ABSTRACT
Lactococcus lactis subsp. lactis bv. diacetylactis BGBU1-4 produces a novel bacteriocin, lactolisterin BU, with strong antimicrobial activity against many species of Gram-positive bacteria, including important food spoilage and foodborne pathogens, such as Listeria monocytogenes, Staphylococcus aureus, Bacillus spp., and streptococci. Lactolisterin BU was extracted from the cell surface of BGBU1-4 by 2-propanol and purified to homogeneity by C18 solid-phase extraction and reversed-phase high-performance liquid chromatography. The molecular mass of the purified lactolisterin BU was 5,160.94 Da, and an internal fragment, AVSWAWQH, as determined by N-terminal sequencing, showed low-level similarity to existing antimicrobial peptides. Curing and transformation experiments revealed the presence of a corresponding bacteriocin operon on the smallest plasmid, pBU6 (6.2 kb), of strain BGBU1-4. Analysis of the bacteriocin operon revealed a leaderless bacteriocin of 43 amino acids that exhibited similarity to bacteriocin BHT-B (63%) from Streptococcus ratti, a bacteriocin with analogy to aureocin A.
IMPORTANCE Lactolisterin BU, a broad-spectrum leaderless bacteriocin produced by L. lactis subsp. lactis bv. diacetylactis BGBU1-4, expresses strong antimicrobial activity against food spoilage and foodborne pathogens, such as Listeria monocytogenes, Staphylococcus aureus, Bacillus spp., and streptococci. Lactolisterin BU showed the highest similarity to aureocin-like bacteriocins produced by different bacteria. The operon for synthesis is located on the smallest plasmid, pBU6 (6.2 kb), of strain BGBU1-4, indicating possible horizontal transfer among producers.
KEYWORDS: bacteriocin, lactolisterin BU, antilisterial activity
INTRODUCTION
Bacteria have the ability to produce an extraordinary array of different antagonistic compounds. These include bacteriocins, described as ribosomally synthesized hydrophobic peptides (1, 2) usually active against bacteria closely related to the producer. In addition, some bacteriocins have broader inhibitory spectra and show activity against medically important pathogens and food spoilage bacteria (3). On the basis of their structures, mechanisms of action, and biochemical and genetic characteristics, bacteriocins from lactic acid bacteria (LAB) are generally classified into two different groups: class I bacteriocins (lantibiotics) contain unusual amino acids, such as lanthionine and dehydrated amino acids, as a result of posttranslational modifications, and class II bacteriocins consist of unmodified peptides or peptides with minor modifications. Furthermore, class II bacteriocins are subdivided into four subclasses: pediocin-like bacteriocins (class IIa), two-peptide bacteriocins (class IIb), cyclic bacteriocins (class IIc), and linear non-pediocin-like bacteriocins (class IId) (4). The bacteriocins produced by LAB have been intensively explored from a fundamental perspective for their potential applications as food preservatives and, more recently, in veterinary and human medicine as possible alternatives to antibiotics.
The positive properties of bacteriocins that make them suitable for application in the food industry are their lack of activity against and toxicity for eukaryotic cells and their sensitivity to digestive proteases. The result of the latter is that they have little influence on the gut microbiota. The application of bacteriocins in the food industry (nisin and pediocin PA-1 are commercially available for use as food preservatives) provides many benefits; for example, they may be used as replacements for chemical preservatives, and their use allows the intensity of heat treatment to be reduced, resulting in food that is more naturally preserved and that has better sensorial and nutritional properties. Furthermore, bacteriocins are relatively thermostable, and some of them can retain antimicrobial activity following pasteurization or sterilization. Some bacteriocins also have a broad spectrum of antimicrobial activity, so they can be used in foods as an effective method to extend the shelf life and to control foodborne pathogens, such as Staphylococcus aureus and Listeria monocytogenes (5, 6). L. monocytogenes is of particular concern since it is the causative agent of listeriosis, a relatively rare disease with high fatality rates in Europe (12%) and in the United States (25%), and can traverse the intestinal, placental, and blood-brain barriers in humans. Because of that, in most European countries and in the United States, there are zero-tolerance standards for the presence of L. monocytogenes in ready-to-eat (RTE) food (7–10).
Traditional fermented foods, such as cheeses produced from raw milk, are a rich ecological niche from which bacteriocin-producing LAB can be isolated (11). The indigenous LAB isolated from white brined cheeses from Serbia are good candidates for screening for antimicrobial substances, as they are well adapted to the microbial environments in cheese and could therefore be the source of novel properties (12).
Aureocins are a new group of leaderless class II bacteriocins with a broad spectrum of activity that were first isolated from Staphylococcus aureus. Aureocins act bactericidally on sensitive cells, causing rapid lysis (13–15). According to their structure, they can be classified into two main groups: multipeptide aureocins (aureocin 70-like aureocins) and one-peptide aureocins (aureocin A53-like aureocins).
In a previous study, it was demonstrated that the crude extract obtained from the cell-free supernatant of the natural isolate Lactococcus lactis subsp. lactis bv. diacetylactis BGBU1-4 inhibited growth and biofilm formation and reduced the 24-h-old biofilms of coagulase-negative staphylococci and Listeria monocytogenes clinical isolates (16). The objective of this work was to purify and biochemically and genetically characterize the broad-spectrum bacteriocin lactolisterin BU, produced by L. lactis subsp. lactis bv. diacetylactis BGBU1-4.
RESULTS
Localization of the genes coding bacteriocin production.
The spectrum of activity of L. lactis subsp. lactis bv. diacetylactis BGBU1-4 is broad, with the strain inhibiting different strains of Lactococcus, Lactobacillus, Enterococcus species, and some pathogenic strains (16). Standard biochemical methods confirmed the proteinaceous nature of the antimicrobial agent, as it was found to be sensitive to proteinase K and pronase E. In addition, it was active against B464, a mannose phosphotransferase (Man-PTS) deletion mutant derivative of IL1403 (17), suggesting that Man-PTS is not a receptor for its antilisterial activity. To determine the bacteriocin-coding gene location, a plasmid-curing assay was performed. It was interesting to note that three types of plasmid-cured derivatives which differed in their spectra of activity and the sizes of their inhibition zones in agar well diffusion assays were obtained. It was noticed that derivative BGBU1-4/2 showed a reduced zone of inhibition against L. lactis subsp. lactis BGMN1-596 and L. monocytogenes ATCC 19111 compared to that against the parental strain and was sensitive to the parental strain. Derivatives BGBU1-4/29 and BGBU1-4/8 did not show antimicrobial activity against BGMN1-596 or ATCC 19111 and were sensitive to the BGBU1-4 parental strain and the derivative BGBU1-4/2. Plasmid profile analysis showed differences between parental strain BGBU1-4 and its derivatives: derivative BGBU1-4/2 lost plasmids pBU12 and pBU20, and derivative BGBU1-4/8 lost the smallest plasmid (pBU6) and pBU12, while in derivative BGBU1-4/29, three plasmids (pBU6, pBU12, and pBU20) were absent (Fig. 1). These results indicate that strain BGBU1-4 synthesizes at least two bacteriocins that are active against Lactococcus spp. and L. monocytogenes strains and whose genes are carried by plasmids. It is possible to conclude that there is a direct correlation between the presence and absence of plasmid pBU6 and bacteriocin activity and most likely that the operon for the synthesis of the second bacteriocin is located on plasmid pBU12.
FIG 1.
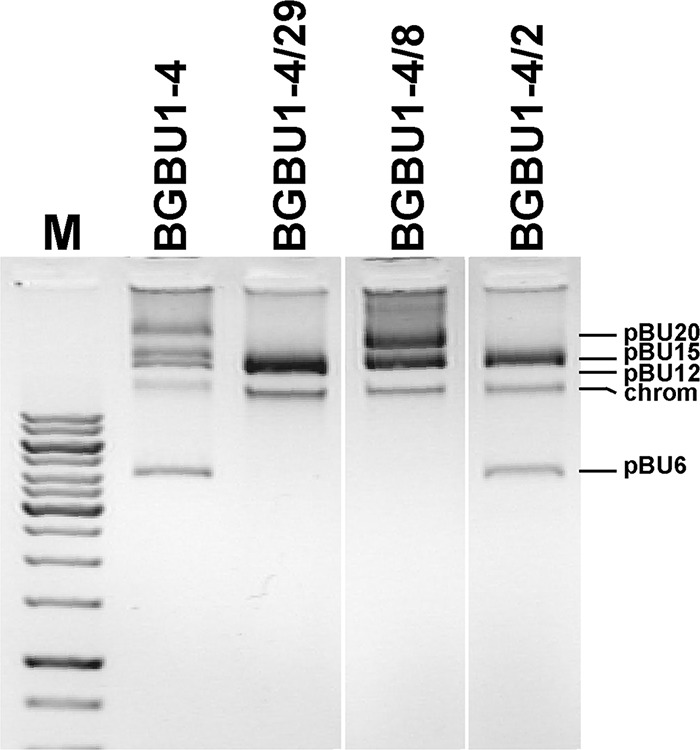
Plasmid profile analysis of parental strain L. lactis subsp. lactis bv. diacetylactis BGBU1-4 and its cured derivatives on a 1% agarose gel. Lane M, the upper part of the GeneRuler 1-kb DNA ladder (from top to bottom, 10, 8, 6, 5, 4, 3.5, 3, 2.5, 2, 1.5, 1, and 0.75 kb; Thermo Fisher Scientific). chrom, the position of chromosomal DNA. Only plasmids of the selected cured derivatives are shown and were taken from different positions of the gel.
Purification and identification of the bacteriocin(s).
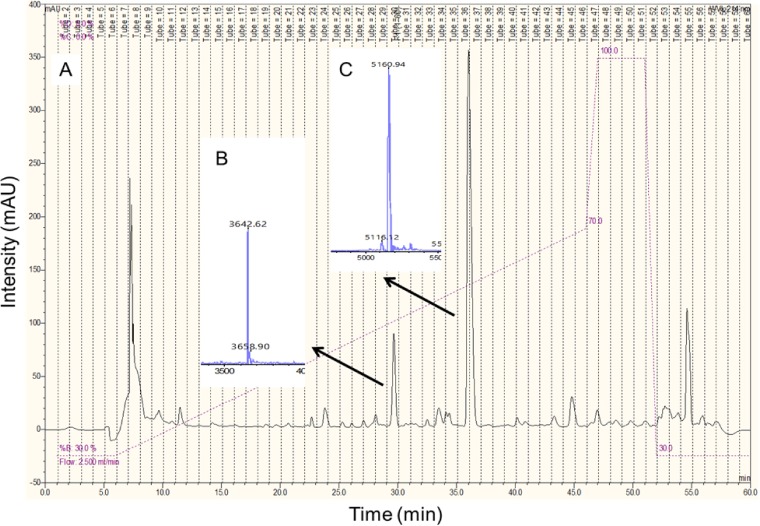
The bacteriocin(s) produced by strain BGBU1-4 was purified by reversed-phase (RP) high-performance liquid chromatography (HPLC), and the molecular mass of the active peptides was determined by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MS). The RP-HPLC chromatogram showed dominant peaks (fractions 30 and 37) (Fig. 2) that were active against L. lactis subsp. lactis BGMN1-596 and L. monocytogenes ATCC 19111. Mass spectrometry analysis determined the molecular mass of fraction 30 to be 3,642.62 Da and that of fraction 37 to be 5,160.94 Da. As fraction 37 was the most active, it was selected for further characterization.
FIG 2.
Reverse-phase high-performance liquid chromatography (RP-HPLC) chromatogram (A) and matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry data (B, C). Arrows, locations of the antimicrobial peptides. mAU, milli-absorbance units.
N-terminal sequencing of protein fraction 37.
N-terminal sequencing of the native peptide in fractions 30 and 37 failed, most probably due to a blocked N terminus. N-terminal sequencing was performed two times, without success. To circumvent this, native peptides were digested with trypsin, and N-terminal sequencing of the internal fragments was done. An internal 1,112.61-Da fragment of the protein from fraction 37 revealed the amino acid sequence AVSWAWQH, which corresponds to lactolisterin BU residues 16 to 23 (see Fig. 4), while N-terminal sequencing of the peptides from fraction 30 failed two times and work was continued only with fraction 37 (lactolisterin BU). Lactolisterin BU is a leaderless peptide, and consequently, the N-terminal amino acid is formylmethionine rather than methionine, as there was no cleavage of the leader peptide. The formyl group of formylmethionine blocks the alpha carbon of the amino acid, making it inaccessible to phenylisothiocyanate (PITC), the reagent used in N-terminal sequencing (18). MALDI-TOF MS was also used to confirm the presence of formylmethionine. Addition of a formyl group resulted in a 28-Da increase in mass, which was in good agreement with the 29-Da mass difference observed when the mass of the native peptide (5,160.94 Da) was compared with the theoretical mass (5,131.67 Da).
FIG 4.
Alignment of the lactolisterin BU (GenBank accession no. SDR48784) amino acid sequence with the amino acid sequences of homologous bacteriocins: lacticin Q from Lactococcus lactis QU5 (GenBank accession no. BAF57910.1), lacticin Z from Lactococcus lactis QU14 (GenBank accession no. BAF75975.1), bacteriocin BHT-B from Streptococcus ratti (GenBank accession no. DQ145753.1), aureocin A53 from Staphylococcus aureus (GenBank accession no. AAN71834), and an aureocin A53-like bacteriocin from Corynebacterium jeikeium (GenBank accession no. WP_010976360). Highlighted residues indicate conservation in at least five of the peptide sequences, while an asterisk indicates completely conserved residues. An x corresponds to unconserved residues, periods indicate amino acids belonging to similar groups, and colons indicate amino acids belonging to the same group.
Heterologous expression of the bacteriocin in Lactococcus lactis subsp. cremoris MG7284.
Plasmid curing indicated that the genes for the synthesis and immunity of bacteriocins in strain L. lactis subsp. lactis bv. diacetylactis BGBU1-4 are located on plasmids. The smallest plasmid from strain BGBU1-4, named pBU6 (6.2 kb), was isolated and used for transformation of the bacteriocin nonproducer Lactococcus lactis subsp. cremoris strain MG7284. GM17 (M17 medium supplemented with d-glucose) agar plates containing lactolisterin BU at a concentration of 1.34 μM were used to select for a bacteriocin-resistant transformant. The transformants obtained were designated MG7284/pBU6 and were used for further purification of lactolisterin BU. Plasmid profile analysis of the MG7284/pBU6 transformants revealed that they all possessed the smallest 6.2-kb plasmid and were found to be active against indicator strains BGMN1-596, ATCC 19111, and B464, confirming that Man-PTS is not a receptor for its antilisterial activity (see Fig. S1 in the supplemental material).
Analysis of plasmid pBU6.
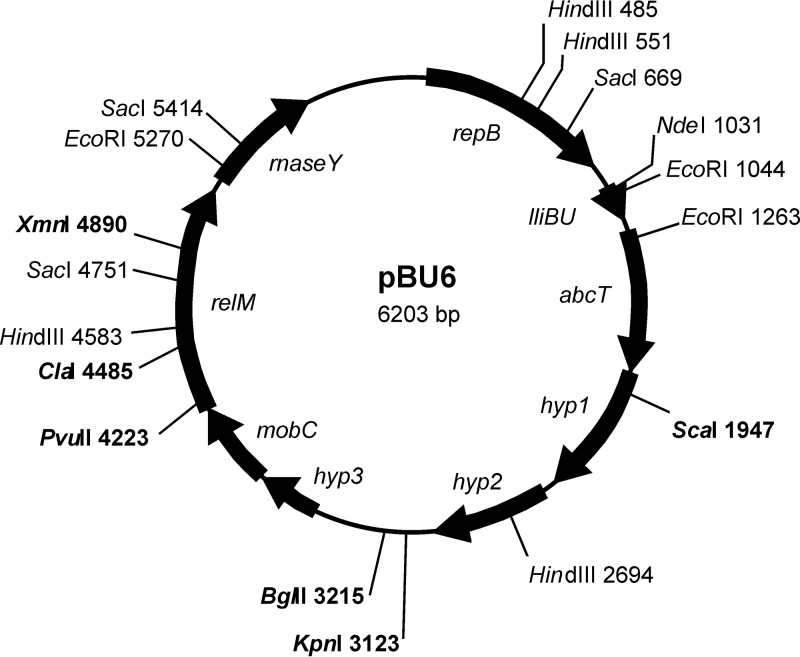
Plasmid pBU6 was sequenced in its entirety. Analysis of the plasmid pBU6 sequence revealed that it is a small rolling-circle replicating (RCR) plasmid which contains nine open reading frames (ORFs): repB, lliBU, abcT, hyp1, hyp2, hyp3, mobC, relM, and rnaseY (Fig. 3; Table 1).
FIG 3.
Circular restriction map of plasmid pBU6. Only relevant restriction sites and their positions are indicated; unique restriction sites are indicated in bold. The positions and orientations of the genes are indicated by arrows.
TABLE 1.
Results of comparison of sequences of proteins carried by plasmid pBU6 with sequences in Entrez protein database by BLAST analysis
| ORF (no. of amino acids) | Positionsa | Proteins with the highest identity (GenBank accession no.) | Predicted domain(s) or superfamily in encoded ORF | Organism | Amino acid sequence identity (%) |
|---|---|---|---|---|---|
| RepB (278) | 80–916 | RepB (WP_011117039.1) | Rep_3 (pfam01051) | Lactobacillus fermentum | 90 |
| 80–916 | RepB (WP_032951507.1) | Rep_3 (pfam01051) | Lactococcus lactis | 85 | |
| LliBU (43) | 1034–1165 | Aureocin-like bacteriocin (AAZ76605.1) | Bacteriocin_Iii (pfam11758) | Streptococcus ratti | 63 |
| AbcT (212) | 1230–1868 | Sugar ABC transporter ATP-binding protein (WP_003089811.1) | ABC_DR_subfamily_A (CD03230) | Streptococcus ratti | 62 |
| Hyp1 (212) | 1861–2449 | Hypothetical protein (WP_003089809.1) | Streptococcus ratti | 42 | |
| Hyp2 (169) | 2503–3012 | Hypothetical protein (AAZ76608.1) | Streptococcus ratti | 43 | |
| Hyp3 (92) | 3531–3809 | Hypothetical protein (WP_027822861.1) | Lactobacillus plantarum | 51 | |
| MobC (127) | 3829–4212 | Mobilization protein C (WP_010729194.1) | MobC (pfam05713) | Enterococcus faecium | 41 |
| 3829–4212 | Hypothetical protein (WP_011117545.1) | MobC (pfam05713) | Enterococcus faecalis | 55 | |
| RelM (333) | 4194–5195 | Relaxase/mobilization nuclease (WP_010730379.1) | Relaxase (pfam03432) | Enterococcus faecium | 51 |
| RNaseY (169) | 5219–5728 | RNase Y (WP_017865030.1) | Phosphodiesterase (PRK12704) | Lactococcus lactis | 29 |
Positions of the gene on plasmid pBU6.
In silico analysis revealed that the amino acid sequence of the RepB protein shows a high degree of similarity to the amino acid sequences of the RepB proteins of Lactobacillus fermentum (GenBank accession no. WP_011117039.1) and Lactococcus lactis (GenBank accession no. WP_032951507.1). The N-terminal sequencing results of the peptide digest of fraction 37 identified lliBU to be the structural gene, encoding a 43-amino acid (aa) peptide which is responsible for the production of the bacteriocin lactolisterin BU. Lactolisterin BU is a leaderless bacteriocin rich in the amino acids glycine and tryptophan (each of which comprises 11.6% of the bacteriocin) with a pI of 10.16. Interestingly, the amino acid sequence of this protein shows the highest degree of similarity with that of bacteriocin BHT-B from Streptococcus ratti (63%; GenBank accession no. AAZ76605.1 [19]) and other aureocin A-like bacteriocins (Fig. 4).
An ORF downstream of the lliBU gene designated abcT (212 aa) encodes a protein similar to the sugar ABC transporter ATP binding protein from Streptococcus ratti (62%; GenBank accession no. WP_003089811.1). One or more of the next three genes, hyp1 (212 aa), hyp2 (169 aa), and hyp3 (92 aa), may encode a protein(s) that plays a role in producer immunity. The three genes mobC (127 aa), relM (333 aa), and rnaseY (169 aa) encode proteins similar to mobilization protein C (GenBank accession no. WP_010729194.1), relaxase/mobilization nuclease (GenBank accession no. WP_010730379.1), and RNase Y (GenBank accession no. WP_017865030.1), respectively.
Biochemical characterization of lactolisterin BU.
The antimicrobial activity of purified lactolisterin BU was unchanged after heat treatment at 60°C, 80°C, and 100°C for 15 and 30 min compared with that after no heat treatment, showing MIC values of 0.67 μM against L. lactis subsp. lactis BGMN1-596 and 1.34 μM against L. monocytogenes ATCC 19111.
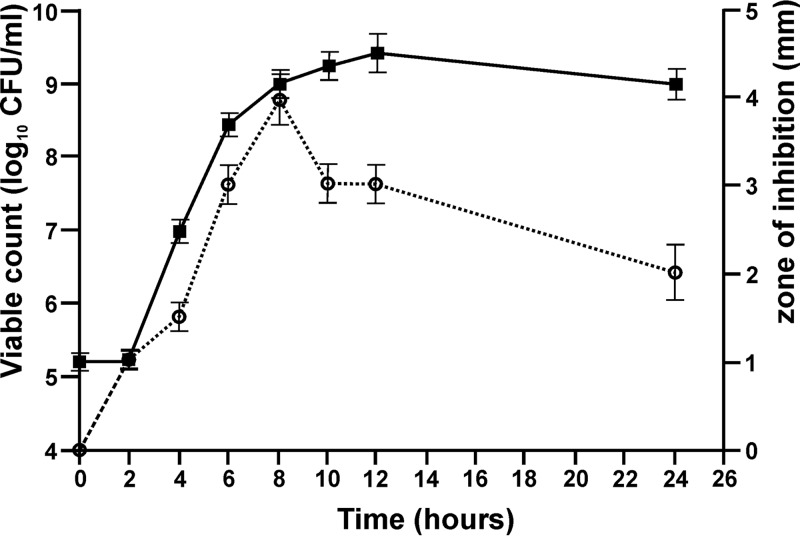
The growth kinetics and antimicrobial activity of transformant MG7284/pBU6 are shown in Fig. 5. Detectable levels of bacteriocin activity were recorded after 2 h of growth at 30°C. The maximum bacteriocin activity of the MG7284/pBU6 transformant was achieved at the end of exponential phase, while its activity against indicator strain Listeria monocytogenes ATCC 19111 was lower after 24 h of growth (Fig. 5).
FIG 5.
Lactolisterin BU levels in culture relative to the cell density of Lactococcus lactis subsp. cremoris MG7284/pBU6 tested on L. monocytogenes ATCC 19111. Filled squares, bacterial growth determined by measurement of the number of CFU per milliliter; circles, the corresponding bacteriocin activity determined by the area of the zone of inhibition. Error bars represent the standard deviations from three independent experiments.
Purified lactolisterin BU was active against various Gram-positive bacteria, with the MICs being in the micromolar range. Lactolisterin BU showed relatively strong activity against Listeria monocytogenes ATCC 19111, as the MIC value of the bacteriocin was 1.34 μM. In addition, lactolisterin BU showed very strong antimicrobial activity against Staphylococcus aureus ATCC 25923, Streptococcus pyogenes A2941, and Streptococcus pneumoniae P156 (Table 2). It is interesting that purified lactolisterin BU exhibited antimicrobial activity against lactolisterin BU producers (parental strain BGBU1-4, a cured derivative BGBU1-4/2, and the MG7284/pBU6 transformant), with the MIC values being 1.34 μM (Table 2).
TABLE 2.
Antimicrobial spectra of purified lactolisterin BU
| Indicator strain | MIC (μM) |
|---|---|
| Lactococcus lactis subsp. lactis BGMN1-596 | 0.67 |
| Lactococcus lactis subsp. lactis BGBU1-4 | 1.34 |
| Lactococcus lactis subsp. lactis BGBU1-4/2 | 1.34 |
| Lactococcus lactis subsp. cremoris MG7284/pBU6 | 1.34 |
| Lactobacillus casei BGHN14 | 0.67 |
| Listeria monocytogenes ATCC 19111 | 1.34 |
| Staphylococcus aureus ATCC 25923 | 0.67 |
| Enterococcus faecalis ATCC 29212 | 1.34 |
| Enterococcus faecalis BGZLS10-27 | 1.34 |
| Bacillus subtilis subsp. subtilis ATCC 23857 | 5.375 |
| Bacillus cereus ATCC 11778 | 5.375 |
| Streptococcus pyogenes | 0.67 |
| Streptococcus pneumoniae | 0.67 |
| Escherichia coli H7:O157 ATCC 35150 | NAa |
| Pseudomonas aeruginosa ATCC 27853 | NA |
| Salmonella Typhimurium ATCC 14028 | NA |
| Salmonella Enteritidis ATCC 13076 | NA |
NA, no activity.
Mode of action of lactolisterin BU.
The results of analysis of L. monocytogenes ATCC 19111 growth in culture (starting with different numbers of cells) in the presence of different concentrations of lactolisterin BU strongly indicated that the effect of lactolisterin BU on growth is not growth phase dependent. The strongest lysis of bacteria was obtained when the cells were in the early logarithmic phase. With the increase in the number of cells, the effect of lactolisterin BU on the cessation of growth of sensitive bacteria gradually decreased (when it was used at a concentration of 1.34 μM, the MIC), but at higher concentrations (4.02 and 13.4 μM), it exhibited strong inhibition of the growth of bacteria at all stages of growth (Fig. S2), indicating that it can successfully be used against pathogens and food contaminants that are in the stationary phase.
An attempt to isolate a lactolisterin BU-resistant mutant.
In our previous experiences in obtaining mutants resistant to bacteriocins, we used two approaches: selection of spontaneous mutants and mutagenesis with N-methyl-N′-nitro-N-nitrosoguanidine (20). We have successfully isolated mutants by the use of both approaches, but using mutagenesis, we isolated greater numbers of mutants that showed greater diversity. First, we tried to isolate spontaneous mutants by spreading 500 μl of 10-fold-concentrated overnight cultures of sensitive strains on GM17 petri dishes containing lactolisterin BU at concentrations of 1.34 μM and 2.68 μM (1 and 2 times the MIC values, respectively). No mutant resistant to lactolisterin BU was obtained, indicating that the mechanism of action is possibly different from that of the bacteriocins studied previously or that a mutation leading to resistance is lethal (that is, the target protein is essential). In order to confirm the impossibility of obtaining mutants resistant to lactolisterin BU, banks of mutants of three sensitive strains (L. lactis subsp. lactis BGMN1-596, L. lactis subsp. cremoris MG7284, and Enterococcus faecalis BGZLS10-27) were constructed using N-methyl-N′-nitro-N-nitrosoguanidine (which increases the chance of getting more mutations in each of the genes) and used to isolate resistant mutants by spreading of aliquots on selective GM17 petri dishes containing lactolisterin BU. We did not manage to select a mutant resistant to lactolisterin BU from the mutant banks after three attempts for each strain, confirming that the treatment that we applied did not yield a mutant resistant to lactolisterin BU or it is very difficult to isolate such a mutant.
DISCUSSION
Lactic acid bacteria are found in different ecological niches, and as a result of their efforts to adapt and survive, they produce various secondary metabolites, among which are bacteriocins. Although bacteriocins have been studied for almost 7 decades, researchers still find them interesting due to their potential applications. In the last decade, this field has broadened because of their possible use instead of and/or in synergy with antibiotics to overcome the immense problem of increases in the prevalence of antibiotic-resistant bacteria (4). Natural isolates from traditionally prepared food products are a tremendous source of highly diverse, unique metabolites. These isolates come from harsh environments and contain genes that are usually lost in industrial strains. Lactococci commonly produce more than one bacteriocin, and the Lactococcus lactis subsp. lactis bv. diacetylactis BGBU1-4 strain produces at least two bacteriocins (21–23). The crude extract from the cell-free supernatant of strain BGBU1-4 inhibited growth and reduced the 24-h-old biofilms formed by clinical isolates of Listeria monocytogenes and coagulase-negative Staphylococcus spp., while the prevention of biofilm formation was demonstrated for L. monocytogenes clinical isolates (16). The genes for the production of the bacteriocin lactolisterin BU were found to be located on plasmid pBU6, which is not unusual (22–25), but the results of assays with plasmid-cured derivatives of strain BGBU1-4 suggested that its broad-spectrum antimicrobial activity is a consequence of the production of at least two bacteriocins. Characterization of the antilisterial bacteriocin purified by HPLC revealed that the ORF lliBU on pBU6 encodes the lactolisterin BU structural gene. In addition, sequence analysis of lactolisterin BU, a 43-amino-acid peptide, showed that its highest similarity was with BHT-B, a class II bacteriocin from Streptococcus ratti. Although lactolisterin BU showed some characteristics of class IIa bacteriocins (antilisterial activity and the absence of unusual amino acids), it does not possess the highly conserved motif T-G-N-G-V/L generally found in pediocin-like bacteriocins with antilisterial activity and so cannot be classified as a class IIa bacteriocin (26–28). Therefore, it is classified as a class IId leaderless bacteriocin and can be compared to LsbB, lacticin Q, lacticin Z, and aureocins A70 and A53, bacteriocins known to be synthesized and exported without a leader sequence (14, 15, 23, 29, 30). A comparative analysis of the amino acid sequences has shown that lactolisterin BU shares a conserved region, AKYGXKAV, with the majority of known aureocin A53-like bacteriocins (Fig. 4). It can be assumed, considering the variation in their lengths and primary structures, that the region AKYGXKAV is responsible for the activity of aureocin A53-like bacteriocins. It is interesting that lactolisterin BU shows a higher degree of identity with the BHT-B bacteriocin from Streptococcus ratti (63%) than with lacticin Q and lacticin Z (33.9%), isolated from lactococci. The antilisterial activity of lactolisterin BU is not mediated by interaction with Man-PTS like that of other class II bacteriocins, as it showed antimicrobial activity against mutant B464 (17).
Biochemical characterization showed that lactolisterin BU, like other bacteriocins, is sensitive to proteolytic enzymes, is relatively thermostable, and is produced at maximum levels in early stationary phase (31–33). Lactolisterin BU has potential as a food preservative due to its strong antimicrobial activity (it is active at micromolar concentrations) against many species of Gram-positive bacteria, including important food spoilage and foodborne pathogens, such as Listeria monocytogenes, Staphylococcus aureus, Bacillus spp., and Streptococcus spp. Additionally, since the production of many food products involves exposure to high temperatures, the relative thermostability of lactolisterin BU is another desirable feature for its application in the food industry. The most desirable characteristic of lactolisterin BU for its use in controlling contaminants of food or pathogens is its inability to induce resistance, which is most likely the result of a specific mechanism of action or because it targets an essential molecule. Genes for bacteriocin production are often located on plasmids, enabling horizontal gene transfer between genera, and it is expected that rearrangements during transfer result in novel peptides. It is assumed that a similar scenario happened with lactolisterin BU. The presence of a highly conserved region between lactococcal bacteriocin lactolisterin BU, bacteriocin BHT-B from Streptococcus ratti, aureocin A53 from Staphylococcus aureus, and an aureocin A53-like bacteriocin from Corynebacterium jeikeium indicates a common origin of the bacteriocin operon. It is interesting that the homologous bacteriocin operon is present in such a wide variety of genera, which indicates that bacteriocin production confers an advantage to the carrier. The greater similarity between lactolisterin BU and aureocins from other bacteria than between lactolisterin BU and lactococcal bacteriocins (lacticin Q and Z) indicates that these lactococcal bacteriocins followed a different evolutionary pathway.
This work provides insight into a new and unusual class II lactococcal bacteriocin, lactolisterin BU. Its genetic and biochemical characteristics, spectrum of activity, protease sensitivity, thermostability, and inability to induce or the very rare occurrence of the induction of resistance recommend lactolisterin BU as a good candidate to be a safe, cheap, and natural food preservative. Further experiments examining the ability of lactolisterin BU to prevent L. monocytogenes development in products obtained from raw milk are ongoing.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Table 3. The bacteriocin producer L. lactis subsp. lactis bv. diacetylactis BGBU1-4 was isolated from a 3-day-old traditional semihard cheese made from mixed cow (20%) and sheep (80%) milk (34). The cheese was produced without the use of starter cultures in a household in the village of Buzina, located on the mountain Beljanica in eastern Serbia. Lactococcal strains were grown in M17 medium (Merck GmbH, Darmstadt, Germany) supplemented with d-glucose (0.5%, wt/vol) (GM17) at 30°C. Lactobacillus strains were grown in MRS medium (Merck GmbH, Darmstadt, Germany). Nonlactococcal indicator strains were grown aerobically in Luria-Bertani (LB) broth at 37°C. Streptococcus strains were grown in brain heart infusion (BHI) medium (Oxoid, Basingstoke, Hampshire, England) at 37°C in atmosphere of 5% CO2. Solid medium and soft agar were made by adding 1.5% or 0.7% (wt/vol) agar (Torlak, Belgrade, Serbia) to the liquid medium, respectively.
TABLE 3.
Strains used in this studya
| Strain | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Lactococcus lactis | ||
| BGBU1-4 | Bacteriocin (lactolisterin BU) producer | 34 |
| BGBU1-4/2 | Derivative of BGBU1-4; Bac+ Bacs | This work |
| BGBU1-4/8 | Derivative of BGBU1-4; Bac+ Bacs | This work |
| BGBU1-4/29 | Derivative of BGBU1-4; Bac− Bacs | This work |
| BGMN1-596 | Bac− Bacs | 20 |
| MG7284 | Bac− Bacs | 44 |
| MG7284/pBU6 | MG7284 transformed with plasmid pBU6 (producer of lactolisterin BU) | This work |
| B464 | Man-PTS deletion mutant of strain IL1403 | 17 |
| Lactobacillus casei BGHN14 | Bac− Bacs | 45 |
| Listeria monocytogenes | ATCC 19111 | |
| Staphylococcus aureus | ATCC 25923 | |
| Enterococcus faecalis | ATCC 29212 | |
| Enterococcus faecalis BGZLS10-27 | 46 | |
| Bacillus cereus | ATCC 11778 | |
| Bacillus subtilis subsp. subtilis | ATCC 23857 | |
| Streptococcus pyogenes A2941 | Pasteur Laboratory, Belgrade, Serbia | |
| Streptococcus pneumoniae P156 | Pasteur Laboratory, Belgrade, Serbia | |
| Escherichia coli H7:O157 | ATCC 35150 | |
| Pseudomonas aeruginosa | ATCC 27853 | |
| Salmonella enterica serovar Typhimurium | ATCC 14028 | |
| Salmonella enterica serovar Enteritidis | ATCC 13076 |
Bac+, bacteriocin producer; Bac−, non-bacteriocin producer; Bacs, sensitivity to lactolisterin BU; ATCC, American Type Culture Collection, Manassas, VA, USA.
Spectrum and kinetics of bacteriocin activity.
The agar well diffusion assay was used to determine the spectrum of antibacterial activity of strain BGBU1-4 (33). Each indicator strain was inoculated into appropriate soft agar, and wells (diameter, 5 mm) were made in the plate. The wells were filled with 50 μl of sample, and plates were incubated under appropriate conditions for the respective indicator strain (Table 2). After 24 h of incubation, the plates were examined for the presence of inhibition zones. A clear zone of inhibition around the wells was taken as evidence of bacteriocin production.
To monitor the kinetics of bacteriocin production and activity, 100 ml of fresh preheated GM17 broth was inoculated with an overnight culture (1%, vol/vol) and incubated at 30°C. Samples were taken at 0, 2, 4, 6, 8, 10, 12, and 24 h. Bacteriocin activity was determined by the area of the zone of inhibition. L. monocytogenes ATCC 19111 and L. lactis subsp. lactis BGMN1-596 were used as indicator strains.
Genetic characterization. (i) Plasmid-curing experiments.
Plasmid-curing assays were done by growing cells of the BGBU1-4 bacterial strain in the presence of novobiocin at sublethal temperatures as described previously (35). Preheated GM17 broth (42°C) containing novobiocin (5 μg/ml) was inoculated with 103 cells per ml. After 2 h of incubation, the cells were collected by centrifugation and resuspended in the same volume of fresh preheated novobiocin containing GM17 broth to avoid the killing effect of bacteriocin on the cured cells. This step was repeated four times, and endpoint aliquots (0.1 ml) were plated onto GM17 agar plates, which were then incubated at 30°C for 48 h.
(ii) Molecular methods.
Derivatives obtained following the plasmid-curing experiments and transformants were confirmed using pulsed-field gel electrophoresis (PFGE) as described previously by Kojic et al. (23). For isolation of total DNA from lactococci, a modified version of the method described by Hopwood et al. (36) was used. Plasmid DNA from lactococci was isolated by the modified method previously described by O'Sullivan and Klaenhammer (37). Plasmids were introduced into lactococci by electroporation using an electroporator (Eppendorf, Hamburg, Germany) (38). Plasmid DNA was sequenced by the Macrogen Sequencing Service (Macrogen Europe, Amsterdam, The Netherlands). Nucleotide sequences were analyzed using the BLAST algorithm. The functions of the proteins encoded by genes carried by the pBU6 plasmid were attributed on the basis of the homology of their sequences with those of known proteins in the Entrez Protein BLAST database, which were compared by use of the BLAST algorithm.
Purification of the bacteriocin.
Lactolisterin BU was purified from the cells according to the method of Rea et al. (39) with the following modifications. Briefly, the cell pellet from a 2-liter culture grown in tryptone-yeast extract broth was resuspended in 250 ml of 70% (vol/vol) 2-propanol, 0.1% (vol/vol) trifluoroacetic acid (TFA) per liter of broth and stirred at room temperature for 3 to 4 h. A sample was centrifuged at 8,280 × g for 20 min, and the cell supernatant was retained for purification. The 2-propanol was evaporated using a rotary evaporator (Buchi Labortechnik AG, Flawil, Switzerland), and the sample was applied to a 5-g (20-ml) Strata C18-E SPE column (Phenomenex, Cheshire, UK) preequilibrated with methanol and water. The column was washed with 40 ml of 30% (vol/vol) ethanol, and the bacteriocin was eluted with 40 ml of 70% (vol/vol) 2-propanol, 0.1% (vol/vol) TFA. An aliquot of the 70% 2-propanol, 0.1% TFA eluent from the cell pellet passed on the C18-E SPE column was concentrated using rotary evaporation before separation of the peptides using RP-HPLC. Aliquots of approximately 4 ml were applied to a Phenomenex (Phenomenex, Macclesfield, Cheshire, UK) Proteo Jupiter (RP)-HPLC column (250 by 10.0 mm; particle size, 4 μm; 90 Å) that had previously been equilibrated with 25% acetonitrile, 0.1% TFA. Peptides were eluted in a gradient of 30% acetonitrile containing 0.1% TFA to 70% acetonitrile containing 0.1% TFA over 40 min with buffer A (Milli-Q water containing 0.1% TFA) and buffer B (90% acetonitrile containing 0.1% TFA), and the flow rate was 2.5 ml/min. Fractions were collected at 1-min intervals and assayed on Lactococcus lactis BGMN1-596 and Listeria monocytogenes ATCC 19111 indicator plates.
MALDI-TOF mass spectrometry was performed on fractions exhibiting positive inhibitory activity, using an Axima TOF2 MALDI-TOF mass spectrometer (Shimadzu Biotech, Manchester, UK), as described by Mills et al. (40).
N-terminal sequencing of protein from fractions 30 and 37.
Protein fractions 30 and 37, which showed antimicrobial activity, were sent to the Department of Molecular and Biomedical Sciences, Jozef Stefan Institute (Ljubljana, Slovenia), for N-terminal sequencing by Edman degradation. N-terminal sequencing of the native peptides failed due to the presence of an N-terminal formylmethionine, so the peptides were digested with trypsin and the trypsin fragments were sequenced on the second attempt.
Determination of MICs.
The MIC of the lactolisterin BU was determined using the broth microdilution method proposed by Steinberg et al. (41). The microdilution testing assay used a mixture of the indicator strains (Tables 2 and 3) and increasing concentrations of the bacteriocin lactolisterin BU. Microdilution testing with in-house-prepared panels was performed following the methods of the Clinical and Laboratory Standards Institute (42). Indicator strains were diluted to a 0.5 McFarland standard, from which 20 μl was distributed to the wells of a clear 96-well flat-bottom microtiter plate. The concentration of pure lactolisterin BU was determined by the spectroscopic method using a theoretical extinction coefficient calculated from the peptide sequence, as described by Blanusa et al. (43). Lactolisterin BU (47 μM) was 2-fold serially diluted to give a dilution series with concentrations ranging from 43 μM to 0.67 μM. The microtiter plates were incubated under appropriate conditions for 24 h, and the optical density at 595 nm (OD595) was recorded at 30-min intervals (Infinite M200pro; Tecan, Switzerland). The values obtained were used to illustrate the antimicrobial activity of the bacteriocin lactolisterin BU. Control wells contained appropriate medium (blanks) and an untreated culture. All experiments were done in triplicate.
Effect of heat treatment on lactolisterin BU activity.
Purified lactolisterin BU dissolved in water (concentration, 47 μM) was incubated at 60°C, 80°C, and 100°C for 15 min and 30 min. After the treatments, antimicrobial activity was determined using the broth microdilution method proposed by Steinberg et al. (41) as described above. The indicator strains were L. monocytogenes ATCC 19111 and L. lactis subsp. lactis BGMN1-596; untreated purified bacteriocin was used as a control. All experiments were done in triplicate.
Mode of action of lactolisterin BU.
To analyze the effect of lactolisterin BU on the growth of the sensitive strain, purified lactolisterin BU (three different concentrations, 1.34 μM [the MIC], 4.02 μM [a concentration 3 times higher than the MIC], and 13.4 μM [a concentration 10 times higher than the MIC], were used) was added to the cultures inoculated with different numbers of L. monocytogenes ATCC 19111 bacteria (3 × 107, 3.3 × 108, and 2 × 109 cells/ml) in microtiter plates. Before the addition of bacteriocin, diluted bacterial cultures were incubated for 1 h at the optimal growth temperature to refresh the cells. The bacterial growth was monitored by measurement of the OD595, which was recorded at 30-min intervals (Infinite M200pro; Tecan, Switzerland) and by determination of the number of viable bacterial cells (number of CFU) at every hour of growth. Control wells contained appropriate medium (blanks) and untreated cultures. All experiments were done in triplicate.
Mutagenesis of sensitive strains with N-methyl-N′-nitro-N-nitrosoguanidine.
Cells of sensitive strains (L. lactis subsp. lactis BGMN1-596, L. lactis subsp. cremoris MG7284, and Enterococcus faecalis BGZLS10-27) from mid-logarithmic-growth phase (OD600, ∼0.6 to 1) were harvested by centrifugation at 10,000 × g for 10 min at 4°C and washed two times in the same volume of 100 mM sodium phosphate buffer (pH 7). Tenfold-concentrated cells in phosphate buffer were exposed to different concentrations of N-methyl-N′-nitro-N-nitrosoguanidine (0, 25, 50, 100, and 200 μg/ml in phosphate buffer) for 1 h at 30°C in the dark. After treatment, the cells were washed two times with a 10-fold volume of phosphate buffer and, finally, resuspended in the same volume of GM17. Cultures were grown for 1 h at 30°C in the dark in order to recover cells, and cell survival was determined by plating 10-fold dilutions on GM17 plates and incubation at 30°C for 2 days. Stabilized mutated cells were stored at −80°C in glycerol (final concentration, 15%) until use.
Accession number(s).
The sequence of plasmid pBU6 has been submitted to the European Nucleotide Archive and may be found under accession no. LT629305.
Supplementary Material
ACKNOWLEDGMENT
Financial support for the conduct of the research was provided by the Ministry of Education, Science and Technological Development of the Republic of Serbia (grant no. 173019).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01519-17.
REFERENCES
- 1.Montesinos E. 2007. Antimicrobial peptides and plant disease control. FEMS Microbiol Lett 270:1–11. doi: 10.1111/j.1574-6968.2007.00683.x. [DOI] [PubMed] [Google Scholar]
- 2.Keymanesh K, Soltani S, Sardari S. 2009. Application of antimicrobial peptides in agriculture and food industry. World J Microbiol Biotechnol 25:933–944. doi: 10.1007/s11274-009-9984-7. [DOI] [Google Scholar]
- 3.Nes IF. 2011. History, current knowledge, and future directions on bacteriocin research in lactic acid bacteria, p 3–12. In Drider D, Rebuffat S (ed), Prokaryotic antimicrobial peptides—from genes to applications. Springer Science & Business Media, New York, NY. [Google Scholar]
- 4.Cotter PD, Ross RP, Hill C. 2013. Bacteriocins—a viable alternative to antibiotics? Nat Rev Microbiol 11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 5.Acuña L, Corbalan NS, Fernandez-No IC, Morero RD, Barros-Velazquez J, Bellomio A. 2015. Inhibitory effect of the hybrid bacteriocin Ent35-MccV on the growth of Escherichia coli and Listeria monocytogenes in model and food systems. Food Bioprocess Technol 8:1063–1075. doi: 10.1007/s11947-015-1469-0. [DOI] [Google Scholar]
- 6.Thomas LV, Ingram RE, Bevis HE, Davies EA, Milne CF, Delves-Broughton J. 2002. Effective use of nisin to control Bacillus and Clostridium spoilage of a pasteurized mashed potato product. J Food Prot 65:1580–1585. doi: 10.4315/0362-028X-65.10.1580. [DOI] [PubMed] [Google Scholar]
- 7.European Food Safety Authority. 2011. EU summary report on zoonoses, zoonotic agents and food-borne outbreaks 2011. EFSA J 11:3129 https://www.efsa.europa.eu/en/efsajournal/pub/3129. [Google Scholar]
- 8.WHO. 2014. Food and health in Europe: a new basis for action. WHO, Geneva, Switzerland. [Google Scholar]
- 9.Schlech WF, Acheson D. 2000. Foodborne listeriosis. Clin Infect Dis 31:770–775. doi: 10.1086/314008. [DOI] [PubMed] [Google Scholar]
- 10.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. 1999. Food-related illness and death in the United States. Emerg Infect Dis 5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cocolin L, Foschino R, Comi G, Graziafortina M. 2007. Description of the bacteriocins produced by two strains of Enterococcus faecium isolated from Italian goat milk. Food Microbiol 24:752–758. doi: 10.1016/j.fm.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Topisirovic L, Kojic M, Fira D, Golic N, Strahinic I, Lozo J. 2006. Potential of lactic acid bacteria isolated from specific natural niches in food production and preservation. Int J Food Microbiol 112:230–235. doi: 10.1016/j.ijfoodmicro.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Netz DJA, Bastos MDCDF, Sahl H-G. 2002. Mode of action of the antimicrobial peptide aureocin A53 from Staphylococcus aureus. Appl Environ Microbiol 68:5274–5280. doi: 10.1128/AEM.68.11.5274-5280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Netz DJA, Pohl R, Beck-Sickinger AG, Selmer T, Pierik AJ, Bastos MDCDF, Sahl H-G. 2002. Biochemical characterisation and genetic analysis of aureocin A53, a new, atypical bacteriocin from Staphylococcus aureus. J Mol Biol 319:745–756. doi: 10.1016/S0022-2836(02)00368-6. [DOI] [PubMed] [Google Scholar]
- 15.Netz DJA, Sahl H-G, Marcolino R, dos Santos Nascimento J, de Oliveira SS, Soares MB, Bastos MDCDF, Marcolino R. 2001. Molecular characterisation of aureocin A70, a multi-peptide bacteriocin isolated from Staphylococcus aureus. J Mol Biol 311:939–949. [DOI] [PubMed] [Google Scholar]
- 16.Cirkovic I, Bozic DD, Draganic V, Lozo J, Beric T, Kojic M, Arsic B, Garalejic E, Djukic S, Stankovic S. 2016. Licheniocin 50.2 and bacteriocins from Lactococcus lactis subsp. lactis biovar diacetylactis BGBU1-4 inhibit biofilms of coagulase negative staphylococci and Listeria monocytogenes clinical isolates. PLoS One 11:e0167995. doi: 10.1371/journal.pone.0167995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diep DB, Skaugen M, Salehian Z, Holo H, Nes IF. 2007. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc Natl Acad Sci U S A 104:2384–2389. doi: 10.1073/pnas.0608775104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyatake N, Kamo M, Satake K, Uchiyama Y, Tsugita A. 1993. Removal of N-terminal formyl groups and deblocking of pyrrolidone carboxylic acid of proteins with anhydrous hydrazine vapor. Eur J Biochem 212:785–789. doi: 10.1111/j.1432-1033.1993.tb17719.x. [DOI] [PubMed] [Google Scholar]
- 19.Hyink O, Balakrishnan M, Tagg JR. 2005. Streptococcus rattus strain BHT produces both a class I two-component lantibiotic and a class II bacteriocin. FEMS Microbiol Lett 252:235–241. doi: 10.1016/j.femsle.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Uzelac G, Kojic M, Lozo J, Aleksandrzak-Piekarczyk T, Gabrielsen C, Kristensen T, Nes IF, Diep DB, Topisirovic L. 2013. A Zn-dependent metallopeptidase is responsible for sensitivity to LsbB, a class II leaderless bacteriocin of Lactococcus lactis subsp. lactis BGMN1-5. J Bacteriol 195:5614–5621. doi: 10.1128/JB.00859-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Belkum MJ, Hayema BJ, Geis A, Kok J, Venema G. 1989. Cloning of two bacteriocin genes from a lactococcal bacteriocin plasmid. Appl Environ Microbiol 55:1187–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirkovic N, Polovic N, Vukotic G, Jovcic B, Miljkovic M, Radulovic Z, Diep DB, Kojic M. 2016. Lactococcus lactis LMG2081 produces two bacteriocins, a nonlantibiotic and a novel lantibiotic. Appl Environ Microbiol 82:2555–2562. doi: 10.1128/AEM.03988-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kojic M, Strahinic I, Fira D, Jovcic B, Topisirovic L. 2006. Plasmid content and bacteriocin production by five strains of Lactococcus lactis isolated from semi-hard homemade cheese. Can J Microbiol 52:1110–1120. doi: 10.1139/w06-072. [DOI] [PubMed] [Google Scholar]
- 24.Mirkovic N, Radulovic Z, Uzelac G, Lozo J, Obradovic D, Topisirovic L, Kojic M. 2015. Isolation and characterization of bacteriocin and aggregation-promoting factor production in Lactococcus lactis ssp. lactis BGBM50 strain. Food Technol Biotechnol 53:237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ansari A, Siddiqui NN, Ghani M, Aman A, Qader SA. 2015. Plasmid borne BAC-IB17: localization of a potential antibacterial positive marker (Bac+) encoded broad inhibitory spectrum bacteriocin. Pak J Pharm Sci 28:1331–1335. [PubMed] [Google Scholar]
- 26.Kalmokoff ML, Banerjee SK, Cyr T, Hefford MA, Gleeson T. 2001. Identification of a new plasmid-encoded sec-dependent bacteriocin produced by Listeria innocua 743. Appl Environ Microbiol 67:4041–4047. doi: 10.1128/AEM.67.9.4041-4047.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Kwaadsteniet M, Fraser T, Van Reenen CA, Dicks LMT. 2006. Bacteriocin T8, a novel class IIa sec-dependent bacteriocin produced by Enterococcus faecium T8, isolated from vaginal secretions of children infected with human immunodeficiency virus. Appl Environ Microbiol 72:4761–4766. doi: 10.1128/AEM.00436-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altena K, Guder A, Cramer C, Bierbaum G. 2000. Biosynthesis of the lantibiotic mersacidin: organization of a type B lantibiotic gene cluster. Appl Environ Microbiol 66:2565–2571. doi: 10.1128/AEM.66.6.2565-2571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwatani S, Zendo T, Yoneyama F, Nakayama J, Sonomoto K. 2007. Characterization and structure analysis of a novel bacteriocin, lacticin Z, produced by Lactococcus lactis QU 14. Biosci Biotechnol Biochem 71:1984–1992. doi: 10.1271/bbb.70169. [DOI] [PubMed] [Google Scholar]
- 30.Fujita K, Ichimasa S, Zendo T, Koga S, Yoneyama F, Nakayama J, Sonomoto K. 2007. Structural analysis and characterization of lacticin Q, a novel bacteriocin belonging to a new family of unmodified bacteriocins of Gram-positive bacteria. Appl Environ Microbiol 73:2871–2877. doi: 10.1128/AEM.02286-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ribeiro SC, O'Connor PM, Ross RP, Stanton C, Silva CCG. 2016. An anti-listerial Lactococcus lactis strain isolated from Azorean Pico cheese produces lacticin 481. Int Dairy J 63:18–28. doi: 10.1016/j.idairyj.2016.07.017. [DOI] [Google Scholar]
- 32.Lozo J, Jovcic B, Kojic M, Dalgalarrondo M, Chobert J-M, Haertlé T, Topisirovic L. 2007. Molecular characterization of a novel bacteriocin and an unusually large aggregation factor of Lactobacillus paracasei subsp. paracasei BGSJ2-8, a natural isolate from homemade cheese. Curr Microbiol 55:266–271. doi: 10.1007/s00284-007-0159-1. [DOI] [PubMed] [Google Scholar]
- 33.Lozo J, Vukasinovic M, Strahinic I, Topisirovic L. 2004. Characterization and antimicrobial activity of bacteriocin 217 produced by natural isolate Lactobacillus paracasei subsp. paracasei BGBUK2-16. J Food Prot 67:2727–2734. doi: 10.4315/0362-028X-67.12.2727. [DOI] [PubMed] [Google Scholar]
- 34.Golić N, Cadež N, Terzić-Vidojević A, Suranská H, Beganović J, Lozo J, Kos B, Sušković J, Raspor P, Topisirović L. 2013. Evaluation of lactic acid bacteria and yeast diversity in traditional white pickled and fresh soft cheeses from the mountain regions of Serbia and lowland regions of Croatia. Int J Food Microbiol 166:294–300. doi: 10.1016/j.ijfoodmicro.2013.05.032. [DOI] [PubMed] [Google Scholar]
- 35.Kojic M, Strahinic I, Topisirovic L. 2005. Proteinase PI and lactococcin A genes are located on the largest plasmid in Lactococcus lactis subsp. lactis bv. diacetylactis S50. Can J Microbiol 51:305–314. doi: 10.1139/w05-009. [DOI] [PubMed] [Google Scholar]
- 36.Hopwood DA, Bibb MJ, Chater KF, Kieser T, Bruton CJ, Kieser HM, Lydiate CM, Smith CP, Ward JM, Schrempf H. 1985. Genetic manipulation of Streptomyces—a laboratory manual. The John Innes Foundation, Norwich, United Kingdom. [Google Scholar]
- 37.O'Sullivan DJ, Klaenhammer TR. 1993. Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus spp. Appl Environ Microbiol 59:2730–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holo H, Nes IF. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol 55:3119–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rea MC, Clayton E, O'Connor PM, Shanahan F, Kiely B, Ross RP, Hill C. 2007. Antimicrobial activity of lacticin 3147 against clinical Clostridium difficile strains. J Med Microbiol 56:940–946. doi: 10.1099/jmm.0.47085-0. [DOI] [PubMed] [Google Scholar]
- 40.Mills S, Serrano LM, Griffin C, O'Connor PM, Schaad G, Bruining C, Hill C, Ross RP, Meijer WC. 2011. Inhibitory activity of Lactobacillus plantarum LMG P-26358 against Listeria innocua when used as an adjunct starter in the manufacture of cheese. Microb Cell Fact 10(Suppl 1):S7. doi: 10.1186/1475-2859-10-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steinberg DA, Hurst MA, Fujii CA, Kung AH, Ho JF, Cheng FC, Loury DJ, Fiddes JC. 1997. Protegrin-1: a broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrob Agents Chemother 41:1738–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement. CLSI document M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 43.Blanusa M, Perovic I, Popovic M, Polovic N, Burazer L, Milovanovic M, Gavrovic-Jankulovic M, Jankov R, Cirkovic Velickovic T. 2007. Quantification of Art v 1 and Act c 1 being major allergens of mugwort pollen and kiwi fruit extracts in mass-units by ion-exchange HPLC-UV method. J Chromatogr B Analyt Technol Biomed Life Sci 857:188–194. doi: 10.1016/j.jchromb.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 44.Gasson MJ. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol 154:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kojic M, Fira D, Banina A, Topisirovic L. 1991. Characterization of the cell wall-bound proteinase of Lactobacillus casei HN14. Appl Environ Microbiol 57:1753–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valenzuela AS, ben Omar N, Abriouel H, Lopez RL, Veljovic K, Canamero MM, Topisirovic MKL, Galvez A. 2009. Virulence factors, antibiotic resistance, and bacteriocins in enterococci from artisan foods of animal origin. Food Control 20:381–385. doi: 10.1016/j.foodcont.2008.06.004. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.