ABSTRACT
Salmonella enterica can utilize fructose-asparagine (F-Asn) as a source of carbon and nitrogen. This capability has been attributed to five genes in the fra locus. Previously, we determined that mutations in fraB (deglycase), fraD (kinase), or fraA (transporter) eliminated the ability of Salmonella to grow on F-Asn, while a mutation in fraE allowed partial growth. We hypothesized that FraE, a putative periplasmic fructose-asparaginase, converts F-Asn to NH4+ and fructose-aspartate (F-Asp). FraA could then transport F-Asp into the cytoplasm for subsequent catabolism. Here, we report that growth of the fraE mutant on F-Asn is caused by a partially redundant activity provided by AnsB, a periplasmic asparaginase. Indeed, a fraE ansB double mutant is unable to grow on F-Asn. Moreover, biochemical assays using periplasmic extracts of mutants that express only FraE or AnsB confirmed that each of these enzymes converts F-Asn to F-Asp and NH4+. However, FraE does not contribute to growth on asparagine. We tested and confirmed the hypothesis that a fraE ansB mutant can grow on F-Asp, while mutants lacking fraA, fraD, or fraB cannot. This finding provides strong evidence that FraA transports F-Asp but not F-Asn from the periplasm to the cytoplasm. Previously, we determined that F-Asn is toxic to a fraB mutant due to the accumulation of the FraB substrate, 6-phosphofructose-aspartate (6-P-F-Asp). Here, we found that, as expected, a fraB mutant is also inhibited by F-Asp. Collectively, these findings contribute to a better understanding of F-Asn utilization by Salmonella.
IMPORTANCE Salmonella is able to utilize fructose-asparagine (F-Asn) as a nutrient. We recently reported that the disruption of a deglycase enzyme in the F-Asn utilization pathway inhibits the growth of Salmonella in mice and recognized this pathway as a novel and specific drug target. Here, we characterize the first step in the pathway wherein FraE hydrolyzes F-Asn to release NH4+ and F-Asp in the periplasm of the cell. A fraE mutant continues to grow slowly on F-Asn due to asparaginase activity encoded by ansB.
KEYWORDS: fructose-asparagine, fructose-aspartate, fraE, ansB, ansA, Salmonella, asparaginase, Amadori product
INTRODUCTION
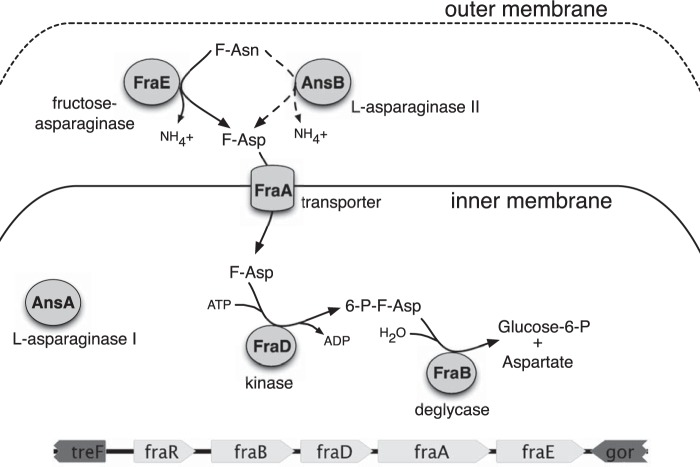
Salmonella enterica is a significant source of serious foodborne infections globally (1–4). In our previous studies, we discovered from a genetic screen that mutants lacking the fra locus were highly attenuated in several mouse models of gastroenteritis (5). This locus contains five genes that confer the ability to utilize fructose-asparagine (F-Asn) as a sole source of carbon and nitrogen (5, 6). Four of the genes are part of the putative fraBDAE operon. The fifth gene, fraR, encodes a GntR-type transcription factor. Our working model is that FraE, a putative asparaginase, converts F-Asn to fructose-aspartate (F-Asp) and NH4+ in the periplasm (Fig. 1). F-Asp is then transported into the cytoplasm through FraA, a Dcu-type transporter. The FraD kinase then phosphorylates F-Asp to yield 6-phosphofructose-aspartate (6-P-F-Asp), which is cleaved by the FraB deglycase to yield the common metabolic intermediates glucose-6-phosphate and aspartate.
FIG 1.
Proposed pathway for fructose-asparagine utilization.
There is growing evidence to validate this model of F-Asn utilization. First, a global proteomic analysis of Salmonella subcellular compartments placed FraE and FraB in the periplasmic and cytoplasmic fractions, respectively (7). Second, we have demonstrated the biochemical activities of FraB and FraD (6, 8). Last, the mutation of fraB causes the accumulation of 6-P-F-Asp, which is toxic to the cell and is responsible for the phenotype of reduced fitness in mice (6). This finding highlights the potential of FraB as a drug target and is a motivation to conduct studies to fully understand the regulation and reactions of the F-Asn utilization pathway. Here, we investigate the putative asparaginase encoded by the fraE gene and provide evidence that FraE converts F-Asn to F-Asp. Interestingly, a Salmonella mutant lacking fraE can grow slowly on F-Asn as a source of carbon and nitrogen, a finding that led us to uncover the partial redundancy afforded by the periplasmic AnsB asparaginase.
RESULTS
The ansB gene contributes to growth of a Salmonella fraE mutant on F-Asn.
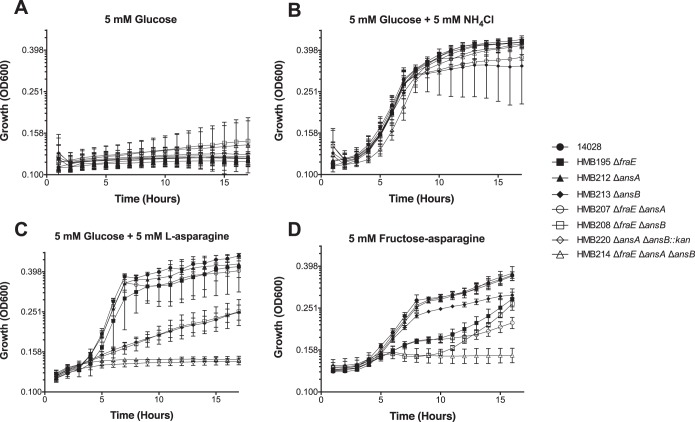
During our studies of the fra locus of Salmonella, we found that a fraE mutant was only partially defective for growth on F-Asn as the sole carbon source (6). Two other asparaginases (AnsA and AnsB) mediate nitrogen assimilation and contribute to virulence (9–11). Because AnsA is cytoplasmic and AnsB is periplasmic (12, 13), we postulated that AnsB was contributing to the growth of a fraE mutant on F-Asn. To test this hypothesis, we constructed double and triple mutants lacking combinations of ansA, ansB, and fraE and grew them with F-Asn as the sole source of carbon and nitrogen (Fig. 2) or just the carbon source (Fig. 3). A fraE mutant was partially defective for growth on F-Asn, while mutants lacking ansA or ansB were not. However, the addition of an ansB mutation to the fraE mutant background eliminates growth on F-Asn for about 11 h (Fig. 2D, or 7 h in Fig. 3C), at which time the cultures begin to resume growth, albeit weakly. We postulate that this late increase is due to ansA contributing to the growth of Salmonella on cell debris. Such a premise is supported by our observation that a triple mutant lacking fraE, ansB, and ansA does not resume growth at late time points (Fig. 2D and 3C). Additionally, a fraE ansA mutant grows slower at later time points than a fraE mutant. Both of these observations provide support for the hypothesis that AnsA is contributing to growth at late time points. Collectively, these results indicate that FraE is the asparaginase homolog primarily responsible for the growth on F-Asn. In the absence of fraE, the periplasmic asparaginase AnsB can partially fulfill this role.
FIG 2.
Growth of asparaginase mutants on different carbon and nitrogen sources. The bacterial strains indicated were grown on M9 minimal medium lacking ammonium chloride and supplemented with the following carbon and nitrogen sources: 5 mM glucose only (no nitrogen source) (A), 5 mM glucose with 5 mM ammonium chloride (B), 5 mM glucose plus 5 mM l-asparagine (C), and 5 mM F-Asn only (D). All data points are the means from triplicate cultures measured on two different occasions (6 total points). Error bars represent standard deviations.
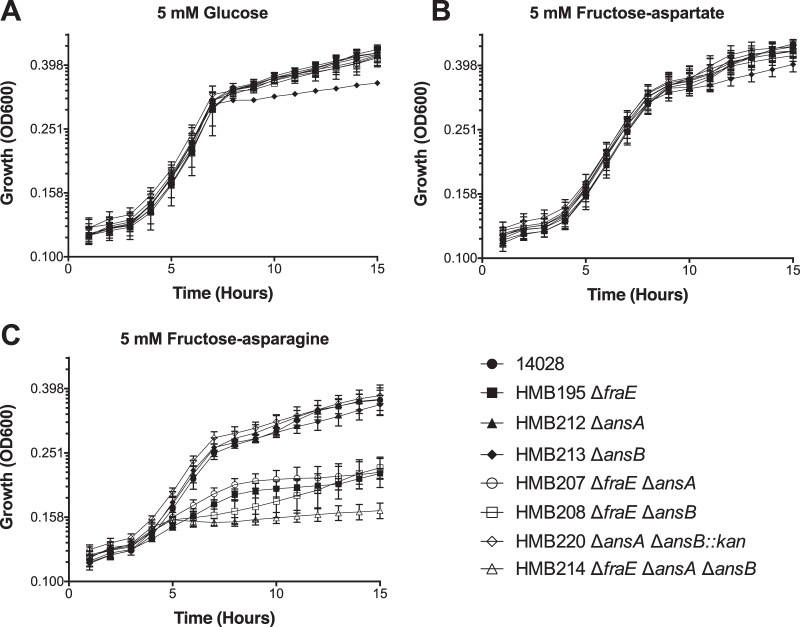
FIG 3.
Growth of asparaginase mutants on different carbon sources. The bacterial strains indicated were grown on M9 minimal medium with 19 mM ammonium chloride and supplemented with the following carbon sources: 5 mM glucose (A), 5 mM F-Asp (B), and 5 mM F-Asn (C). All data points are the means from triplicate cultures measured on three different occasions (9 total points). Error bars represent standard deviations.
FraE and AnsB are the only sources of fructose-asparaginase activity in the periplasm.
To further investigate the ability of the FraE and AnsB enzymes to release NH4+ from F-Asn, we grew cells overnight in LB broth and then subcultured them 1:100 into fresh LB medium containing 5 mM F-Asn. A 6-h growth was used to induce the fra locus. These conditions are different from those used as described above for the growth curve experiments, largely to fulfill the objective of harvesting live cells for biochemical assays even if there is poor or no growth on F-Asn. The cells were fractionated, and the periplasmic fractions were tested for their ability to cleave F-Asn through the use of a glutamate dehydrogenase (GDH)-based coupled assay that directly measures NH4+ (Table 2). The periplasmic contents of a fraE ansB double mutant or a fraE ansB ansA triple mutant had no fructose-asparaginase activity, demonstrating that fraE and ansB are the only two enzymes capable of this activity. In the periplasmic contents of an ansA ansB mutant or an ansA fraE mutant, each had fructose-asparaginase activity, demonstrating that the presence of either FraE or AnsB results in cleavage of F-Asn in vitro (Table 2). The specific activity of the ansB mutant was slightly higher than that of the fraE mutant.
TABLE 2.
Fructose-asparaginase activity in periplasmic subcellular fractionsa
| Salmonella strain | ansA | ansB | fraE | Fructose-asparaginase sp act (102 U/mg) |
|---|---|---|---|---|
| 14028 | ✓ | ✓ | ✓ | 25.8 ± 1.2 |
| HMB195 | ✓ | ✓ | X | 12.0 ± 2.9 |
| HMB207 | X | ✓ | X | 14.1 ± 1.8 |
| HMB208 | ✓ | X | X | 1.9 ± 0.3 |
| HMB212 | X | ✓ | ✓ | 26.8 ± 0.9 |
| HMB213 | ✓ | X | ✓ | 17.7 ± 1.3 |
| HMB220 | X | X | ✓ | 18.7 ± 1.5 |
| HMB214 | X | X | X | 0.85 ± 0.1 |
Shown are the mean values and the standard errors calculated from two independent measurements; ✓ and X refer to the presence and absence of the corresponding locus, respectively.
FraA transports F-Asp, not F-Asn.
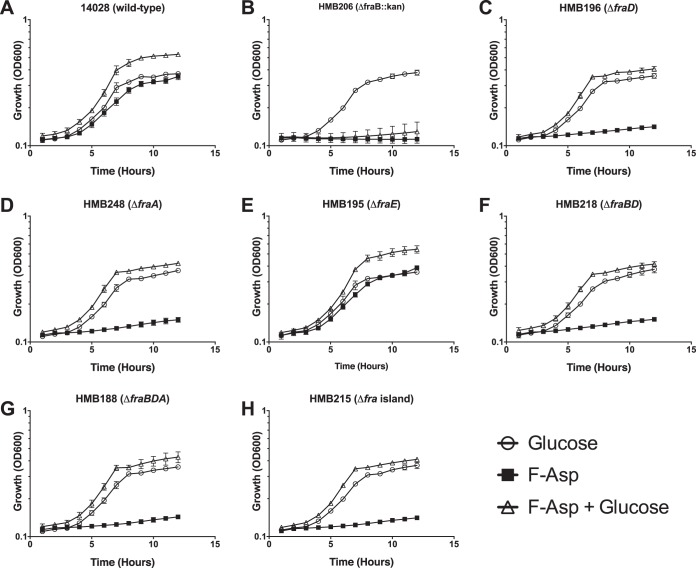
All of the single, double, and triple asparaginase mutants were able to grow on F-Asp, while mutants lacking fraA, fraB, fraD, fraBD, fraBDA, or the entire fra locus (fraR fraBDAE) could not (Fig. 3B and 4). This result is consistent with the conversion of F-Asn to F-Asp in the periplasm and the role of FraA in transporting F-Asp but not F-Asn into the cytoplasm (Fig. 1).
FIG 4.
Growth of fra mutants on F-Asp. The bacterial strains indicated were grown on M9 minimal medium with 19 mM ammonium chloride and supplemented with the following carbon sources: 5 mM glucose, 5 mM F-Asp, or 5 mM F-Asp and 5 mM glucose. All data points are the means from triplicate cultures measured on two different occasions (6 total points). Error bars represent standard deviations.
Both F-Asp and F-Asn are toxic to a fraB mutant.
Previously, we observed that F-Asn is toxic to a fraB mutant (6). This toxicity is due to the accumulation of 6-P-F-Asp. The toxicity can be observed as a lack of growth in the presence of F-Asn, even when glucose is present (6). Based on our model of F-Asn utilization, F-Asp should also be toxic to a fraB mutant. We tested and confirmed this hypothesis (Fig. 4). As expected, this toxicity is independent of fraE, because F-Asp can be transported into the cell without the activity of FraE in the periplasm. However, the toxicity is dependent upon fraA and fraD, which are required for uptake of F-Asp and conversion to 6-P-F-Asp, respectively.
The fraE gene does not contribute to growth of Salmonella on asparagine.
While AnsB was able to contribute to F-Asn metabolism, we sought to test the hypothesis that FraE could contribute to asparagine metabolism. All of the asparaginase mutants were therefore grown with asparagine as the sole source of nitrogen (Fig. 2C). A single mutant lacking fraE or ansA was not defective for growth on asparagine. However, a single mutant lacking ansB was partially defective. A complete loss of growth was observed with an ansA ansB double mutant, consistent with previous reports (9). Adding a fraE mutation to either the ansB mutant strain or the ansA ansB mutant strain had no additional effect. These results indicate that ansA and ansB contribute to asparagine metabolism, while fraE does not.
DISCUSSION
The fra locus of Salmonella is located between the gor and treF genes. The gor and treF genes are present in Escherichia coli, but the fra locus is not. Instead, E. coli contains the gad acid fitness island in this location; thus, fra appears to be a horizontal acquisition (5). A fraB mutant of Salmonella is extremely attenuated in the intestinal tract of inflamed mice due to the accumulation of the FraB substrate, 6-P-F-Asp, during growth on F-Asn (6). This makes FraB an attractive species-specific drug target. We previously determined that mutations in fraB, fraD, and fraA eliminate the ability of Salmonella to grow on F-Asn, while a mutation in fraE does not (6). Here, we have determined that the growth of a fraE mutant on F-Asn is primarily due to the redundancy provided by the periplasmic asparaginase, AnsB (Fig. 2 and 3). However, the converse does not hold: the fraE gene did not contribute to asparagine metabolism (Fig. 2C). Biochemical evidence for the ability of FraE and AnsB to cleave F-Asn was obtained by assaying the periplasmic content of the wild type and a panel of mutants (Table 2). Importantly, the periplasmic extract of a fraE ansB mutant had no activity in this assay, demonstrating that there are no other sources of fructose-asparaginase activity.
Despite the weaker growth of the fraE mutant in minimal medium containing F-Asn than that of the ansB mutant (Fig. 2), our in vitro assays indicate roughly similar specific activities for FraE and AnsB after growth in LB medium containing F-Asn (Table 2). These findings can be rationalized by considering at least two possibilities. First, if FraE and FraA (the transporter) are somehow physically coupled for compartmentalization in vivo, it is conceivable that AnsB is unable to fulfill this specific requirement despite its catalytic ability to convert F-Asn to F-Asp. Second, the relative expression levels of AnsB and FraE may be different in the minimal medium used for growth curves from those in the LB medium used to grow the cells for enzymatic assays. This conjecture is supported by a previous report that ansB of E. coli is upregulated when grown anaerobically in the presence of amino acids (13). While there is no information about the regulation of fraE or ansB in Salmonella, the amino acids in LB could engender an increase in AnsB activity. Further studies on the regulation of AnsB and FraE would be informative.
The latitude in substrate recognition of AnsB may be important for regulation of the fra locus. If the products of F-Asn catabolism are required to induce gene expression of the fraBDAE operon, this secondary activity of AnsB to convert F-Asn to F-Asp may play a role in initiating the pathway; such a hypothesis is testable.
Mutation of a cytoplasmic asparaginase gene, ansA, revealed that AnsA did not contribute to F-Asn metabolism, except modestly at late time points, when it may be contributing to growth on substrates released from dying cells. Consistent with the role of FraE and AnsB in converting F-Asn to F-Asp in the periplasm, the fraE ansB double mutant was able to grow on F-Asp, while fraA, fraD, and fraB mutants were not (Fig. 3B and 4). These results are also consistent with the proposed function of FraA being a transporter of F-Asp but not F-Asn (Fig. 1). A fraB mutant was not able to grow on F-Asp, even in the presence of glucose, which is certainly due to the accumulation of 6-P-F-Asp, as is the case during growth on F-Asn (6). Mutants lacking fraD, fraA, fraBD, fraBDA, or the entire fra locus were able to grow on glucose in the presence of F-Asp, consistent with the lack of 6-P-F-Asp accumulation (Fig. 4).
MATERIALS AND METHODS
Strains and media.
All bacterial strains and plasmids are listed in Table 1. Bacteria were routinely grown in Luria-Bertani (LB) broth (Fisher Bioreagents) or on LB agar plates containing 1.5% (wt/vol) agar (Fisher Bioreagents). For growth studies using defined carbon sources, we employed M9 minimal medium containing 1× M9 salts, 2 mM MgSO4, 0.1 mM CaCl2, 0.01 mM thiamine, and trace metals (14, 15). For the growth assays using defined nitrogen sources, we used M9 minimal medium lacking NH4Cl. As needed, kanamycin (50 μg/ml) was added to the media.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Genotype or descriptiona | Reference, source, or construction |
|---|---|---|
| Strains | ||
| 14028 | Wild-type Salmonella enterica subspecies enterica serovar Typhimurium | ATCC |
| HMB106 | 14028 ΔansA80::Kan | Lambda Red mutation of ansA made using PCR primers BA3004 and BA3005 |
| HMB107 | 14028 ΔansB80::Kan | Lambda Red mutation of ansB made using PCR primers BA3006 and BA3007 |
| HMB174 | 14028 ΔansA80::Kan | P22 transduction of ΔansA80::Kan from HMB106 into a clean 14028 background |
| HMB175 | 14028 ΔansB80::Kan | P22 transduction of ΔansB80::Kan from HMB107 into a clean 14028 background |
| HMB188 | 14028 ΔfraBDA80 | 6 |
| HMB195 | 14028 ΔfraE4 | 6 |
| HMB196 | 14028 ΔfraD4 | 6 |
| HMB203 | 14028 ΔfraE4 ΔansA80::Kan | P22 transduction of ΔansA80::Kan from HMB106 into HMB195 |
| HMB204 | 14028 ΔfraE4 ΔansB80::Kan | P22 transduction of ΔansB80::Kan from HMB107 into HMB195 |
| HMB206 | 14028 ΔfraB80::Kan | 6 |
| HMB207 | 14028 ΔfraE4 ΔansA80 | Antibiotic cassette in HMB203 was flipped out using pCP20 |
| HMB208 | 14028 ΔfraE4 ΔansB80 | Antibiotic cassette in HMB204 was flipped out using pCP20 |
| HMB210 | 14028 ΔfraE4 ΔansA80 ΔansB80::Kan | P22 transduction of ΔansB80::Kan from HMB107 into HMB207 |
| HMB212 | 14028 ΔansA80 | Antibiotic cassette in HMB174 was flipped out using pCP20 |
| HMB213 | 14028 ΔansB80 | Antibiotic cassette in HMB175 was flipped out using pCP20 |
| HMB214 | 14028 ΔfraE4 ΔansA80 ΔansB80 | Antibiotic cassette in HMB210 was flipped out using pCP20 |
| HMB215 | 14028 Δfra80 | 6 |
| HMB218 | 14028 ΔfraBD81 | 6 |
| HMB220 | 14028 ΔansA80 ΔansB80::Kan | P22 transduction of ΔansB80::Kan from HMB107 into HMB212 |
| HMB248 | 14028 ΔfraA4 | 6 |
| Plasmids | ||
| pKD46 | PBAD gam bet exo pSC101 oriTS (Ampr) | 16 |
| pKD4 | FRT-kan-FRT oriR6K (Ampr) | 16 |
| pCP20 | cI857 λPR flp pSC101 oriTS (Ampr Camr) | 20 |
Ampr, ampicillin resistance; Camr, chloramphenicol resistance.
Construction of mutants.
Lambda Red mutagenesis was used to insert antibiotic resistance genes into sites of interest, as previously described (16). In each case, all but the first 10 and last 10 codons of the gene of interest were deleted. The antibiotic resistance genes were flanked by FLP recombination target (FRT) sites and amplified from the plasmid template pKD4 (16). The primers used for the ansA mutation were BA3004 (5′-GAACATTATCATGCAAAAGAAATCAATTTACGTTGCCTATGTGTAGGCTGGAGCTGCTTC-3′) and BA3005 (5′-GGGATTTTTTAATCATCCGGCGTCAATTCACCACGCAACATATGAATATCCTCCTTAG-3′). For the ansB mutation, the primers were BA3006 (5′-GAGATATAACATGGAGTTTTTCAGGAAAACGGCATTAGCTGTGTAGGCTGGAGCTGCTTC-3′) and BA3007 (5′-GGCATTATCTTTAATACTGATTGAACATCGTCTGGATCTGCATATGAATATCCTCCTTAG-3′). The correct location for insertion of the antibiotic resistance gene was confirmed by PCR. Each mutation was transduced into a clean 14028 background using phage P22HTint. The antibiotic cassette was then removed by electroporation of pCP20 (ampicillin resistance [Ampr]), which encodes FLP recombinase, into the strain and plating on LB with ampicillin at 30°C. Single colonies were streaked onto LB agar and incubated at 42°C to cure the strain of pCP20. PCR was used to verify loss of the antibiotic resistance cassette using primers upstream and downstream of the target gene. Colonies with the correct PCR product were also screened for loss of the antibiotic resistance gene and pCP20.
Synthesis of F-Asp and ammonia-free F-Asn.
F-Asp and F-Asn were synthesized and purified as described elsewhere (17). In the case of F-Asn, the final preparation from this synthesis still contains between 0.1 and 0.2 mol of ammonium ion per mole (i.e., 85% zwitterion and 15% ammonium salt). Most of the residual ammonia was removed by treatment with 4-Å molecular sieves as follows: 0.3 g of F-Asn was dissolved in 5 ml of methanol. One and one-half grams of 600-mesh sieves (Strem Chemicals, Newburyport, MA) was added and the suspension stirred for about an hour at 22°C. The mixture was filtered by gravity and washed twice with methanol. The procedure was repeated with a fresh batch of sieves, and the methanol was then removed by rotary evaporation. Analysis for ammonia in a Conway diffusion apparatus (18) revealed that the ammonium ion content had been reduced to about 0.003 mol per mole F-Asn. This value was also independently confirmed using the enzymatic assay described below.
Growth assays.
Growth curves were performed using clear flat-bottom 96-well plates. In each well, 198-μl aliquots of medium were inoculated with 2 μl of overnight cultures that had been washed twice with sterile water. A Breathe-Easy membrane film (Diversified Biotech) was placed over the 96-well plate. Growth at 37°C was measured using hourly measurements of the optical density at 600 nm (OD600), with shaking for 5 s prior to each reading, in the SpectraMax M5 microplate reader (Molecular Devices) and the SoftMax Pro 6.1 software.
Preparation of periplasmic extracts.
Salmonella periplasmic extracts were prepared using essentially the protocol described by Brown et al. (7), except with minor modifications as described below. Salmonella was grown in 5 ml LB for 16 h at 37°C with shaking. The cells were washed with sterile water, subcultured 1:100 in 5 ml of fresh LB supplemented with 5 mM F-Asn, and grown for 6 h at 37°C with shaking. The cells were centrifuged at 5,000 × g for 10 min at 4°C, and the cell pellets were washed with 1.5 ml of 50 mM Tris-HCl (pH 8). The cell pellets were resuspended in 1.5 ml of spheroplasting buffer (50 mM Tris-HCl [pH 8], 250 mM sucrose, 2.5 mM EDTA) and were incubated at 22°C for 5 min. Following incubation, the cell suspension was centrifuged at 11,500 × g for 10 min at 4°C. The cell pellets were resuspended in 300 μl of ice-cold 5 mM MgSO4 and kept on ice for 10 min, with intermittent manual mixing. This suspension was subjected to centrifugation (11,500 × g, 10 min, 4°C) to yield a supernatant containing the soluble periplasmic fraction. Prior to enzymatic assays, 300 μl of the periplasmic fraction was supplemented with 0.1 mg/ml bovine serum albumin (BSA) and dialyzed against 20 mM Tris (pH 8) and 5 mM MgSO4 at 4°C, with two changes over 90 min. The periplasmic extract obtained after dialysis was used in the activity assays, as described below. To calculate the specific activities, the protein content in the dialysates was determined using the Bradford assay (19), with BSA serving as the standard.
Enzyme assays.
For the measurement of fructose-asparaginase activity in periplasmic extracts, we used a glutamate dehydrogenase (GDH)-based coupled assay that measures NH4+. All assays were performed at 37°C and in a 105-μl reaction mixture. Each periplasmic extract (20 μl) was added to an 84-μl assay mixture (0.38 mM F-Asn, 2.3 mM α-ketoglutarate, and 0.15 mM NADPH in the buffer provided by Sigma, the GDH assay kit supplier) that had been preincubated for 4 min at 37°C. The reaction was initiated by the addition of 0.5 U of GDH (1 μl, 525 U/ml), which had been preincubated separately for 4 min at 37°C. Approximately 100 μl from this reaction mixture was quickly transferred to 96 well-white flat-bottom polystyrene microplates (Corning), and absorbance at 340 nm was monitored using a SpectraMax M5 (Molecular Devices) microplate reader (integration time of 1,000 ms, settle time of 300 ms).
The NADPH utilized by GDH during amination was measured by monitoring the decrease in Abs340 and taken as a direct readout of the ammonia generated by the action of either FraE or AnsB on F-Asn. Linear regression (Excel) analysis of NADPH consumed as a function of time was used to calculate the initial velocity (0.987 ≤ r2 ≥ 0.998). A standard curve was generated using different concentrations of ammonium ion standard (5.9 to 47 μM in assay buffer) provided with the kit and used as a reference for calculating the ammonia produced by fructose-asparaginase activities in the periplasmic extracts. Blanks for the ammonia standard curve included all the assay components except GDH, and slopes obtained in the blank reactions were subtracted from the test readings to obtain the final measurement. Blanks for the fructose-asparaginase assay lacked F-Asn and allowed a direct measure of residual ammonia in the periplasmic extract that was being tested. One unit of activity is defined as the amount of enzyme catalyzing the formation of 1 μmol ammonia/min. The mean values for the reported specific activities (Table 2) were calculated from the results from two independent assays.
ACKNOWLEDGMENTS
We thank Jikang Wu and Vicki Wysocki (OSU) for mass spectrometry analyses that helped establish the purity of the Amadori compounds synthesized for this study.
This work was supported by NIH NIAID grant 1R01AI116119.
REFERENCES
- 1.Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Döpfer D, Fazil A, Fischer-Walker CL, Hald T, Hall AJ, Keddy KH, Lake RJ, Lanata CF, Torgerson PR, Havelaar AH, Angulo FJ. 2015. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 12:e1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pires SM, Fischer-Walker CL, Lanata CF, Devleesschauwer B, Hall AJ, Kirk MD, Duarte ASR, Black RE, Angulo FJ. 2015. Aetiology-specific estimates of the global and regional incidence and mortality of diarrhoeal diseases commonly transmitted through food. PLoS One 10:e0142927. doi: 10.1371/journal.pone.0142927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acácio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 4.Scallan E, Mahon BE, Hoekstra RM, Griffin PM. 2013. Estimates of illnesses, hospitalizations and deaths caused by major bacterial enteric pathogens in young children in the United States. Pediatr Infect Dis J 32:217–221. [DOI] [PubMed] [Google Scholar]
- 5.Ali MM, Newsom DL, Gonzalez JF, Sabag-Daigle A, Stahl C, Steidley B, Dubena J, Dyszel JL, Smith JN, Dieye Y, Arsenescu R, Boyaka PN, Krakowka S, Romeo T, Behrman EJ, White P, Ahmer BMM. 2014. Fructose-asparagine is a primary nutrient during growth of Salmonella in the inflamed intestine. PLoS Pathog 10:e1004209. doi: 10.1371/journal.ppat.1004209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabag-Daigle A, Blunk HM, Sengupta A, Wu J, Bogard AJ, Ali MM, Stahl C, Wysocki VH, Gopalan V, Behrman EJ, Ahmer BMM. 2016. A metabolic intermediate of the fructose-asparagine utilization pathway inhibits growth of a Salmonella fraB mutant. Sci Rep 6:28117. doi: 10.1038/srep28117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown RN, Sanford JA, Park JH, Deatherage BL, Champion BL, Smith RD, Heffron F, Adkins JN. 2012. A comprehensive subcellular proteomic survey of Salmonella grown under phagosome-mimicking versus standard laboratory conditions. Int J Proteomics 2012:123076. doi: 10.1155/2012/123076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biswas PK, Behrman EJ, Gopalan V. 2017. Characterization of a Salmonella sugar kinase essential for utilization of fructose-asparagine. Biochem Cell Biol 95:304–309. doi: 10.1139/bcb-2016-0138. [DOI] [PubMed] [Google Scholar]
- 9.McLaughlin PA, McClelland M, Yang H-J, Porwollik S, Bogomolnaya L, Chen J-S, Andrews-Polymenis H, van der Velden AWM. 2017. Contribution of asparagine catabolism to Salmonella virulence. Infect Immun 85:e00740-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torres A, Luke JD, Kullas AL, Kapilashrami K, Botbol Y, Koller A, Tonge PJ, Chen EI, Macian F, van der Velden AWM. 2016. Asparagine deprivation mediated by Salmonella asparaginase causes suppression of activation-induced T cell metabolic reprogramming. J Leukoc Biol 99:387–398. doi: 10.1189/jlb.4A0615-252R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kullas AL, McClelland M, Yang H-J, Tam JW, Torres A, Porwollik S, Mena P, McPhee JB, Bogomolnaya L, Andrews-Polymenis H, van der Velden AWM. 2012. l-Asparaginase II produced by Salmonella Typhimurium inhibits t cell responses and mediates virulence. Cell Host Microbe 12:791–798. doi: 10.1016/j.chom.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell HA, Mashburn LT, Boyse EA, Old LJ. 1967. Two l-asparaginases from Escherichia coli B. Their separation, purification, and antitumor activity. Biochemistry 6:721–730. doi: 10.1021/bi00855a011. [DOI] [PubMed] [Google Scholar]
- 13.Cedar H, Schwartz JH. 1968. Production of l-asparaginase II by Escherichia coli. J Bacteriol 96:2043–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price-Carter M, Tingey J, Bobik TA, Roth JR. 2001. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J Bacteriol 183:2463–2475. doi: 10.1128/JB.183.8.2463-2475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 16.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen AL, Behrman EJ. 2016. Synthesis of 6-phosphofructose aspartic acid and some related Amadori compounds. Carbohydr Res 431:1–5. doi: 10.1016/j.carres.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conway EJ. 1963. Chapter X. Microdiffusion analysis and volumetric error. Chemical Publishing Co, Inc., New York, NY. [Google Scholar]
- 19.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 20.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-A. [DOI] [PubMed] [Google Scholar]