ABSTRACT
Group A streptococci (GAS) are highly prevalent human pathogens whose primary ecological niche is the superficial epithelial layers of the throat and/or skin. Many GAS strains with a strong tendency to cause pharyngitis are distinct from strains that tend to cause impetigo; thus, genetic differences between them may confer host tissue-specific virulence. In this study, the FbaA surface protein gene was found to be present in most skin specialist strains but largely absent from a genetically related subset of pharyngitis isolates. In an ΔfbaA mutant constructed in the impetigo strain Alab49, loss of FbaA resulted in a slight but significant decrease in GAS fitness in a humanized mouse model of impetigo; the ΔfbaA mutant also exhibited decreased survival in whole human blood due to phagocytosis. In assays with highly sensitive outcome measures, Alab49ΔfbaA was compared to other isogenic mutants lacking virulence genes known to be disproportionately associated with classical skin strains. FbaA and PAM (i.e., the M53 protein) had additive effects in promoting GAS survival in whole blood. The pilus adhesin tip protein Cpa promoted Alab49 survival in whole blood and appears to fully account for the antiphagocytic effect attributable to pili. The finding that numerous skin strain-associated virulence factors make slight but significant contributions to virulence underscores the incremental contributions to fitness of individual surface protein genes and the multifactorial nature of GAS-host interactions.
KEYWORDS: Streptococcus pyogenes, group A streptococci, impetigo, phagocytosis, surface proteins
INTRODUCTION
Group A streptococcus (GAS; Streptococcus pyogenes) is a global human pathogen that causes ∼750 million infections per year (1, 2). The epithelia of the throat and skin are the primary ecological niches and reservoirs for GAS. It is well established that some GAS strains have a strong predilection for causing infection only at the throat (pharyngitis), whereas other strains have a high tendency to cause superficial infection only at the skin (impetigo). The determinants of tissue site preference for GAS infection and their relative contributions to fitness and virulence are not fully understood.
The key sources of structural and functional diversity of GAS exoproteins include the genes within the emm and FCT regions, which map on the chromosome approximately equidistant from the origin of replication, but on opposite sides. Several statistically significant associations between emm region genes (encoding M protein and other surface and secreted proteins), FCT region genes (encoding surface pili and other adhesins), and clinical associations with impetigo versus pharyngitis have been made (reviewed in reference 3). Although there are >200 emm types, the content and arrangement of emm and emm-like genes can be organized into the following three emm pattern groupings that closely correspond to the primary tissue site of infection: throat specialists (emm patterns A to C [emm pattern A-C]), skin specialists (emm pattern D), and generalists (emm pattern E). The specialized plasminogen-binding M protein (PAM) is exclusive to the emm pattern D skin specialists, as is a lineage of the plasminogen activator streptokinase, which is encoded by the ska locus that lies within the greater emm region (4, 5). Throat and skin specialist strains also display significant differences in the content of FCT region genes (6). The contributions of several impetigo-associated genes to virulence have been validated experimentally (7–12).
In this report, the relationship between the less-well-studied GAS surface protein gene fbaA and skin versus throat specialist strains is defined, and the effect of FbaA on the virulence of an impetigo strain is quantified. The impact of FbaA on GAS pathogenesis is compared to those of two other cell surface protein genes that are disproportionately associated with classical skin strains.
RESULTS
Distribution of fbaA in the GAS population.
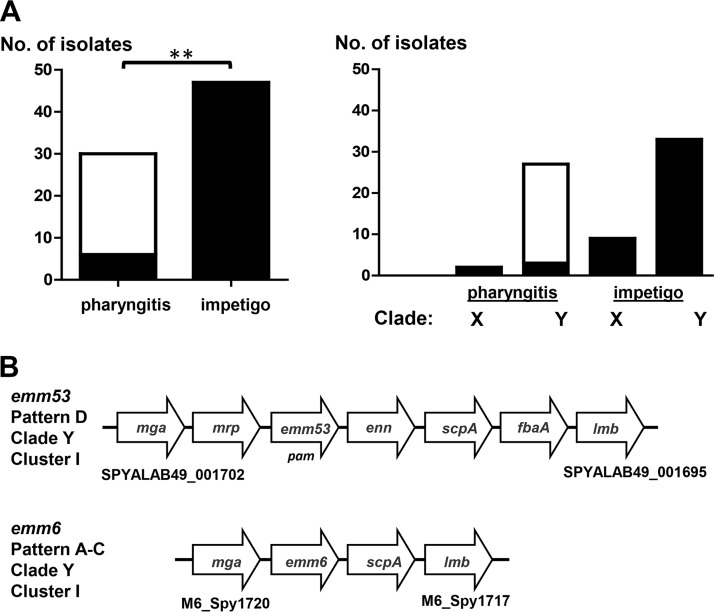
Since fbaA is an accessory gene as opposed to a core gene, its relative distribution among GAS strains may reveal biologically important associations that, in turn, may provide clues to the role of FbaA in GAS disease. For a genetically diverse set of 85 GAS strains representing 85 distinct emm types, the data show that the majority (86%) harbor fbaA (Table 1). GAS strains lacking fbaA are concentrated in the cluster I group (10 of 12 [83%] fbaA-negative strains) (Table 1). Cluster I is one of two major groupings of genetically distinct GAS strains, defined by the presence versus absence of numerous accessory gene loci (13). Furthermore, cluster I strains are almost exclusively of emm pattern A-C or emm pattern D (i.e., throat or skin specialists, respectively), whereby the vast majority of emm pattern D strains belong to cluster I, as does a clinically important subset of emm pattern A-C strains (13). Strikingly, 80% of cluster I emm pattern A-C strains are fbaA deficient, in contrast to <4% of cluster I pattern D strains (P < 0.0001; Fisher's exact test) (Table 1). The fbaA chromosomal regions of a classical skin strain (Alab49) (emm53; emm pattern D) and a throat strain (emm6; emm pattern A-C), both of which are assigned to the cluster I grouping, are illustrated in Fig. 1B.
TABLE 1.
Distribution of fbaA among genotype-defined subpopulations of GAS
| Clustera | emm pattern group | No. of isolates | No. of fbaA- positive isolatesb | No. of fbaA-negative isolates | % fbaA-positive isolatese | Comparator group | P valuec |
|---|---|---|---|---|---|---|---|
| I and II combined | All | 85d | 73 | 12 | 85.9 | ||
| I | All | 41 | 31 | 10 | 75.6 | Cluster II, all | 0.012 |
| II | All | 44 | 42 | 2 | 95.5 | ||
| I | A-C | 10 | 2 | 8 | 20.0 | Cluster I, pattern D | <0.0001 |
| D | 29 | 28 | 1 | 96.6 | |||
| E | 2 | 1 | 1 | NA | |||
| II | A-C | 3 | 1 | 2 | NA | ||
| D | 3 | 3 | 0 | NA | |||
| E | 38 | 38 | 0 | 100 | |||
| I and II combined | A-C | 17 | 4 | 13 | 23.5 | Cluster I/II, pattern D | <0.0001 |
| D | 33 | 32 | 1 | 97.0 | Cluster I/II, pattern E | NS | |
| E | 43 | 42 | 1 | 97.7 | Cluster I/II, pattern A-C | <0.0001 |
Assignment is based on accessory gene haplotypes, as described previously (13).
Determined by PCR amplification of genomic DNA.
By Fisher's exact test (two-tailed). NS, not significant.
All 85 GAS isolates harbor distinct emm types.
NA, not applicable.
FIG 1.
Distribution of fbaA among cluster I pharyngitis and impetigo isolates. (A) fbaA-negative (open) and fbaA-positive (filled) isolates. **, P < 0.01 (Fisher's exact test, two-tailed). (B) Chromosomal maps of fbaA and flanking genes for representative impetigo (Alab49; emm53, emm pattern D, clade Y, cluster I, fbaA positive) and pharyngitis (MGAS10394; emm6, emm pattern A-C, clade Y, cluster I, fbaA negative) isolates. Genes and intergenic regions are not drawn to scale (13, 39).
To better assess clinical relevance, a sample of 77 GAS isolates recovered from known cases of pharyngitis or impetigo and having emm types represented by the GAS strains belonging to the cluster I group (13) was analyzed for the presence or absence of fbaA. This collection is a “convenience” sample selected from in-house GAS strains; together, the 77 organisms exhibit high genetic diversity, representing 28 of the 41 emm types assigned to cluster I. Strikingly, the data show that only 20% of the cluster I pharyngitis isolates harbor fbaA, compared to 100% of the impetigo isolates, a distinction that has high statistical significance (Fig. 1A, left panel). Thus, an important subset of pharyngitis isolates which are otherwise highly similar to classical emm pattern D impetigo strains in their content of accessory genes (13) can be distinguished from those skin strains by a relative lack of fbaA.
emm type assignments can be stratified further according to emm phylogenetic clade (X or Y), which is based on amino acid sequence alignment of the entire surface-exposed portion of the M protein (14). The majority (78%) of GAS isolates in the set of 77 cluster I GAS isolates harbor clade Y emm types. Unlike clade Y impetigo isolates, nearly all clade Y pharyngitis isolates (89%) lack fbaA (Fig. 1A, right panel). Included among the pharyngitis isolates lacking fbaA are the highly prevalent clade Y emm types 3, 5, 6, and 18 (data not shown), which rank high among the emm types most often recovered in association with acute rheumatic fever (15). Taken together, the findings on the distribution of fbaA within the GAS population provided the basis for a testable hypothesis, i.e., that FbaA is a critical virulence factor for superficial GAS infection at the skin.
Bacterial cell surface expression of FbaA.
Immunofluorescence assay of whole bacterial cells was used to evaluate the expression of FbaA on the cell surface of the cluster I classical skin strain Alab49 (emm53; clade Y, emm pattern D). The data show that FbaA was expressed in a growth phase-dependent manner, whereupon FbaA was present during logarithmic growth but absent in stationary phase (Table 2; see Fig. S4B and C in the supplemental material). Loss of surface FbaA during stationary phase can be explained by its degradation via the secreted cysteine protease SpeB, as evidenced by the recovery of FbaA cell surface expression following bacterial growth in the presence of the cysteine protease inhibitor E64 (Fig. S4D) and the presence of FbaA on the surface of the Alab49ΔspeB isogenic mutant (Fig. S4H).
TABLE 2.
Summary of immunofluorescence staining of whole bacterial cells
| Alab49 construct | Growth phase | E64 cysteine protease inhibitor | Antiserum | Immunofluorescence staining scorea |
|---|---|---|---|---|
| WT | Logarithmic | Absent | Preimmune | 0 |
| Logarithmic | Absent | Anti-rFbaA | 3+ | |
| Stationary | Absent | Anti-rFbaA | 0 | |
| Stationary | Present | Anti-rFbaA | 3+ | |
| ΔfbaA | Stationary | Present | Anti-rFbaA | 0 |
| ΔfbaAΔpam | Stationary | Present | Anti-rFbaA | 0 |
| Δmga | Stationary | Present | Anti-rFbaA | 0 |
| ΔspeB | Stationary | Absent | Anti-rFbaA | 3+ |
0, no staining; 3+, strong staining.
In strain Alab49, transcription of fbaA was activated by the global transcriptional regulator Mga, whereby the activation effect was ∼4-fold greater during logarithmic-phase growth than during stationary phase (Table 3), confirming previous findings (9). As expected, cell surface expression of FbaA in strain Alab49 was also dependent on Mga, as evidenced by the lack of detectable FbaA on Alab49Δmga following its growth in E64 (Fig. S4G). As a control, loss of Mga was shown to have no measurable effect on the SpeB activity phenotype during cultivation on Columbia agar (Table S3).
TABLE 3.
Effects of genetic deletions on RNA transcript levels of fbaA and flanking genes
| Alab49 construct | Growth phasea | RNA transcript level relative to that in Alab49WTb |
|||
|---|---|---|---|---|---|
| pam | scpA | fbaA | lmb | ||
| ΔfbaA | Logarithmic | ND | 1.59 ± 0.076 | <0.001 | 1.12 ± 0.84 |
| ΔfbaAΔpam | Logarithmic | ND | 1.22 ± 1.06 | <0.001 | 0.78 ± 0.43 |
| Δmga | Logarithmic | 0.012 ± 0.011 | 0.045 ± 0.049 | 0.043 ± 0.027 | ND |
| Stationary | 0.096 ± 0.134 | ND | 0.164 ± 0.171 | ND | |
OD600 values for logarithmic cultures ranged from 0.448 to 0.482.
Based on two or more independent RNA preparations analyzed by qRT-PCR. ND, not determined.
Two mutant constructs with inactivated fbaA genes were characterized for cell surface expression of FbaA. As expected, FbaA was absent from the cell surface of Alab49ΔfbaA and Alab49ΔfbaAΔpam mutants following their growth in E64-containing culture medium (Fig. S4E and F). Importantly, the ΔfbaA mutants showed no evidence of transcriptional polar effects on transcription of an upstream (scpA) or downstream (lmb) gene (Table 3).
Contributions of fbaA and pam to impetigo.
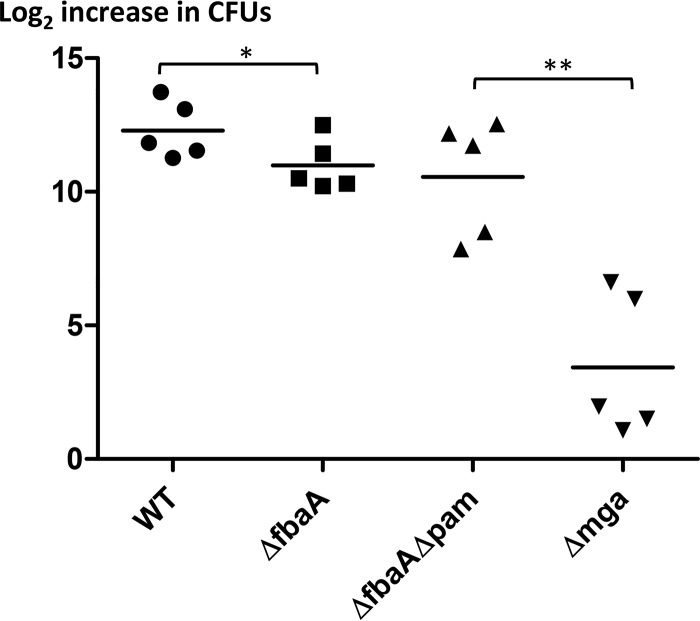
The wild-type Alab49 strain (Alab49WT) was directly compared to its ΔfbaA isogenic mutant for relative fitness and virulence in a humanized experimental model of impetigo. In each test experiment, bacteria grown to mid-logarithmic phase were inoculated onto two grafts of a single hu-skin-SCID mouse (SCID mouse engrafted with human skin), whereby each graft received ∼2,200 CFU; this inoculum size is likely a close parallel to the infective dose encountered during natural human infections. The data show that, by 6 days, there was a mean average of 12.295 population doublings for Alab49WT, compared to 10.988 population doublings for the Alab49ΔfbaA mutant; the difference in the relative change in CFU between the two organisms is statistically significant (Fig. 2) (t = 0.035; two-tailed paired t test). The data correspond to a relative growth index for the ΔfbaA mutant that is only 39% of that of Alab49WT (Table 4). Although FbaA makes a statistically significant contribution to GAS fitness and virulence at the skin, its overall effect is rather modest, and Alab49 is still able to thrive in the absence of FbaA.
FIG 2.
Bacterial population doubling in an experimental model of impetigo. Pairs of human skin grafts on doubly engrafted hu-skin-SCID mice (n = 5 animals per group) were infected with either Alab49WT (●; mean average inoculum, 2,266 ± 321 CFU) and Alab49ΔfbaA (■; inoculum, 2,109 ± 628 CFU) or the ΔfbaAΔpam (▲; inoculum, 1,258 ± 160 CFU) and Δmga (▼; inoculum 1,255 ± 422 CFU) mutants. Paired t test (two-tailed) values for differences in bacterial population doubling are shown as follows: *, t ≤ 0.05; and **, t < 0.01. Differences in inoculum doses were not statistically significant between graft inoculum pairs (paired t test) (data not shown).
TABLE 4.
Relative growth indices of Alab49 and Mga regulon mutants under different experimental conditions
| Test organism | Comparator organism | Relative growth index |
||
|---|---|---|---|---|
| Human skin graft on SCID mouse (6 days)a | Rotation in whole human blood (3 h) | Rotation in human plasma (3 h) | ||
| ΔfbaA | WT | 0.394 | 0.464 | 1.146 |
| Δpam | WT | NDb | 0.215 | 1.065 |
| ΔfbaAΔpam | WT | 0.462 | 0.107 | 0.929 |
| Δmga | WT | 0.005 | 0.028 | 1.068 |
| ΔfbaAΔpam | 0.012 | 0.123 | NDb | |
The correlation coefficient between bacterial growth indices for infected skin grafts and those for rotation in whole blood (excluding the Δpam construct) is nonsignificant (Pearson r = 0.5390).
ND, not determined.
Previous studies showed that loss of either pam (12) or mga (9) results in a significant loss of virulence at the skin for GAS strain Alab49. Mga is a positive regulator of pam and fbaA transcription in Alab49 (Table 3) (9). The plasminogen-binding M protein (PAM) is disproportionately associated with the emm pattern D skin specialist strains (4). In this study, the Alab49Δmga mutant was directly compared to an Alab49ΔfbaAΔpam double mutant for virulence in the hu-skin-SCID mouse model of impetigo. The data show a mean average of only 3.433 population doublings for the Δmga mutant, compared to 10.558 population doublings for the ΔfbaAΔpam double mutant; the difference in net reproductive growth at the skin between these two genetic constructs is highly significant (Fig. 2) (t = 0.0014; two-tailed paired t test). Mean average inoculum doses were slightly lower (∼1,250 CFU) than those used for the group of mice infected with both Alab49WT and the ΔfbaA mutant.
For experiments in which Alab49 constructs were used to infect different sets of mice, each graft could be treated as an independent experiment, and the unpaired t test was used for statistical analysis. The data show that the differences in CFU between Alab49Δmga and either Alab49WT or Alab49ΔfbaA at 6 days postinoculation were highly significant (Fig. 2) (t = 0.0010 and 0.0019, respectively; two-tailed unpaired t test with Welch's correction for unequal variance). In contrast, population doubling at the skin for the Alab49ΔfbaAΔpam construct was not significantly different from that for either the ΔfbaA mutant or Alab49WT. In fact, the increases in population size for Alab49ΔfbaA and Alab49ΔfbaAΔpam were virtually identical for the two groups of mice (10.998 and 10.558 population doublings, respectively). On applying the unpaired t test to Alab49WT versus the ΔfbaA mutant, the difference in population doubling was not statistically significant (t = 0.0819).
In summary, significant differences between the Alab49ΔfbaA single-gene mutant and the Alab49ΔfbaAΔpam double-gene mutant were not evident in terms of bacterial growth at the superficial layers of the skin at 6 days postinoculation. Furthermore, only modest (but statistically significant by the paired t test) differences were detected between Alab49WT and Alab49ΔfbaA. The findings point to a critical role for additional, unidentified Mga-regulated genes in fitness and infectivity at the skin.
Roles of fbaA and pam in bacterial survival and growth in human blood.
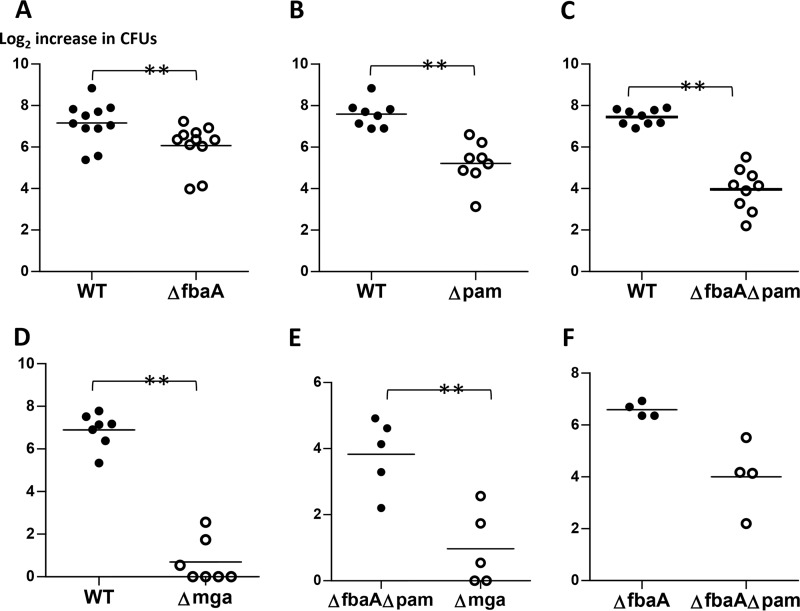
The hu-skin-SCID mouse model measures a complex series of host-pathogen interplays during infection, including GAS interactions with extravasated mouse neutrophils at the epidermis (11, 16, 17). The Lancefield bactericidal assay was used to measure the resistance of Alab49WT and its isogenic mutants to killing via opsonophagocytosis in whole human blood. Experiments comparing Alab49WT to the ΔfbaA isogenic mutant indicated that the contribution of FbaA to the antiphagocytic effect was highly significant (Fig. 3A), whereby the number of population doublings averaged 6.073 and 7.156 for Alab49ΔfbaA and the WT, respectively (t < 0.01; two-tailed paired t test). Over the 3-h period, the growth index of the Alab49ΔfbaA mutant was 46.4% of the value for the WT (Table 4), indicating that even without FbaA, Alab49 can undergo substantial reproductive growth in whole blood, albeit with a reduction in relative fitness.
FIG 3.
Bacterial population doubling in whole blood. Pairs of Alab49WT and/or isogenic mutant strains were rotated in whole human blood for 3 h (bactericidal assay), and the increase in bacterial population size relative to the inoculum size was measured. The mean average value for the log2 increase in CFU is indicated for each sample group (bar); a net loss of CFU was given a score of zero. Paired t test (two-tailed) values for differences in the number of bacterial population doublings are shown as follows: **, t < 0.01. Differences in inoculum doses were not statistically significant between bacterial pairs (two-tailed paired t test) (data not shown); each set of comparisons (A to F) employed 2 to 4 different blood donors. The difference between groups in panel F (ΔfbaA versus ΔfbaAΔpam) is also highly significant (two-tailed paired t test); however, the sample size is smaller than the minimum (i.e., 5 pairs) that is recommended for implementing this statistical test.
Compared to FbaA, PAM (M53 protein) had an even more marked effect on resistance to opsonophagocytosis (Fig. 3B), whereby the Alab49Δpam construct exhibited a growth index that was 21.5% of the value for the WT over the 3-h assay (Table 4). Compared to the single-gene mutants, the double-gene mutant Alab49ΔfbaAΔpam was even more susceptible to opsonophagocytosis (Fig. 3C), with a growth index that was 10.7% of that of Alab49WT (Table 4). The hypothesis that PAM and FbaA most likely have additive effects is supported by their relative growth index values: the product of 0.464 (for the ΔfbaA mutant relative to the WT) and 0.215 (for the Δpam mutant relative to the WT) equates to 0.100 (predicted), close to the actual value of 0.107 (Table 4).
The mean average number of population doublings in whole human blood was 5.222 for the Alab49Δpam construct and 3.958 for the Alab49ΔfbaAΔpam construct (Fig. 3B and C). The population doubling values equate to net population doubling times of 29.6, 34.5, and 45.5 min for Alab49ΔfbaA, Alab49Δpam, and Alab49ΔfbaAΔpam, respectively, over the 3 h of incubation (Fig. 3A to C); in contrast, the population doubling times of the comparator strain Alab49WT were shorter, ranging from 23.7 to 25.1 min. As an experimental control to rule out intrinsic growth defects, Alab49WT and each isogenic mutant were rotated in human plasma for 3 h; the data show that all constructs displayed nearly equivalent growth (Table S3). In summary, the antiphagocytic effects of FbaA and PAM contribute to the fitness of impetigo strain Alab49 during rotation in whole human blood.
Bactericidal assays testing the Alab49Δmga mutant showed a very large loss in the mean average number of population doublings (Fig. 3D). The substantially lower growth index value for the Δmga construct than that for the Alab49ΔfbaAΔpam double mutant (Fig. 3E; Table 4) indicates that other Mga-regulated genes, in addition to the FbaA and PAM genes, make a significant contribution to the antiphagocytic phenotype of Alab49. Transcriptomics reveals that Mga can either activate or repress the transcription of >10% of genes in a given GAS strain (18). Relative to Alab49WT, Alab49Δmga displayed decreased transcription of several genes mapping to the emm region (mrp, pam, enn, scpA, and fbaA), as demonstrated by quantitative reverse transcription-PCR (qRT-PCR) (Table 3; Table S4). Transcomplemention of the Δmga mutant with a plasmid harboring mga plus its upstream promoter region (Δmga::pMga667) led to an approximate restoration of WT levels of RNA transcripts for the surface protein genes tested (Table S4).
In summary, loss of FbaA reduced Alab49 survival in whole blood ∼2-fold, whereas loss of PAM reduced Alab49 survival ∼5-fold, based on relative growth indices. The concurrent loss of FbaA and PAM reduced Alab49 survival ∼10-fold (Fig. 3; Table 4). Loss of Mga reduced Alab49 survival ∼35-fold, which is indicative of a role for additional, unidentified Mga-regulated genes conferring a net effect of resistance to opsonophagocytosis. These findings roughly follow the trend for population doubling on infected human skin grafts, which undergo extensive infiltration by mouse neutrophils; however, the correlation between relative growth indices for the two different experimental models, with four pairwise comparisons (Table 4), is not statistically significant.
Role of FCT region genes in bacterial growth in human blood.
Isogenic mutants with defects in pilus formation were previously shown to exhibit highly significant losses in growth and virulence in the hu-skin-SCID mouse model of impetigo compared to the growth and virulence of Alab49WT following inoculation of logarithmic-phase cultures (7, 8). The gene encoding the (purported) pilus tip adhesin Cpa is disproportionately associated with emm pattern D skin specialist strains compared to emm pattern A-C throat specialist strains (6). Thus, it was of interest to determine whether the loss in virulence during skin infection by FCT region mutants correlated with a diminished antiphagocytic effect in whole blood.
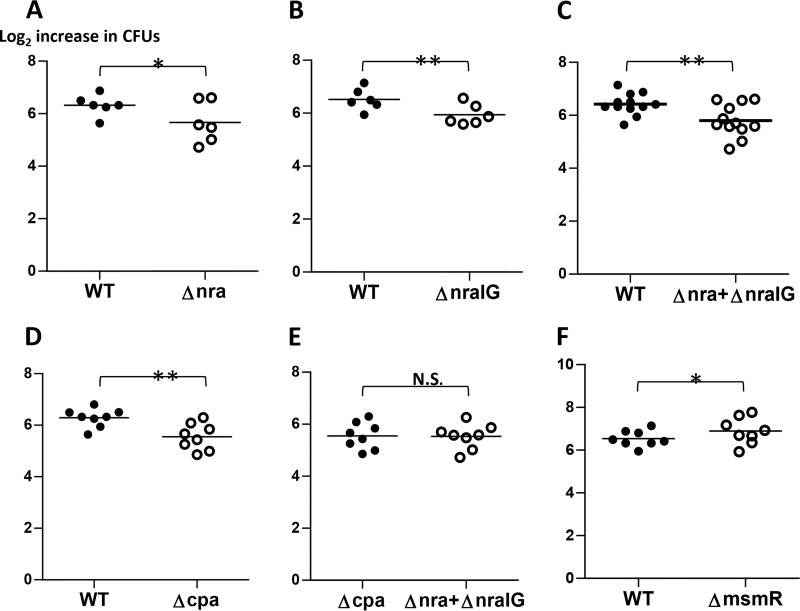
Deletion of the nra gene (Alab49Δnra), which encodes a transcriptional activator of pilus gene expression in strain Alab49, resulted in a statistically significant reduction of bacterial growth in human blood (Fig. 4A; Table 5). The loss in bacterial survival was of a similar magnitude for the Alab49ΔnraIG isogenic mutant (Fig. 4B and C), which lacks both nra and the nra-cpa intergenic region (10), which contains promoter sites for transcription of both genes (Fig. S3). Deletion of the gene encoding the (putative) pilus adhesin protein Cpa (Alab49Δcpa mutant) also resulted in a highly significant decrease in survival in the bactericidal assay (Fig. 4D). Overall, both the Nra and Cpa effects were somewhat modest, with growth indices that were ∼65% that of Alab49WT (Table 5). Thus, mutagenesis of either nra or cpa leaves most of the bacterium's antiphagocytic properties intact. Direct comparison of the Alab49Δnra and Alab49ΔnraIG mutants to the Alab49Δcpa mutant (Fig. 4E) revealed no apparent difference in bacterial growth, providing strong support that Cpa alone accounts for (nearly) all of the antiphagocytic effect conferred by Nra-regulated genes (8). The observed reduction in the antiphagocytic effect (Fig. 4A to D) may provide a (partial) explanation for the reduced virulence of Alab49Δnra and Alab49Δcpa mutants in the humanized mouse model of impetigo (7, 8).
FIG 4.
Bacterial population doubling in whole blood for FCT region mutants. Pairs of Alab49WT and/or isogenic mutant strains were rotated in whole human blood for 3 h (bactericidal assay), and the increase in bacterial population size relative to the inoculum size was measured. The mean average value for the log2 increase in CFU is indicated for each sample group (bar). Paired t test (two-tailed) values for differences in the number of bacterial population doublings are shown as follows: *, t < 0.05; **, t < 0.01; and N.S., not significant. Differences in inoculum doses were not statistically significant between bacterial pairs (two-tailed paired t test) (data not shown); each set of comparisons (A to F) employed 2 to 4 different blood donors.
TABLE 5.
Relative growth indices of Alab49 and FCT region mutants in the bactericidal assay
| Test organism | Comparator organism | Relative growth index |
|
|---|---|---|---|
| Rotation in whole human blood (3 h) | Rotation in human plasma (3 h)a | ||
| Δnra | WT | 0.698 | ND |
| ΔnraIG | WT | 0.668 | 0.898 |
| Δnra + ΔnraIG combined | WT | 0.682 | ND |
| Δcpa | WT | 0.620 | 1.013 |
| ΔmsmR | WT | 1.341 | 0.906 |
| Δnra + ΔnraIG combined | Δcpa | 0.976 | ND |
See Table S5 in the supplemental material. ND, not determined.
MsmR is a second transcriptional regulator encoded within the FCT region of Alab49. In this GAS strain, MsmR acts as both an activator of the (putative) fibronectin (Fn)-binding protein gene prtF2 and a repressor of nra (and cpa) transcription (8, 10), and it possibly affects the transcription of numerous other genes as well (19). Compared directly to Alab49WT, the isogenic Alab49ΔmsmR mutant exhibited a statistically significant increase in population doubling (Fig. 4F; Table 5), consistent with the idea that increased pilus expression by Alab49ΔmsmR may lead to a higher survival rate in whole blood. Overall, the effect of cpa loss on the antiphagocytic effect (∼1.6-fold reduction in growth index) (Table 5) was slightly less than that observed for loss of fbaA (∼2.2-fold reduction) (Table 4).
DISCUSSION
The classical skin strains of GAS differ from classical throat strains in both their epidemiological associations and content of accessory genes (reviewed in reference 3). The presence of fbaA among all cluster I impetigo isolates tested and its absence from 80% of cluster I pharyngitis isolates make FbaA a strong candidate for conferring tissue specificity of infection at the skin for many cluster I organisms. The cluster I organisms are defined by their sharing of numerous other accessory genes (13), and therefore the differential distribution of fbaA within cluster I may contribute to the throat versus skin preference phenotypes. The humanized mouse model of impetigo provides support for the hypothesis that FbaA enhances GAS fitness at the skin, although the magnitude of the effect is rather modest. Nonetheless, even a small fitness advantage can contribute to the shaping of larger epidemiological trends.
The humanized mouse model captures numerous pathogen-host interactions (reviewed in reference 11), including bacterial colonization and reproductive growth, the latter of which may be constrained by the bacterium's ability to acquire nutrients and resist innate host defenses. The small but significant contribution of FbaA to GAS survival and growth in whole human blood may reflect an ability to resist opsonophagocytosis by extravasated mouse polymorphonuclear leukocytes during colonization of the superficial epidermal layer in the hu-skin-SCID mouse model (17). However, the antiphagocytic effect attributed to FbaA in Alab49 pales in comparison to that of the M53 protein (i.e., PAM), which in turn is overshadowed by the collective effect of Mga-regulated gene products.
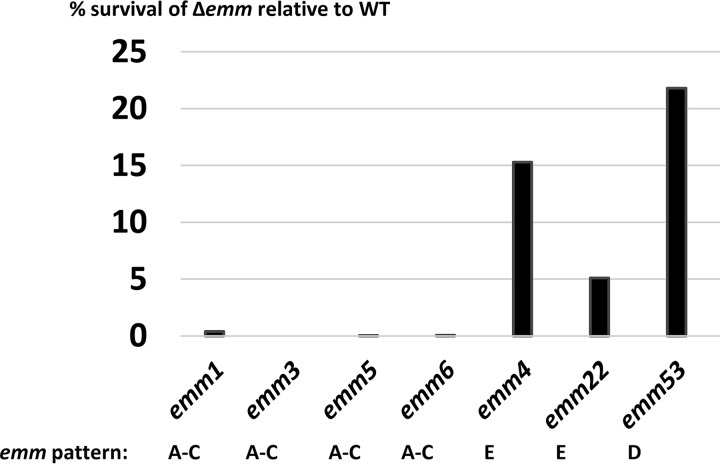
Most studies measuring the contribution of M protein to blocking of phagocytosis have employed emm pattern A-C and E strains, the so-called throat specialists and generalists, respectively. With similarly designed bactericidal assays and calculations, the % GAS survival in whole blood following emm gene deletion relative to that of the WT was <1% for pattern A-C strains of four different emm types (M1, M3, M5, and M6) (20–22) (Fig. 5). For emm pattern E strains of M types 4 and 22, the survival of Δemm mutants relative to that of the WT was 15.4 and 5.1%, respectively (23, 24). All values are lower than the ∼22% survival for Alab49Δpam (i.e., Alab49Δemm53) observed in the present study (Fig. 5). Using slightly different calculations (i.e., using median values in place of means), the survival of Δemm mutants relative to that of the WT is 2.0 and 17.7%, respectively, for pattern E strains of M types 2 and 49, respectively, and 18.2% for an emm95 pattern D strain (previously known as 64/14 and renamed SS1343) (25). Considering all reported values combined, the survival rates for Δemm mutants are 0.13% ± 0.2%, 10% ± 7.6%, and 20% ± 2.5% for emm pattern groups A-C (throat specialists), E (generalists), and D (skin specialists), respectively. This trend may be relevant to epidemiological findings showing that, of the three emm pattern groups, pattern A-C strain emm types (followed by pattern E types) are most often associated with invasive disease in each of six distinct global regions examined (26). The bactericidal assay with whole human blood may be a highly relevant experimental model for invasive GAS disease (e.g., bacteremia).
FIG 5.
Meta-analysis of GAS survival in whole blood following emm deletion. The mean average % survival relative to that of the wild type is shown for several Δemm mutants of different M types, based on the findings of several other investigators (20–24). Each study used whole human blood and logarithmic-phase GAS rotated for 3 h at 37°C, although inoculum doses may have differed slightly; the % survival calculations were made in almost exactly the same way as that described for the Alab49 (emm53) strain from this report.
Pandiripally et al. (27) reported the effects of M1 protein (and FbaA) on opsonophagocytosis of GAS in the bactericidal assay; however, precise % survival values were not reported, so direct comparisons to the Alab49 data presented in Fig. 5 were not attempted. By extrapolation of values from bar graphs, survival of the Δemm1 mutant appears to be (very) roughly 10% (27).
Studies of an FbaA variant derived from an M protein type 1 (M1) strain showed that FbaA1 is a multifunctional cell surface protein that binds fibronectin (Fn) and the complement regulators factor H (FH) and factor H-like 1 (FHL-1) (28, 29). FHL-1 allows GAS to resist opsonophagocytosis (27) and also facilitates epithelial cell invasion (28) in an FbaA1-dependent manner. The overall amino acid sequence identity between FbaA1 (MGAS5005) and FbaA53 (Alab49) is 88% following alignment using MUSCLE (data not shown). FbaA from an invasive M type 53 GAS strain (AP53) displays 100% amino acid sequence identity with FbaA53 from Alab49, and an AP53ΔfbaA mutant exhibits reduced binding of FH (30). Although FH and FHL-1 binding to FbaA from Alab49 (FbaA53) was not measured in this study, two regions of FH and FHL-1 binding have been mapped for FbaA1 and can be compared as follows: the 16-amino-acid sequence of region I has 87.5% identity to FbaA53, and the 18-amino-acid sequence of region II is 100% identical to that of FbaA53 (28; data not shown). FbaA53 also has a highly homologous COOH-terminal proline-rich repeat domain responsible for Fn binding in its FbaA1 counterpart (31). Thus, it is expected, though not directly proven, that FbaA53 from Alab49 also binds Fn and complement regulators and may function in a manner similar to that of FbaA1. M1 strains are distinguished from many classic throat strains in that they harbor an fbaA gene. Conceivably, the propensity of M1 GAS to cause skin and soft tissue infections in temperate regions (26, 32) partially overlaps the virulence strategy used by classic impetigo strains in causing superficial skin infections.
Both the M53 protein (i.e., PAM) and FbaA are positively regulated by Mga and contribute to the virulence of strain Alab49 at the skin and to its survival in whole blood. However, loss of Mga leads to a decrease in virulence, to well below the level of the Alab49ΔfbaAΔpam double mutant. Additional Mga-regulated genes in the Alab49 strain include those that encode the surface proteins SclA (collagen-like) (9) and ScpA (C5a peptidase) and the M-like proteins Mrp and Enn. After 3 h of rotation in whole human blood, the number of surviving Alab49Δmga organisms was 8-fold lower than that for the Alab49ΔfbaAΔpam double mutant, whereas 6 days after inoculation of the human skin graft, the net growth of the Alab49ΔfbaAΔpam mutant exceeded that of the Δmga mutant by almost 100-fold. Thus, another, unspecified Mga-regulated gene product(s) makes a substantial contribution to the fitness of the Alab49 strain under pathological conditions. SclA, Mrp, and Enn are structurally diverse, and Mrp and Enn are absent from most classical throat strains. Whether the functional features of these surface proteins coordinate with the biological activities of PAM and FbaA to yield skin tissue specificity of infection remains an open possibility, although any of the Mga-regulated genes—including many that remain unidentified in this study—may have a contributory effect.
Other studies have reported that the GAS pilus alters the susceptibility of GAS to phagocytosis in whole human blood by either increased bacterial killing (M1T1 strain; FCT-2, emm pattern A-C) or decreased bacterial killing (M2T2 strain; FCT-6, emm pattern E) (33, 34). The emm pattern D strain Alab49 expresses the FCT-3 form of pili (T serotype T3/13/B) (7). In the present study, loss of pili in Alab49 due to mutation of the transcriptional activator Nra gene or the pilus tip adhesion Cpa gene yielded nearly equivalent levels of decreased survival in whole human blood; the decrease in survival relative to that of Alab49WT was slight but significant. The slight but significant increase in survival following loss of MsmR parallels increased pilus production, although other MsmR-regulated genes may contribute to the effect.
The accessory gene content of classic skin strains (i.e., emm pattern D) is similar to that of a highly prevalent subset of classic throat strains (i.e., cluster I, emm pattern A-C), wherein the differential presence/absence of fbaA is tightly coupled to the two distinct clinical phenotypes. Thus, gain/loss of fbaA may facilitate a transition in tissue site preference for infection. Consistent with this hypothesis is the slight loss of fitness at the skin for the Alab49ΔfbaA mutant. Thus, FbaA and other emm pattern D-specific gene products, such as PAM, and possibly Cpa (7) and/or the streptokinase lineage 2b form (5, 12), may each contribute incrementally to shaping the phenotype for preferred infection at the skin. The absence of any one of the impetigo-associated signature genes may lead to a lower fitness that reduces transmissibility ever so slightly and, over time, that variant genotype may inch toward extinction from the global GAS population.
MATERIALS AND METHODS
Bacterial culture.
GAS was cultivated in Todd-Hewitt broth supplemented with 1% yeast extract (THYB). Mid-logarithmic-phase GAS cultures were grown to the optical density at 600 nm (OD600) specified for each assay; stationary-phase cultures were cultures grown for 16 h. Escherichia coli used for cloning and recombinant polypeptide expression was grown at 37°C in Luria broth (LB) with shaking. For selection of GAS transformants harboring the antibiotic resistance genes aphA3 and aad9, kanamycin was used at 500 μg/ml and spectinomycin was used at 200 μg/ml, respectively. GAS harboring pDCerm was grown in the presence of 5 or 10 μg/ml of erythromycin.
PCR-based screening.
PCR products were generated using the primers listed in Table S1 in the supplemental material. The genomic (gDNA) and plasmid DNAs used as templates were prepared as previously described (7). Bacterial strains that underwent PCR-based screening included many from previous reports (13, 35–37).
Bacterial mutants.
Several targeted gene replacements were made in impetigo strain Alab49 (M type 53). The fbaA gene was inactivated in both Alab49WT and an Alab49Δpam mutant (i.e., Δemm53) (12) to generate the Alab49ΔfbaA single-gene mutant and the Alab49ΔfbaAΔpam double-gene mutant, respectively (Fig. S1). Mutagenesis was achieved by allelic exchange following transformation of bacteria with purified linear DNA containing the spectinomycin resistance gene (aad9) flanked by sequences upstream and downstream of the target gene, as previously described (7), using the oligonucleotide primers listed in Table S1. The linear DNA cassette was generated using CloneAmp high-fidelity polymerase (Clontech, Mountain View, CA), and electroporation (2.5 kV, 200 Ω, 25 μF) (Micropulser instrument; Bio-Rad, Hercules, CA) of glycine-grown competent GAS cells in 10% glycerol yielded a time constant of ∼5 to 6 ms, which is critical for yielding transformants in strain Alab49 by this method. Cells underwent selection for antibiotic resistance, and the transformants were evaluated for replacement of the target fbaA gene by PCR-based mapping and nucleotide sequence determination; growth curves for the Alab49-derived mutants were identical (Fig. S2). Additional mutants used in this study, i.e., Alab49Δmga, Alab49Δnra, Alab49Δcpa, and Alab49ΔmsmR, were constructed and characterized as previously described (7–10) (Fig. S1 and S3). Alab49Δmga::pMga667 is a transcomplemented strain with pDCerm containing mga plus the 667-bp upstream autoregulatory region, as previously described (9); Alab49Δmga::pEmpty is a transcomplemented strain with pDCerm lacking an insert. The Alab49ΔnraIG mutant, lacking the nra gene plus the entire nra-cpa intergenic region (Fig. S3), was constructed by allelic exchange with a linear DNA cassette containing the aphA3 gene (conferring kanamycin resistance) and generated according to previously described methods (8), using the primers listed in Table S1. Controls for caseinolytic digestion of mutant constructs are presented in Table S2.
Immunofluorescence assay of bacterial cells.
GAS cells were grown in THYB, with or without the cysteine proteinase inhibitor E64 (25 μM; Sigma-Aldrich, St. Louis, MO), for 24 h at 30°C. The 24-h cultures were subsequently diluted 100-fold in fresh THYB, in the presence or absence of 25 μM E64, and grown at 30°C to either the mid-logarithmic or stationary phase, and cells were harvested by centrifugation and washed in phosphate-buffered saline (PBS). To block nonspecific binding of immunoglobulins to Alab49 via cell surface IgG-binding proteins, GAS cells were suspended in a 1/10 volume of blocking solution (PBS, pH 7.4, with 50 μg/ml purified human IgG Fc fragment and 10% goat serum) and incubated at room temperature for 1 h. Cell suspensions were subsequently split into equal volumes and centrifuged, and bacterial pellets were suspended in preimmune or hyperimmune rabbit serum at a 1:1,000 dilution; these mixtures were incubated at room temperature for 1 h and then washed with ice-cold PBS. Next, the bacterial cells were stained with secondary antibody by incubation for 1 h with Alexa Fluor 488-labeled goat anti-rabbit IgG F(ab′)2 (Life Technologies, Carlsbad, CA) at 1:250 in PBS and then were washed thoroughly. Wet mounts were viewed on a Zeiss Axiovert 200 inverted motorized microscope with a 100× oil immersion objective, and images were recorded using a Zeiss Axiocam MRm camera; ≥3 fields per sample were visualized and recorded.
Hyperimmune rabbit serum.
A recombinant polypeptide corresponding to the predicted mature form of FbaA (rFbaA) of Alab49 was used to generate FbaA-specific antiserum. The fbaA gene of Alab49, excluding the portions encoding the predicted signal peptide and the LPXTG motif and cell anchoring domain, was amplified by PCR from gDNA by use of LongAmp Taq polymerase and the oligonucleotide primers listed in Table S1. The fbaA-derived PCR product was cloned into the E. coli plasmid expression vector pGEX-3X (GE Healthcare Life Sciences, Pittsburgh, PA), and the recombinant plasmid was used to transform Star BL21(DE3) competent E. coli cells. rFbaA was expressed as a fusion with an NH2-terminal glutathione S-transferase (GST) tag, and rFbaA was affinity purified using a GST SpinTrap column. FbaA-specific antiserum was generated by intradermal injection of a rabbit (New Zealand White) with 400 μg of purified rFbaA emulsified in TiterMax Gold adjuvant (Sigma-Aldrich, St. Louis, MO). Three booster immunizations, each consisting of 400 μg of rFbaA in TiterMax Gold, were administered at 4-week intervals; serum was collected 7 to 10 days after each immunization. All animal experimentation was approved by the New York Medical College Institutional Animal Care and Use Committee.
RNA transcript levels.
Total cellular RNAs were extracted from mid-logarithmic- and stationary-phase GAS cultures; cDNA was prepared from 1 μg of purified RNA as previously described (8). For qRT-PCR, oligonucleotide primers (Table S1) were designed to amplify a sequence of ∼100 bp at the 5′ end of the target gene; samples in each qRT-PCR run were tested in triplicate. Expression of the target gene was normalized to that of the housekeeping gene recA or to both recA and gyr, and relative RNA transcript levels were measured using the 2−(ΔΔCT) method (8). Each gene was analyzed in ≥2 independent experiments, and the mean average fold change was calculated.
Casein digestion assay.
For characterization of mutants, SpeB protease activity was measured on Columbia agar plates (0.5× Columbia agar [Sigma-Aldrich, St. Louis, MO], 1.5% Bacto agar, 3% skim milk powder). Columbia agar was inoculated via stabs with single GAS colonies and incubated for 48 h at 37°C; the diameter of the zone of clearance (i.e., casein digestion) around the inoculation site was measured in millimeters. The cysteine protease inhibitor E64 (25 μM in agar) and Alab49ΔspeB (7, 16) were included as controls.
Impetigo model.
A SCID mouse model engrafted with human skin (i.e., hu-skin-SCID mice) was used in an experimental model of GAS impetigo (11, 17). For inoculation, a 0.002× dilution of a mid-logarithmic-phase (OD600, ∼0.350) GAS culture grown at 37°C in 50 μl of THYB was spotted onto bandage pads and delivered to freshly scratched (nonbleeding) human skin grafts on the backs of C.B-17-scid mice (Taconic Biosciences, Inc., Germantown, NY). In paired experiments, two skin grafts present on a single mouse each received a bacterial inoculum at the same time; in unpaired experiments, each skin graft was treated as an individual experiment. The number of CFU in each GAS inoculum was calculated via serial dilutions on blood agar plates, performed in duplicate. Six days after inoculation with GAS, human skin grafts were surgically removed, weighed, placed in sterile saline, and vigorously vortexed to release the bacteria; the number of CFU associated with each biopsied skin graft was calculated following (duplicate) serial dilutions of the released organisms. The number of bacterial population doublings was calculated as the log2 of the ratio of the average mean CFU in skin biopsy specimens to the average mean CFU in the inoculum. The relative bacterial growth index at the skin was defined as the ratio of the average mean CFU at 6 days postinoculation/inoculum CFU for the test strain to the average mean CFU at 6 days postinoculation/inoculum CFU for the comparator strain.
Phagocytosis (bactericidal) assay.
Growth of GAS in human whole blood was measured using a modified version of the Lancefield bactericidal assay (12, 38). Heparinized human blood was kept on ice and used for bactericidal assays within 2 h of collection; blood was collected in accordance with a protocol approved by the New York Medical College Institutional Review Board for Human Subjects. For the bactericidal assay, 400 μl of heparinized blood was mixed with GAS (in 100 μl THYB) and incubated at 35°C for 3 h with rotation. GAS inoculums were based on dilutions of 1:8 and 1:2 of a 10,000-fold dilution of early-logarithmic-phase cultures (OD600, ∼0.150); actual inoculums (as determined by serial dilution on blood agar plates, in duplicate, and CFU counts) ranged from ∼50 to 200 CFU. Alab49WT and its isogenic mutants were each tested with ≥2 different blood donors. GAS constructs were also tested for the ability to grow in human plasma to rule out possible bacterial growth defects; plasma was collected from the heparinized human blood samples by centrifugation. The number of bacterial population doublings was calculated as the log2 of the ratio of the average mean CFU after 3 h of incubation in whole blood or plasma to the average mean CFU in the inoculum. The relative bacterial growth index was defined as the ratio of the average mean CFU after 3 h of incubation in whole blood (or plasma)/inoculum CFU for the test strain to the average mean CFU after 3 h of incubation in whole blood (or plasma)/inoculum CFU for the comparator strain. The paired t test was performed for pairs of organisms incubated in identical whole-blood or plasma samples.
Statistical analysis.
Statistical significance was measured using GraphPad for paired or unpaired Student's t test (two-tailed) and Fisher's exact test (two-tailed). Correlation analysis was performed by calculating Pearson's correlation coefficient.
Supplementary Material
ACKNOWLEDGMENTS
We are especially grateful to Yoann Le Breton and Kevin McIver for helpful advice on transformation protocols. We also thank the numerous investigators who provided many of the GAS strains used in this study, which included isolates originating from Robert Tanz and Stanford Shulman (Ann & Robert H. Lurie Children's Hospital of Chicago), the Menzies School of Health Research (Darwin, Australia), and the CDC (Atlanta, GA).
This work was supported by a bridge award from NYMC-Touro and by NIH grants AI-053826 and AI-065572.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00374-17.
REFERENCES
- 1.Carapetis JR. 2007. Rheumatic heart disease in developing countries. N Engl J Med 357:439–441. doi: 10.1056/NEJMp078039. [DOI] [PubMed] [Google Scholar]
- 2.Carapetis JR, Steer AC, Mulholland EK, Weber M. 2005. The global burden of group A streptococcal diseases. Lancet Infect Dis 5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 3.Bessen DE, McShan WM, Nguyen SV, Shetty A, Agrawal S, Tettelin H. 2015. Molecular epidemiology and genomics of group A streptococcus. Infect Genet Evol 33:393–418. doi: 10.1016/j.meegid.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Svensson MD, Sjöbring U, Bessen DE. 1999. Selective distribution of a high-affinity plasminogen binding site among group A streptococci associated with impetigo. Infect Immun 67:3915–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalia A, Bessen DE. 2004. Natural selection and evolution of streptococcal virulence genes involved in tissue-specific adaptations. J Bacteriol 186:110–121. doi: 10.1128/JB.186.1.110-121.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kratovac Z, Manoharan A, Luo F, Lizano S, Bessen DE. 2007. Population genetics and linkage analysis of loci within the FCT region of Streptococcus pyogenes. J Bacteriol 189:1299–1310. doi: 10.1128/JB.01301-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lizano S, Luo F, Bessen DE. 2007. Role of streptococcal T antigens in superficial skin infection. J Bacteriol 189:1426–1434. doi: 10.1128/JB.01179-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo F, Lizano S, Bessen DE. 2008. Heterogeneity in the polarity of Nra regulatory effects on streptococcal pilus gene transcription and virulence. Infect Immun 76:2490–2497. doi: 10.1128/IAI.01567-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo F, Lizano S, Banik S, Zhang H, Bessen DE. 2008. Role of Mga in group A streptococcal infection at the skin epithelium. Microb Pathog 45:217–224. doi: 10.1016/j.micpath.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lizano S, Luo F, Tengra FK, Bessen DE. 2008. Impact of orthologous gene replacement on the circuitry governing pilus gene transcription in streptococci. PLoS One 3:e3450. doi: 10.1371/journal.pone.0003450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bessen DE, Lizano S. 2010. Tissue tropisms in group A streptococcal infections. Future Microbiol 5:623–638. doi: 10.2217/fmb.10.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Svensson MD, Sjobring U, Luo F, Bessen DE. 2002. Roles of the plasminogen activator streptokinase and the plasminogen-associated M protein in an experimental model for streptococcal impetigo. Microbiology 148:3933–3945. doi: 10.1099/00221287-148-12-3933. [DOI] [PubMed] [Google Scholar]
- 13.Bessen DE, Kumar N, Hall GS, Riley DR, Luo F, Lizano S, Ford CN, McShan WM, Nguyen SV, Dunning Hotopp JC, Tettelin H. 2011. Whole-genome association study on tissue tropism phenotypes in group A streptococcus. J Bacteriol 193:6651–6663. doi: 10.1128/JB.05263-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanderson-Smith M, De Oliveira DM, Guglielmini J, McMillan DJ, Vu T, Holien JK, Henningham A, Steer AC, Bessen DE, Dale JB, Curtis N, Beall BW, Walker MJ, Parker MW, Carapetis JR, Van Melderen L, Sriprakash KS, Smeesters PR, M Protein Study Group. 2014. A systematic and functional classification of Streptococcus pyogenes that serves as a new tool for molecular typing and vaccine development. J Infect Dis 210:1325–1338. doi: 10.1093/infdis/jiu260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shulman ST, Stollerman G, Beall B, Dale JB, Tanz RR. 2006. Temporal changes in streptococcal M protein types and the near-disappearance of acute rheumatic fever in the United States. Clin Infect Dis 42:441–447. doi: 10.1086/499812. [DOI] [PubMed] [Google Scholar]
- 16.Svensson MD, Scaramuzzino DA, Sjobring U, Olsen A, Frank C, Bessen DE. 2000. Role for a secreted cysteine proteinase in the establishment of host tissue tropism by group A streptococci. Mol Microbiol 38:242–253. doi: 10.1046/j.1365-2958.2000.02144.x. [DOI] [PubMed] [Google Scholar]
- 17.Scaramuzzino DA, McNiff JM, Bessen DE. 2000. Humanized in vivo model for streptococcal impetigo. Infect Immun 68:2880–2887. doi: 10.1128/IAI.68.5.2880-2887.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hondorp ER, McIver KS. 2007. The Mga virulence regulon: infection where the grass is greener. Mol Microbiol 66:1056–1065. doi: 10.1111/j.1365-2958.2007.06006.x. [DOI] [PubMed] [Google Scholar]
- 19.Nakata M, Podbielski A, Kreikemeyer B. 2005. MsmR, a specific positive regulator of the Streptococcus pyogenes FCT pathogenicity region and cytolysin-mediated translocation system genes. Mol Microbiol 57:786–803. doi: 10.1111/j.1365-2958.2005.04730.x. [DOI] [PubMed] [Google Scholar]
- 20.Gustafsson MC, Lannergard J, Nilsson OR, Kristensen BM, Olsen JE, Harris CL, Ufret-Vincenty RL, Stalhammar-Carlemalm M, Lindahl G. 2013. Factor H binds to the hypervariable region of many Streptococcus pyogenes M proteins but does not promote phagocytosis resistance or acute virulence. PLoS Pathog 9:e1003323. doi: 10.1371/journal.ppat.1003323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Casal J, Caparon MG, Scott JR. 1992. Introduction of the emm6 gene into an emm-deleted strain of Streptococcus pyogenes restores its ability to resist phagocytosis. Res Microbiol 143:549–558. doi: 10.1016/0923-2508(92)90112-2. [DOI] [PubMed] [Google Scholar]
- 22.Kotarsky H, Thern A, Lindahl G, Sjobring U. 2000. Strain-specific restriction of the antiphagocytic property of group A streptococcal M proteins. Infect Immun 68:107–112. doi: 10.1128/IAI.68.1.107-112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlsson F, Berggard K, Stalhammar-Carlemalm M, Lindahl G. 2003. Evasion of phagocytosis through cooperation between two ligand-binding regions in Streptococcus pyogenes M protein. J Exp Med 198:1057–1068. doi: 10.1084/jem.20030543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Courtney HS, Hasty DL, Dale JB. 2006. Anti-phagocytic mechanisms of Streptococcus pyogenes: binding of fibrinogen to M-related protein. Mol Microbiol 59:936–947. doi: 10.1111/j.1365-2958.2005.04977.x. [DOI] [PubMed] [Google Scholar]
- 25.Podbielski A, Schnitzler N, Beyhs P, Boyle M. 1996. M-related protein (Mrp) contributes to group A streptococcal resistance to phagocytosis by human granulocytes. Mol Microbiol 19:429–441. doi: 10.1046/j.1365-2958.1996.377910.x. [DOI] [PubMed] [Google Scholar]
- 26.Steer AC, Law I, Matatolu L, Beall BW, Carapetis JR. 2009. Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect Dis 9:611–616. doi: 10.1016/S1473-3099(09)70178-1. [DOI] [PubMed] [Google Scholar]
- 27.Pandiripally V, Gregory E, Cue D. 2002. Acquisition of regulators of complement activation by Streptococcus pyogenes serotype M1. Infect Immun 70:6206–6214. doi: 10.1128/IAI.70.11.6206-6214.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pandiripally V, Wei L, Skerka C, Zipfel PF, Cue D. 2003. Recruitment of complement factor H-like protein 1 promotes intracellular invasion by group A streptococci. Infect Immun 71:7119–7128. doi: 10.1128/IAI.71.12.7119-7128.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terao Y, Kawabata S, Nakata M, Nakagawa I, Hamada S. 2002. Molecular characterization of a novel fibronectin-binding protein of Streptococcus pyogenes strains isolated from toxic shock-like syndrome patients. J Biol Chem 277:47428–47435. doi: 10.1074/jbc.M209133200. [DOI] [PubMed] [Google Scholar]
- 30.Agrahari G, Liang Z, Mayfield JA, Balsara RD, Ploplis VA, Castellino FJ. 2013. Complement-mediated opsonization of invasive group A Streptococcus pyogenes strain AP53 is regulated by the bacterial two-component cluster of virulence responder/sensor (CovRS) system. J Biol Chem 288:27494–27504. doi: 10.1074/jbc.M113.494864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terao Y, Okamoto S, Kataoka K, Hamada S, Kawabata S. 2005. Protective immunity against Streptococcus pyogenes challenge in mice after immunization with fibronectin-binding protein. J Infect Dis 192:2081–2091. doi: 10.1086/498162. [DOI] [PubMed] [Google Scholar]
- 32.O'Loughlin RE, Roberson A, Cieslak PR, Lynfield R, Gershman K, Craig A, Albanese BA, Farley MM, Barrett NL, Spina NL, Beall B, Harrison LH, Reingold A, Van Beneden C. 2007. The epidemiology of invasive group A streptococcal infection and potential vaccine implications: United States, 2000–2004. Clin Infect Dis 45:853–862. doi: 10.1086/521264. [DOI] [PubMed] [Google Scholar]
- 33.Tsai JC, Loh JM, Clow F, Lorenz N, Proft T. 2017. The group A Streptococcus serotype M2 pilus plays a role in host cell adhesion and immune evasion. Mol Microbiol 103:282–298. doi: 10.1111/mmi.13556. [DOI] [PubMed] [Google Scholar]
- 34.Crotty Alexander LE, Maisey HC, Timmer AM, Rooijakkers SH, Gallo RL, von Kockritz-Blickwede M, Nizet V. 2010. M1T1 group A streptococcal pili promote epithelial colonization but diminish systemic virulence through neutrophil extracellular entrapment. J Mol Med (Berl) 88:371–381. doi: 10.1007/s00109-009-0566-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGregor KF, Spratt BG, Kalia A, Bennett A, Bilek N, Beall B, Bessen DE. 2004. Multilocus sequence typing of Streptococcus pyogenes representing most known emm types and distinctions among subpopulation genetic structures. J Bacteriol 186:4285–4294. doi: 10.1128/JB.186.13.4285-4294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGregor KF, Bilek N, Bennett A, Kalia A, Beall B, Carapetis JR, Currie BJ, Sriprakash KS, Spratt BG, Bessen DE. 2004. Group A streptococci from a remote community have novel multilocus genotypes but share emm types and housekeeping alleles with isolates from worldwide sources. J Infect Dis 189:717–723. doi: 10.1086/381452. [DOI] [PubMed] [Google Scholar]
- 37.Bessen DE, Carapetis JR, Beall B, Katz R, Hibble M, Currie BJ, Collingridge T, Izzo MW, Scaramuzzino DA, Sriprakash KS. 2000. Contrasting molecular epidemiology of group A streptococci causing tropical and nontropical infections of the skin and throat. J Infect Dis 182:1109–1116. doi: 10.1086/315842. [DOI] [PubMed] [Google Scholar]
- 38.Lancefield RC. 1957. Differentiation of group A streptococci with a common R antigen into three serological types, with special reference to the bactericidal test. J Exp Med 106:525–544. doi: 10.1084/jem.106.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banks DJ, Porcella SF, Barbian KD, Beres SB, Philips LE, Voyich JM, DeLeo FR, Martin JM, Somerville GA, Musser JM. 2004. Progress toward characterization of the group A streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. J Infect Dis 190:727–738. doi: 10.1086/422697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.