ABSTRACT
This study was aimed at improving the functional attributes and shelf life of burrata cheese by using protective lactobacilli (Lactobacillus plantarum LPAL and Lactobacillus rhamnosus LRB), fructooligosaccharides, and inulin. Six burrata cheeses were made using (i) the traditional protocol (control), (ii) the addition of 0.5% fructooligosaccharides and inulin (DF cheese), (iii) protective lactobacilli in milk alone (PL cheese), (iv) protective lactobacilli in milk and governing liquid (2PL cheese), (v) protective lactobacilli in milk and dietary fibers (DF_PL cheese), and (vi) protective lactobacilli in milk and governing liquid and dietary fibers (DF_2PL cheese). As expected, DF, DF_PL, and DF_2PL cheeses showed 1.5% of total fibers. Burrata cheeses produced by adding protective lactobacilli only in milk (PL and DF_PL cheeses) showed the lowest acidification during cheese making and storage. Lactic and acetic acids and ethanol were found at the lowest concentrations in these samples. Analyses of cultivable microbiota and the microbiome showed that protective lactobacilli reduced the house microbiota components (e.g., Streptococcus thermophilus, Lactococcus lactis, and Leuconostoc lactis) during cheese making and storage. Protective lactobacilli slowed the growth of staphylococci, coliforms, and Pseudomonas spp., especially in early storage. According to the different microbiome assemblies, burrata samples differed in peptide profiles and the levels of free amino acids. As shown by a sensory analysis, the addition of protective lactobacilli in milk improved the flavor and increased the shelf life of burrata cheese. In comparison to cheeses made using protective cultures only in milk, the shelf lives of those containing cultures also in the governing liquid were not further prolonged and they received lower acceptability scores by the panelists.
IMPORTANCE This study provides more in-depth knowledge of the microbiome of burrata cheese and the set-up for a novel biotechnology using prebiotic dietary fibers and protective probiotic Lactobacillus plantarum LPAL and Lactobacillus rhamnosus LRB in milk. The biotechnology proposed in this study should be considered a useful tool to improve the functional value of burrata cheese. The use of protective lactobacilli in milk enhanced the flavor formation and shelf life of burrata cheese.
KEYWORDS: burrata cheese, protective lactobacilli, dietary fibers, microbiome
INTRODUCTION
Burrata, a creamy cheese, is a specialty of Southern Italy, especially in the regions of Apulia, Campania, and Basilicata, and it is included on the list of traditional agri-food products (“prodotto agroalimentare tradizionale” [PAT]) (1). Recently, Burrata di Andria cheese obtained the EU protected geographical indication (2). Burrata cheese is manufactured using pasteurized cow's milk, which is mixed with the acidified serum of the previous day's cheese making so that the initial equilibrated pH is adjusted to 6.1 ± 0.10, and calf rennet (3). The final product consists of a double structure, which is composed of a “bag” made of mozzarella paste and an inner core called “stracciatella.” The bag is produced by using mozzarella curd obtained by the use of chemically acidified milk without a starter culture and stretching in hot water (80 to 90°C) (4). The pasta filata is molded to form an open hollow cheese sphere. The stracciatella, made by mixing cream with mozzarella cheese strips, is used to fill the hollow sphere, which is closed manually. Burrata cheese is salted in brine for a few minutes and cooled in water at 4°C (5). Part of the burrata cheese is exported, but this is limited by its short shelf life (6). Indeed, due to its naturally poor competitive microbiota, relatively high values of activity water (aw), and mildly acidic pH, various microorganisms, especially bacteria, may grow in burrata cheese, thus causing various shelf life-limiting spoilages (e.g., loss of elasticity and discolorations) (3). In addition, different ingredients are used depending on the producers (e.g., whey cream produced by spontaneous rising or ultrahigh temperature centrifuged cream and cheeses produced at artisanal or industrial levels) (7). These factors result in cheeses that differ in shape, sensory and safety features, and consequently, the shelf life (4, 8). Psychrotrophic microorganisms (Pseudomonas spp. and Enterobacteriaceae) are the main spoiling agents of burrata cheeses (4, 9, 10). One of the approaches used to prevent the growth of undesirable microorganisms and prolong the shelf life of fresh cheese is the application of biopreservatives (i.e., nisin or protective cultures) (11–16). Protective cultures are live microorganisms that are deliberately added to food items to inhibit the growth of undesired (spoiling and/or pathogenic) microorganisms, without negatively affecting the sensorial quality of food (11). To date, no studies have reported the use of protective cultures for increasing the shelf life of burrata cheese.
The shelf lives of foods are affected by changes in their sensory features, including the structural characteristics (17). Conte et al. (3) showed that the sensory shelf life of burrata cheese was directly affected by modifying the consistency. Dietary fibers improve the sensory characteristics, such as taste and texture of dairy products (18, 19). Fructooligosaccharides (FOS) are dietary fibers that potentially have beneficial effects on human health (19). Among them, inulin is used as a bulking agent that is able to replace fat, modify cheese texture, and improve the sensorial quality of the cheese (20). Previously, the positive effects of dietary fibers on the shelf lives of soft dairy products, such as some fresh cheeses and ice creams, were described (19, 21–23). In addition, positive effects on the human gut microbiota and health from the intake of dietary fibers have been described (19, 24). The European Food Safety Authority (EFSA) authorized the use of the following health claim for inulin from chicory. Inulin “contributes to normal bowel function by increasing stool frequency” if ingested via food at a daily intake of 12 g (25). The use of dietary fibers and protective cultures could represent a new natural biotechnological tool to improve the sensory quality, functional features, and the shelf life of burrata cheeses.
The current study was aimed to improve the functional quality and the shelf life of burrata cheese using FOS, inulin, and protective probiotic lactobacilli. The approaches integrated biochemical, microbiological, and sensorial analyses of cheeses.
RESULTS
Compositional and physicochemical analyses.
After 1 day of manufacturing, burrata cheeses without added protective cultures or fibers (control) and those with protective lactobacilli added in milk (PL cheese) or in milk and governing liquid (2PL cheese) had the following gross chemical composition (wt/wt): moisture, 65.7 ± 0.9%; total carbohydrates, 3.4 ± 0.2%; proteins, 13.5 ± 0.5%; fat, 16.5 ± 0.6%; ashes, 0.60 ± 0.02%; and sodium chloride, 0.30 ± 0.04%. Burrata cheeses supplemented with (i) FOS and inulin (DF), (ii) FOS, inulin, and protective lactobacilli in milk (DF_PL cheese), and (iii) FOS, inulin, and protective lactobacilli in milk and governing liquid (DF_2PL cheese) differed (P < 0.05) from the others in terms of moisture (66.9 ± 0.5%), proteins (12.3 ± 0.3%), fat (15.2% ± 0.2%), and fibers (1.55 ± 0.04%). After 1 day of manufacture, the pH values ranged between ca. 6.55 (cheeses with protective cultures added) and ca. 6.35 (DF cheese). No acidification was found in any of the cheeses after 6 days of storage at 4°C. pH values decreased during 16 days of storage, especially for DF_2PL and 2PL cheeses (see Table S1 in the supplemental material). The lowest decreases in pH after 16 days were found for DF_PL and PL cheeses (pH 6.43 and 6.25, respectively). Overall, the values of aw did not vary significantly (P > 0.05) among the different cheeses or during storage. DF_PL and PL cheeses showed the highest concentrations of residual lactose after 16 days of storage (see Table S2). Galactose was detected only in burrata cheeses supplemented with dietary fibers, and concentrations decreased during storage. Lactic acid was detected in trace amounts until 8 days of storage and strongly increased at 16 days, especially for DF_2PL and 2PL cheeses (Table S2). Acetic acid and ethanol were detected only at 8 and 16 days of storage at 4°C. Acetic acid was not detected in DF_PL and PL cheeses.
Cultivable microbiota of burrata cheeses.
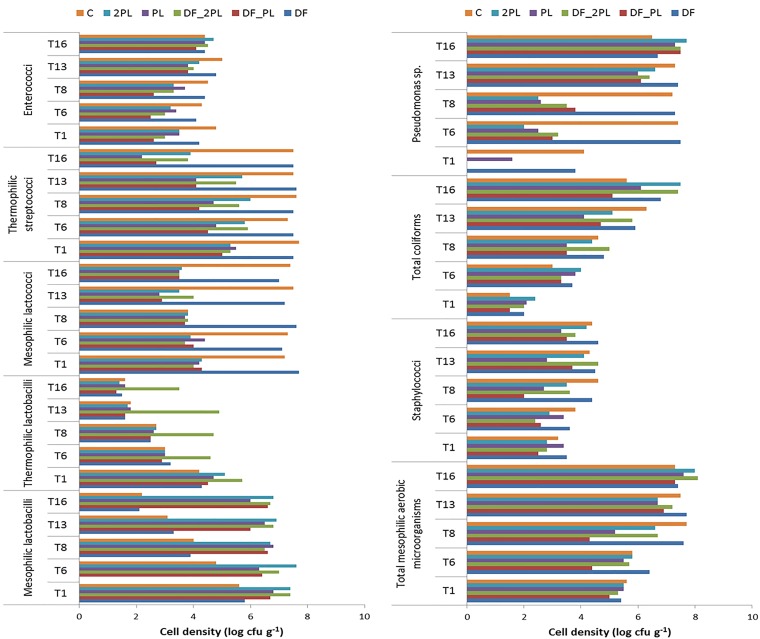
Mesophilic lactobacilli ranged from ca. 5.6 log (DF and control cheeses) to 7.4 (DF_2PL and 2PL) log CFU · g−1 (Fig. 1). During 16 days of storage at 4°C, the greatest decreases were found for DF and control cheeses. Compared to mesophilic lactobacilli, thermophilic lactobacilli were found at lower levels in the control and DF samples. After 16 days of storage, they decreased to 1.3 log to 1.6 log CFU · g−1 in all cheeses. The only exception was DF_2PL, showing a final cell density of 3.5 log CFU · g−1. At 1 day of storage, mesophilic lactococci and thermophilic streptococci were found at the highest cell densities in the control and DF cheeses. Their levels were constant during the 16 days of storage. Compared to that of the control, cheeses containing protective lactobacilli showed a decrease in cell viability of mesophilic lactococci and thermophilic streptococci during storage. Specifically, mesophilic lactococci were found at the lowest cell density in DF_PL and PL cheeses after 13 days. Among the cheeses containing protective lactobacilli, DF_2PL and 2PL samples showed the highest density of thermophilic streptococci during storage.
FIG 1.
Cell densities of microbial groups in the burrata cheeses supplemented with fructooligosaccharides (FOS) and inulin (DF), protective cultures in milk with FOS and inulin (DF_PL), protective cultures in milk and governing liquid with FOS and inulin (DF_2PL), protective cultures in milk (PL), and protective cultures in milk and governing liquid (2PL) after 1 (T1), 6 (T6), 8 (T8), 13 (T13), and 16 (T16) days of storage at 4°C. Control (C) is burrata cheese without FOS, inulin, or protective culture.
After 1 day, the amounts of enterococci were larger in the control and DF cheeses than in the cheeses containing protective lactobacilli. Increases in the amounts of enterococci were found for all cheeses containing protective lactobacilli at 13 and 16 days of storage. The total amounts of mesophilic aerobic microorganisms increased during storage at 4°C, reaching the highest after 8 (control and DF cheeses) and 16 (cheeses containing protective lactobacilli) days. Except for in PL cheese, staphylococci increased during storage at 4°C, reaching the maximum cell densities after 8 (DF and control cheeses) and 13 or 16 (all the other cheeses) days. Staphylococcus aureus was never detected. Coliforms increased during storage to ca. 4 log (DF_PL and PL cheeses) and 5 log CFU · g−1 (all the other cheeses). For all the cheeses, fecal coliforms were less than 2 log CFU · g−1 (data not shown).
Pseudomonas was found at the highest levels in control and DF cheeses after 1 day of storage. The level increased during storage at 4°C, reaching the maximum cell densities after 6 (control and DF samples) and 13 or 16 (all the other cheeses) days.
Burrata cheese microbiome.
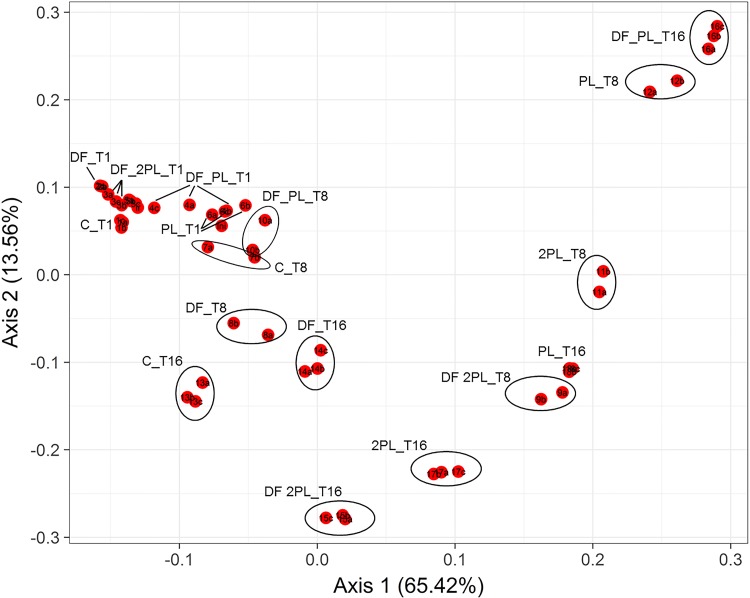
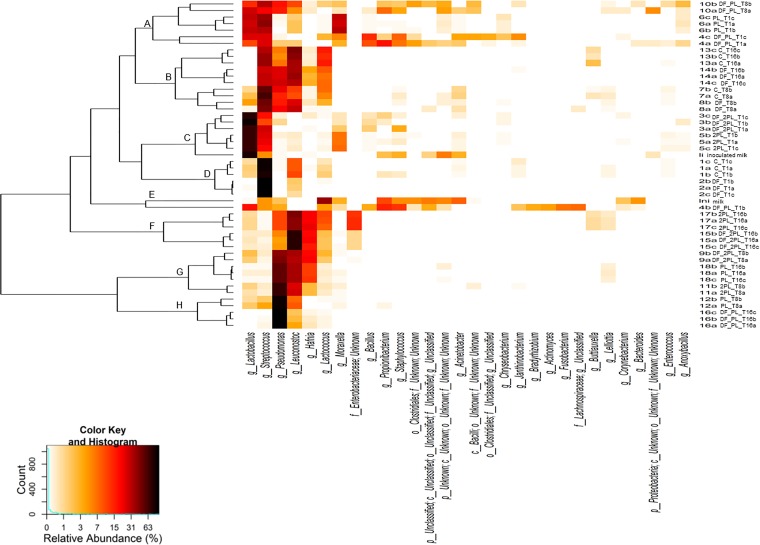
The addition of dietary fibers and protective bacteria affected α-diversity (see Table S3) and beta-diversity (Fig. 2) indices after 1, 8, and 16 days of storage at 4°C. After 1 day, the number of operational taxonomic units (OTU) ranged from ca. 15 to 64 (Table S3). Compared to that of the control, the numbers of OTU were lowest in all cheeses with added dietary fibers (DF, DF_PL, and DF_2PL cheeses) after 1 and 16 days of storage. Specific differences were found between cheeses for Chao and Shannon indices. The difference in the cheese microbiome assembly was further confirmed using phylogeny-based beta-diversity measures. The microbiomes of different samples were clearly differentiated after 8 days of storage based on bacterial lineage-specific principal-coordinate analysis with a weighted UniFrac distance matrix (Fig. 2). Based on the composition of the most abundant 30 genera, samples were grouped based on weighted UniFrac distances (Fig. 3). DF and control samples were grouped together (cluster D) and were characterized by the highest relative abundance of the genus Streptococcus. However, the microbiome changed during storage, and control and DF cheeses were grouped together in another cluster (B) showing large relative amounts of Streptococcus, Pseudomonas, Leuconostoc, and Lactococcus. According to the double inoculum of L. rhamnosus LRB and L. plantarum LPAL, DF_2PL, and 2PL cheeses after 1 day of storage were grouped in a cluster (C) showing the largest relative amount of the genus Lactobacillus. The double inoculum of protective bacteria drove a different microbiome, and DF_2PL and 2PL cheeses grouped together (cluster F) after 16 days of storage.
FIG 2.
Principal-coordinate analysis (PCoA) based on the weighted UniFrac analysis of all 16S rRNA gene sequences of bacteria found in the burrata cheeses supplemented with fructooligosaccharides (FOS) and inulin (DF), protective cultures in milk with FOS and inulin (DF_PL), protective cultures in milk and governing liquid with FOS and inulin (DF_2PL), protective cultures in milk (PL), and protective cultures in milk and governing liquid (2PL), after 1 (T1), 8 (T8), and 16 (T16) days of storage at 4°C. Control (C) is burrata cheese without FOS, inulin, or protective culture.
FIG 3.
Heatmap of relative abundances of the 30 most dominant bacterial genera found in the burrata cheeses supplemented with fructooligosaccharides (FOS) and inulin (DF), protective cultures in milk with FOS and inulin (DF_PL), protective cultures in milk and governing liquid with FOS and inulin (DF_2PL), protective cultures in milk (PL), and protective cultures in milk and governing liquid (2PL), after 1 (T1), 8 (T8), and 16 (T16) days of storage at 4°C. Control is burrata cheese without FOS, inulin, or protective culture. Samples are sorted based on weighted UniFrac distances. The color key defines the percentages of OTU in the samples.
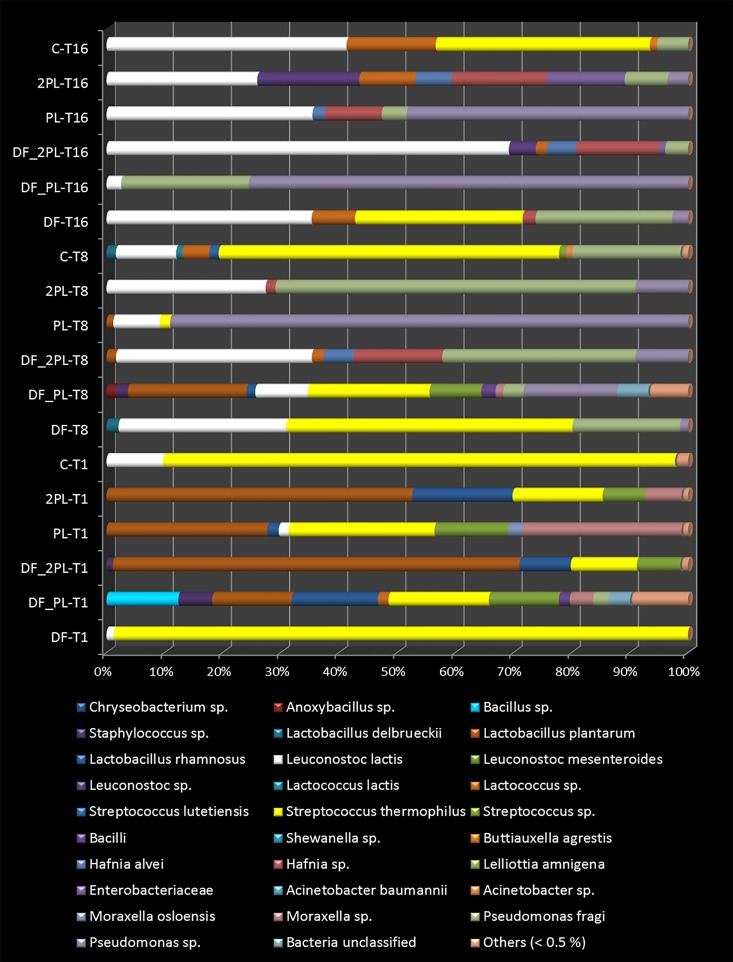
At the species level, L. plantarum and L. rhamnosus were detected in burrata cheeses with added protective bacteria until 16 days of storage (Fig. 4). They were absent in both control and DF cheeses at all time points assayed. Lactobacillus delbrueckii was always found in DF and control cheeses, while it was never detected in the other cheeses after 8 and/or 16 days of storage. The detection value of Leuconostoc lactis was the highest in control cheese after 1 day and increased during storage, especially for DF_2PL cheese. After 16 days of storage, the lowest value was found for DF_PL cheese. Lactococcus lactis was detected in all samples after 1 day of storage, but it was not detected after 16 days in DF_PL and PL cheeses. The abundance of Lactococcus spp. increased during 16 days of storage, and the lowest relative abundance was for DF_PL and PL cheeses. S. thermophilus dominated the microbiome of DF and control cheeses, but it was inhibited in the other cheeses. Streptococcus lutetiensis and a Streptococcus sp. were detected in all samples after 1 day of storage. S. lutetiensis was not detected in cheeses containing protective bacteria after 8 and 16 days of storage. The Streptococcus sp. decreased during storage and after 16 days, and the highest value found was for the control cheese.
FIG 4.
Relative abundance (%) of OTU assigned to the highest possible taxonomic level found as significantly (P < 0.05) different in the burrata cheeses supplemented with fructooligosaccharides (FOS) and inulin (DF), protective cultures in milk with FOS and inulin (DF_PL), protective cultures in milk and governing liquid with FOS and inulin (DF_2PL), protective cultures in milk (PL), and protective cultures in milk and governing liquid (2PL) after 1 (T1), 8 (T8), and 16 (T16) days of storage at 4°C. Control (C) is burrata cheese without FOS, inulin, or protective culture.
Compared to the control, FOS- and inulin-enriched cheese (DF) showed no OTU belonging to an Anoxybacillus sp. (Fig. 4), Staphylococcus epidermidis (see Table S4), a Janthinobacterium sp., Hafnia alvei, Moraxella osloensis, Pseudomonas fragi, Pseudomonas hibiscicola, or a Pseudomonas sp. after 1 day of storage. Except for H. alvei and Pseudomonas species, all these OTU decreased during storage. An Anoxybacillus sp. and S. epidermidis were not detected after 16 days. Similar trends were also found for Propionibacterium acnes, a Chryseobacterium sp., Bacillus sporothermodurans, a Bacillus sp., a Staphylococcus sp., and Acinetobacter baumannii. An Acinetobacter sp. also decreased during storage and, after 16 days, was detected only in 2PL and control samples. Different trends were found for H. alvei, P. fragi, and a Pseudomonas sp. that increased during storage in all cheeses. The largest relative amounts of P. fragi and a Pseudomonas sp. were found after 8 days in 2PL and PL cheeses, respectively.
OTU correlations were investigated considering genus-level taxonomic assignments (see Fig. S4 and S5), with significant correlations considered at a false discovery rate (FDR) of < 0.05. At 1 day of storage, Lactobacillus was negatively correlated with all the other genera, except for Moraxella (Fig. S4). At 16 days of storage, this genus was negatively correlated only with Streptococcus and Pseudomonas (Fig. S5). Similar results were found also for Leuconostoc.
Assessment of proteolysis and concentration of free amino acids.
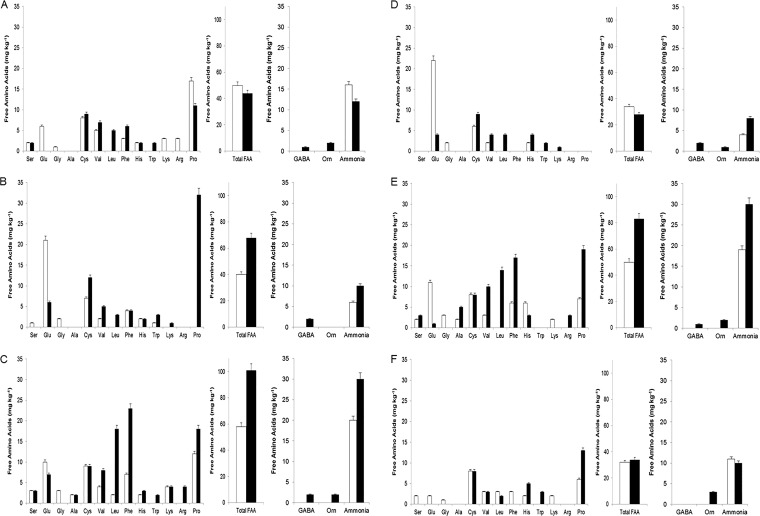
Urea-PAGE profiles of pH 4.6-insoluble N fractions showed very reduced hydrolysis of caseins throughout storage without statistical (P > 0.05) differences among the cheeses (see Fig. S2). Peptide profiles of the pH 4.6-soluble N fractions showed specific differences between cheeses at both 1 and 16 days of storage (Fig. 5). Cheeses also differed for the free amino acid (FAA) profiles (Fig. 6). After 1 day of storage, control and PL cheeses showed the lowest levels of total FAA. Glu, Gly, Cys, Val, and His were found in all cheeses. Except for PL cheese, Ser and Phe were also detected in all the cheeses. During storage, the total FAAs increased for DF_PL, DF_2PL, and 2PL cheeses, while decreases were found for DF and PL cheeses. With few exceptions, Val, Leu, Phe, Trp, and Pro strongly increased during storage. On the contrary, Glu decreased and one of its metabolites (γ-aminobutyric acid [GABA]) increased. Regarding FAA-derived compounds, ammonia and ornithine (orn) also increased in most of the cheeses during storage. Correlations between genera and metabolites were found (see Fig. S6). Overall, at 1 and 16 days of storage, Lactobacillus was positively correlated with all the amino acids except Trp. At 16 days of storage, a positive correlation was found for Leuconostoc and acetic acid.
FIG 5.
RP-FPLC peptide profiles of the pH 4.6-soluble fraction of the burrata cheeses supplemented with fructooligosaccharides (FOS) and inulin (DF) (A), protective cultures in milk with FOS and inulin (DF_PL) (B), protective cultures in milk and governing liquid with FOS and inulin (DF_2PL) (C), protective cultures in milk (PL) (D), and protective cultures in milk and governing liquid (2PL) (E) after 1 (continuous lines) and 16 (dotted lines) days of storage at 4°C. (F) Control is burrata cheese without FOS, inulin, or protective culture.
FIG 6.
Concentrations of individual and total free amino acids (FAA), γ-aminobutyric acid (GABA), ornithine, and ammonia in the pH 4.6-soluble fractions of the burrata cheeses supplemented with fructooligosaccharides (FOS) and inulin (DF) (A), protective cultures in milk with FOS and inulin (DF_PL) (B), protective cultures in milk and governing liquid with FOS and inulin (DF_2PL) (C), protective cultures in milk (PL) (D), and protective cultures in milk and governing liquid (2PL) (E) after 1 (white bars) and 16 (black bars) days of storage at 4°C. (F) Control is burrata cheese without FOS, inulin and protective culture.
Sensory analysis.
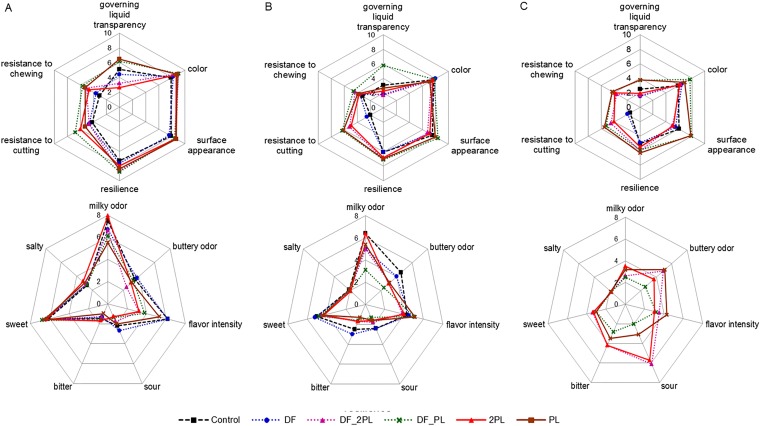
The sensory characteristics of the six burrata cheeses were determined during 16 days of storage at 4°C (Fig. 7; see also Fig. S3). Most of the attributes (especially “resilience,” “milky odor,” and “sweet flavor”) decreased over time on average. However, sour and bitter flavors increased during the shelf life. At 1 day of storage, significant differences (P < 0.05) were found for the governing liquid transparency, the resistance to chewing and cutting, milky and buttery odors, flavor intensity, and sourness. Specifically, the clearest governing liquid was found for DF_PL and PL cheeses, whereas DF_2PL and 2PL cheeses were characterized by the lowest transparency of the governing liquid. Overall, the transparency of governing liquid and the resistance to cutting and chewing were the highest for DF_PL and PL cheeses during further storage. Control and DF cheeses showed the strongest decreases of resistance to chewing and, especially, cutting. The scores for color and surface appearance were not significantly different (P > 0.05) until 8 days. However, at 13 and 16 days of storage, DF_PL and PL cheeses received the highest scores for both these parameters. Milky odor at the highest score was found for control and 2PL cheeses (until 8 days) and for 2PL and PL cheeses (13 and 16 days). Based on texture, sourness, and bitterness scores, DF and control cheeses were not further tasted. At the end of storage, only DF_PL and PL cheeses were judged with sufficient scores. Indeed, the other cheeses (DF_2PL and 2PL) showed excessive sourness and bitterness.
FIG 7.
Sensory analysis of the burrata cheeses supplemented with fructooligosaccharides (FOS) and inulin (DF), protective cultures in milk with FOS and inulin (DF_PL), protective cultures in milk and governing liquid with FOS and inulin (DF_2PL), protective cultures in milk (PL), and protective cultures in milk and governing liquid (2PL), after 1 (A), 8 (B) and 16 (C) days of storage at 4°C. Control is burrata cheese without FOS, inulin, or protective culture.
DISCUSSION
This study used probiotic protective Lactobacillus strains and prebiotic dietary fibers (FOS and inulin) to improve the functional attributes (26) and shelf life of burrata cheese. Burrata, fresh pasta filata cheese with cream, is rapidly spreading in Europe and the United States, but it has a very high caloric content and is characterized by a high risk of microbial contaminations with a short shelf life (10, 27). Recently, a prototype of reduced-fat polyunsaturated fatty acids (PUFA)-enriched burrata cheese was described (6) but without market development. FOS lower blood cholesterol (28) and glucose (29), as well as the risk of cardiovascular disease, diabetes, hypertension, obesity, and gastrointestinal disorders, and improve lipid metabolism (30–32). FOS and inulin drive both the gut microbiota composition and the related metabolome, including the synthesis of short-chain fatty acids (SCFA), whose positive effects on human health are well known (24). Using approaches similar to those previously described for other cheeses (11, 14, 33, 34), protective probiotic L. plantarum LPAL and L. rhamnosus LRB (35, 36) were applied to inhibit undesired bacteria with or without the addition of FOS and inulin. Overall, the addition of inulin and FOS and the use of protective strains modified the microbiome assembly of burrata cheese. During cheese making and 16 days of storage, burrata cheeses showed different microbiomes and biochemical and sensory features. Stochastic (e.g., dispersal), temporal, and deterministic (e.g., biotic and abiotic factors) drivers shape not only the microbiome assembly but, most relevantly, the microbiome functionalities (37–39). According to previous findings (40, 41), during cheese making and storage, the house microbiota components (including Firmicutes, such as Streptococcus, and Proteobacteria, such as Pseudomonas, etc.) are transferred to the burrata samples. The general paradigm is that generalists with redundant metabolic traits mainly assemble stochastically, while specialists for selected metabolisms are mainly determined by the food components and environmental drivers (38). Thus, the final microbiome results from both deterministic drivers and functional traits of microbes, which co-occur or disappear, depending on their capacity to thrive or not in such an environment (38, 42). Well-adapted cheese genera Streptococcus (mainly S. thermophilus) and Lactococcus (L. lactis) dominated the microbiome of burrata cheeses produced traditionally or with the addition of dietary fibers, and accordingly, they acidified the burrata during cheese making. This negatively affects the sensory properties (especially mild taste) of fresh dairy products (11) such as burrata cheese. Previously, it was found that mesophilic (L. lactis) and thermophilic (S. thermophilus, L. delbrueckii, and L. helveticus) lactic acid bacteria are contaminants of chemically acidified mozzarella and burrata cheeses (43, 44). First, this study showed that protective lactobacilli drive the microbial communities, also reducing the Streptococcus and Lactococcus contamination during cheese making. Compared to the control, the addition of dietary fibers and protective lactobacilli in milk strongly reduced L. lactis, which is involved in the spoilage of fresh products (45). Based on culture-dependent methods, protective lactobacilli slowed the growth of staphylococci, coliforms, and Pseudomonas spp., especially in early storage. The bacterial groups that were inhibited are all recognized as pathogens/spoiling bacteria of fresh cheeses that reduce the shelf lives of the products (45, 46). Human and animal pathogenic species were not detected. Pseudomonas fragi and another Pseudomonas sp. increased in all the cheeses after 8 days of storage. Partial inhibition of Pseudomonas species by protective bacteria was also described for ricotta fresca (11). Previously, P. fragi and Pseudomonas spp. were the only Proteobacteria detected in burrata cheeses (44). This study also identified Hafnia spp. and Hafnia alvei as a component of the burrata cheese microbiome. H. alvei is not conventionally used in food processing but influences the synthesis of sulfur compounds in cheese (47). The double inoculum of protective L. rhamnosus LRB and L. plantarum LPAL resulted in a specifically different microbiome during storage.
According to the different microbiome assemblies, burrata samples differed in peptide profiles and levels of free amino acids (FAA). Previously, it was reported that probiotic lactobacilli affect the proteolysis of mozzarella and other cheese varieties depending on the strains (48–50). Microorganisms, including probiotic lactobacilli, metabolize FAA to obtain ATP and other compounds to survive under hostile abiotic conditions, such as acid and cold stresses (51, 52). The decarboxylation of acid substrates (such as glutamic acid) into a neutral compound (GABA), by consuming an H+, increases the intracellular pH (53, 54). The catabolism of FAA by deaminase pathways producing ammonia is another common mechanism of probiotic lactobacilli for survival under acid and/or starvation stress (51, 53). The synthesis/liberation of FAA constitutes a key factor impacting the flavor of cheeses, and FAA catabolism, by forming volatile organic compounds, influences taste and aroma (55). As shown by the sensory analysis, the addition of protective lactobacilli improved the flavor of the burrata cheeses. Overall, no body/texture or bitter defects were found in cheeses containing dietary fibers and protective lactobacilli. The use of protective lactobacilli strains increased the shelf life of burrata cheese by 3 days. Compared to cheeses made using a protective culture only in the milk, those also containing cultures in the governing liquid did not have a prolonged shelf life and received lower acceptability scores by the panelists.
This study provided an in-depth knowledge on the microbiome of burrata cheese and set up a novel biotechnology using prebiotic dietary fibers and probiotic protective L. plantarum and L. rhamnosus in milk. Although further studies are needed to show the in vivo probiotic and prebiotic effects, the biotechnology proposed in this study should be considered a useful tool to improve the functional value of burrata cheese. The use of protective L. plantarum and L. rhamnosus in milk during cheese making enhanced the flavor formation and shelf life of burrata cheese.
MATERIALS AND METHODS
Protective cultures and dietary fibers.
Commercially available freeze-dried probiotic Lactobacillus plantarum LPAL (Lyofast LPAL) and Lactobacillus rhamnosus LRB (Lyofast LRB) were purchased from Clerici-Sacco S.r.l. (Cadorago, Italy) (35). L. plantarum LPAL is a bacteriocin-producing strain that can be used as a protective culture for cheese to inhibit undesired bacteria (e.g., Listeria spp. and the pathogens Clostridium difficile and Clostridium perfringens) (35). Similar to L. rhamnosus GG, the strain LRB showed four bacteriocin loci (56) and it is used as a protective culture in various food and feed items, because it inhibits undesired microorganisms. Powdered fructooligosaccharides ([FOS] 95.9% fiber) and inulin (93.0% dietary fiber) extracted from chicory roots were purchased from Farmalabor S.r.l. (Canosa di Puglia, Italy).
Manufacture of burrata cheese.
Six types of burrata cheese were manufactured at the industrial plant Ignalat (in a vat of ∼200 liters), located in Noci (Bari, Italy) (see Fig. S1 in the supplemental material), namely, cheese (i) supplemented with FOS and inulin (DF cheese); (ii) supplemented with FOS, inulin, and protective lactobacilli in milk (DF_PL cheese); (iii) supplemented with FOS, inulin, and protective lactobacilli in milk and governing liquid (DF_2PL cheese); (iv) supplemented with protective lactobacilli in milk (PL cheese); (v) supplemented with protective lactobacilli in milk and governing liquid (2PL_cheese); and (vi) without protective cultures and fibers (control). Pasteurized cows' milk was acidified with 1.45 g · liter−1 lactic acid. Freeze-dried protective lactobacilli were added to milk (final cell density for each strain of ca. 7 log CFU · g−1), except for the DF and control cheeses. Thirty minutes after the addition of liquid calf rennet, coagulation took place. The coagulum was first cut, held under whey at 37°C for 2 h, and then reduced to particles of 1.5 to 2 cm in diameter. The curd was stretched (ca. 65°C for 5 min) in hot (80°C) water after it reached a pH of 5.7. The filling consisted of ultra-high-processing-temperature (UHT) milk cream (65%) and strips (frayed stretched curd pieces, 35%). FOS (0.8% of the total cheese weight) and inulin (0.8% of the total cheese weight) were dissolved in the filling only for the burrata cheeses supplemented with fibers (DF, DF_PL, and DF_2PL cheeses). The filling was inserted in the stretched curd, and burrata cheese was given its final shape. After being hardened by dipping in cold water, cheeses (single piece weighing ca. 100 g) were singly packaged in plastic tubs containing governing liquid. Only for DF_2PL and 2PL cheeses, protective lactobacilli were added to the governing liquid (final cell density of ca. 7 log CFU · g−1). The tubs were sealed and stored at 4°C for 16 days. Each type of burrata cheese was manufactured in triplicate.
Compositional and physicochemical analyses.
Burrata cheeses were analyzed for the concentrations of total carbohydrates (57), proteins (58), fats (59), and sodium chloride (60). Moisture and ashes were determined according to Association of Official Analytical Chemists International (AOAC) official methods 923.10 (61) and 923.03 (62), respectively. The pH was determined by direct insertion of a Foodtrode (Hamilton, Bonaduz, Switzerland) electrode. Water activity (aw) was determined at 25°C by the Aqualab Dew Point 4TE water activity meter (Decagon Devices Inc., USA). Concentrations of glucose, lactose, galactose, lactic and acetic acids, and ethanol were determined by high-pressure liquid chromatography (HPLC) using an Äkta purifier system (GE Healthcare Biosciences, Uppsala, Sweden) equipped with a 300-mm 7.8-mm-internal-diameter cation exchange column (Aminex HPX-87H, Bio-Rad Laboratories) and a Perkin-Elmer 200a refractive index detector (Perkin-Elmer Corp., Waltham, MA). Elution was carried out isocratically at 60°C, with a flow rate of 0.3 ml/min and using H2SO4 as the mobile phase (63).
Cultivable microbiota.
Microbiological analyses were carried out as previously described (48), using culture media and supplements purchased from Oxoid. Ten grams of cheese was homogenized with 90 ml of sterile saline (NaCl, 9 g · liter−1) in a 400P bag mixer (3 min of treatment). Total amounts of mesophilic aerobic microorganisms were determined using plate count agar after incubating at 30°C. Presumptive mesophilic and thermophilic lactobacilli were enumerated using MRS agar plates incubated at 30°C and 45°C, respectively. Presumptive mesophilic and thermophilic cocci were enumerated using lactose M17 agar plates incubated at the same temperatures as above. Enterococci were counted after inoculating, by the spreading technique, plates of Slanetz and Bartley agar and incubating at 37°C. Staphylococci were determined using Baird Parker agar supplemented with egg yolk tellurite and were inoculated by the spreading technique and incubated at 37°C. Total and fecal coliforms were plate counted on violet red bile glucose agar (VRBGA) after incubating plates at 37°C and 45°C, respectively. Plates of Pseudomonas agar supplemented with cetrimide, fucidin, and cephalosporin (CFC supplement) were spread inoculated with 0.1 ml of diluted sample and used to enumerate Pseudomonas spp. after incubating at 30°C. All plates were incubated for 48 h, except for VRBGA and Pseudomonas agar, which were incubated for 24 h.
Extraction of total bacterial genomic RNA and reverse transcription.
Forty-five milliliters of saline solution was added to 5 g of burrata cheese and homogenized for 3 min. Homogenates were centrifuged (1,000 × g for 5 min at 4°C) and the supernatants were recovered, manually defatted, and centrifuged (5,000 × g for 15 min at 4°C). The pellets were suspended in 1 ml of saline solution and further centrifuged (21,600 × g for 1 min at 4°C). After discarding the supernatants, the pellets were used for the extraction of RNA. Total RNA was extracted using the RiboPure-bacteria kit (Ambion RNA, Life Technologies Co., Carlsbad, CA), according to the manufacturer's instructions (64). The concentrations of extracted RNA were determined by spectrophotometric determination (Nanodrop ND-1000; Thermo Fisher Scientific Inc.). The purified RNA (100 ng) was mixed with a random hexamer primer mix, deoxynucleoside triphosphates (dNTPs), RNase inhibitor, and reverse transcriptase (Tetro cDNA synthesis kit; Bioline USA Inc., Taunton, MA) and incubated (25°C for 10 min, 45°C for 30 min, and 85°C for 5 min) to obtain cDNA, according to the manufacturer's instructions (65). cDNA from three cheese-making trials was pooled, dried using a vacuum centrifuge (SpeedVac Concentrator SPD121P; Thermo Scientific), and used as the template for 16S metagenetics.
Analysis of bacterial diversity.
16S metagenetics was carried out at RTLGenomics (Lubbock, TX) using the Illumina MiSeq platform. A fragment of the 16S rRNA gene for analysis of the diversity inside the domain of Bacteria was amplified using the primers 28F (GAGTTTGATCNTGGCTCAG) (66) and 519R (GTNTTACNGCGGCKGCTG) (67). PCR and sequencing analyses were carried out according to the protocol of RTLGenomics.
The sequenced reads were processed through denoising and chimera detection. Specifically, denoising was performed by (i) merging the forward and reverse reads using the PEAR Illumina paired-end read merger (68); (ii) grouping reads (having an average quality higher than 25) using the USEARCH (69) algorithm (prefix dereplication) into clusters (4% dissimilarity among sequences of the same cluster), so that each sequence of a length shorter than the centroid sequence must be a 100% match to the centroid sequence for the length of the sequence; and (iii) selecting operational taxonomic units (OTU) using the UPARSE OTU selection algorithm (70). Following denoising, the selected OTU were chimera checked using the UCHIME software (71). Specifically, each trimmed read was mapped to its corresponding nonchimeric cluster using the USEARCH global alignment algorithm (69). Each sequence in a cluster was then aligned to the consensus sequence. Each sequence was corrected base by base to remove noise. An analysis of bacterial diversity was finally performed by running the centroid sequences from each cluster against the USEARCH algorithm, using a database of high quality sequences derived from the NCBI. Lastly, the outputs were analyzed using an internally developed python program that assigns taxonomic information to each sequence.
The percentages of each of the bacterial OTU were analyzed individually for each sample, providing relative abundance information among the samples based on the relative numbers of reads within each (72). Alpha diversity (Chao 1 richness and Shannon diversity indices) was calculated using QIIME (73–75).
Assessment of proteolysis and concentration of free amino acids.
The pH 4.6-soluble and -insoluble extracts of burrata cheeses were obtained as described by Kuchroo and Fox (76). The pH 4.6-insoluble fractions were analyzed by urea-PAGE using a Protean II xi vertical slab gel unit (Bio-Rad Laboratories) and the stacking gel system. Gels were stained by the method described by Blakesley and Boezi (77) with Coomassie brilliant blue G250, and the color background was faded by continuous washing in distilled water. Gel images were acquired by a flatbed scanner (Expression 11000 XL; Epson). The peptide profiles of the pH 4.6-soluble fractions were determined by reverse-phase (RP) chromatography with a Resource RPC 3-ml column using an Äkta fast protein liquid chromatography (FPLC) system (GE Healthcare Biosciences). Concentrations of total and individual free amino acids (FAA) in the pH 4.6-soluble extract were determined using the Biochrom 30 amino acid analyzer (Biochrom LTD, Cambridge Science Park, England) as previously described (78).
Sensory analysis.
The sensory analysis of burrata cheese was carried out using the descriptive model described by Coppola et al. (79) as modified by De Angelis et al. (78). Ten volunteers (5 male and 5 female; mean age, 30 years [range, 20 to 40 years]) were recruited from the laboratory staff. Three introductory sensory training sessions were held for discussing the sensory attributes with the panelists. Before the sensory evaluation, cheeses were taken out of the refrigerator 1 h before serving and served at room temperature under normal (daylight) illumination. Each cheese (two pieces per thesis), identified by a code number, was given to each panelist on a single tray. Samples were served in a random order and evaluated in two replicates by all panelists. The quality attributes evaluated were governing liquid transparency, color, surface appearance, resilience, resistance to cutting, resistance to chewing, milky odor, buttery odor, flavor intensity, sourness, bitterness, sweetness, and salty taste. Each sensory trait was rated with a score from 0 (lowest) to 10 (highest).
Statistical analyses.
Data were subjected to one-way analysis of variance (ANOVA), and pair-wise comparisons of treatment means were achieved by Tukey's test at a P value of < 0.05, using the statistical software Statistica v. 7.0 for Windows. Multivariate differences among burrata cheeses were estimated by the permutational multivariate analysis of variance using the distance matrix function in ADONIS (80). For ADONIS, distances among samples first were calculated using the weighted UniFrac, and then an ANOVA-like simulation was conducted to test for group differences. Spearman correlations for OTU, as well as between OTU and metabolite concentrations, were computed using Statistica v. 7.0 and elaborated through PermutMatrix software.
Accession number(s).
The 16S rRNA gene sequences are available in the Sequence Read Archive of NCBI (accession number PRJNA392015).
Supplementary Material
ACKNOWLEDGMENTS
F.M. supervised the biochemical and microbiological analyses, A.C. supervised sensory evaluation, M.A.D.N. revised the manuscript, M.G. codesigned the study and revised the manuscript, and M.D.A. codesigned the study, directed the experimental phases, and wrote the manuscript.
This work was supported by Apulian Region Project no. QCBRAJ6 (Biotecnologie degli alimenti per l'innovazione e la competitività delle principali filiere regionali: estensione della conservabilità e aspetti funzionali—BIOTECA).
We thank the industrial plant Ignalat, located in Noci, Bari (Apulia region), Italy, for experimental cheese-making trials. We thank Farmalabor (Canosa di Puglia, BT, Italy) for providing scientific support in the use of FOS and inulin. We also thank Sabino Formiglio, Cristina Costa, and Anna Lattanzi for technical assistance.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01494-17.
REFERENCES
- 1.Italian Parliament. 20 June 2014. No. 141. The Official Gazette of the Italian Republic. Istituto Poligrafico e Zecca dello Stato, Rome, Italy. [Google Scholar]
- 2.European Commission. 2 December 2016. Regulations. Commission implementing regulation (EU) 2016/2103. L 327/16. Official Journal of the European Union. European Union, Brussels, Belgium. [Google Scholar]
- 3.Conte A, Brescia I, Del Nobile MA. 2011. Lysozyme/EDTA disodium salt and modified-atmosphere packaging to prolong the shelf life of burrata cheese. J Dairy Sci 94:5289–5297. doi: 10.3168/jds.2010-3961. [DOI] [PubMed] [Google Scholar]
- 4.Rea S, Marino L, Stocchi R, Branciari R, Loschi AR, Miraglia D, Ranucci D. 2016. Differences in chemical, physical and microbiological characteristics of Italian burrata cheeses made in artisanal and industrial plants of Apulia Region. Ital J Food Saf 5:e5879. doi: 10.4081/ijfs.2016.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salvadori del Prato O. 2001. Trattato di tecnologia casearia. Calderoni ed Agricole, Bologna, Italy. [Google Scholar]
- 6.Trani A, Gambacorta G, Gomes TF, Loizzo P, Cassone A, Faccia M. 2016. Production and characterisation of reduced-fat and PUFA-enriched Burrata cheese. J Dairy Res 83:236–241. doi: 10.1017/S0022029916000078. [DOI] [PubMed] [Google Scholar]
- 7.Faccia M, Gammariello AD, Conte A, Del Nobile MA. 2013. Pasta filata cheeses: advances in processing and preservation, p 251–260. In Preedy VR, Patel V, Watson RR (ed), Handbook of cheese in health: production, nutrition and medical sciences. Academic Publishers, Wageningen, The Netherlands. [Google Scholar]
- 8.Tantillo MG. 2007. I prodotti tradizionali pugliesi: burrata. Infosei, Bari, Italy. [Google Scholar]
- 9.Pintado ME, Macedo AC, Malcata FX. 2001. Review: technology, chemistry and microbiology of whey cheeses. Food Sci Technol Int 7:105–116. doi: 10.1177/108201320100700202. [DOI] [Google Scholar]
- 10.Tirloni E, Stella S, Bernardi C. 2014. Concerns about the microbiological quality of traditional raw milk cheeses: a worldwide issue. Int J Health Anim Sci Food Saf 1:24–31. [Google Scholar]
- 11.Spanu C, Scarano C, Piras F, Spanu V, Pala C, Casti D, Lamon S, Cossu F, Ibba M, Nieddu G, De Santis EPL. 2017. Testing commercial biopreservative against spoilage microorganisms in MAP packed Ricotta fresca cheese. Food Microbiol 66:72–76. doi: 10.1016/j.fm.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 12.Sobrino-Lopez A, Martín-Belloso O. 2008. Use of nisin and other bacteriocins for preservation of dairy products. Int Dairy J 18:329–343. doi: 10.1016/j.idairyj.2007.11.009. [DOI] [Google Scholar]
- 13.Elsser-Gravesen D, Elsser-Gravesen A. 2013. Biopreservatives, p 29–49. In Zorn H, Czermak P (ed), Biotechnology of food and feed additives. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 14.Grattepanche F, Miescher-Schwenninger S, Meile L, Lacroix C. 2008. Recent developments in cheese cultures with protective and probiotic functionalities. Dairy Sci Technol 88:421–444. doi: 10.1051/dst:2008013. [DOI] [Google Scholar]
- 15.Coelho MC, Silva CCG, Ribeiro SC, Dapkevicius MLNE, Rosa HJD. 2014. Control of Listeria monocytogenes in fresh cheese using protective lactic acid bacteria. Int J Food Microbiol 191:53–59. doi: 10.1016/j.ijfoodmicro.2014.08.029. [DOI] [PubMed] [Google Scholar]
- 16.Mills S, Griffin C, O'Connor PM, Serrano LM, Meijer WC, Hill C, Ross RP. 2017. A multibacteriocin cheese starter system comprising nisin and lacticin 3147 in Lactococcus lactis, in combination with plantaricin from Lactobacillus plantarum. Appl Environ Microbiol 83:e00799-17. doi: 10.1128/AEM.00799-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giménez A, Ares F, Ares G. 2012. Sensory shelf-life estimation: a review of current methodological approaches. Food Res Int 49:311–325. doi: 10.1016/j.foodres.2012.07.008. [DOI] [Google Scholar]
- 18.Alnemr TM, El-Razek AMA, Hasan HMA, Massoud MI. 2013. Improving of Karish cheese by using enhanced technological texturizing inulin. Alex J Agr Res 58:173–181. [Google Scholar]
- 19.Karimi R, Azizi MH, Ghasemlou M, Vaziri M. 2015. Application of inulin in cheese as prebiotic, fat replacer and texturizer: a review. Carbohydr Polym 119:85–100. doi: 10.1016/j.carbpol.2014.11.029. [DOI] [PubMed] [Google Scholar]
- 20.Meyer D, Bayarri S, Tárrega A, Costell E. 2011. Inulin as texture modifier in dairy products. Food Hydrocoll 25:1881–1890. doi: 10.1016/j.foodhyd.2011.04.012. [DOI] [Google Scholar]
- 21.Cardarelli HR, Buriti FCA, de Castro IA, Saad SMI. 2008. Inulin and oligofructose improve sensory quality and increase the probiotic viable count in potentially symbiotic petit-suisse cheese. Lebenson Wiss Technol 41:1037–1046. doi: 10.1016/j.lwt.2007.07.001. [DOI] [Google Scholar]
- 22.Araujo EA, de Carvalho AF, Leandro ES, Furtado MM, de Moraes CA. 2010. Development of a symbiotic cottage cheese added with Lactobacillus delbrueckii UFV H2b20 and inulin. J Funct Foods 2:85–89. doi: 10.1016/j.jff.2009.12.002. [DOI] [Google Scholar]
- 23.Ferrão LL, Silva EB, Silva HLA, Silva R, Mollakhalili N, Granato D, Freitas MQ, Silva MC, Raices RSL, Padilha MC, Zacarchenco PB, Barbosa MIMJ, Mortazavian AM, Cruz AG. 2016. Strategies to develop healthier processed cheeses: reduction of sodium and fat contents and use of prebiotics. Food Res Int 86:93–102. doi: 10.1016/j.foodres.2016.04.034. [DOI] [Google Scholar]
- 24.Ríos-Covián D, Ruas-Madiedo P, Margolles A, Gueimonde M, de los Reyes-Gavilán CG, Salazar N. 2016. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol 7:e185. doi: 10.3389/fmicb.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.European Commission. 7 December 2015. Commission regulation (EU) 2015/2314. L328 European Union, Brussels, Belgium. [Google Scholar]
- 26.Vijaya Kumar B, Vijayendra SV, Reddy OV. 2015. Trends in dairy and non-dairy probiotic products—a review. J Food Sci Technol 52:6112–6124. doi: 10.1007/s13197-015-1795-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dambrosio A, Quaglia NC, Saracino M, Malcangi M, Montagna C, Quinto M, Lorusso V, Normanno G. 2013. Microbiological quality of burrata cheese produced in Puglia region, southern Italy. J Food Prot 76:1981–1984. doi: 10.4315/0362-028X.JFP-13-067. [DOI] [PubMed] [Google Scholar]
- 28.Costa GT, Abreu GC, Guimarães AB, Vasconcelos PR, Guimarães SB. 2015. Fructo-oligosaccharide effects on serum cholesterol levels. An overview. Acta Cir Bras 30:366–370. doi: 10.1590/S0102-865020150050000009. [DOI] [PubMed] [Google Scholar]
- 29.Costa GT, Guimarães SB, Sampaio HA. 2012. Fructo-oligosaccharide effects on blood glucose: an overview. Acta Cir Bras 27:279–282. doi: 10.1590/S0102-86502012000300013. [DOI] [PubMed] [Google Scholar]
- 30.Anderson JW, Baird P, Davis RH Jr, Ferreri S, Knudtson M, Koraym A, Waters V, Williams CL. 2009. Health benefits of dietary fiber. Nutr Rev 67:188–205. doi: 10.1111/j.1753-4887.2009.00189.x. [DOI] [PubMed] [Google Scholar]
- 31.Raninen K, Lappi J, Mykkänen H, Poutanen K. 2011. Dietary fiber type reflects physiological functionality: comparison of grain fiber, inulin, and polydextrose. Nutr Rev 69:9–21. doi: 10.1111/j.1753-4887.2010.00358.x. [DOI] [PubMed] [Google Scholar]
- 32.Williams CM, Jackson KG. 2002. Inulin and oligofructose: effects on lipid metabolism from human studies. Br J Nutr 87:S261–S264. doi: 10.1079/BJN/2002546. [DOI] [PubMed] [Google Scholar]
- 33.Aljewicz M, Cichosz G. 2015. Protective effects of Lactobacillus cultures in Dutch-type cheese-like products. Lebenson Wiss Technol 63:52–56. doi: 10.1016/j.lwt.2015.03.054. [DOI] [Google Scholar]
- 34.Arqués Juan L, Rodríguez E, Langa S, Landete JM, Medina M. 2015. Antimicrobial activity of lactic acid bacteria in dairy products and gut: effect on pathogens. Biomed Res Int 2015:584183. doi: 10.1155/2015/584183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schoster A, Kokotovic B, Permin A, Pedersen PD, Dal Bello F, Guardabassi L. 2013. In vitro inhibition of Clostridium difficile and Clostridium perfringens by commercial probiotic strains. Anaerobe 20:36–41. doi: 10.1016/j.anaerobe.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 36.Nyanzi R, Jooste PJ, Abu JO, Beukes EM. 2010. Consumer acceptability of a synbiotic version of the maize beverage mageu. Dev South Afr 27:447–463. doi: 10.1080/0376835X.2010.498955. [DOI] [Google Scholar]
- 37.Shafquat A, Joice R, Simmons S, Huttenhower C. 2014. Functional and phylogenetic assembly of microbial communities in the human microbiome. Trends Microbiol 22:261–266. doi: 10.1016/j.tim.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolfe BE, Dutton RJ. 2015. Fermented foods as experimentally tractable microbial ecosystems. Cell 16:49–55. doi: 10.1016/j.cell.2015.02.034. [DOI] [PubMed] [Google Scholar]
- 39.De Filippis F, Genovese A, Ferranti P, Gilbert JA, Ercolini D. 2016. Metatranscriptomics reveals temperature-driven functional changes in microbiome impacting cheese maturation rate. Sci Rep 6:21871. doi: 10.1038/srep21871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bokulich NA, Mills DA. 2013. Facility-specific “house” microbiome drives microbial landscapes of artisan cheesemaking plants. Appl Environ Microbiol 79:5214–5223. doi: 10.1128/AEM.00934-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calasso M, Ercolini D, Mancini L, Stellato G, Minervini F, Di Cagno R, De Angelis M, Gobbetti M. 2016. Relationships among house, rind and core microbiotas during manufacture of traditional Italian cheeses at the same dairy plant. Food Microbiol 54:115–126. doi: 10.1016/j.fm.2015.10.008. [DOI] [Google Scholar]
- 42.De Filippis F, Parente E, Ercolini D. 2017. Metagenomics insights into food fermentations. Microb Biotechnol 10:91–102. doi: 10.1111/1751-7915.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guidone A, Zotta T, Matera A, Ricciardi A, De Filippis F, Ercolini D, Parente E. 2016. The microbiota of high-moisture mozzarella cheese produced with different acidification methods. Int J Food Microbiol 216:9–17. doi: 10.1016/j.ijfoodmicro.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 44.Garofalo C, Bancalari E, Milanović V, Cardinali F, Osimani A, Savo Sardaro ML, Bottari B, Bernini V, Aquilanti L, Clementi F, Neviani E, Gatti M. 2017. Study of the bacterial diversity of foods: PCR-DGGE versus LH-PCR. Int J Food Microbiol 242:24–36. doi: 10.1016/j.ijfoodmicro.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 45.Remenant B, Jaffrès E, Dousset X, Pilet M-F, Zagorec M. 2015. Bacterial spoilers of food: behavior, fitness and functional properties. Food Microbiol 45:45–53. doi: 10.1016/j.fm.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 46.Baruzzi F, Lagonigro R, Quintieri L, Morea M, Caputo L. 2012. Occurrence of non-lactic acid bacteria populations involved in protein hydrolysis of cold-stored high moisture mozzarella cheese. Food Microbiol 30:37–44. doi: 10.1016/j.fm.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 47.Irlinger F, Yung SA, Sarthou A-S, Delbès-Paus C, Montel M-C, Coton E, Coton M, Helinck S. 2012. Ecological and aromatic impact of two Gram-negative bacteria (Psychrobacter celer and Hafnia alvei) inoculated as part of the whole microbial community of an experimental smear soft cheese. Int J Food Microbiol 153:332–338. doi: 10.1016/j.ijfoodmicro.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 48.Minervini F, Siragusa S, Faccia M, Dal Bello F, Gobbetti M, De Angelis M. 2012. Manufacture of fior di latte cheese by incorporation of probiotic lactobacilli. J Dairy Sci 95:508–520. doi: 10.3168/jds.2011-4150. [DOI] [PubMed] [Google Scholar]
- 49.Ong L, Henriksson A, Shah NP. 2007. Proteolytic pattern and organic acid profiles of probiotic cheddar cheese as influenced by probiotic strains of Lactobacillus acidophilus, Lb. paracasei, Lb. casei or Bifidobacterium sp. Int Dairy J 17:67–78. doi: 10.1016/j.idairyj.2005.12.009. [DOI] [Google Scholar]
- 50.Bergamini CV, Hynes ER, Candioti MC, Zalazar CA. 2009. Multivariate analysis of proteolysis patterns differentiated the impact of six strains of probiotic bacteria on a semi-hard cheese. J Dairy Sci 92:2455–2467. doi: 10.3168/jds.2008-1794. [DOI] [PubMed] [Google Scholar]
- 51.De Angelis M, Gobbetti M. 2004. Environmental stress responses in Lactobacillus: a review. Proteomics 4:106–122. doi: 10.1002/pmic.200300497. [DOI] [PubMed] [Google Scholar]
- 52.De Angelis M, Calasso M, Cavallo N, Di Cagno R, Gobbetti M. 2016. Functional proteomics within the genus Lactobacillus. Proteomics 16:946–962. doi: 10.1002/pmic.201500117. [DOI] [PubMed] [Google Scholar]
- 53.Papadimitriou K, Alegría A, Bron PA, De Angelis M, Gobbetti M, Kleerebezem M, Lemos JA, Linares DM, Ross P, Stanton C, Turroni F, van Sinderen D, Varmanen P, Ventura M, Zúñiga M, Tsakalidou E, Kok J. 2016. Stress physiology of lactic acid bacteria. Microbiol Mol Biol Rev 80:837–890. doi: 10.1128/MMBR.00076-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siragusa S, De Angelis M, Di Cagno R, Rizzello CG, Coda R, Gobbetti M. 2007. Synthesis of gamma-aminobutyric acid by lactic acid bacteria isolated from a variety of Italian cheeses. Appl Environ Microbiol 73:7283–7290. doi: 10.1128/AEM.01064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De Angelis M, De Pasquale I, Gobbetti M. 2013. Catabolism of free amino acids by lactic acid bacteria during cheese ripening, p 67–96. In Randazzo C, Caggia C, Neviani E (ed), Cheese ripening: quality, safety and health. Nova Science Publishers, Inc., Hauppauge, NY. [Google Scholar]
- 56.Biswas S, Biswas I. 2016. Complete genome sequence of Lactobacillus rhamnosus strain LRB. Genome Announc 4:e01208-16. doi: 10.1128/genomeA.01208-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.James CS. 1995. Analytical chemistry of foods, 1st ed Chapman and Hall, New York, NY. [Google Scholar]
- 58.International Dairy Federation. 1964. Determination of the protein content of processed cheeses products. Standard 25. International Dairy Federation, Brussels, Belgium. [Google Scholar]
- 59.Institute for Industrial Research and Standards. 1955. Determination of the percentage of fat in cheese. Irish standard 69. Institute for Industrial Research and Standards, Dublin, Ireland. [Google Scholar]
- 60.Fox PF. 1963. Potentiometric determination of salt in cheese. J Dairy Sci 46:744–745. doi: 10.3168/jds.S0022-0302(63)89134-1. [DOI] [Google Scholar]
- 61.Association of Official Analytical Chemists. 2016. AOAC: method 923.10 In Horwitz W. (ed), Official methods of analysis of Association of Official Analytical Chemists International, 20th ed AOAC Press, Rockville, MD. [Google Scholar]
- 62.Association of Official Analytical Chemists. 2016. AOAC: method 923.03 In Horwitz W. (ed), Official methods of analysis of Association of Official Analytical Chemists International, 20th ed AOAC Press, Rockville, MD. [Google Scholar]
- 63.Zeppa G, Conterno L, Gerbi V. 2001. Determination of organic acids, sugars, diacetyl, and acetoin in cheese by high-performance liquid chromatography. J Agric Food Chem 49:2722–2726. doi: 10.1021/jf0009403. [DOI] [PubMed] [Google Scholar]
- 64.See MJ, Staggs SE, Dubey JP, Villegas EN. 2012. Evaluation of four RNA extraction methods for gene expression analyses of Cryptosporidium parvum and Toxoplasma gondii oocysts. J Microbiol Methods 89:185–192. doi: 10.1016/j.mimet.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 65.Gowen CM, Fong SS. 2010. Genome-scale metabolic model integrated with RNAseq data to identify metabolic states of Clostridium thermocellum. Biotechnol J 7:759–767. doi: 10.1002/biot.201000084. [DOI] [PubMed] [Google Scholar]
- 66.Handl S, Dowd SE, Garcia-Mazcorro JF, Steiner JM, Suchodolski JS. 2011. Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiol Ecol 76:301–310. doi: 10.1111/j.1574-6941.2011.01058.x. [DOI] [PubMed] [Google Scholar]
- 67.Rhoads DD, Wolcott RD, Sun Y, Dowd SE. 2012. Comparison of culture and molecular identification of bacteria in chronic wounds. Int J Mol Sci 13:2535–2550. doi: 10.3390/ijms13032535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang J, Kobert K, Flouri T, Stamatakis A. 2014. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 70.Edgar RC. 2013. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 71.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Andreotti R, Pérez de León A, Dowd SE, Guerrero FD, Bendele KG, Scoles G. 2011. Assessment of bacterial diversity in the cattle tick Rhipicephalus (Boophilus) microplus through tag-encoded pyrosequencing. BMC Microbiol 11:e6. doi: 10.1186/1471-2180-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shannon CE, Weaver W. 1949. The mathematical theory of communication. University of Illinois Press, Urbana, IL. [Google Scholar]
- 74.Chao A, Bunge J. 2002. Estimating the number of species in a stochastic abundance model. Biometrics 58:531–539. doi: 10.1111/j.0006-341X.2002.00531.x. [DOI] [PubMed] [Google Scholar]
- 75.Suchodolski JS, Dowd SE, Wilke V, Steiner JM, Jergens AE. 2012. 16S rRNA gene pyrosequencing reveals bacterial dysbiosis in the duodenum of dogs with idiopathic inflammatory bowel disease. PLoS One 7:e39333. doi: 10.1371/journal.pone.0039333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kuchroo CN, Fox PF. 1982. Soluble nitrogen in cheddar cheese: comparison of extraction procedures. Milchweissnschaft 37:331–335. [Google Scholar]
- 77.Blakesley RW, Boezi JA. 1977. A new staining technique for proteins in polyacrylamide gels using Coomassie brilliant blue G250. Anal Biochem 82:580–582. doi: 10.1016/0003-2697(77)90197-X. [DOI] [PubMed] [Google Scholar]
- 78.De Angelis M, de Candia S, Calasso MP, Faccia M, Guinee TP, Simonetti MC, Gobbetti M. 2008. Selection and use of autochthonous multiple strain cultures for the manufacture of high-moisture traditional mozzarella cheese. Int J Food Microbiol 125:123–132. doi: 10.1016/j.ijfoodmicro.2008.03.043. [DOI] [PubMed] [Google Scholar]
- 79.Coppola S, Villani F, Coppola R, Parente E. 1990. Comparison of different starter systems for water-buffalo mozzarella cheese manufacture. Lait 70:411–423. doi: 10.1051/lait:19905-631. [DOI] [Google Scholar]
- 80.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. 2016. Vegan: community ecology package. R package version 2-4-0. https://cran.r-project.org/web/packages/vegan/index.html Accessed 23 June 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.