ABSTRACT
Biofilms are microbial communities that inhabit various surfaces and are surrounded by extracellular matrices (ECMs). Clinical microbiologists have shown that the majority of chronic infections are caused by biofilms, following the introduction of the first biofilm infection model by J. W. Costerton and colleagues (J. Lam, R. Chan, K. Lam, and J. W. Costerton, Infect Immun 28:546–556, 1980). However, treatments for chronic biofilm infections are still limited to surgical removal of the infected sites. Pseudomonas aeruginosa and Enterococcus faecalis are two frequently identified bacterial species in biofilm infections; nevertheless, the interactions between these two species, especially during biofilm growth, are not clearly understood. In this study, we observed phenotypic changes in a dual-species biofilm of P. aeruginosa and E. faecalis, including a dramatic increase in biofilm matrix thickness. For clear elucidation of the spatial distribution of the dual-species biofilm, P. aeruginosa and E. faecalis were labeled with red and green fluorescence, respectively. E. faecalis was located at the lower part of the dual-species biofilm, while P. aeruginosa developed a structured biofilm on the upper part. Mutants with altered exopolysaccharide (EPS) productions were constructed in order to determine the molecular basis for the synergistic effect of the dual-species biofilm. Increased biofilm matrix thickness was associated with EPSs, not extracellular DNA. In particular, Pel and Psl contributed to interspecies and intraspecies interactions, respectively, in the dual-species P. aeruginosa and E. faecalis biofilm. Accordingly, targeting Pel and Psl might be an effective part of eradicating P. aeruginosa polymicrobial biofilms.
IMPORTANCE Chronic infection is a serious problem in the medical field. Scientists have observed that chronic infections are closely associated with biofilms, and the vast majority of infection-causing biofilms are polymicrobial. Many studies have reported that microbes in polymicrobial biofilms interact with each other and that the bacterial interactions result in elevated virulence, in terms of factors, such as infectivity and antibiotic resistance. Pseudomonas aeruginosa and Enterococcus faecalis are frequently isolated pathogens in chronic biofilm infections. Nevertheless, while both bacteria are known to be agents of numerous nosocomial infections and can cause serious diseases, interactions between the bacteria in biofilms have rarely been examined. In this investigation, we aimed to characterize P. aeruginosa and E. faecalis dual-species biofilms and to determine the molecular factors that cause synergistic effects, especially on the matrix thickening of the biofilm. We suspect that our findings will contribute to the development of more efficient methods for eradicating polymicrobial biofilm infections.
KEYWORDS: Enterococcus faecalis, Pseudomonas aeruginosa, biofilms, polymicrobial
INTRODUCTION
Biofilms are communities of microbes that dwell on surfaces and are surrounded by extracellular matrices (ECMs) (1). The major characteristics of biofilms are high resistance to antibiotics and other various stresses, a high rate of horizontal gene transfer (HGT), and differential gene expression patterns relative to the planktonic state (1). Biofilms became a popular concept in microbiology approximately 3 decades ago; however, microbiologists still face many difficulties in studying and managing biofilms. Biofilms exist on most surfaces and cause serious problems in the medical field as infectious agents and reservoirs for many pathogens (2, 3). Chronic infections are caused by bacteria in a biofilm mode of growth (3). Due to biofilm characteristics, chronic biofilm infections are very difficult to eradicate (4). There are two major categories of biofilm infection: those associated with medically implanted devices (5–9), and those directly associated with tissues, such as chronic otitis media, dental plaque, endocarditis, lung infections in cystic fibrosis patients, urinary tract infections, and chronic wound infections (4, 10–13). The vast majority of biofilm infections contain more than one species of bacterium, fungus, or other microbe (13).
Several studies have investigated polymicrobial biofilms and have identified beneficial and synergistic interactions between microbes in a biofilm. The interactions in a polymicrobial biofilm affect its overall function, physiology, or surroundings, which enhance resistance or virulence (10, 13, 14). For example, many microbes exist in dental plaque and undergo spatiotemporal interactions, wherein one bacterial species attaches to the tooth surface and alters the surroundings to fit the next bacterial species (15). Staphylococcus aureus has been shown to increase infectivity, biofilm development, and antibiotic resistance when grown with Candida albicans in serum (14). Also, Pseudomonas aeruginosa has been shown to enhance virulence when incubated with Gram-positive bacteria (16).
Pseudomonas species are Gram-negative bacilli and are ubiquitous in the environment; some cause disease in both animals and plants. Among the Pseudomonas species, Pseudomonas aeruginosa is a common human opportunistic pathogen and can cause serious infections. P. aeruginosa is also a very common causative agent of health care-associated infections (HAIs) and the second most common cause of ventilator-associated pneumonia (VAP) in the United States. P. aeruginosa is known to produce various virulence factors, and the expression of these virulence factors is regulated by complex signal transduction systems in response to changes in the surrounding environment, such as biofilm formation (17).
Enterococci are Gram-positive cocci and opportunistic pathogens that are frequently isolated in the normal flora of the human gastrointestinal tract, oral cavity, and female genital tract. Enterococci have been reported to readily adhere to various medical devices and produce biofilms (18). Among enterococcal species, Enterococcus faecalis is the most common nosocomial pathogen and typically causes urinary tract infections, peritonitis, bacteremia, infections in abscesses, decubiti, and foot ulcers, and endocarditis. The pathogen is responsible for approximately 90% of all enterococcus-related HAIs (18–20). Furthermore, because of its ability to resist various antibiotics, E. faecalis causes serious problems in clinical areas (21).
P. aeruginosa and E. faecalis share many characteristics and niches and have been found together in clinical samples from humans (9, 15, 20). Even though several studies have investigated synergism in polymicrobial biofilms, only a few studies on polymicrobial biofilms with P. aeruginosa and E. faecalis have been undertaken. For instance, there is evidence that P. aeruginosa uses peptidoglycan molecules of Gram-positive bacteria as a signal to produce more virulence factors and antimicrobials, which damages the host and alters the microbiome compositions (22). Also, a study showed that pyelonephritis caused by P. aeruginosa was exacerbated by coinfection with E. faecalis (23). In the present research, synergistic effects of biofilm development were detected, including enhanced matrix thickness in P. aeruginosa and E. faecalis dual-species biofilms. Since this synergistic effect of the dual-species biofilm might enhance its virulence, the molecular elements for the synergistic effects were investigated. In order to investigate the dual-species biofilm, we constructed mutants with altered exopolysaccharide (EPS) production and several fluorescent strains. We also described the characteristics of the dual-species P. aeruginosa and E. faecalis biofilm using various techniques, such as viscosity measurement, a crystal violet assay, and confocal laser scanning microscopy. In light of our results, we suggest that the increased matrix thickness of the dual-species biofilm was due, in part, to the expression of Pel and Psl EPSs by P. aeruginosa. In conclusion, we suggest a simple model for dual-species P. aeruginosa and E. faecalis biofilm development and possible molecules that could be targeted for more effective eradication of P. aeruginosa polymicrobial biofilm infections.
RESULTS
Coculture and biofilm development of P. aeruginosa and E. faecalis.
Wild-type P. aeruginosa PAO1 and E. faecalis were grown together in brain heart infusion broth (BHIB) medium. Each bacterial species was differentiated via colony morphology (Fig. 1A). Planktonic growth experiments of individual bacterial samples and cocultured samples were conducted to determine if one bacterial species affected the growth of the other bacterial species. The CFU per milliliter values were used to plot the data in lieu of optical density, due to interference from the excessive production of extracellular materials (Fig. 1B and C). Each bacterial species presented very similar growth patterns and generated similar numbers of bacterial cells during growth (Fig. 1B). Cocultured samples presented a growth pattern almost identical to that of individually grown cultures (Fig. 1C). The results of the growth experiment indicated that P. aeruginosa and E. faecalis did not affect each other during planktonic growth.
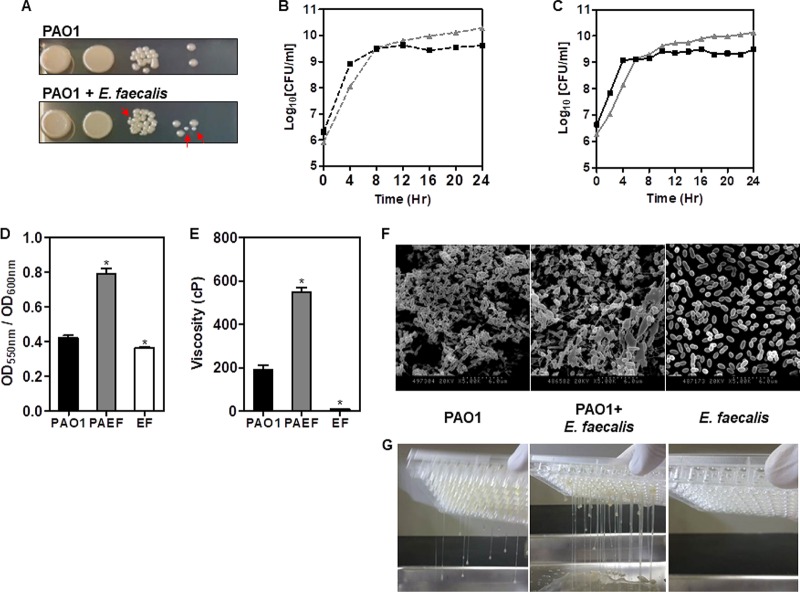
FIG 1.
Polymicrobial biofilm tests of P. aeruginosa and E. faecalis. (A) Colony observation of cocultured P. aeruginosa and E. faecalis (red arrows). The growth experiment of P. aeruginosa (gray triangle) and E. faecalis (black square), grown individually (B) or together (C), in a shaking incubator at 37°C. The viable count assay was used to determine the CFU per milliliter. (D) The CV biofilm assay of the monospecies and the dual-species biofilms. *, P < 0.001 versus the biofilm levels of PAO1 and PAO1 plus E. faecalis (PAEF) or E. faecalis (EF). (E) Viscosity measurements of the mono- and dual-species biofilms. Viscosity appears in units of centipoise (cP). *, P < 0.0001 versus viscosity of PAO1 and PAEF or EF. (F) SEM images of the mono- and dual-species biofilms of P. aeruginosa and/or E. faecalis. The magnifications are ×5,000. (G) Images of the mono- and dual-species biofilms of P. aeruginosa and/or E. faecalis dripping out of the 96-well plates. Detailed video clips are provided in the movies in the supplemental material.
Dual-species biofilm development with P. aeruginosa and E. faecalis was conducted to determine if interactions occur between the two bacterial species during biofilm development. First, the amount of biofilm formation was measured using the standard crystal violet (CV) assay (Fig. 1D). The data indicated that the dual-species biofilm showed enhanced biofilm formation compared to the monospecies biofilms (Fig. 1D). During biofilm development, an interesting biofilm phenotype was observed. The biofilm exhibited a highly sticky phenotype when the two bacterial species were incubated together (see the movies in the supplemental material). Thus, the viscosities of the biofilms were measured using a viscosity meter, revealing a significantly higher viscosity for the dual-species biofilm than for the monospecies biofilms (Fig. 1E). Scanning electron microscope (SEM) images also revealed corresponding biofilm phenotypes (Fig. 1F). The SEM images of P. aeruginosa biofilms showed ECMs with encased and connected bacterial cells, whereas the E. faecalis biofilm SEM images showed no ECMs, with barely any structures. The dual-species biofilm, however, revealed significantly greater amounts of ECM than connected cells of both the same and different species. The images of the corresponding biofilms dripping out of the 96-well plates are displayed in Fig. 1G and Movie S1. The images demonstrate a stickier phenotype of the dual-species biofilm than the monospecies biofilms.
Spatial composition of the dual-species biofilm.
It is very important to distinguish bacterial strains in polymicrobial biofilm investigation in order to determine the accurate distribution of each species in the biofilm architecture. To distinguish P. aeruginosa and E. faecalis under confocal laser scanning microscope (CLSM), each bacterium was inserted with different fluorescent plasmids: pME-dsRED for P. aeruginosa, and pMV158GFP for E. faecalis. Both bacterial strains harboring recombinant fluorescence plasmids were incubated in appropriate medium, with BHIB, gentamicin (5 μg/ml), and tetracycline (1 μg/ml), to prevent the loss of plasmids during growth; the medium did not affect the growth or biofilm formation of the bacteria (data not shown). Both monospecies bacterial cultures presented fluorescence, with red for P. aeruginosa pME-dsRED (PAO1/red) and green for E. faecalis pMV158GFP (EF/green) (Fig. S2A and B). The two bacterial species were clearly distinguishable in the dual-species culture (Fig. S2C). Growth experiments were conducted to confirm that the growth pattern was the same as that with strains lacking fluorescence constructs. The results showed that fluorescent bacteria had growth patterns identical to those of nonfluorescent bacteria (Fig. S3A to C) and that the biofilm development pattern was also identical (Fig. S3D).
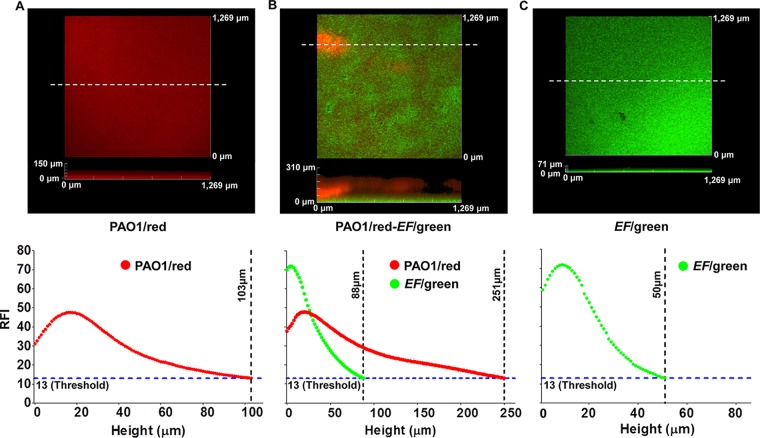
The biofilms of PAO1/red and EF/green were analyzed by CLSM and ImageJ program analyses. The Z-stack images of each biofilm were captured by CLSM, and the fluorescence intensities of each focal plane were analyzed using ImageJ (Fig. 2). Biofilm in CLSM observations disappeared around a relative fluorescence intensity of 13 in all biofilm samples. Thus, the relative fluorescence intensity of 13 was considered the threshold for biofilm detection. The CLSM images revealed a flat PAO1/red biofilm with a height of 103 μm (Fig. 2A, bottom view). Fluorescence intensity demonstrated that most of the biofilm could be detected around 15 to 20 μm, gradually decreasing thereafter (Fig. 2A). The EF/green biofilm also exhibited a flat biofilm, although the height of the biofilm was around 50 μm (Fig. 2C, bottom view), much thinner than the PAO1/red biofilm. In addition, the fluorescence intensity decreased more rapidly than in the PAO1/red biofilm (Fig. 2C). The dual-species biofilm exhibited a striking difference in the spatial distribution of each species along the height of the biofilm. The Z-stack images showed a flat EF/green biofilm at the bottom of the polymicrobial biofilm with a cluster-structured biofilm of PAO1/red on top of it. The fluorescence intensity measurements showed a stronger EF/green fluorescence relative to PAO1/red fluorescence at or below the height of 30 μm, with the PAO1/red fluorescence intensity more dominant than the EF/green at planes higher than 30 μm (Fig. 2B).
FIG 2.
CLSM image analysis of mono- and dual-species PAO1/red and EF/green biofilms. CLSM images and their relative fluorescent intensity measures along the heights of biofilms of PAO1/red (A), PAO1/red and EF/green (B), and EF/green (C). White dotted lines indicate the locations of sagittal sections for height representation of the biofilms. The relative fluorescence intensity (RFI) of 13 was considered the threshold for biofilm detection. The biofilms were incubated at 37°C for 48 h. The relative fluorescence intensities were measured using the ImageJ program.
Effect of extracellular DNA on biofilm development.
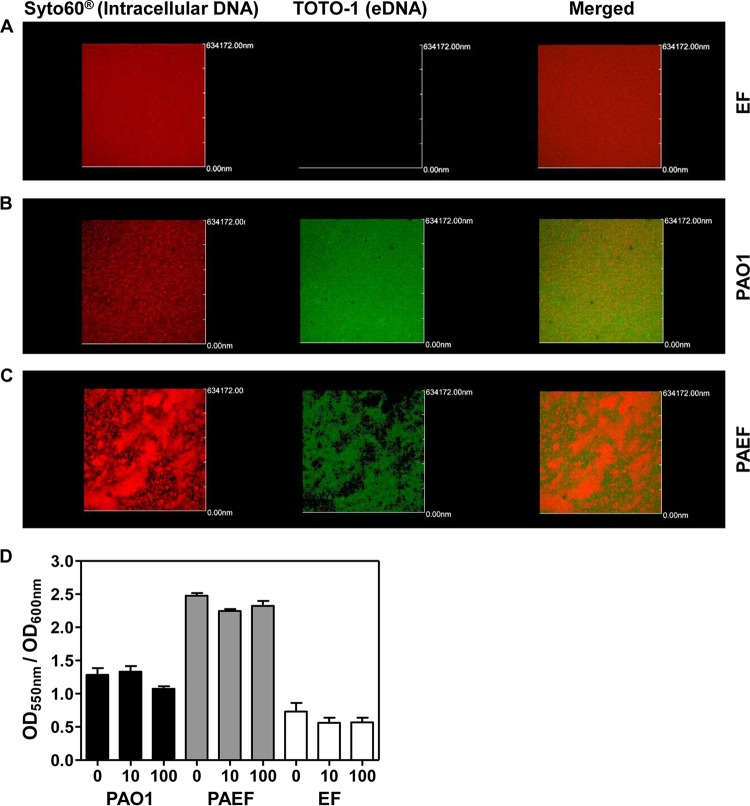
The dual-species PAO1 and E. faecalis biofilm produced a significantly thicker matrix that was not detected in monospecies biofilms (Fig. 1E). In order to determine which component of the ECM contributed to the enhancement of matrix thickness in the dual-species biofilm, we sought to assess the presence of extracellular DNA (eDNA), which has been determined to be a major ECM component (24, 25). To achieve this goal, we stained biofilms with TOTO-1, a DNA-specific green fluorescent dye that does not penetrate live bacterial cells. Green fluorescent signals were not detected in the E. faecalis monospecies biofilm, demonstrating that no eDNA was present inside the biofilm (Fig. 3A and S4A). In contrast, strong fluorescent signals were detected in the PAO1 monospecies biofilm (Fig. 3B and S4B). Of note, a slightly lower level of eDNA-specific signaling was observed in the dual-species biofilm (Fig. 3C and S4C). Images revealed that E. faecalis biofilm produced very small amounts of eDNA. On the other hand, PAO1 produced large amounts of eDNA in its biofilm. The dual-species biofilm presented larger amounts of eDNA than the mono-E. faecalis biofilm but smaller amounts than the mono-PAO1 biofilm (Fig. S4). Furthermore, a CV biofilm assay indicated that the biofilm integrity was not affected when 32-h-old mature biofilms were treated with DNase I for 2 h (Fig. 3D). These results suggest that eDNA is not likely responsible for the matrix thickening in the dual-species biofilm.
FIG 3.
Extracellular DNA detection and DNase I treatment of mono- and dual-species P. aeruginosa and E. faecalis biofilms. The monospecies biofilms of E. faecalis (A), P. aeruginosa (B), and the dual-species biofilm of E. faecalis and P. aeruginosa (C) were stained for intracellular DNA (Syto60) and extracellular DNA (TOTO-1). Wavelengths of 652 nm and 514 nm were used for Syto60 and TOTO-1, respectively. (D) Crystal violet biofilm assay results when two different concentrations of DNase I (10 μg/ml and 100 μg/ml) were used to treat 32-h-old mature biofilms.
Effects of P. aeruginosa EPSs on the formation of the thicker matrix of the dual-species biofilm.
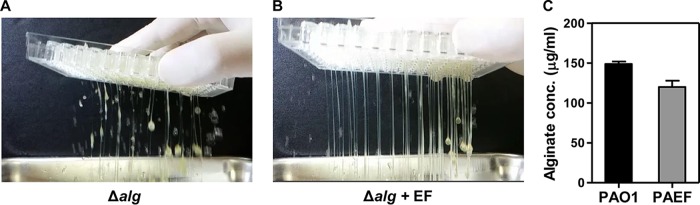
We next examined whether the matrix thickening of the dual-species biofilm is caused by overproduction of EPS molecules. First, we tested the role of alginate, an EPS molecule produced by mucoid P. aeruginosa strains (26). Although alginate is not a major component of nonmucoid PAO1 biofilms (27), we postulated that alginate production might be stimulated during dual-species biofilm formation. An alginate-negative PAO1 mutant, the Δalg mutant, displayed levels of biofilm matrix thickening comparable to that of the PAO1 biofilm when it formed a monospecies biofilm (Fig. 4A and Movie S2). Importantly, the dual-species biofilm of the Δalg mutant plus E. faecalis presented the same matrix thickness as that observed in the PAO1 plus E. faecalis biofilms (Fig. 4B and Movie S3). These results suggest that alginate has no significant effect on the matrix thickening of the dual-species biofilm. Consistent with these results, dual-species biofilms did not exhibit any increase in alginate production (Fig. 4C). Alginate production was only slightly decreased in the PAO1 plus E. faecalis dual-species biofilm, compared with the PAO1 biofilm (Fig. 4C).
FIG 4.
Alginate is not responsible for the elevated biofilm matrix thickness in the dual-species biofilms. The matrix thickness of biofilms was tested by flipping the biofilms upside down. Biofilms of PAO1 Δalg mutant of its own (A) and with E. faecalis (B) were formed and tested. (C) Quantification of alginate production in PAO1 monospecies and PAO1 plus E. faecalis dual-species biofilms. *, P < 0.05 versus the alginate levels of PAO1 and PAO1 plus E. faecalis (PAEF).
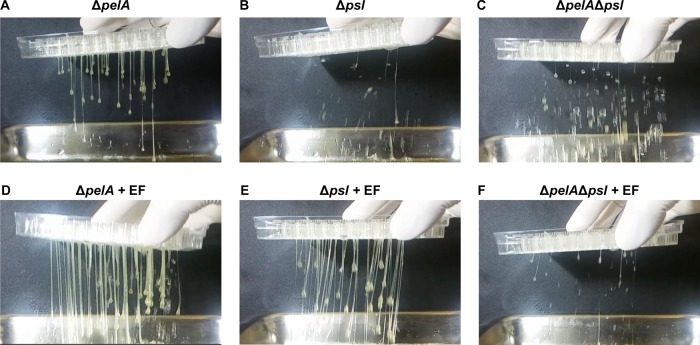
Other EPSs, Pel and Psl, are also known to be important for biofilm development for P. aeruginosa (28). Herein, Pel-negative PAO1 ΔpelA biofilm produced a similarly thicker phenotype relative to the PAO1 biofilm (Fig. 5A and Movie S4). In contrast, a Psl-negative Δpsl mutant strain showed no matrix thickness on its biofilm (Fig. 5B and Movie S6), demonstrating that Psl is critically involved in matrix thickening in PAO1 monospecies biofilm. The dual-species biofilm of the ΔpelA mutant plus E. faecalis showed a significantly increased biofilm matrix thickness, while the Δpsl mutant plus E. faecalis biofilm displayed a matrix thickness that recovered to normal levels (Fig. 5D and E, and Movies S5 and S7). These results suggest that the production of Psl and Pel is stimulated by the presence of E. faecalis. As expected, a PAO1 mutant defective in both Pel and Psl production, the ΔpelA Δpsl mutant, appeared to have no matrix thickness in both the mono- and dual-species biofilms (Fig. 5C and F, and Movies S8 and S9).
FIG 5.
Role of Psl and Pel in the matrix thickening of the dual-species biofilms. The matrix thickness of biofilms was tested by flipping the biofilms upside down. PAO1 ΔpelA (A and D), Δpsl (B and E), and ΔpelA Δpsl mutants (C and F) were grown as monospecies biofilms (A to C) or together with EF (D to F) as dual-species biofilms.
Psl stimulates intraspecies PAO1-PAO1 interactions, while Pel is involved in interspecies PAO1-E. faecalis interactions.
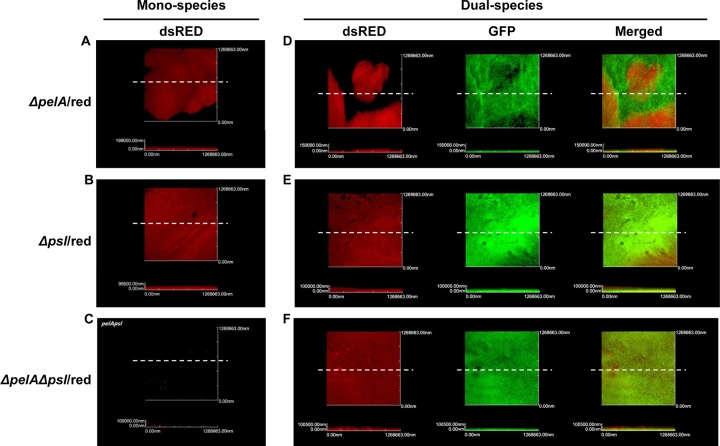
Our results described above indicate that both Psl and Pel are involved in matrix thickening in biofilms. Thus, we aimed to explore the effect of EPS deficiencies on the dual-species biofilm formation. The red fluorescent ΔpelA and Δpsl mutants presented with normal biofilm structures (Fig. 6A and B). The ΔpelA Δpsl double mutant, however, showed a dramatic decrease in its ability to produce biofilm (Fig. 6C). When each of these mutants was coculture with green fluorescent E. faecalis, different patterns of biofilm formation were observed. In the ΔpelA mutant plus E. faecalis dual-species biofilm, ΔpelA mutant cells and E. faecalis were spatially segregated, yielding a structurally distinct biofilm (Fig. 6D). Psl production appeared to be increased in the ΔpelA mutant plus E. faecalis dual-species biofilm (Fig. 5A and D), while Pel was absent in this particular biofilm. Thus, the results shown in Fig. 6D indicate that Psl is likely involved in PAO1-PAO1 intraspecies interactions. In contrast, the Δpsl mutant, when grown together with E. faecalis, produced a biofilm in which the two organisms were mixed quite evenly, suggesting that Pel is more involved in interspecies interactions between PAO1 and EF (Fig. 6E). The ΔpelA Δpsl mutant plus E. faecalis coculture biofilm presented with red fluorescent ΔpelA Δpsl mutant cells that were not detected in its own monospecies biofilm (Fig. 6C and F), suggesting that the biofilm-forming capability of the ΔpelA Δpsl mutant can be restored in the presence of EF, even though the biofilm still exhibited no matrix thickness, as demonstrated in Fig. 5F. Together, these results suggest that Pel and Psl play distinct roles during the formation of dual-species biofilm.
FIG 6.
Biofilms of ΔpelA/red mutant, Δpsl/red mutant, and/or EF/green. The CLSM images of monospecies ΔpelA/red mutant (A), Δpsl/red mutant (B), ΔpelAΔpsl/red mutant (C), the dual-species EF/green and ΔpelA/red mutant (D), Δpsl/red mutant (E), and ΔpelAΔpsl/red mutant (F). White dotted lines indicate the locations of sagittal sections for height representation of the biofilms.
qRT-PCR analysis of Psl and Pel production in mono- and dual-species biofilms.
Quantitative real-time PCR (qRT-PCR) of pslA and pelB of the monospecies PAO1 biofilm and the dual-species biofilm was performed to quantify the expression levels of the two genes at different time points. The qRT-PCR data revealed that both pslA and pelB expression levels were increased in the dual-species biofilms (Fig. 7). The transcription levels of pslA were about 2-fold higher in the dual-species biofilm than in the PAO1 monospecies biofilm in the early and mature stages of biofilm development (Fig. 7A and B). The transcription levels of pelB were also higher in the dual-species biofilms than the monospecies biofilms (Fig. 7C and D). Interestingly, pelB was expressed much more in the early stage of biofilm development (approximately 30-fold that in the monospecies biofilm) than in the mature stage (approximately 6-fold that in the monospecies biofilm). The data confirm that Psl and Pel are expressed more in the dual-species biofilms and support the notion that Pel is closely related to interspecies interactions between P. aeruginosa and E. faecalis.
FIG 7.
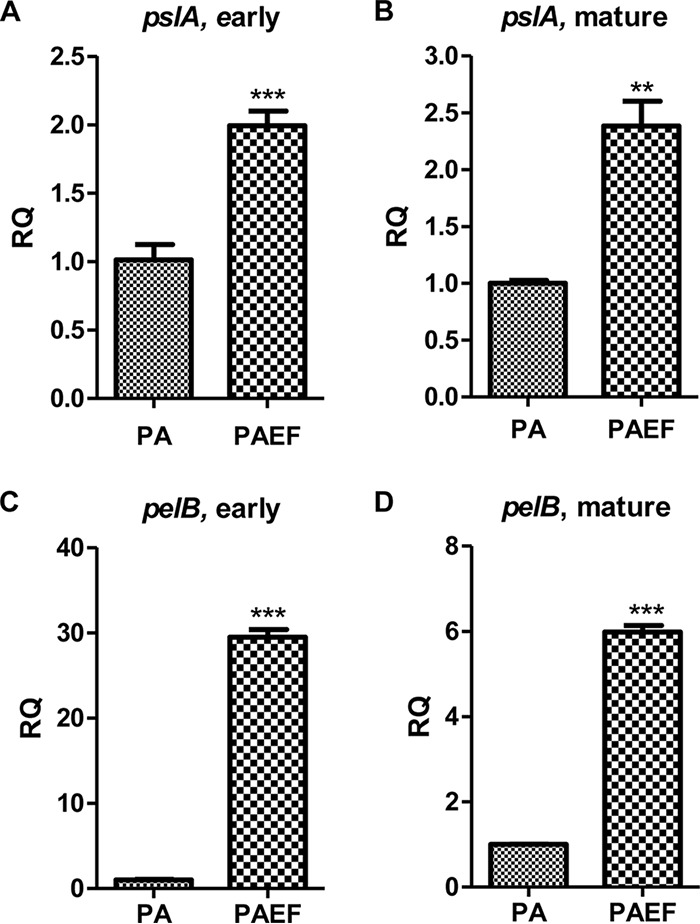
Quantitative real-time PCR analysis of pslA and pelB expression. The qRT-PCR of PAO1 monospecies (the reference samples; PA) and PAO1 plus E. faecalis dual-species (the experimental samples; PAEF) biofilms for pslA (A and B) and pelB (C and D) genes. Gene expression was measured at the early (A and C) and mature (B and D) stages of the biofilm developments. RQ represents the relative quantitation value for genetic expression. **, P < 0.001 versus gene expression of PAO1 and PAO1 plus E. faecalis biofilms. ***, P < 0.0001 versus gene expression of PAO1 and PAO1 plus E. faecalis biofilms.
DISCUSSION
Most chronic infections have been shown to be associated with biofilm infections. However, the eradication of biofilm infections is still extremely difficult, and effective treatments other than surgical removal have not yet been found. One of the major reasons for the difficulties in biofilm infection treatment is that the majority of biofilm infections do not consist of a single bacterial species (13). Biofilms typically consist of multiple species of microbes, and the interactions between the microbes in biofilms make these biofilm infections more virulent and more resistant (10, 20, 29). In this investigation, we focused on characterizing and understanding a P. aeruginosa and E. faecalis dual-species biofilm, as the two species are some of the major causative agents of chronic biofilm infections (4, 20, 30).
P. aeruginosa and E. faecalis belong to two different bacterial groups, Gram-negative and Gram-positive bacteria, respectively, and they cause biofilm infections in similar areas of the human body, such as the urinary tract and wounds (8, 11, 21, 30, 31). Preliminary experiments on the coculturability of P. aeruginosa and E. faecalis showed that P. aeruginosa and E. faecalis did not affect each other's growth under planktonic culture conditions (Fig. 1B and C). Interestingly, the CV biofilm assay showed enhanced biofilm formation (Fig. 1D), and biofilm matrix thickness was enhanced substantially in the dual-species biofilm compared to the monospecies biofilms (Fig. 1E and the movies in the supplemental material). SEM images of the biofilms indicated that there was increased production in the ECM in the dual-species biofilm (Fig. 1F). According to these observations, we discerned that these two bacterial species interact with each other and have synergistic effects on biofilm development.
In our investigation, the dual-species biofilm presented with a more structured PAO1/red biofilm than the monospecies biofilm on top of a flat EF/green biofilm layer (Fig. 2). This spatial distribution of the dual-species biofilm might be due to the ability to grow under a lack of oxygen. Even though both bacterial species are facultative anaerobes, E. faecalis is able to grow using fermentation pathways, while P. aeruginosa cannot ferment and requires nitrogen oxides as an alternative electron acceptor for anaerobic growth (18, 32). Therefore, it would be more efficient for P. aeruginosa to reside on top of E. faecalis in the dual-species biofilm, as there is more oxygen available. The dual-species biofilms were prepared with a 24-h interval between the inoculation times of each bacterium. Regardless of which bacterial species was inoculated first, the biofilms presented the same spatial distribution (data not shown).
One of the major components of the ECM is eDNA, which is known to play a role in the adhesion of bacterial cells on surfaces with repulsive electronic charge and structure stability (24, 25). Accordingly, we observed eDNA in the mono- and dual-species P. aeruginosa and E. faecalis biofilms. The E. faecalis biofilm presented no eDNA (Fig. 3A). On the other hand, the P. aeruginosa biofilm was covered with a substantial amount of eDNA (Fig. 3B). Notwithstanding, the production and distribution of eDNA in the dual-species biofilm were not exactly as we expected: first, the amount of eDNA was not increased in the dual-species biofilm compared to that in the P. aeruginosa monospecies biofilm (Fig. 3C). Second, the distribution of eDNA in the dual-species biofilm was mainly present outside structured biofilms, which may indicate that the eDNA acts less in biofilm structure formation of P. aeruginosa and that the eDNA was present due to the P. aeruginosa. Furthermore, regardless of when and how much DNase I was used, it did not significantly affect the dual-species biofilm (Fig. 3D). According to these results, we deduced that eDNA is not an essential component of the ECM in dual-species biofilm formation.
Another major component of the ECM is EPS. Herein, P. aeruginosa mutants defective in the production of each known EPS molecule, alginate, Pel, and Psl, were constructed, and matrix thickening of the biofilms with these mutants and E. faecalis was observed (Fig. 4 and 5, and the movies in the supplemental material). In biofilms of each P. aeruginosa EPS gene knockout, alginate and Psl seemed to be necessary to obtain thickening of the matrix of the P. aeruginosa biofilm; however, the ΔpelA Δpsl mutant biofilm showed that alginate alone did not express the matrix thickening of P. aeruginosa biofilms (Fig. 5C and Movie S8). The ΔpelA Δpsl mutant plus E. faecalis biofilm showed that alginate is not significantly involved in interspecies interactions with E. faecalis for matrix thickening (Fig. 5F and Movie S9). The Δalg mutant plus E. faecalis biofilm results indicated that both Pel and Psl are necessary for increased matrix thickness (Fig. 4B). Meanwhile, the ΔpelA mutant plus E. faecalis and Δpsl mutant plus E. faecalis biofilms showed that Psl is associated more with the matrix thickness of monospecies biofilm and bacterial surface adhesion and that Pel is more related to interspecies interactions, since the matrix thickness of the Δpsl mutant plus E. faecalis biofilm recovered to normal levels (Fig. 5E and Movie S7). Even though alginate did not seem to be directly involved in matrix thickening, alginate is known to be an important factor for virulence and antibiotic resistance in P. aeruginosa biofilms (33, 34). Therefore, we evaluated alginate production in the mono- and dual-species biofilms via the alginate assay. The results showed no enhancement of alginate in the dual-species biofilm (Fig. 4).
The CLSM images of The PAO1 ΔpelA mutant exhibiting red fluorescence (ΔpelA/red) and PAO1 Δpsl mutant exhibiting red fluorescence (Δpsl/red) with EF/green biofilms indicated that Psl is involved in the interaction between P. aeruginosa cells to form a structured biofilm and that Pel has a greater role in interspecies interactions (Fig. 6). The data also confirmed that P. aeruginosa loses the ability to form biofilm when both Pel and Psl are missing (Fig. 6C). However, the ΔpelA Δpsl mutant plus E. faecalis biofilm showed that ΔpelA Δpsl/red exists within the EF/green biofilm, indicating that E. faecalis has the ability to weakly interact with P. aeruginosa (Fig. 6F). Nevertheless, ΔpelA Δpsl mutant plus E. faecalis biofilm did not present any matrix thickness, suggesting that the matrix thickening of the dual-species biofilm is solely due to Pel and Psl (Fig. 5). qRT-PCR analysis also revealed increased production of Psl and Pel in the dual-species biofilms (Fig. 7). The significant increase in Pel production might indicate that Pel plays an important role in the interspecies interaction of P. aeruginosa and E. faecalis. A limitation of the study, however, was that it was difficult to distinguish between Pel, Psl, and alginate in these biofilms. Different approaches are needed to precisely distinguish the distribution of each EPS. Furthermore, Chew et al. reported similar results for a P. aeruginosa and S. aureus mixed biofilm (35). This may suggest that the results that we obtained reflect general characteristics of P. aeruginosa and Gram-positive coccus mixed biofilms.
In conclusion, P. aeruginosa and E. faecalis can form a dual-species biofilm and have a distinct spatial distribution within the biofilm. E. faecalis tends to locate at the bottom of the biofilm, and P. aeruginosa forms a more structured biofilm on the top of the E. faecalis biofilm. However, the species were not completely separated, as the biofilms did contain cells of the other species when observed under higher magnifications (data not shown). The dual-species biofilm had much thicker ECM than its monospecies counterparts, potentially contributing to the increase in virulence of polymicrobial biofilm infections. For examples, the thicker ECM can protect the dual-species biofilms from antimicrobial reagents, host immune responses, and other environmental stresses. The Psl polysaccharide can inhibit opsonization, which resulted in a decrease in reactive oxygen species (ROS) production by neutrophils (36). As a result of the increased ECM, the biofilm infection can become chronic. This phenotype was mainly due to components of the ECM, particularly Pel and Psl, rather than eDNA and alginate (Fig. 8). Therefore, targeting Pel and Psl of P. aeruginosa might help eradicate polymicrobial infections more effectively. Furthermore, the results of biofilm formation experiments with supernatants or fixed cells of E. faecalis under planktonic or static culture conditions indicated that cell-to-cell contact with live E. faecalis is essential for the expression of the thicker matrix phenotype of the dual-species biofilm (data not shown). This indicates that there should be a mechanism for live E. faecalis recognition on the P. aeruginosa membrane, and the identification of said mechanism will likely contribute to the development of a method with which to eradicate polymicrobial biofilms with P. aeruginosa. Further investigation of the matrix-thickening factors of the dual-species biofilm with E. faecalis is warranted. Investigation using E. faecalis forward genetic screening is in progress.
FIG 8.
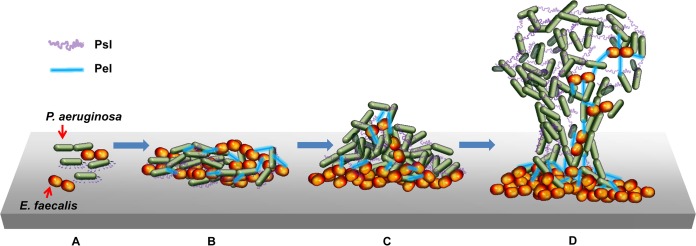
Scheme of the dual-species P. aeruginosa and E. faecalis biofilm development. (A) P. aeruginosa and E. faecalis attach to a surface without spatial separation. (B) During the early stages of dual-species biofilm formation, the attached cells proliferate and start producing EPS, Pel (blue straight line), and Psl (purple curved line). (C) P. aeruginosa and E. faecalis start to exhibit distinct spatial distributions. (D) In the maturation stage of the dual-species biofilm, P. aeruginosa forms a structured biofilm on top of the E. faecalis biofilm: Psl is associated with the surface and between P. aeruginosa, while Pel is associated with interspecies interactions between P. aeruginosa and E. faecalis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. aeruginosa and E. faecalis were used as the model bacteria for this polymicrobial biofilm study. P. aeruginosa PAO1 (wild type) was used as the basis for all mutant P. aeruginosa strains used in this investigation. E. faecalis 12448 (Korean Culture Collection of Microorganism [KCCM]) was used for nonfluorescent E. faecalis experiments. E. faecalis OG1RF harboring pMV158GFP (EF/green) and pAMβ1 were used for the fluorescent E. faecalis experiments. P. aeruginosa harboring the pME plasmid vector that was transcriptionally fused with a dsRED gene after GFP promoter (pME-dsRED) was constructed in the lab (PAO1/red). The E. faecalis pMV158GFP plasmid possessed a tetracycline resistance (Tetr) marker, and P. aeruginosa pME-dsRED contained a gentamicin resistance (Gmr) marker. The bacterial strains and plasmids used are described in Table 1. P. aeruginosa strains lacking plasmid were grown in Luria-Bertani (LB) medium at 37°C. For selection purposes, P. aeruginosa harboring pME-dsRED (PAO1/red) was grown on LB medium with 100 μg/ml gentamicin. E. faecalis was grown on brain heart infusion broth (BHIB) medium, and E. faecalis pMV158GFP (EF/green) was grown on BHIB medium with tetracycline (1 μg/ml) and erythromycin (1 μg/ml) to maintain the plasmids.
TABLE 1.
Bacterial strains and plasmids used in this investigation
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Bacterial strains | ||
| PAO1 | Prototype P. aeruginosa laboratory strain | ATCC 10145 |
| Δalg mutant | PAO1 with in-frame deletion of alg operon (PA3540–PA3548) | This study |
| ΔpelA mutant | PAO1 with in-frame deletion of PA3064 gene | This study |
| Δpsl mutant | PAO1 with in-frame deletion of psl operon (PA2231–PA2242) | This study |
| ΔpelA Δpsl mutant | PAO1 with in-frame deletion of PA3064 gene and psl operon | This study |
| PAO1/red | PAO1 harboring pME-dsRED | This study |
| Δalg/red | Δalg mutant harboring pME-dsRED | This study |
| ΔpelA/red | ΔpelA mutant harboring pME-dsRED | This study |
| Δpsl/red | Δpsl mutant harboring pME-dsRED | This study |
| ΔpelA Δpsl/red | ΔpelA Δpsl mutant harboring pME-dsRED | This study |
| E. faecalis 12448 | E. faecalis type strain (ATCC 19433) | Korean Culture Collection of Microorganisms (KCCM) |
| EF/green | E. faecalis OG1RF strain, harboring pMV158GFP and pAMβ1 | 41 |
| Plasmids | ||
| pME-dsRED | Transcriptional fusion of GFP promoter with a gene encoding DsRed, Gmr | This study |
| pMV158GFP | pMV158, harbors the GFP gene under the control of the PM-inducible promoter, Tetr | 41 |
| pAMβ1 | Auxiliary plasmid for pMV158GFP transport, Emr | 41 |
GFP, green fluorescent protein; Gmr, gentamicin resistance; Tetr, tetracycline resistance; Emr, erythromycin resistance.
Construction of the alg, pelA, psl, and pelA psl clean-deletion mutants.
The Δalg, ΔpelA, Δpsl, and ΔpelA Δpsl mutants were constructed by allelic replacement (37). Briefly, approximately 600 bp of flanking sequences at both ends of each gene or operon were amplified by PCR with primers (Table 2). The upstream reverse primers and the downstream forward primers were complementary to each other. Thus, the 3′ end of the upstream sequence and the 5′ end of the downstream sequence were annealed together during PCR amplification. The ΔpelA Δpsl double mutant was constructed by performing the allelic replacement twice. The deletions of the alg, pelA, and psl operon genes were confirmed by PCR and DNA sequencing.
TABLE 2.
Primers used in this study
| Mutation/gene target by primer type | Flanking region | Direction | Primer sequence (5′–3′)a | Restriction enzyme |
|---|---|---|---|---|
| Primers for clean deletion | ||||
| alg | Left | Forward | ACCTTGAGCTCGCATGGGTCGAAGATTAAGG | SacI |
| Left | Reverse | CGTTAATGAGGTGGCCGTATAAGTCGAAGTAGAGCTGCGC | ||
| Right | Forward | GCGCAGCTCTACTTCGACTTATACGGCCACCTCATTAACG | ||
| Right | Reverse | TAGAGGAGCTCTCTGCAATGGCTGGTTGTAG | SacI | |
| psl | Left | Forward | ACCTTGCATGCGGGCTGGTACATCCAGAAGA | SphI |
| Left | Reverse | TCGTCGATAGTGGCTTTGTGAGCATTCCGACAAGGAGC | ||
| Right | Forward | GCTCCTTGTCGGAATGCTCACAAAGCCACTATCGACGA | ||
| Right | Reverse | TAGAGGCATGCGCATCGACCTGAAAATCCTC | SphI | |
| pelA | Left | Forward | ACCTTGAGCTCCGATCATCCTCGGCTTTCT | SacI |
| Left | Reverse | CAAAACCTGTCGCGTAGTGGTAATCGCTCATCCACAGC | ||
| Right | Forward | GCTGTGGATGAGCGATTACCACTACGCGACAGGTTTTG | ||
| Right | Reverse | TAGAGGAGCTCCGCTGGGCATGAATACTTCT | SacI | |
| qRT-PCR primers | ||||
| pelA | Forward | GGT GAT TAT GTT CCA GGC ACT | ||
| Reverse | GGT GAA CCA GAA GAT CAC CA | |||
| pslB | Forward | TGG CTG ACC TTC AAC AGC GA | ||
| Reverse | TGC TCG AAG TCA CCG AGC TT | |||
| 16S rRNA | Forward | CTT ACG GCC AGG GCT ACA CA | ||
| Reverse | GTA CAA GGC CCG GGA ACG TA |
Restriction enzyme recognition sites are underlined.
Biofilm preparation.
Biofilms were prepared in accordance with the method described by O'Toole (38). Briefly, a static biofilm was prepared by inoculating subcultured bacterial strains (1:100 dilution) in BHIB. BHIB was used for both P. aeruginosa strain and E. faecalis strain biofilms: E. faecalis strains presented retarded growth in LB medium, whereas P. aeruginosa showed similar growth in LB and BHIB media (data not shown). Biofilms for quantification and visual observation were prepared in 96-well plates (HM, South Korea), biofilms for confocal microscopy were prepared in glass-bottom confocal dishes (SPL, South Korea), and biofilms for matrix thickness measurement and extracellular matrix analysis were prepared in 500-ml cell culture flasks (SPL).
Biofilm analysis.
The quantification of biofilms was executed by crystal violet (CV) biofilm assays (38). Also, the CFU of biofilms were measured. The biofilms, grown in 96-well plates, were sonicated with three 10-s pulses at 40 kHz (Branson 8510 ultrasonic cleaner) to disperse the biofilm-associated bacterial cells. Sonicated biofilm samples (100 μl) were harvested and serially diluted for CFU measurements.
Matrix thickness measurements.
Observations by the naked eye were made to assess the matrix thickening of each biofilm. Biofilms were grown in 96-well plates for 48 h and dumped by turning the plates upside down. The thicknesses of biofilms and their ECMs were observed. Briefly, 50 ml of the biofilm was grown in a 500-ml cell culture flask for 48 h at 37°C. Then, the medium was carefully removed by pipetting and rinsing the biofilms with phosphate-buffered saline (PBS) once to remove planktonic cells. The biofilms and their matrices from the surface of the cell culture flask were harvested using cell scrapers. The harvested biofilms and ECMs were transferred to 50-ml conical tubes (HM, South Korea), and viscosity was measured using a viscometer (Brookfield digital viscometer model DV-II) with spindle no. 6 (RV/HA/HB series) at 25°C and 50 rpm. The viscosity measurements were performed three times for each biofilm sample, and average readings thereof were recorded.
CLSM image analysis of biofilms.
Differential interference contrast (DIC) images were acquired using a confocal laser scanning microscope (FV-1000; Olympus Optical Co. Ltd., Japan) equipped with FV10-ASW operating software (version 02.01). For CLSM image analysis, biofilms were grown in cover-glass bottom dishes (SPL, South Korea). After 24 h of biofilm growth, the planktonic portion of the cultures was removed, and the plates were washed with 0.9% saline. Bacterial strains harboring fluorescent plasmids were examined at 488 nm and 594 nm for the pMV158GFP and pME-dsRED plasmids, respectively. Syto60 (Life Technologies) and TOTO-1 (Life Technologies) fluorescent dyes were used for the detection of intracellular DNA and extracellular DNA, respectively. Excitation wavelengths at 652 nm and 514 nm were used for Syto60 and TOTO-1, respectively. Calcofluor white dye was used for EPS detection at 358 nm. For three-dimensional (3D) image analysis, Z-stack images were obtained, and 3D images were reconstructed using the FV10-ASW software. The ImageJ program was used to analyze the fluorescence intensities of the Z-stack images (39).
SEM of biofilms.
The biofilm samples were grown on 12-mm round cover glass and fixed with Karnovsky fixing solution (2% glutaraldehyde, 2% paraformaldehyde, 0.5% CaCl2) for 6 h. The fixed samples were washed with 0.1 M phosphate buffer for 2 h and treated with 1% OsO4 for 2 h. Then, the biofilm samples were dehydrated in an ascending gradual series (50 to ∼100%) of ethanol, infiltrated with isoamyl acetate, and dried (critical point dryer HCP-2; Hitachi, Japan). The dried biofilm samples were coated with gold via ion sputter at 6 mA for 6 min. The samples were observed using field emission scanning electron microscopy (FE-SEM; S-800; Hitachi Ltd., Tokyo, Japan).
Alginate quantification.
Bacterial alginate was indirectly quantified by means of an alginate assay that measures uronic acid levels using a d-mannuronic acid lactone standard curve (40). Briefly, bacterial samples were grown at 37°C for 24 h in both planktonic and biofilm modes of growth. The planktonic cultures were centrifuged to obtain supernatants. The supernatants of the biofilm samples were carefully harvested by pipetting, and the remaining biofilms were scraped for harvesting. The biofilm samples were sonicated, and the resultant supernatants were harvested. The culture supernatants (20 μl) were mixed with 80 μl of distilled water and assayed for alginate quantification (40). One hundred microliters each of alginate standard and sample were mixed thoroughly with 600 μl of the cold sulfuric acid-borate solution in an ice bath. The mixtures were heated to 100°C for 10 min and then rapidly cooled in an ice bath. Ten microliters of 0.1% carbazole solution was added to the mixture and reheated at 100°C for 15 min. The samples were cooled immediately in ice after heating. The absorbance at 525 nm was measured. The optical density at 600 nm (OD600) values of bacterial suspensions were used for normalization.
qRT-PCR analysis of Pel and Psl production in monospecies and dual-species biofilms.
Pel and Psl polysaccharide production was indirectly measured by qRT-PCR analysis of pslA and pelB, the constituents of the psl and pel operons, respectively. The primer sequences used for the qRT-PCR are listed in Table 2. Briefly, PAO1 monospecies biofilms and PAO1 plus E. faecalis dual-species biofilms were harvested at 4 h and 24 h postinoculation. RNA was isolated using an RNeasy minikit (Qiagen). cDNA was prepared using the PrimeScript II first-strand cDNA synthesis kit (TaKaRa Bio, Inc., Japan). qRT-PCR was performed using a SYBR Premix Ex Taq kit (TaKaRa Bio, Inc.) and gene-specific primers. For each sample, three qRT-PCR replicates were performed using the Applied Biosystems 7300 real-time PCR system. The following thermocycling conditions were utilized: 94°C for 5 min, followed by 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min. The level of each gene was normalized to that of the 16S rRNA of P. aeruginosa. The results are expressed relative to the gene expression level obtained with gene-specific primers from the PAO1 monospecies biofilm.
Statistical analysis.
All data are expressed as the mean ± standard deviation (SD). Unpaired Student's t test and one-way analysis of variance (ANOVA) were used to analyze the significance of all comparisons. A P value of <0.05 was considered statistically significant. All experiments were repeated at least three times for reproducibility.
Supplementary Material
ACKNOWLEDGMENTS
E. faecalis OG1RF harboring pMV158GFP (EF/green) and pAMβ1 was kindly provided by Manuel Espinosa Padron (Centro de Investigaciones Biológicas, Consejo Superior de Investigaciones Científicas, Madrid, Spain).
This work was supported by grants from the National Research Foundation of Korea (NRF), funded by the South Korean government (grants 2014R1A2A2A01002861 and 2014R1A4A1008625). This work was also supported by a grant from the Korea Healthcare Technology R&D project, of the Ministry of Health, Welfare, and Family Affairs (grant HI15C0694).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01182-17.
REFERENCES
- 1.O'Toole G, Kaplan HB, Kolter R. 2000. Biofilm formation as microbial development. Annu Rev Microbiol 2000 54:49–79. [DOI] [PubMed] [Google Scholar]
- 2.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 3.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 21:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 4.Clinton A, Carter T. 2015. Chronic wound biofilms: pathogenesis and potential therapies. Lab Med 46:277–284. doi: 10.1309/LMBNSWKUI4JPN7SO. [DOI] [PubMed] [Google Scholar]
- 5.Donlan RM. 2001. Biofilms and device-associated infections. Emerg Infect Dis 7:277–281. doi: 10.3201/eid0702.010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandoe JA, Witherden IR, Cove JH, Heritage J, Wilcox MH. 2003. Correlation between enterococcal biofilm formation in vitro and medical-device-related infection potential in vivo. J Med Microbiol 52:547–550. doi: 10.1099/jmm.0.05201-0. [DOI] [PubMed] [Google Scholar]
- 7.Lleo M, Bonato B, Tafi MC, Caburlotto G, Benedetti D, Canepari P. 2007. Adhesion to medical device materials and biofilm formation capability of some species of enterococci in different physiological states. FEMS Microbiol Lett 274:232–237. doi: 10.1111/j.1574-6968.2007.00836.x. [DOI] [PubMed] [Google Scholar]
- 8.Niveditha S, Pramodhini S, Umadevi S, Kumar S, Stephen S. 2012. The isolation and the biofilm formation of uropathogens in the patients with catheter-associated urinary tract infections (UTIs). J Clin Diagn Res 6:1478–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandes A, Dias M. 2013. The microbiological profiles of infected prosthetic implants with an emphasis on the organisms which form biofilms. J Clin Diagn Res 7:219–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolcott R, Costerton JW, Raoult D, Cutler SJ. 2013. The polymicrobial nature of biofilm infection. Clin Microbiol Infect 19:107–112. doi: 10.1111/j.1469-0691.2012.04001.x. [DOI] [PubMed] [Google Scholar]
- 11.Semedo-Lemsaddek T, Mottola C, Alves-Barroco C, Cavaco-Silva P, Tavares L, Oliveira M. 2016. Characterization of multidrug-resistant diabetic foot ulcer enterococci. Enferm Infecc Microbiol Clin 34:114–116. doi: 10.1016/j.eimc.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 12.Burmølle M, Thomsen TR, Fazli M, Dige I, Christensen L, Homoe P, Tvede M, Nyvad B, Tolker-Nielsen T, Givskov M, Moser C, Kirketerp-Moller K, Johansen HK, Hoiby N, Jensen PO, Sorensen SJ, Bjarnsholt T. 2010. Biofilms in chronic infections–a matter of opportunity–monospecies biofilms in multispecies infections. FEMS Immunol Med Microbiol 59:324–336. doi: 10.1111/j.1574-695X.2010.00714.x. [DOI] [PubMed] [Google Scholar]
- 13.Peters BM, Jabra-Rizk MA, O'May GA, Costerton JW, Shirtliff ME. 2012. Polymicrobial interactions: impact on pathogenesis and human disease. Clin Microbiol Rev 25:193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harriott MM, Noverr MC. 2009. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: effects on antimicrobial resistance. Antimicrob Agents Chemother 53:3914–3922. doi: 10.1128/AAC.00657-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colombo AV, Barbosa GM, Higashi D, di Micheli G, Rodrigues PH, Simionato MR. 2013. Quantitative detection of Staphylococcus aureus, Enterococcus faecalis and Pseudomonas aeruginosa in human oral epithelial cells from subjects with periodontitis and periodontal health. J Med Microbiol 62:1592–1600. doi: 10.1099/jmm.0.055830-0. [DOI] [PubMed] [Google Scholar]
- 16.Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. 2013. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci U S A 110:1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alhazmi A. 2015. Pseudomonas aeruginosa—pathogenesis and pathogenic mechanisms. Int J Biol 7:44–67. doi: 10.5539/ijb.v7n2p44. [DOI] [Google Scholar]
- 18.Fisher K, Phillips C. 2009. The ecology, epidemiology and virulence of Enterococcus. Microbiology 155:1749–1757. doi: 10.1099/mic.0.026385-0. [DOI] [PubMed] [Google Scholar]
- 19.Nallapareddy SR, Singh KV, Sillanpaa J, Garsin DA, Hook M, Erlandsen SL, Murray BE. 2006. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest 116:2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, Rhoads D. 2008. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PLoS One 3:e3326. doi: 10.1371/journal.pone.0003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohamed JA, Huang DB. 2007. Biofilm formation by enterococci. J Med Microbiol 56:1581–1588. doi: 10.1099/jmm.0.47331-0. [DOI] [PubMed] [Google Scholar]
- 22.Korgaonkar AK, Whiteley M. 2011. Pseudomonas aeruginosa enhances production of an antimicrobial in response to N-acetylglucosamine and peptidoglycan. J Bacteriol 193:909–917. doi: 10.1128/JB.01175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuchimori N, Hayashi R, Shino A, Yamazaki T, Okonogi K. 1994. Enterococcus faecalis aggravates pyelonephritis caused by Pseudomonas aeruginosa in experimental ascending mixed urinary tract infection in mice. Infect Immun 62:4534–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 25.Okshevsky M, Meyer RL. 2015. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit Rev Microbiol 41:341–352. doi: 10.3109/1040841X.2013.841639. [DOI] [PubMed] [Google Scholar]
- 26.Yoon SS, Coakley R, Lau GW, Lymar SV, Gaston B, Karabulut AC, Hennigan RF, Hwang SH, Buettner G, Schurr MJ, Mortensen JE, Burns JL, Speert D, Boucher RC, Hassett DJ. 2006. Anaerobic killing of mucoid Pseudomonas aeruginosa by acidified nitrite derivatives under cystic fibrosis airway conditions. J Clin Invest 116:436–446. doi: 10.1172/JCI24684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wozniak DJ, Wyckoff TJ, Starkey M, Keyser R, Azadi P, O'Toole GA, Parsek MR. 2003. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc Natl Acad Sci U S A 100:7907–7912. doi: 10.1073/pnas.1231792100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colvin KM, Gordon VD, Murakami K, Borlee BR, Wozniak DJ, Wong GC, Parsek MR. 2011. The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog 7:e1001264. doi: 10.1371/journal.ppat.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy S, Elgharably H, Sinha M, Ganesh K, Chaney S, Mann E, Miller C, Khanna S, Bergdall VK, Powell HM, Cook CH, Gordillo GM, Wozniak DJ, Sen CK. 2014. Mixed-species biofilm compromises wound healing by disrupting epidermal barrier function. J Pathol 233:331–343. doi: 10.1002/path.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watters C, DeLeon K, Trivedi U, Griswold JA, Lyte M, Hampel KJ, Wargo MJ, Rumbaugh KP. 2013. Pseudomonas aeruginosa biofilms perturb wound resolution and antibiotic tolerance in diabetic mice. Med Microbiol Immunol 202:131–141. doi: 10.1007/s00430-012-0277-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watters C, Everett JA, Haley C, Clinton A, Rumbaugh KP. 2014. Insulin treatment modulates the host immune system to enhance Pseudomonas aeruginosa wound biofilms. Infect Immun 82:92–100. doi: 10.1128/IAI.00651-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma G, Rao S, Bansal A, Dang S, Gupta S, Gabrani R. 2014. Pseudomonas aeruginosa biofilm: potential therapeutic targets. Biologicals 42:1–7. doi: 10.1016/j.biologicals.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Min KB, Lee KM, Oh YT, Yoon SS. 2014. Nonmucoid conversion of mucoid Pseudomonas aeruginosa induced by sulfate-stimulated growth. FEMS Microbiol Lett 360:157–166. doi: 10.1111/1574-6968.12600. [DOI] [PubMed] [Google Scholar]
- 34.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. 2010. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents 35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 35.Chew SC, Kundukad B, Seviour T, van der Maarel JR, Yang L, Rice SA, Doyle P, Kjelleberg S. 2014. Dynamic remodeling of microbial biofilms by functionally distinct exopolysaccharides. mBio 5(4):e01536-14. doi: 10.1128/mBio.01536-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mishra M, Byrd MS, Sergeant S, Azad AK, Parsek MR, McPhail L, Schlesinger LS, Wozniak DJ. 2012. Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell Microbiol 14:95–106. doi: 10.1111/j.1462-5822.2011.01704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee KM, Park Y, Bari W, Yoon MY, Go J, Kim SC, Lee HI, Yoon SS. 2012. Activation of cholera toxin production by anaerobic respiration of trimethylamine N-oxide in Vibrio cholerae. J Biol Chem 287:39742–39752. doi: 10.1074/jbc.M112.394932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Toole GA. 2011. Microtiter dish biofilm formation assay. J Vis Exp 2011(47):e2437. doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abramoff MD, Magalhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int 11:36–42. [Google Scholar]
- 40.Damron FH, Qiu D, Yu HD. 2009. The Pseudomonas aeruginosa sensor kinase KinB negatively controls alginate production through AlgW-dependent MucA proteolysis. J Bacteriol 191:2285–2295. doi: 10.1128/JB.01490-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nieto C, Espinosa M. 2003. Construction of the mobilizable plasmid pMV158GFP, a derivative of pMV158 that carries the gene encoding the green fluorescent protein. Plasmid 49:281–285. doi: 10.1016/S0147-619X(03)00020-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.