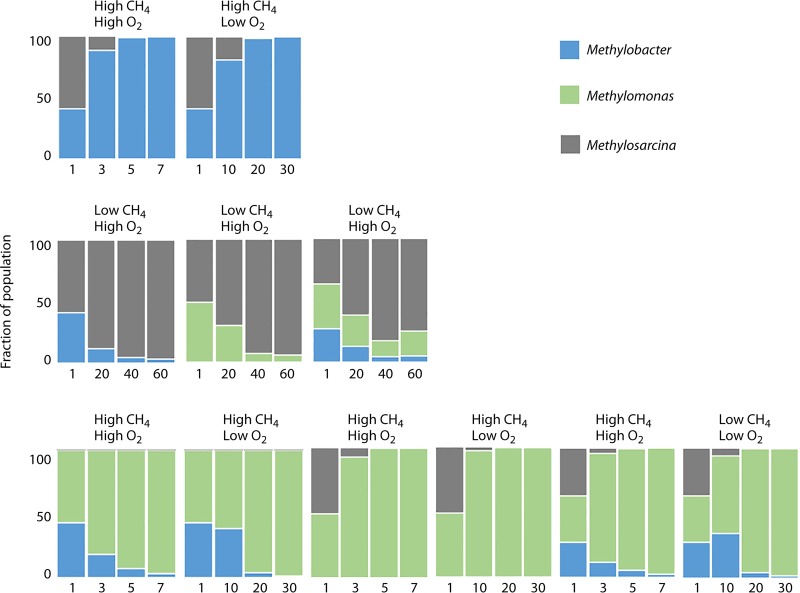
While important insights have been gained by investigating natural methane-oxidizing communities, in situ or in the laboratory, such communities tend to display significant variations between biological replicates (43, 44) (Hernandez and Chistoserdova, unpublished), likely due to the complexity of the communities, with multiple species being involved in competitions within and between different functional guilds; they are also likely affected by the activities of predatory species, phages, and other factors that cannot be controlled for in “wild” experimental settings. One approach to overcoming these obstacles is manipulation of simplified synthetic communities, whose composition is predetermined by the knowledge gained in experiments with natural communities but is limited to a select set of strains, with characterized genomes and physiologies (46). Synthetic community experiments are also a prominent approach for hypothesis testing. For example, the data on cooccurrence of specific functional guilds in the environment and selection for specific functional guilds in methane-fed microcosms contradict the public goods hypothesis (26, 28). To date, two types of synthetic community experiments have been carried out to test this hypothesis. The first type employed a collection of pure culture isolates originating from the same ecological niche, including species that revealed the most competitive behavior in natural microcosms, as described above, and the isolates were incubated with transfers and dilutions designed to select for the most competitive species (47). The second type employed randomly combined species originating from nonrelated environmental niches, and the species were not subjected to competition (48, 49). The outcomes of these experiments were very different. Yu and colleagues (47) employed communities built of 50 pure cultures isolated from Lake Washington sediment (10 methanotrophs and 40 nonmethanotrophs) and incubated them under different regimens with transfers and dilutions, following community dynamics via iTag profiling. They observed that general trends in synthetic community dynamics were similar to trends in natural communities (43, 44). As observed for wild communities, representatives of Methylococcaceae and Methylophilaceae dominated the synthetic communities, while the relative abundances of other methylotrophs steadily declined. As in natural microcosms, Bacteroidetes and Burkholderiales remained relatively abundant over time. However, species dynamics among different methanotrophs were different in synthetic communities. While Methylobacter and Methylosarcina were repeatedly observed in natural communities, Methylomonas species appeared to be most competitive in synthetic communities under most regimens, in accord with their performance and fitness as pure cultures (34). To confirm these results, representatives of Methylomonas, Methylobacter, and Methylosarcina were competed against each other in two- or three-species microcosms; Methylomonas revealed competitive fitness under most regimens, with the exception of a low-methane regimen, under which Methylosarcina prevailed (Fig. 4). These experiments demonstrated certain discrepancies between wild and synthetic microcosm behavior. However, the general trends reminded similar, with Methylococcaceae and Methylophilaceae being the most competitive species, supporting data from DNA SIP experiments as well as data from naturally cooccurring species.
FIG 4.
Dynamics among three methanotroph species in simple synthetic communities, under different conditions. The duration of experiments is shown at the bottom, in days. Based on data from reference 47.
In contrast, Ho and colleagues concluded that interactions among species were not necessarily exclusive and that any randomly selected partner could enhance methanotroph activity, as long as more than one partner was present (“the more the merrier”) (48). In that particular study, growth of the methanotroph did not appear to be stimulated by the partner species, as no changes in cell counts were observed; this observation contradicted a prior report from the same group, in which the growth of methanotrophs was stimulated up to 4-fold by some randomly selected partners (49). These discrepancies highlight the need for further studies, including synthetic community manipulation, toward understanding the patterns and details of syntrophic interactions in aerobic methanotrophy.
THE RARE EARTH ELEMENT SWITCH AND ITS ROLE IN COMMUNITY FUNCTION
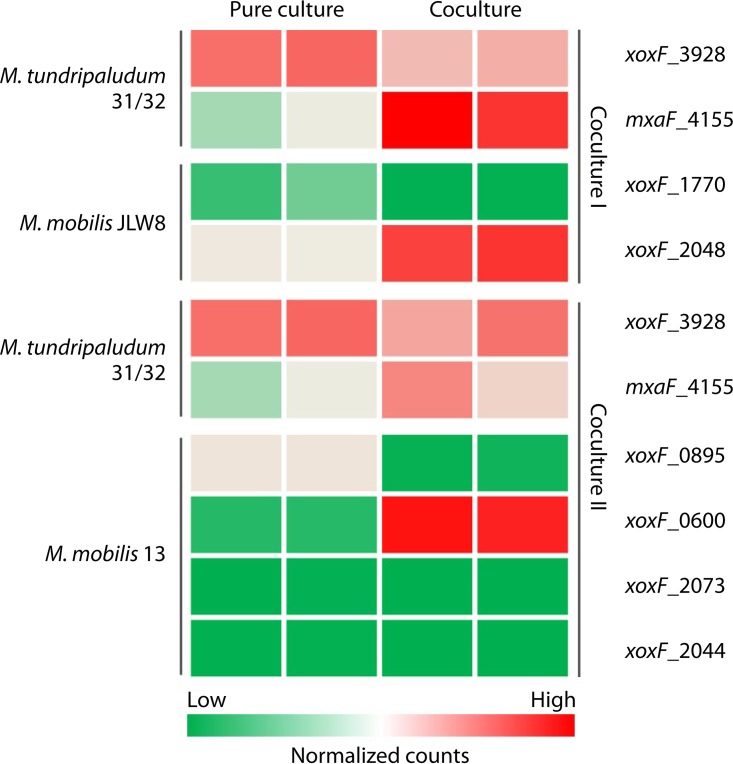
One of the most surprising and potentially very important recent discoveries in the biochemistry of aerobic methane oxidation has been the discovery of the so-called rare Earth element (REE) switch (50–52), a regulatory mechanism that determines the switch between two alternative methanol dehydrogenases (MDHs), i.e., the calcium-dependent enzyme (MxaFI) and the REE-dependent enzyme (XoxF). In itself, demonstration of a biochemical function for REEs is very important, as it overturns the long-held dogma of biological inertia of REEs (53, 54). With respect to methanotrophy and more broadly methylotrophy, this discovery points to the gaps in our understanding of these processes, especially given the fact that expression of the two enzymes is inversely regulated (50–54). What is the significance of the coexistence of these two types of enzymes in a single organism, and what is the physiological significance of the REE switch? In pure cultures of methylotrophs, as so far reported, the switch acts in a straightforward fashion, i.e., when REEs are present, transcription of MxaFI genes goes down and transcription of the XoxF gene(s) goes up (50–52). However, a recent investigation involving a simple, two-species, synthetic community model revealed that the REE switch may function differently in communities versus pure cultures (55). In those experiments, Methylobacter was employed as the methanotroph model and was cocultured separately with two different strains of Methylophilaceae (Methylotenera mobilis JLW8 and Methylotenera mobilis 13) as nonmethanotroph satellite models, and all (co)cultures were supplemented with REE lanthanum. Transcriptomes were sequenced for the cocultures and for each pure culture. From transcriptome analysis, some of the most differentially expressed genes were the genes for alternative MDH enzymes in both Methylobacter and Methylotenera. While xoxF, the gene for the REE-dependent MDH, was expressed at the highest level in pure cultures of Methylobacter, genes for MxaFI, the calcium-dependent MDH, were most highly expressed in cocultures (55) (Fig. 5). In M. mobilis JLW8, which does not encode MxaFI but encodes two different XoxF enzymes, the gene for one (xoxF_1770) was more highly expressed in pure cultures, while the gene for the other (xoxF_2048) was overexpressed in cocultures. In M. mobilis 13, a gene encoding one of the multiple REE-dependent MDH enzymes (xoxF_0600) was overexpressed in cocultures and another gene (xoxF_0895) was overexpressed in pure cultures, while the third xoxF gene (xoxF_2044) and the mxaFI genes were expressed at low levels (55) (Fig. 5). These results suggest that the REE switch acts differently depending on whether an organism is engaged in community functions or is growing as a pure culture. These results also demonstrate that the REE switch acts differently on different xoxF genes when multiple copies are present in the same genome, suggesting a more complex nature for the REE switch than previously appreciated. While the precise roles of (multiple) alternative MDHs in each organism require further investigation, these data clearly point to the importance of methanol metabolism and suggest that methanol must be one of the metabolites that feeds the Methylophilaceae. In support of this finding, a double-knockout mutant of M. mobilis JLW8 that does not produce any functional MDH enzymes and thus is incapable of metabolizing methanol was tested (56). This mutant was not able to establish cocultures with Methylobacter, further supporting methanol as one metabolite shared between Methylococcaceae and Methylophilaceae (55). However, yet more complex biochemistry may be governed by the REE switch, as other REE-dependent enzymes may participate in communal metabolism of methane. Recently, an enzyme (ExaF) only distantly related to either MxaF or XoxF has been described as an REE enzyme, showing broad substrate specificity toward alcohols and aldehydes (57). While homologs of this enzyme are not present in the genomes of the methanotrophs or the Methylophilaceae, they are present in the genomes of some of the organisms representing minor populations in the communities (47) (Z. Yu and L. Chistoserdova, unpublished data). These enzymes may play important roles in primary substrate consumption, competition for REEs, or both, thus presenting another mechanism for community member interdependency.
FIG 5.
Heatmap depicting differential expression of genes for alternative methanol dehydrogenases (xoxF and mxaF) in pure cultures versus cocultures of Methylobacter tundripaludum and Methylotenera mobilis, demonstrating that communal living triggers the rare Earth element switch (55).
CONCLUSIONS AND FUTURE PERSPECTIVES
Methane oxidation as a community function is still a novel concept. In this way, methanotrophs appear to act as “primary producers,” akin to photoautotrophs, which also tend to support diverse communities by releasing carbon as a form of public goods (58, 59) but appear to select for specific partners (60, 61). A recent study that explored species interdependencies in a cyanobacterial consortium, both in the laboratory and in situ, applied the black queen hypothesis (BQH) to explain nonrandom distribution of the public goods in such communities, identifying vitamin B12 as one of the metabolites exchanged between species (61). In accordance with the BQH, species can greatly benefit from a loss of function as long as this function can be fulfilled by helper species, altruistically or in exchange for other goods (62). Vitamin B12 exchange has been previously implicated in maintaining stable cocultures of methanotrophs and nonmethanotrophs (63). However, this does not appear to be the mechanism that selects for Methylobacter-Methylotenera or Methylosarcina-Methylophilus partnerships, as all of these species are capable of growth in minimal media (64, 65) and no support for B12 dependency emerged from comparative transcriptomic analysis (55). However, methanol has been identified as a carbon compound shared between Methylococcaceae and Methylophilaceae, and its metabolism appears to be subject to the REE switch (55). Other metabolites released by the methanotrophs likely include acetate, a product of methane fermentation (28) that feeds the Acidovorax and other Burkholderiales, and polymeric substances that are released by both Methylococcaceae and Methylophilaceae and feed Bacteroidetes. Whether and what the methanotrophs may be gaining in return remains unclear. Interspecies electron transfer may be taking place, contributing to methane activation, but such a mechanism would need experimental validation. Neither Methylococcaceae nor Methylophilaceae encode any multiheme cytochromes that are implicated in such transfers (66, 67). However, such cytochromes were previously characterized from the thermophilic methanotroph Methylococcus capsulatus (68). Both Methylobacter and Methylophilaceae encode and highly express pilus functions (69, 70) (Yu and Chistoserdova, unpublished), which are also implicated in interspecies electron transfer (67, 71). To further investigate the mechanistic details of syntrophic interactions in aerobic methane oxidation, the synthetic community manipulation approach bears promise. However, this approach faces serious methodological challenges. Because most of the microbes involved can survive and thrive on their own as pure cultures (based on their behavior in the laboratory), the choice of experimental conditions (e.g., medium composition, partial pressures of methane and oxygen, and nitrogen sources) conducive to cooperative behavior must be important. Other factors also may be important in controlling species ratios in natural communities, including functional guilds not primarily involved in methane utilization or cometabolism but sporadically influencing community structure, such as predatory species, species harboring predatory plasmids or phages, or free-living phages. Overall, there may be tradeoffs between manipulation of natural communities versus synthetic communities, with the outcomes, in terms of the community structure or the activity of individual species, being somewhat different, as observed in the experiments described above. Ultimately, while some fundamental questions could be addressed through manipulation of very simple synthetic communities, an understanding of the finer details of interspecies interactions might require experiments with communities more precisely reflecting the activities of natural communities. To achieve that goal, synthetic communities should probably be modeled to better reflect natural communities, as observed from manipulation of microcosms initiated with native environmental samples, and manipulations should ideally recreate conditions approximating natural conditions, as much as reasonably possible in laboratory settings. We imagine that an intelligent community design, as opposed to a random selection of species, can be achieved as long as a selection of appropriate model organisms is available, representing different functional guilds. The dynamics of these rationally designed communities can be monitored in real time, using, for example, flow cytometry to distinguish between different cell sizes and shapes (Fig. 6). Each species' signature features, such as differential gene transcription (or protein expression or metabolite production), could then be determined to gain insights into which functions may be important for cooperative behavior, as has already been revealed for the alternative MDH enzymes and the REE switch. Additional points of cross talk between the community partners could then be elucidated via knockout mutant manipulation, which can be carried out on a large scale with high throughput. Such approaches will be most powerful when combined with analyses of purified protein activity levels for select targets identified through omics, as part of community function analysis.
FIG 6.
Schematic of a rationally designed community experiment in which differences in cell size and shape allow real-time monitoring of species dynamics.
ACKNOWLEDGMENTS
This material is based on work supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, under award DE-SC-0016224.
We declare no conflicts of interest.
REFERENCES
- 1.Sherwood Lollar B, Westgate TD, Ward JA, Slater GF, Lacrampe-Couloume G. 2002. Abiogenic formation of alkanes in the Earth's crust as a minor source for global hydrocarbon reservoirs. Nature 416:522–524. doi: 10.1038/416522a. [DOI] [PubMed] [Google Scholar]
- 2.Sherwood Lollar B, Lacrampe-Couloume G, Slater GF, Ward J, Moser DP, Gihring TM, Lin L-H, Onstott TC. 2006. Unravelling abiogenic and biogenic sources of methane in the Earth's deep subsurface. Chem Geol 226:328–339. doi: 10.1016/j.chemgeo.2005.09.027. [DOI] [Google Scholar]
- 3.Singh BK, Bardgett RD, Smith P, Reay DS. 2010. Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat Rev Microbiol 8:779–790. doi: 10.1038/nrmicro2439. [DOI] [PubMed] [Google Scholar]
- 4.Conrad R. 2009. The global methane cycle: recent advances in understanding the microbial processes involved. Environ Microbiol Rep 1:285–292. doi: 10.1111/j.1758-2229.2009.00038.x. [DOI] [PubMed] [Google Scholar]
- 5.Trotsenko YA, Murrell JC. 2008. Metabolic aspects of aerobic obligate methanotrophy. Adv Appl Microbiol 6:183–229. doi: 10.1016/S0065-2164(07)00005-6. [DOI] [PubMed] [Google Scholar]
- 6.Chistoserdova L, Lidstrom ME. 2013. Aerobic methylotrophic prokaryotes, p 267–285. In Rosenberg E, DeLong EF, Thompson F, Lory S, Stackebrandt E (ed), The prokaryotes, 4th ed Springer-Verlag, Berlin, Germany. [Google Scholar]
- 7.Knittel K, Boetius A. 2009. Anaerobic oxidation of methane: progress with an unknown process. Annu Rev Microbiol 63:311–334. doi: 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- 8.Offre P, Spang A, Schleper C. 2013. Archaea in biogeochemical cycles. Annu Rev Microbiol 67:437–457. doi: 10.1146/annurev-micro-092412-155614. [DOI] [PubMed] [Google Scholar]
- 9.Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJ, van Alen T, Luesken F, Wu ML, van de Pas-Schoonen KT, Op den Camp HJ, Janssen-Megens EM, Francoijs KJ, Stunnenberg H, Weissenbach J, Jetten MS, Strous M. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 10.Söhngen NL. 1906. Über Bakterien, welche Methan als Kohlenstoffnahrung und Energiequelle gebrauchen. Zentrabl Bakteriol Parasitenk Infektionskr 15:513–517. [Google Scholar]
- 11.Kaserer H. 1906. Uber die Oxydation des Wasserstoffs und des Methans durch Mikroorganismen. Zentrabl Bakteriol Parasitenk Infektionskr 15:573–576. [Google Scholar]
- 12.Sirajuddin S, Rosenzweig AC. 2015. Enzymatic oxidation of methane. Biochemistry 54:2283–2294. doi: 10.1021/acs.biochem.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chistoserdova L. 2011. Modularity of methylotrophy, revisited. Environ Microbiol 13:2603–2622. doi: 10.1111/j.1462-2920.2011.02464.x. [DOI] [PubMed] [Google Scholar]
- 14.Hallam SJ, Putnam N, Preston CM, Detter JC, Rokhsar D, Richardson PM, DeLong EF. 2004. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305:1457–1462. doi: 10.1126/science.1100025. [DOI] [PubMed] [Google Scholar]
- 15.Meyerdierks A, Kube M, Kostadinov I, Teeling H, Glöckner FO, Reinhardt R, Amann R. 2010. Metagenome and mRNA expression analyses of anaerobic methanotrophic archaea of the ANME-1 group. Environ Microbiol 12:422–439. doi: 10.1111/j.1462-2920.2009.02083.x. [DOI] [PubMed] [Google Scholar]
- 16.Thauer RK. 2011. Anaerobic oxidation of methane with sulfate: on the reversibility of the reactions that are catalyzed by enzymes also involved in methanogenesis from CO2. Curr Opin Microbiol 14:292–299. doi: 10.1016/j.mib.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Hinrichs K-U, Hayes JM, Sylva SP, Brewer PG, DeLong EF. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802–805. doi: 10.1038/19751. [DOI] [PubMed] [Google Scholar]
- 18.Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, Amann R, Jørgensen BB, Witte U, Pfannkuche O. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623–626. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- 19.Orphan VJ, House CH, Hinrichs KU, McKeegan KD, DeLong EF. 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484–487. doi: 10.1126/science.1061338. [DOI] [PubMed] [Google Scholar]
- 20.Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, Yuan Z, Tyson GW. 2013. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500:567–570. doi: 10.1038/nature12375 (Erratum, 501:578, 2013.) [DOI] [PubMed] [Google Scholar]
- 21.Arshad A, Speth DR, de Graaf RM, Op den Camp HJ, Jetten MS, Welte CU. 2015. A metagenomics-based metabolic model of nitrate-dependent anaerobic oxidation of methane by Methanoperedens-like archaea. Front Microbiol 6:1423. doi: 10.3389/fmicb.2015.01423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beal EJ, House CH, Orphan VJ. 2009. Manganese- and iron-dependent marine methane oxidation. Science 325:184–187. doi: 10.1126/science.1169984. [DOI] [PubMed] [Google Scholar]
- 23.Milucka J, Ferdelman TG, Polerecky L, Franzke D, Wegener G, Schmid M, Lieberwirth I, Wagner M, Widdel M, Kuypers MM. 2012. Zero-valent sulphur is a key intermediate in marine methane oxidation. Nature 491:541–546. doi: 10.1038/nature11656. [DOI] [PubMed] [Google Scholar]
- 24.Raghoebarsing AA, Pol A, van de Pas-Schoonen KT, Smolders AJ, Ettwig KF, Rijpstra WI, Schouten S, Damsté JS, Op den Camp HJ, Jetten MS, Strous M. 2006. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918–921. doi: 10.1038/nature04617. [DOI] [PubMed] [Google Scholar]
- 25.Hanson RS, Hanson TE. 1996. Methanotrophic bacteria. Microbiol Rev 60:439–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Modin O, Fukushi K, Yamamoto K. 2007. Denitrification with methane as external carbon source. Water Res 41:2726–2738. doi: 10.1016/j.watres.2007.02.053. [DOI] [PubMed] [Google Scholar]
- 27.Dedysh SN, Dunfield PF. 2014. Cultivation of methanotrophs, p 231–247. In McGenity TJ, Timmis K, Nogales N (ed), Hydrocarbon and lipid microbiology protocols: isolation and cultivation. Springer, Berlin, Germany. [Google Scholar]
- 28.Kalyuzhnaya MG, Yang S, Rozova ON, Smalley NE, Clubb J, Lamb A, Gowda GA, Raftery D, Fu Y, Bringel F, Vuilleumier S, Beck DA, Trotsenko YA, Khmelenina VN, Lidstrom ME. 2013. Highly efficient methane biocatalysis revealed in a methanotrophic bacterium. Nat Commun 4:2785. doi: 10.1038/ncomms3785. [DOI] [PubMed] [Google Scholar]
- 29.Radajewski S, Ineson P, Parekh NR, Murrell JC. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646–649. doi: 10.1038/35001054. [DOI] [PubMed] [Google Scholar]
- 30.Radajewski S, Webster G, Reay DS, Morris SA, Ineson P, Nedwell DB, Prosser JI, Murrell JC. 2002. Identification of active methylotroph populations in an acidic forest soil by stable-isotope probing. Microbiology 148:2331–2342. doi: 10.1099/00221287-148-8-2331. [DOI] [PubMed] [Google Scholar]
- 31.Hutchens E, Radajewski S, Dumont MG, McDonald IR, Murrell JC. 2004. Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ Microbiol 6:111–120. doi: 10.1046/j.1462-2920.2003.00543.x. [DOI] [PubMed] [Google Scholar]
- 32.Kalyuzhnaya MG, Lapidus A, Ivanova N, Copeland AC, McHardy AC, Szeto E, Salamov A, Grigoriev IV, Suciu D, Levine SR, Markowitz VM, Rigoutsos I, Tringe SG, Bruce DC, Richardson PM, Lidstrom ME, Chistoserdova L. 2008. High-resolution metagenomics targets specific functional types in complex microbial communities. Nat Biotechnol 26:1029–1034. doi: 10.1038/nbt.1488. [DOI] [PubMed] [Google Scholar]
- 33.Kuivila KM, Murray JW, Devol AH, Lidstrom ME, Reimers CE. 1988. Methane cycling in the sediments of Lake Washington. Limnol Oceanogr 33:571–581. doi: 10.4319/lo.1988.33.4.0571. [DOI] [Google Scholar]
- 34.Auman AJ, Stolyar S, Costello AM, Lidstrom ME. 2000. Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl Environ Microbiol 66:5259–5266. doi: 10.1128/AEM.66.12.5259-5266.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beck DA, Kalyuzhnaya MG, Malfatti S, Tringe SG, Glavina del Rio T, Ivanova N, Lidstrom ME, Chistoserdova L. 2013. A metagenomic insight into freshwater methane-utilizing communities and evidence for cooperation between the Methylococcaceae and the Methylophilaceae. PeerJ 1:e23. doi: 10.7717/peerj.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martineau C, Whyte LG, Greer CW. 2010. Stable isotope probing analysis of the diversity and activity of methanotrophic bacteria in soils from the Canadian high Arctic. Appl Environ Microbiol 76:5773–5784. doi: 10.1128/AEM.03094-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crevecoeur S, Vincent WF, Comte J, Lovejoy C. 2015. Bacterial community structure across environmental gradients in permafrost thaw ponds: methanotroph-rich ecosystems. Front Microbiol 6:192. doi: 10.3389/fmicb.2015.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crespo-Medina M, Meile CD, Hunter KS, Diercks A-R, Asper VL, Orphan VJ, Tavormina PJ, Nigro LM, Battles JJ, Chanton JP, Shiller AM, Joung D-J, Amon RMW, Bracco A, Montoya JP, Villareal TA, Wood AM, Joye SB. 2014. The rise and fall of methanotrophy following a deepwater oil-well blowout. Nat Geosci 7:423–427. doi: 10.1038/ngeo2156. [DOI] [Google Scholar]
- 39.Rivers AR, Sharma S, Tringe SG, Martin J, Joye SB, Moran MA. 2013. Transcriptional response of bathypelagic marine bacterioplankton to the Deepwater Horizon oil spill. ISME J 7:2315–2329. doi: 10.1038/ismej.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kessler JD, Valentine DL, Redmond MC, Du M, Chan EW, Mendes SD, Quiroz EW, Villanueva CJ, Shusta SS, Werra LM, Yvon-Lewis SA, Weber TC. 2011. A persistent oxygen anomaly reveals the fate of spilled methane in the deep Gulf of Mexico. Science 331:312–315. doi: 10.1126/science.1199697. [DOI] [PubMed] [Google Scholar]
- 41.Lösekann T, Knittel K, Nadalig T, Fuchs B, Niemann H, Boetius A, Amann R. 2007. Diversity and abundance of aerobic and anaerobic methane oxidizers at the Haakon Mosby Mud Volcano, Barents Sea. Appl Environ Microbiol 73:3348–3362. doi: 10.1128/AEM.00016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruff SE, Biddle JF, Teske AP, Knittel K, Boetius A, Ramette A. 2015. Global dispersion and local diversification of the methane seep microbiome. Proc Natl Acad Sci U S A 112:4015–4020. doi: 10.1073/pnas.1421865112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oshkin IY, Beck DA, Lamb AE, Tchesnokova V, Benuska G, McTaggart TL, Kalyuzhnaya MG, Dedysh SN, Lidstrom ME, Chistoserdova L. 2015. Methane-fed microbial microcosms show differential community dynamics and pinpoint taxa involved in communal response. ISME J 9:1119–1129. doi: 10.1038/ismej.2014.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hernandez ME, Beck DA, Lidstrom ME, Chistoserdova L. 2015. Oxygen availability is a major factor in determining the composition of microbial communities involved in methane oxidation. PeerJ 3:e801. doi: 10.7717/peerj.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei XM, He R, Chen M, Su Y, Ma RC. 2016. Conversion of methane-derived carbon and microbial community in enrichment cultures in response to O2 availability. Environ Sci Pollut Res Int 23:7517–7528. doi: 10.1007/s11356-015-6017-y. [DOI] [PubMed] [Google Scholar]
- 46.Grosskopf T, Soyer OS. 2014. Synthetic microbial communities. Curr Opin Microbiol 18:72–77. doi: 10.1016/j.mib.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu Z, Krause SM, Beck DA, Chistoserdova L. 2016. A synthetic ecology perspective: how well does behavior of model organisms in the laboratory predict microbial activities in natural habitats? Front Microbiol 7:946. doi: 10.3389/fmicb.2016.00946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ho A, de Roy K, Thas O, De Neve J, Hoefman S, Vandamme P, Heylen K, Boon N. 2014. The more, the merrier: heterotroph richness stimulates methanotrophic activity. ISME J 8:1945–1948. doi: 10.1038/ismej.2014.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stock M, Hoefman W, Kerckhof F-M, Boon N, De Vos P, De Baets B, Heylen K, Waegeman W. 2013. Exploration and prediction of interactions between methanotrophs and heterotrophs. Res Microbiol 164:1045–1054. doi: 10.1016/j.resmic.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 50.Vu HN, Subuyuj GA, Vijayakumar S, Good NM, Martinez-Gomez NC, Skovran E. 2016. Lanthanide-dependent regulation of methanol oxidation systems in Methylobacterium extorquens AM1 and their contribution to methanol growth. J Bacteriol 198:1250–1259. doi: 10.1128/JB.00937-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chu F, Lidstrom ME. 2016. XoxF acts as the predominant methanol dehydrogenase in the type I methanotroph Methylomicrobium buryatense. J Bacteriol 198:1317–1325. doi: 10.1128/JB.00959-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gu W, Farhan Ul Haque M, DiSpirito AA, Semrau JD. 2016. Uptake and effect of rare Earth elements on gene expression in Methylosinus trichosporium OB3b. FEMS Microbiol Lett 363:fnw129. doi: 10.1093/femsle/fnw129. [DOI] [PubMed] [Google Scholar]
- 53.Chistoserdova L. 2016. Lanthanides: new life metals? World J Microbiol Biotechnol 32:138. doi: 10.1007/s11274-016-2088-2. [DOI] [PubMed] [Google Scholar]
- 54.Martinez-Gomez NC, Vu HN, Skovran E. 2016. Lanthanide chemistry: from coordination in chemical complexes shaping our technology to coordination in enzymes shaping bacterial metabolism. Inorg Chem 55:10083–10089. doi: 10.1021/acs.inorgchem.6b00919. [DOI] [PubMed] [Google Scholar]
- 55.Krause SMB, Johnson T, Samadhi Karunaratne Y, Fu Y, Beck DAC, Chistoserdova L, Lidstrom ME. 2017. Lanthanide-dependent cross-feeding of methane-derived carbon is linked by microbial community interactions. Proc Natl Acad Sci U S A 114:358–363. doi: 10.1073/pnas.1619871114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mustakhimov I, Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L. 2013. Insights into denitrification in Methylotenera mobilis from denitrification pathway and methanol metabolism mutants. J Bacteriol 195:2207–2211. doi: 10.1128/JB.00069-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Good NM, Vu HN, Suriano CJ, Subuyuj GA, Skovran E, Martinez-Gomez NC. 2016. Pyrroloquinoline quinone ethanol dehydrogenase in Methylobacterium extorquens AM1 extends lanthanide-dependent metabolism to multicarbon substrates. J Bacteriol 198:3109–3118. doi: 10.1128/JB.00478-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sher D, Thompson JW, Kashtan N, Croal L, Chisholm SW. 2011. Response of Prochlorococcus ecotypes to co-culture with diverse marine bacteria. ISME J 5:1125–1132. doi: 10.1038/ismej.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hmelo LR, Van Mooy BAS, Mincer TJ. 2012. Characterization of bacterial epibionts on the cyanobacterium Trichodesmium. Aquat Microb Ecol 67:1–14. doi: 10.3354/ame01571. [DOI] [Google Scholar]
- 60.Cole JK, Hutchison JR, Renslow RS, Kim YM, Chrisler WB, Engelmann HE, Dohnalkova AC, Hu D, Metz TO, Fredrickson JK, Lindemann SR. 2014. Phototrophic biofilm assembly in microbial-mat-derived unicyanobacterial consortia: model systems for the study of autotroph-heterotroph interactions. Front Microbiol 5:109. doi: 10.3389/fmicb.2014.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee MD, Walworth NG, McParland EL, Fu FX, Mincer TJ, Levine NM, Hutchins DA, Webb EA. 2017. The Trichodesmium consortium: conserved heterotrophic co-occurrence and genomic signatures of potential interactions. ISME J 11:1813–1824. doi: 10.1038/ismej.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morris J, Lenski RE, Zinser ER. 2012. The Black Queen hypothesis: evolution of dependencies through adaptive gene loss. mBio 3:e00036-12. doi: 10.1128/mBio.00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iguchi H, Yurimoto H, Sakai Y. 2011. Stimulation of methanotrophic growth in cocultures by cobalamin excreted by rhizobia. Appl Environ Microbiol 77:8509–8515. doi: 10.1128/AEM.05834-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beck DA, McTaggart TL, Setboonsarng U, Vorobev A, Kalyuzhnaya MG, Ivanova N, Goodwin L, Woyke T, Lidstrom ME, Chistoserdova L. 2014. The expanded diversity of Methylophilaceae from Lake Washington through cultivation and genomic sequencing of novel ecotypes. PLoS One 9:e102458. doi: 10.1371/journal.pone.0102458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kalyuzhnaya MG, Lamb AE, McTaggart TL, Oshkin IY, Shapiro N, Woyke T, Chistoserdova L. 2015. Draft genomes of gammaproteobacterial methanotrophs isolated from Lake Washington sediment. Genome Announc 3:e00103-15. doi: 10.1128/genomeA.00103-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lovley DR. 2017. Happy together: microbial communities that hook up to swap electrons. ISME J 11:327–336. doi: 10.1038/ismej.2016.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wegener G, Krukenberg V, Riedel D, Tegetmeyer HE, Boetius A. 2015. Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 526:587–590. doi: 10.1038/nature15733. [DOI] [PubMed] [Google Scholar]
- 68.Karlsen OA, Larsen O, Jensen HB. 2011. The copper responding surfaceome of Methylococcus capsulatus Bath. FEMS Microbiol Lett 323:97–104. doi: 10.1111/j.1574-6968.2011.02365.x. [DOI] [PubMed] [Google Scholar]
- 69.Kalyuzhnaya MG, Beck DAC, Suciu D, Pozhitkov A, Lidstrom ME, Chistoserdova L. 2010. Functioning in situ: gene expression in Methylotenera mobilis in its native environment as assessed through transcriptomics. ISME J 4:388–398. doi: 10.1038/ismej.2009.117. [DOI] [PubMed] [Google Scholar]
- 70.Vorobev A, Beck DAC, Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L. 2013. Comparative transcriptomics in three Methylophilaceae species uncover different strategies for environmental adaptation. PeerJ 1:e115. doi: 10.7717/peerj.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Holmes DE, Shrestha PM, Walker DJF, Dang Y, Nevin KP, Woodard TL, Lovley DR. 2017. Metatranscriptomic evidence for direct interspecies electron transfer between Geobacter and Methanothrix species in methanogenic rice paddy soils. Appl Environ Microbiol 83:e00223-17. doi: 10.1128/AEM.00223-17. [DOI] [PMC free article] [PubMed] [Google Scholar]