ABSTRACT
Staphylococcus aureus is a human commensal but also has devastating potential as an opportunistic pathogen. S. aureus bacteremia is often associated with an adverse outcome. To identify potential targets for novel control approaches, we have identified S. aureus components that are required for growth in human blood. An ordered transposon mutant library was screened, and 9 genes involved specifically in hemolysis or growth on human blood agar were identified by comparing the mutants to the parental strain. Three genes (purA, purB, and pabA) were subsequently found to be required for pathogenesis in the zebrafish embryo infection model. The pabA growth defect was specific to the red blood cell component of human blood, showing no difference from the parental strain in growth in human serum, human plasma, or sheep or horse blood. PabA is required in the tetrahydrofolate (THF) biosynthesis pathway. The pabA growth defect was found to be due to a combination of loss of THF-dependent dTMP production by the ThyA enzyme and increased demand for pyrimidines in human blood. Our work highlights pabA and the pyrimidine salvage pathway as potential targets for novel therapeutics and suggests a previously undefined role for a human blood factor in the activity of sulfonamide antibiotics.
KEYWORDS: Staphylococcus aureus, blood, nucleotide
INTRODUCTION
The pathogenicity of the Gram-positive bacterium Staphylococcus aureus requires a multitude of virulence factors that are intricately coordinated and regulated (1, 2). In addition to the more “classic” virulence factors, such as pore-forming toxins and superantigens, fundamental metabolic processes of bacteria are also recognized as a prerequisite for disease. Indeed, the majority of antibiotics act by disrupting essential metabolic processes (3). However, pathogens, including S. aureus, have adapted so as to resist such insults by switching off, or severely reducing, the activity of aspects of metabolism in order to persist in the presence of antibiotics (4, 5).
Microbial fitness during pathogenesis requires efficient utilization of available nutrients. Although the mammalian host is nutrient rich, many nutrients are sequestered as a means of inhibiting pathogen growth, a concept referred to as “nutritional immunity” (6). Strategies for overcoming the nutrient-limited environment in vivo have been well described for S. aureus and other bacteria; they include the upregulation of peptide or amino acid transport mechanisms (7) and of proteins that enable the acquisition of nutrients sequestered by the host (8, 9). De novo biosynthetic pathways are also required to produce essential products not readily available in the environment. Nucleotide biosynthetic pathways have been identified as critical for the proliferation of Gram-positive pathogens on human blood (10), yet detailed studies of the growth requirements of S. aureus are lacking.
To support studies on S. aureus, the Nebraska transposon mutant library (NTML) was recently constructed in the community-acquired methicillin-resistant S. aureus (CA-MRSA) USA300 JE2 strain, deposited in the Network on Antimicrobial Resistance in S. aureus (NARSA) strain repository, and made freely available to registered users (11). This library was created using the mariner-based transposon (bursa aurealis) and employing the same methodology as that of Bae and colleagues (12). To date, the NTML has been used to carry out diverse screens to identify genes involved in S. aureus antibiotic persistence in vitro (13), altered hemolytic activity on rabbit blood agar (11, 14), polymicrobial interactions (15), and hyaluronidase activity (16).
A comprehensive approach to identify genes involved in the growth of S. aureus on human blood was undertaken using the NTML. The genes were then further characterized to analyze their potential roles in human infection. We show that purine biosynthesis is indispensable for growth on human blood and in vivo pathogenicity, using a zebrafish embryo model. In addition, a gene involved in tetrahydrofolate (THF) biosynthesis, pabA, was identified as being required for virulence in vivo and was unable to grow specifically on human blood. The relationship between human blood, a folate-poor environment, and S. aureus pyrimidine salvage pathways was further elucidated.
RESULTS
Screening of an S. aureus Tn library for growth defects on human blood.
The NTML was screened to define gene disruptions leading to alterations in growth and/or hemolysis on agar containing human blood as the only nutrient source (see Materials and Methods). The library was also screened on bovine serum agar and 5% (vol/vol) sheep blood with a Columbia agar base as comparators, in order to determine traits specific to human blood (data not shown). The transposon (Tn) insert for each strain identified in the screen was transduced back into the parent strain (S. aureus JE2), and transductants were rescreened in order to establish that the mutant phenotype was associated with each Tn insertion. Fifteen transductants maintained the altered phenotype, and nine of these (the purB, purA, pabA, atl, murQ, araC, mecA, odhB, and lipA transductants) were selected for further study (Table 1). The remaining six strains had transposon disruptions in genes expected to produce an altered phenotype when grown on human blood agar (agrA, agrB, agrC, hla, saeR, and saeS), confirming the ability of the screen to identify specific phenotypes.
TABLE 1.
Tn library mutants identified as having an altered phenotype on human blood agar
| Categorya | Protein ID | NARSA ID | Protein name | Growth phenotype in: |
Hemolysis phenotype in: |
||
|---|---|---|---|---|---|---|---|
| Human blood | Rabbit blood | 5% human blood + Columbia agar | 5% sheep blood + Columbia agar | ||||
| A1 | SAUSA300_1889 | NE522 | Adenylosuccinate lyase (PurB) | Reduced growth | Reduced growth | Increased hemolysis | Reduced hemolysis |
| SAUSA300_0017 | NE529 | Adenylosuccinate synthetase (PurA) | Reduced growth | Reduced growth | Increased hemolysis | Slightly increased hemolysis | |
| SAUSA300_0698 | NE821 | para-Aminobenzoate synthase, glutamine amidotransferase, component II (PabA) | Highly reduced growth | Slightly reduced growth | —b | — | |
| A2 | SAUSA300_0955 | NE460 | Autolysin (Atl) | Opaque colony | Opaque colony | Increased hemolysis | — |
| SAUSA300_0193 | NE1253 | N-Acetylmuramic acid-6-phosphate etherase (MurQ) | — | — | Increased hemolysis | Increased hemolysis | |
| SAUSA300_2326 | NE1304 | Transcription regulatory protein (AraC) | — | — | Reduced hemolysis | — | |
| SAUSA300_0899 | NE1315 | Adaptor protein (MecA) | — | — | Reduced hemolysis | Slightly reduced hemolysis | |
| SAUSA300_1305 | NE1391 | Dihydrolipoamide succinyltransferase (OdhB) | Slightly reduced growth | Slightly reduced growth | Increased hemolysis | Increased hemolysis | |
| SAUSA300_0320 | NE1775 | Triacylglycerol lipase (LipA) | Slightly reduced growth | — | Increased hemolysis | — | |
| B | SAUSA300_1989 | NE95 | Accessory gene regulator protein B (AgrB) | — | — | — | Reduced hemolysis |
| SAUSA300_1991 | NE873 | Accessory gene regulator protein C (AgrC) | — | — | Slightly reduced hemolysis | No hemolysis | |
| SAUSA300_0690 | NE1296 | Sensor histidine kinase (SaeS) | — | — | — | No hemolysis | |
| SAUSA300_1058 | NE1354 | Alpha-hemolysin (Hla) | — | — | — | No hemolysis | |
| SAUSA300_1992 | NE1532 | Accessory gene regulator protein A (AgrA) | — | — | Reduced hemolysis | No hemolysis | |
| SAUSA300_0691 | NE1622 | DNA-binding response regulator (SaeR) | — | — | — | No hemolysis | |
A1, strains with a defect in growth on human blood agar, which were investigated further; A2, strains with altered hemolysis on human blood agar, which were investigated further; B, strains expected to show a hemolysis phenotype, which were not explored further.
—, no difference from the control strain, JE2.
Phenotypic characterization of growth-defective mutants in vivo.
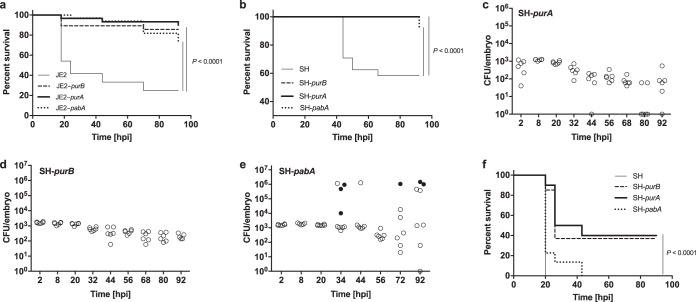
In order to define genes for further study, the pathogenicity of the nine transduced strains in the JE2 background was assessed using the zebrafish embryo model of systemic S. aureus infection (17). The atl, murQ, araC, mecA, odhB, and lipA transductants did not show altered killing in this model (see Fig. S1a and b in the supplemental material). However, three of the strains, harboring Tn inserts in the purA, purB, and pabA genes (referred to here as JE2-purA, JE2-purB, and JE2-pabA), showed significant attenuation in the zebrafish model (P < 0.0001) (Fig. 1a). To confirm that the reduced pathogenicity was not strain specific, Tn inserts containing the purA, purB, and pabA genes were transduced into another strain background, S. aureus SH1000. These strains (referred to here as SH-purA, SH-purB, and SH-pabA) also showed significant attenuation in the zebrafish embryo model (P < 0.0001) (Fig. 1b). In vivo growth analysis demonstrated that SH-purA and SH-purB were unable to replicate within zebrafish embryos, and the numbers of bacteria recovered were lower than the inoculated dose (Fig. 1c and d). This is in stark contrast to the bacterial kinetics observed when the parental S. aureus strain is injected at the same dose as that published previously (Fig. S1c) (17). SH-pabA retained limited capacity to replicate and to cause host death in the zebrafish model (Fig. 1e). Using a knockdown approach to deplete zebrafish myeloid cells (morpholino-mediated pu.1 knockdown), SH-pabA was restored to a level of virulence similar to that of the parental strain, but with a slight temporal delay (Fig. 1f). By 20 h postinfection (hpi), all embryos injected with the parent strain, and 80% of SH-pabA-injected embryos, had succumbed. The remaining SH-pabA-injected embryos died over the following 24 h. In myeloid cell-depleted zebrafish, SH-purA and SH-purB caused the death of approximately two-thirds of subjects injected, significantly less than the parent strain (P < 0.0001). The pu.1 knockdown approach causes a temporary delay in phagocytic cell development, and as expected, no further host death was observed after 40 h, a time point at which phagocyte production would recover (18, 19).
FIG 1.
In vivo characterization of S. aureus strains in the zebrafish embryo model of infection with reduced growth in human blood in vitro. (a) Survival curves of fish injected with wild-type S. aureus JE2 or with JE2-purA, JE2-purB, or JE2-pabA (1,500 CFU each). (b) Survival curves of fish injected with wild-type S. aureus SH1000 (SH) or with SH1000 purA, SH1000 purB, or SH1000 pabA (1,500 CFU each). (c to e) Growth of S. aureus mutants within embryos after injection with 1,500 CFU of SH1000 purA (c), SH1000 purB (d), or SH1000 pabA (e). Open circles, live embryos; filled circles, dead embryos. (f) Survival curves of pu.1 knockdown fish injected with wild-type S. aureus SH1000, SH1000 purA, SH1000 purB, or SH1000 pabA (1,500 CFU).
Purine biosynthesis is required for growth in blood.
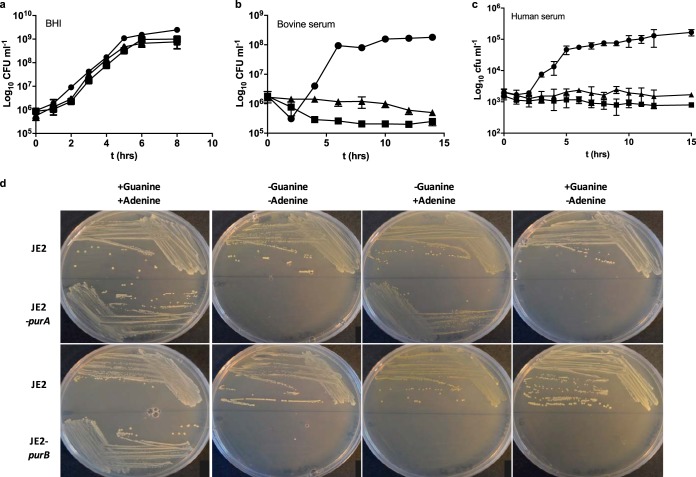
Analysis of the purA and purB genes (20, 21) demonstrated that purA and purB code for enzymes in the purine biosynthesis pathways (adenylosuccinate synthase and adenylosuccinate lyase, respectively) (see Fig. S2 in the supplemental material). In vitro, JE2-purA and JE2-purB showed reduced growth on human blood and bovine serum agar plates but growth similar to that of the parent strain on 5% (vol/vol) sheep blood, which contained a rich nutrient base (data not shown). Growth assays of JE2-purA and JE2-purB were also conducted in liquid media (brain heart infusion [BHI], bovine serum, or human serum) (Fig. 2a to c). Growth was comparable to that of the parent only in the nutrient-rich BHI medium, matching that seen in the initial NTML screen. This finding suggested that the reduced-growth phenotype was due to a requirement for a nutrient not readily available in human blood or in human or bovine serum. Analysis of the purine biosynthesis pathway suggested that both strains should require adenine for growth, while in addition to adenine, JE2-purB should also require guanine (or inosine). Chemically defined medium (CDM) analysis confirmed that the growth of JE2-purA was dependent on the presence of adenine, while the growth of JE2-purB was dependent on adenine and guanine (Table 2; Fig. 2d). The addition of 20 μg ml−1 adenine and 20 μg ml−1 inosine restored the growth of each pur mutant, to levels similar to that obtained for the parent strain (data not shown). Biochemical complementation of the purA and purB mutants was not successful in the zebrafish infection model, likely due to poor diffusion of nucleobases into zebrafish embryos (data not shown). The importance of purine biosynthesis pathway enzymes in disease has been well characterized (22, 23).
FIG 2.
The S. aureus purA and purB mutants require exogenous purines for growth. (a to c) Strains were grown in BHI (a), bovine serum (b), or human serum (c). Data are from three independent repeats; error bars represent standard errors. Circles, JE2; squares, JE2-purB; triangles, JE2-purA. (d) Growth of strains on CDM agar plates with or without adenine (20 μg ml−1) and guanine (20 μg ml−1) after 24 h of aerobic incubation at 37°C.
TABLE 2.
Growth analysis of JE2-purA and JE2-purB on solid media in the presence or absence of adenine and guanine
| Strain | Growtha on chemically defined medium |
|||
|---|---|---|---|---|
| With adenine and guanineb | Without adenine or guanine | With adenine, without guanine | Without adenine, with guanine | |
| JE2 | + | + | + | + |
| purB mutant | + | − | − | − |
| purA mutant | + | − | + | − |
+, growth; −, no growth.
The concentration of adenine or guanine, when present, was 20 μg ml−1.
pabA is required for virulence in the murine sepsis model and for growth in human blood.
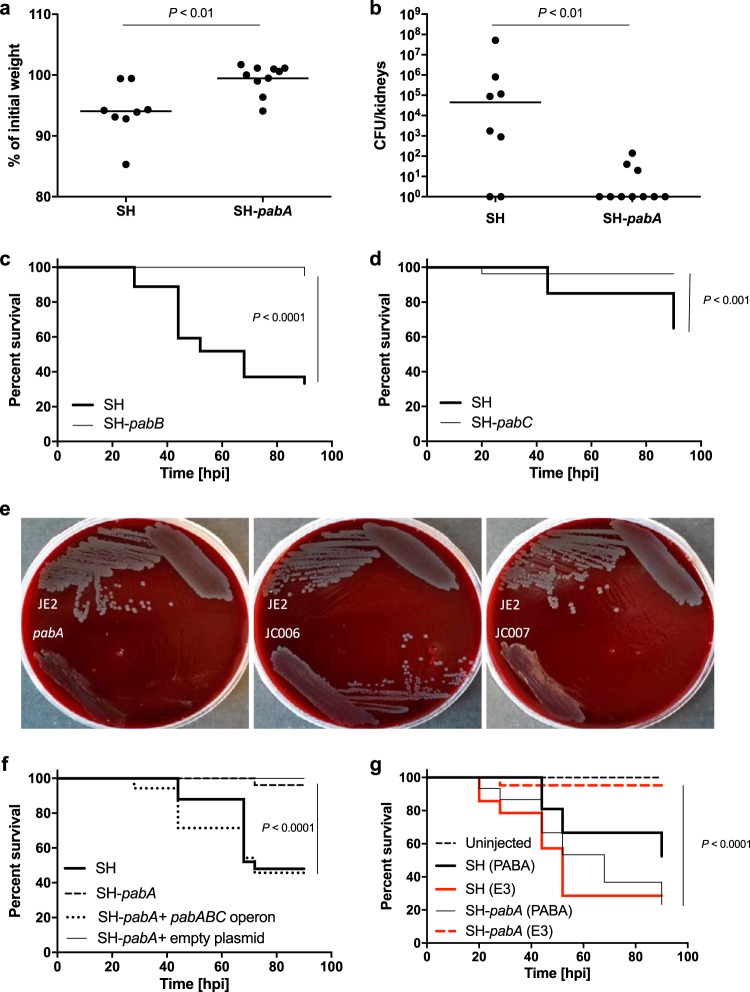
In a mouse sepsis model, mice injected with S. aureus SH-pabA (4 × 107 CFU) lost significantly less weight than those receiving the parent strain (2 × 107 CFU) (Fig. 3a). Bacterial numbers were also significantly lower in kidneys harvested from mice injected with SH-pabA (Fig. 3b) (P < 0.01).
FIG 3.
The pabABC operon is required for pathogenesis. (a and b) Female BALB/c mice (n = 10) were injected i.v. with 2 × 107 CFU S. aureus SH1000 (SH) or 4 × 107 CFU S. aureus SH1000 pabA (SH-pabA). Weight loss (a) and CFU counts in the kidneys (b) were measured after 3 days. (c) Survival curves of fish injected with S. aureus SH1000 (1,500 CFU) or S. aureus SH1000 pabB. (d) Survival curves of fish injected with S. aureus SH1000 (1,500 CFU) or S. aureus SH1000 pabC. (e) Growth of the parent strain (JE2), the pabA mutant, the genetically complemented pabA strain (with pJC002 integrated) (strain JC006), or a control integrated strain (with the empty pKASBAR plasmid in the pabA mutant) (strain JC007) on unsupplemented human blood agar (30%, vol/vol). Plates were incubated aerobically at 37°C for 48 h. (f) Survival curves of fish injected with 1,500 CFU of S. aureus SH1000, S. aureus SH1000 pabA, S. aureus SH1000 pabA plus the pabABC operon (with pJC002 integrated) (strain JC010), or S. aureus SH1000 pabA with an empty plasmid only (pKASBAR) (strain JC011). (g) Survival curves of fish injected with 1,500 CFU of S. aureus SH1000 or S. aureus SH1000 pabA followed by immediate immersion in either unsupplemented E3 medium (red) or E3 medium supplemented with 7 μg ml−1 PABA (black). Uninjected fish were included as controls under each condition.
The pabA Tn mutant was found to have a unique growth phenotype in the initial screen. Growth was highly reduced on 30% (vol/vol) human blood but only slightly reduced on 30% (vol/vol) rabbit blood (Table 1). However, the pabA mutant grew well on both sheep and horse blood agar (30% [vol/vol]), demonstrating that the phenotype was species specific (data not shown). In addition, the pabA mutant demonstrated good growth on 50% (vol/vol) human serum or plasma agar (see Fig. S3 in the supplemental material). To ascertain if the amount of human plasma in 30% (vol/vol) whole blood agar was too small to support growth, the pabA mutant was compared to the parent strain on agar in which the plasma concentration was increased up to 50% (vol/vol). At the lower concentrations of 10% (vol/vol) and 15% (vol/vol) (15% being the approximate concentration of plasma in 30% [vol/vol] blood agar), the growth of the pabA mutant was poor but comparable to that of JE2. Therefore, the reduced growth of the pabA mutant on human blood was not a result of lower plasma levels in human blood agar (data not shown).
PabA is an enzyme required for tetrahydrofolate (THF) synthesis (para-aminobenzoate synthetase component II) (20), and pabA is found in an operon with pabB and pabC, which is responsible for the synthesis of the folate pathway intermediate 4-aminobenzoic acid (para-aminobenzoic acid [PABA]) (20). Strains from the NTML harboring a Tn disrupting pabB or pabC were transduced into the SH1000 background and were also found to be attenuated in the zebrafish infection model (Fig. 3c and d) (P < 0.001). Genetic complementation of the pab operon restored JE2-pabA growth on human blood (Fig. 3e) and SH-pabA virulence in the zebrafish model (Fig. 3f).
Reduced growth on human blood could be due to a lack of nutrients that are required by a strain lacking THF. The end product of the folate pathway, THF, acts as a single-carbon donor/acceptor in glycine/serine interconversion, vitamin B5 synthesis, methionine synthesis, purine synthesis, N-formylmethionine-tRNA charging, glycine cleavage, and dTMP synthesis (see Fig. S4 in the supplemental material). To further characterize pabA, different media were used to interrogate the mechanism underpinning the lack of growth on human blood. With liquid culture, the growth of the pabA mutant was comparable to that of the wild type in BHI, bovine serum, and human serum (data not shown), suggesting that the reduced-growth phenotype was specific to blood. When CDM lacking purines, serine, and glycine was used as the base, only the addition of purines, serine, and glycine together could restore the growth yield of the mutant to parental levels (as measured by the maximum optical density at 600 nm [OD600] reached) (data not shown). Biochemical complementation with the same supplements did not restore the growth of the pabA mutant on human blood, nor did the addition of folic acid. However, the addition of PABA fully complemented growth (see Fig. S5a in the supplemental material), as would be expected based on similar work done with Lactococcus lactis (24). Immersion of SH-pabA-injected zebrafish embryos in E3 medium containing PABA restored virulence in vivo (Fig. 3g) (P < 0.0001).
Pyrimidine salvage pathways are required to bypass pabA.
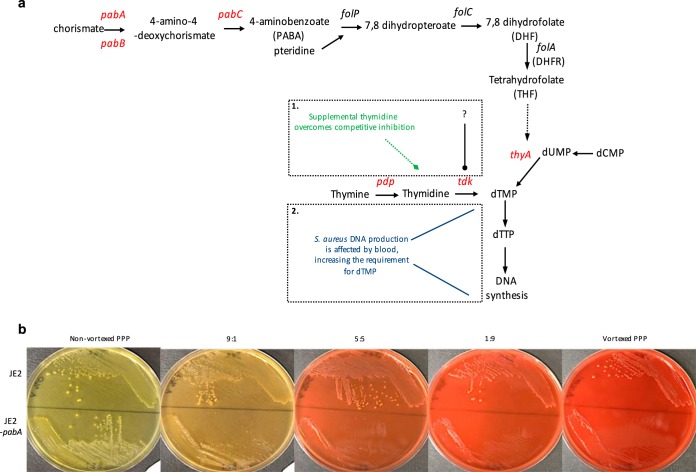
dTMP is synthesized via a THF-dependent route, or via an alternative nucleotide salvage pathway requiring thymine or thymidine (Fig. 4a). A combination of glycine, serine, and purines could not restore the growth of the pabA mutant on human blood; however, the addition of pyrimidines (thymine) supported growth to the extent of the parent strain, JE2 (Fig. S5a in the supplemental material). The crucial role of pyrimidines in bacterial survival under conditions of folate deprivation has been reported previously (25, 26). Neither pyrimidines nor folic acid could restore the pathogenicity of the pabA mutant in the zebrafish embryo model (data not shown).
FIG 4.
Folate biosynthesis pathway and effect of lysed RBCs on the growth of S. aureus pabA. (a) The folate biosynthesis and pyrimidine nucleotide salvage pathways (20, 21). Possible hypotheses for poor pabA growth on human blood are shown: either S. aureus Tdk is the target of competitive inhibition by human blood (frame 1) or increased dTMP demand necessitates supplemental thymidine in S. aureus (frame 2). (b) Growth of S. aureus JE2 or JE2-pabA on nonvortexed human PPP or with a decreasing ratio of vortexed to nonvortexed agar. Plates were incubated aerobically at 37°C for 48 h.
Double mutants defective in pabA and one of the pyrimidine nucleotide salvage pathway genes—pdp, tdk, or the thymidine transporter gene, nupC—were constructed in order to assess the roles of these genes in pabA growth. The growth of all three double mutants was reduced on human blood but could be complemented with PABA (Fig. S5b in the supplemental material). The addition of thymine or thymidine to blood could complement all the mutants except for the pabA tdk double mutant. This highlighted the fact that pyrimidine salvage pathways are required to bypass the deficit of pabA. If an inhibitory factor in blood was responsible for preventing the growth of the pabA mutant, Tdk was the likely target. Unexpectedly, the pabA pdp double mutant was complemented by thymine, and the pabA nupC mutant was complemented by thymidine. This suggests that thymine can be converted to thymidine independently of Pdp and that an alternative thymidine transporter to NupC is available in S. aureus. Two remaining putative pyrimidine transporters have been identified in S. aureus and have not yet been investigated (27).
Investigating a nucleotide salvage pathway inhibitory component in human blood.
The nucleotide salvage pathway appears to provide enough dTMP (later converted to dTTP) for the DNA synthesis and growth of the pabA mutant on human plasma or serum, but not on human blood, unless thymine or thymidine is added. This suggested that either a factor in whole blood competitively inhibits the nucleotide salvage pathway enzymes or growth on human blood leads to an increased requirement for dTMP, which cannot be met without increasing the thymine or thymidine concentration (Fig. 4a). To hone in on an inhibitory factor, different components of blood were assessed for their abilities to replicate the poor-growth phenotype of the pabA mutant seen on whole human blood. The growth of JE2-pabA was comparable to that of JE2 on platelet-rich plasma (PRP) and on PRP that had been vortexed to disrupt platelets (data not shown). Similarly, the growth of the parent strain and that of the mutant were equivalent when white blood cells (WBCs), either intact or lysed, were added to platelet-poor plasma (PPP). Vortexing of whole human blood followed by centrifugation produces red, rather than straw-colored, plasma, indicating red blood cell (RBC) lysis. Plasma from vortexed blood was mixed with PPP to give a 9:1 ratio of nonvortexed to vortexed plasma, decreasing incrementally to a ratio of 1:9. At the lowest ratio of nonvortexed to vortexed plasma, the growth of JE2-pabA was greatly reduced (Fig. 4b). This suggested that there is a potent inhibitor of the growth of the pabA mutant in the RBC component of human blood.
Hemoglobin or heme was deemed a likely candidate for the inhibitory factor. Hemoglobin, a complex of four heme groups, is the most abundant hemoprotein in humans. Heme is an iron-containing ring structure, and usage of heme as an iron source can be toxic to bacteria due to its active redox potential (28). Although the underlying mechanisms are not fully understood, it has been reported that heme-induced monooxygenase-like activity can cause direct DNA damage (28, 29). In S. aureus, heme is extracted from hemoglobin and is transported into the cell by the iron-regulated surface determinant (Isd) system (30). Toxicity induced by the liberation of iron from heme by S. aureus is reduced by the two-component heme-regulated transporter (hrtAB). Heme is also transported into S. aureus by the ABC transporter HtsABC, which requires the extraction of heme from hemoglobin by the Isd system (30). Both transport systems are upregulated under low Fe conditions by alleviation of the negative regulator Fur. However, supplementation of human blood agar with an alternative Fe source (ammonium ferrous sulfate) did not support the growth of JE2-pabA on human blood (data not shown). In addition, lyophilized bovine hemin, bovine hemoglobin, and human hemoglobin failed to prevent the growth of the pabA mutant on plasma (data not shown).
S. aureus growth in human blood requires increased levels of pyrimidines.
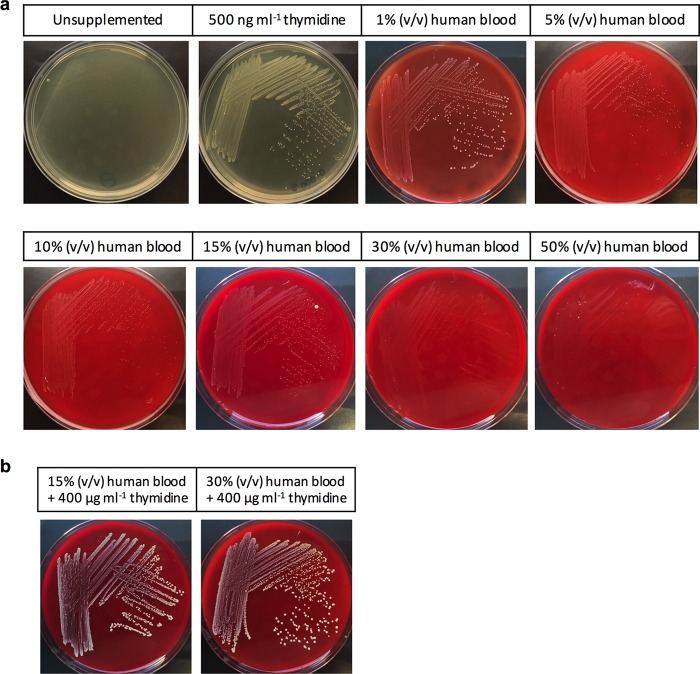
Rather than an inhibitory factor in blood preventing the growth of the pabA mutant, it is possible that human blood leads to an increased requirement for dTMP, which cannot be met in a folate-deficient mutant reliant solely on the pyrimidine salvage pathway (Fig. 4a). Thymidylate synthase (encoded by thyA) is highly conserved, requiring THF as a cofactor for the conversion of dUMP to dTMP, an essential step in DNA synthesis. To maintain viability, thyA mutants can utilize extracellular thymidine via pyrimidine salvage pathways (31) and thus cannot grow in vitro on media lacking pyrimidines, such as Mueller-Hinton (MH) agar or human blood (27). To determine if human blood increases the demand for pyrimidines, a minimal permissive concentration of thymidine to allow thyA mutant growth (500 ng ml−1) was added to MH agar (Fig. 5a). As the concentration of added human blood increased, ranging from 1 to 50% (vol/vol), the growth of the thyA mutant was increasingly inhibited, suggesting that, as with the pabA mutant, pyrimidine requirements are elevated by human blood. This was further confirmed by the addition of a higher concentration of thymidine (400 μg ml−1), which allowed biochemical complementation of thyA (Fig. 5b).
FIG 5.
Increased concentrations of thymidine are required for S. aureus growth on human blood. (a) Growth of S. aureus SH-thyA on MH agar. The medium either was left unsupplemented (top left) or contained a permissive concentration of thymidine (500 ng ml−1). Increasing concentrations of human blood, ranging from 1 to 50% (vol/vol), were added to the MH agar base, which contained thymidine (500 ng ml−1). Plates were incubated aerobically at 37°C for 24 h. (b) At concentrations of human blood causing reduced S. aureus SH-thyA growth, biochemical complementation was achieved by the addition of 400 μg ml−1 thymidine. Plates were incubated aerobically at 37°C for 24 h.
In the host environment, when innate immune cells encounter bacteria, reactive oxygen species (ROS), such as superoxide and nitric oxide, are generated (32). Bacteria have developed sophisticated mechanisms to resist such oxidative stress. Although heme acquisition is a necessity for S. aureus survival in vivo, we hypothesized that heme causes oxidative stress on bacteria, increasing dTTP requirements for DNA repair, and that the pabA mutant would be less able to compensate than the parent strain. To test this hypothesis, the pabA mutation was transduced into a strain unable to acquire heme due to disrupted Isd and heme transport systems, LS1 ΔisdE ΔhtsA (33). The triple mutant (LS1 ΔisdE ΔhtsA pabA) was inoculated onto human blood agar in order to determine whether the removal of potential heme toxicity would restore the growth of the pabA mutant. No growth was observed for the pabA or ΔisdE ΔhtsA pabA strain on unsupplemented blood agar, but both strains displayed good growth in the presence of exogenous pyrimidines (data not shown). However, it has been demonstrated that in the absence of functional heme transport and Isd systems, S. aureus can still acquire heme, by a third, as yet unknown heme transport mechanism (33).
In the presence of sulfonamide antibiotics, nucleotide salvage pathways are required for S. aureus growth in blood.
The use of folate-antagonistic sulfonamide antibiotics, such as trimethoprim, against S. aureus leads to the loss of THF synthesis and, like the pabA mutation, a dependence on the pyrimidine nucleotide salvage pathway for dTMP synthesis. The activity of this class of antibiotics can be reversed by providing enough thymidine to bypass the requirement for the THF-dependent dTMP synthesis pathway (34). The reversal of trimethoprim activity against JE2, JE2-pabA, and JE2-tdk by pyrimidine was assessed by growth on human, sheep, or horse blood agar. On human blood, thymidine reversed the activity of trimethoprim against JE2, and JE2-pabA growth was restored in the presence of thymidine; however, trimethoprim was active against JE2-tdk in the presence or absence of thymidine (Table 3). Similar results were obtained on horse blood. On sheep blood, trimethoprim was inactive against JE2 and JE2-pabA, likely due to a higher pyrimidine concentration in sheep blood (35), demonstrating that the JE2-pabA phenotype on human blood may also be due to differences in the pyrimidine content of blood. As with human and horse blood, JE2-tdk was inhibited by trimethoprim on sheep blood, and the addition of thymidine could not reverse this inhibition, since the nucleotide salvage pathways are prohibited.
TABLE 3.
Trimethoprim MICs for the S. aureus parent strain, JE2-pabA, and JE2-tdk on various media
| Strain | Trimethoprim MIC (mg/liter) for the indicated strain on the following mediuma: |
|||||||
|---|---|---|---|---|---|---|---|---|
| BHI |
Human blood |
Sheep blood |
Horse blood |
|||||
| −T | +T | −T | +T | −T | +T | −T | +T | |
| Parent | 1 | >32 | 0.75 | >32 | >32 | >32 | 1 | >32 |
| pabA mutant | 1 | >32 | >32 | >32 | >32 | >0.002 | >32 | |
| tdk mutant | 0.25 | 0.25 | 0.75 | 0.75 | 0.5 | 0.5 | 1 | 1 |
−T, no exogenous thymidine added, +T, thymidine added at 400 μg ml−1.
DISCUSSION
In order to identify novel pathogenicity determinants, an ordered library of transposon mutants was screened for gene disruptions causing growth and hemolysis defects on agar containing human blood as the only nutrient source. This screen identified purA, purB, and pabA as required for growth on human blood. The purA and purB genes are part of the de novo biosynthetic pathway for purines, and pabA is involved in folate synthesis. All three mutations were found to lead to significant attenuation in the zebrafish model of systemic infection, confirming the important role of these genes in pathogenesis.
In a study using a microarray-based system to detail the nonessential genes involved in the growth of Escherichia coli, Salmonella enterica, and Bacillus anthracis in human serum, the majority of mutants identified were involved in purine or pyrimidine biosynthesis (10). This suggests a scarcity of nucleotides in vivo, which bacteria counteract by being equipped with energetically costly metabolic pathways permitting de novo synthesis. Similarly, in our study, the ability of purine biosynthesis mutants to grow in nutrient-rich media suggested that purA and purB mutants require nutrients not readily available in human serum, whole blood, or the live zebrafish.
The reduced growth of S. aureus pabA in vitro was intriguing, because it was specific to human blood; normal growth was seen on blood components (serum, plasma), horse blood, and sheep blood. PabA is required for the production of PABA, an essential intermediate in the synthesis of THF. A pabB mutant of Streptococcus pneumoniae has been used as an attenuated strain for vaccine research, highlighting the importance of this pathway in the development of prophylactic strategies (36). The use of liquid CDM and solid CDM agar showed that purines, glycine, and serine were required for growth by the pabA mutant in excess of the amounts required by the parental strain. However, when assessed on human blood agar, growth inhibition could not be rescued with any compound except pyrimidines, suggesting that all other factors necessary to bypass the lack of THF are present in serum, plasma, and whole blood. The concept of “thymineless death” has been introduced previously and demonstrates the fundamental importance of pyrimidines in bacterial survival, over and above the other downstream effectors of THF (25). The addition of thymidine to human blood permitted the growth of the pabA mutant (Fig. S5a in the supplemental material). Human blood is known to have a lower thymidine content than the blood of other animals (35). However, the growth of the mutant on other thymidine-poor media (e.g., CDM, horse blood) suggested that thymidine deficiency per se was not solely responsible for the growth phenotype.
In the absence of THF-dependent thyA activity, pyrimidine salvage pathways are essential for the conversion of thymidine to dTMP (via Tdk), which is necessary for DNA replication. Both the pabA and the thyA mutant rely on these salvage pathways to provide a permissive amount of thymidine and, therefore, dTTP, allowing them to remain viable. It is difficult to tease apart exactly how human blood subverts this process, and we hypothesized that Tdk was the target of competitive inhibition in the pabA mutant, given that supplemental thymidine restored the growth of the pabA mutant, and a genetic knockout of pabA and tdk eliminated this biochemical complementation. Double knockouts of pabA with the gene responsible for the conversion of thymine to thymidine (pabA pdp) or with a pyrimidine transporter gene (pabA nupC) had no effect on biochemical complementation. Furthermore, the growth of the pabA mutant was reduced on human plasma supplemented with lysed RBC products. Since excess heme is toxic to S. aureus (28), heme and related molecules were ruled out as Tdk inhibitory factors. Tdk is a zinc-requiring enzyme that is purported to be required for transcriptional regulation (37). Zinc sequestration by human blood and other potential inhibitory factors should be investigated in future work (38).
Although the exact mechanism has yet to be elucidated, it is clear that human blood, or a component thereof, leads to an increased demand for dTMP, which cannot be met in a THF-deficient mutant; hence, exogenous thymidine or thymine is necessary to support the growth of the mutant specifically on human blood.
Finally, little is known about the clinical prevalence or relevance of pabA mutations. Trimethoprim is used in the control of S. aureus infections, and long-standing treatment can lead to failure due to the development of antibiotic resistance (39). In this context, thyA mutations are usually observed in the resistant subpopulation, and such mutations cause the formation of thymidine-dependent small-colony variants (SCVs), which rely on pyrimidine salvage pathways (via Pdp and Tdk) (40). However, sulfonamide antibiotics remain bactericidal unless a thymidine-rich environment exists, such as damaged host tissues, which allow S. aureus to utilize pyrimidine salvage pathways and thus survive (41). The work presented here suggests that the activity of sulfonamide drugs results from the inhibition of THF coupled with reduced activity of the pyrimidine salvage pathways and/or an increased demand for dTMP imparted by human blood. The identification of metabolic pathways important for host-pathogen interactions provides novel avenues to be explored in the effort to combat antibiotic-resistant pathogens.
MATERIALS AND METHODS
Ethics statement.
Zebrafish embryos less than 5 days postfertilization (dpf) are not protected under the Animals (Scientific Procedures) Act 1986, but all zebrafish work was carried out according to the details set out in Project License PPL 40/3574. Murine work was carried out according to UK law in the Animals (Scientific Procedures) Act 1986, under Project License PPL 40/3699. Human blood was obtained from healthy volunteers in compliance with the guidelines of the South Sheffield Research Ethics Committee (STH13927).
Bacterial strains, plasmids, and growth conditions.
The Nebraska transposon mutagenesis library (11) was acquired from the Network on Antimicrobial Resistance in S. aureus (NARSA) strain repository, now available from BEI Resources (www.beiresources.org/), and was used for screening experiments. Originally in the USA300 LAC JE2 background, mutations were transduced back into JE2 or SH1000 as required. All other strains, and the plasmids used in this study, are listed in Table 4. S. aureus strains were routinely grown in brain heart infusion (BHI) medium at 37°C with aeration at 250 rpm, unless otherwise stated. Mueller-Hinton agar (Oxoid) was used as a thymidine-poor medium where stated. E. coli strains were grown in Luria-Bertani medium at 37°C with aeration at 250 rpm. Agar at 1.5% (wt/vol) was added for solid media. Antibiotics were added as required. For MIC determination, a bacterial colony was inoculated into 2 ml sterile distilled water (dH2O) and was spread onto an agar plate using a sterile swab (Oxoid). Trimethoprim Etests (bioMérieux) were applied to the solid medium surface using tweezers, and incubation was carried out overnight at 37°C.
TABLE 4.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype/markers | Source or reference |
|---|---|---|
| Strains | ||
| S. aureus strains | ||
| SH1000 | Functional rsbU+ derivative of 8325-4 | 51 |
| RN4220 | Restriction-negative, modification-positive strain | 52 |
| USA300 JE2 | USA300 LAC strain cured of plasmids p01 and p03 | 11 |
| SJF4669 | SH-pabA::spc pdp::ery | This study |
| SJF4670 | SH-pabA::spc nupC::ery | This study |
| SJF4671 | SH-thyA::ery | 27 |
| SJF4678 | SH-pabA::spc tdk::ery | This study |
| JC006 | JE2-pabA, pJC002 inserted at lipase; pabA+ Eryr Linr Tetr | This study |
| JC007 | JE2-pabA, pKASBAR inserted at lipase; Eryr Linr Tetr | This study |
| JC010 | SH-pabA, pJC002 inserted at lipase; pabA+ Eryr Linr Tetr | This study |
| JC011 | SH-pabA, pKASBAR inserted at lipase; Eryr Linr Tetr | This study |
| LS1 | Spontaneous murine arthritis isolate | 53 |
| LS1 ΔisdE ΔhtsA | LS1 derivative; ΔisdE ΔhtsA | 33 |
| E. coli TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80 lacZΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara-leu)7697 galK rpsL (Strr) endA1 nupG | Invitrogen |
| Plasmids | ||
| pKASBAR | Hybrid vector of pCL84 and pUC18 for integration into S. aureus lipase gene (geh), attP; Tetr (S. aureus); Specr (E. coli) | 46 |
| pJC002 | pKASBAR containing the pab operon, with pabA, pabB, and pabC, and upstream control elements; Tetr | This study |
The chemically defined medium used in this study has been described previously (42). The following components were dissolved in 1 liter of H2O: Na2HPO4·2H2O, 7 g; KH2PO4, 3 g; l-aspartic acid, 0.15 g; l-alanine, 0.1 g; l-arginine, 0.1 g; l-cysteine, 0.05 g; glycine, 0.1 g; l-glutamic acid, 0.15 g; l-histidine, 0.1 g; l-isoleucine, 0.15 g; l-lysine, 0.1 g; l-leucine, 0.15 g; l-methionine, 0.1 g; l-phenylalanine, 0.1 g; l-proline, 0.15 g; l-serine, 0.1 g; l-threonine, 0.15 g; l-tryptophan, 0.1 g; l-tyrosine, 0.1 g; l-valine, 0.15 g; biotin, 0.02 g; pyridoxal HCl, 0.8 g; nicotinic acid, 0.4 g; pyridoxamine di-HCl, 0.8 g; d-pantothenic acid, 0.4 g; riboflavin, 0.4 g; thiamine HCl, 0.4 g; adenine sulfate, 0.02 g; guanine HCl, 0.02 g; CaCl2·6H2O, 0.01 g; (NH4)2Fe(SO4)2·6H2O, 0.006 g; glucose, 10 g; MgSO4·7H2O, 0.5g. Inosine was used at 20 µg/ml.
Human, or other animal, blood or blood components were added to agar at various concentrations, as required. Venous blood was collected from healthy volunteers following informed consent. For plasma preparation, blood was centrifuged at 270 × g for 20 min in 50-ml Falcon tubes. The upper, platelet-rich phase was collected and was either used directly as platelet-rich plasma (PRP) or centrifuged again at 1,155 × g for 30 min to yield platelet-poor plasma (PPP). Plasma was stored at −20°C. Animal blood and blood products were purchased from Thermo Scientific or Sigma and were stored at 4°C. Bovine hemin or hemoglobin, human hemoglobin, thymine, thymidine, glycine, serine, PABA, or folic acid was added to media as and when required.
Genetic manipulation.
Electroporation was used to transform S. aureus RN4220 and E. coli using previously published methods (43, 44). All S. aureus transduction experiments were carried out with ϕ11 as described previously (45).
For genetic complementation of SH-pabA and JE2-pabA, Phusion polymerase (NEB) was used to amplify the pab operon from S. aureus SH1000 genomic DNA, using primers containing appropriate restriction sites (forward, ATAATAGGGCCCATTGTA-CTGTCTTGACCACCACT; reverse, ATAATACTCGAGATACGTATACAAGAATTAA-CAACAGCA). The PCR product was inserted into pKASBAR (46), a plasmid encoding an attP site. Using this attP site, bacteriophage DNA can integrate into the S. aureus genome at the attB site, in the presence of an integrase (47). The attB site is located at the glycerol ester hydrolase (geh) gene, so integration can be verified by loss of lipase activity. For such genetic manipulation, the integrase is provided by an additional helper plasmid, pYL112Δ19, propagated in the S. aureus recipient strain, RN4220. The insert was then transduced from RN4220 into pabA and control strains.
To prepare double mutants within Tn insertions, the “toolkit” for switching antibiotic resistance within NTML strains was used as published previously (48). Tn inserts in pdp, nupC, and tdk genes, with alternate antibiotic resistance markers, were transduced into the pabA mutant as listed in Table 4.
Strains LS1 and LS1 ΔisdE ΔhtsA were kindly provided by Sean Nair (University College London). pabA was transduced into both strains, and successful transductants were confirmed by PCR.
Transposon library screen.
The NTML was grown for 18 h at 37°C in 96-well microtiter dishes. Using a 96-pin replicator (Boekel Industries), the contents of each well were transferred to BHI agar, BHI agar plus erythromycin (10 μg/ml) and lincomycin (25 μg/ml), 30% (vol/vol) human blood agar, 50% (vol/vol) bovine serum agar, and 5% (vol/vol) sheep blood, plus Columbia agar base in rectangular OmniTray plates (Nunc). Human blood and bovine serum plates were incubated for 48 h at 37°C; all other plates were incubated for 18 h at 37°C, with an additional 4 h at 4°C for sheep blood plates, to ensure efficient hemolysis. Phenotypes were determined by comparison of each spot (colony size and hemolysis zone) to the surrounding spots on the plate.
Zebrafish model.
Zebrafish embryos, strain London wild type (LWT), were maintained in E3 medium at 28°C by following standard protocols (17). Embryos were bred in the aquarium facilities at the University of Sheffield. Microinjection of embryos was performed as described previously (17). Individual infected embryos were kept in 100 μl E3 medium, and survival was assessed over 90 h. For in vivo complementation experiments, compounds were dissolved in E3 medium and were buffered to a pH of 6.5 to 7.5. Immediately following injections, embryos were placed in appropriate compound solutions. Further compound solution was added in the embryo washing step. Ninety-six-well microtiter plates were placed in a plastic box, with damp paper, to reduce evaporation during incubation.
pu.1-antisense morpholino-modified oligonucleotides (49) were injected into zebrafish embryos using the method described previously (17). Bacteria were recovered from infected embryos at 12-h intervals. Individual embryos were transferred to microcentrifuge tubes and were homogenized using a PreCellys 24-Dual homogenizer (Peqlab). Bacterial numbers were then determined by serial dilution in phosphate-buffered saline (PBS) and plating onto BHI agar.
Murine infection model.
Female BALB/c mice were purchased from Charles River Laboratories (Margate, UK) and were maintained by standard husbandry techniques at the University of Sheffield (Biological Services). Bacteria were washed in endotoxin-free PBS (Sigma), and 100 μl (2 × 107 to 4 × 107 CFU) was injected intravenously (i.v.) into the tail vein. Serial dilutions of culture were prepared to confirm the injection CFU. Mice were monitored and were sacrificed at 72 hpi. Mouse organs were individually homogenized in PBS and, after serial dilution, plated onto BHI agar supplemented with antibiotics as needed for bacterial enumeration.
Statistical analysis.
Sample sizes were predetermined for mouse (n = 10) and zebrafish (n = 20) experiments based on previous experimental data (50). All zebrafish experimental results are representative of 2 experiments unless otherwise stated. For zebrafish embryo survival experiments, the Kaplan-Meier method was employed. Survival curves were compared using the log rank (Mantel-Cox) test. For bacterial count comparisons in murine experiments, the Mann-Whitney U test was used. Statistical analysis was performed using Prism, version 6.0 (GraphPad), and a P value of <0.05 was considered significant.
Supplementary Material
ACKNOWLEDGMENTS
We have no conflicts of interest to declare.
This work was funded by the Wellcome Trust (grants 099957/Z/12/Z and 089981) and the University of Sheffield 2022 Futures program. We are grateful for the use of the Bateson Centre aquarium and Biological Services Unit at the University of Sheffield.
We thank Kenneth Bayles for provision of the NARSA mutant library, Daria Shamarina for technical assistance with the murine work, Sean Nair for providing us with the LS1 ΔisdE ΔhtsA strain, and Barbara Kahl for providing SH1000ΔthyA.
Author contributions for this study were as follows. J.C. and E.B. performed and analyzed the experiments. L.R.P., S.A.R., M.K.W., and S.J.F. contributed to the study design and data analysis. J.C., E.B., and S.J.F. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00337-17.
REFERENCES
- 1.Gordon RJ, Lowy FD. 2008. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis 46:S350–S359. doi: 10.1086/533591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Powers ME, Wardenburg JB. 2014. Igniting the fire: Staphylococcus aureus virulence factors in the pathogenesis of sepsis. PLoS Pathog 10:e1003871. doi: 10.1371/journal.ppat.1003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol 186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lechner S, Lewis K, Bertram R. 2012. Staphylococcus aureus persisters tolerant to bactericidal antibiotics. J Mol Microbiol Biotechnol 22:235–244. doi: 10.1159/000342449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassat JE, Skaar EP. 2013. Iron in infection and immunity. Cell Host Microbe 13:509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coulter SN, Schwan WR, Ng EYW, Langhorne MH, Ritchie HD, Westbrock-Wadman S, Hufnagle WO, Folger KR, Bayer AS, Stover CK. 1998. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol Microbiol 30:393–404. doi: 10.1046/j.1365-2958.1998.01075.x. [DOI] [PubMed] [Google Scholar]
- 8.Hammer ND, Skaar EP. 2012. The impact of metal sequestration on Staphylococcus aureus metabolism. Curr Opin Microbiol 15:10–14. doi: 10.1016/j.mib.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinberg ED. 1974. Iron and susceptibility to infectious disease. Science 184:952–956. doi: 10.1126/science.184.4140.952. [DOI] [PubMed] [Google Scholar]
- 10.Samant S, Lee H, Ghassemi M, Chen J, Cook JL, Mankin AS, Neyfakh AA. 2008. Nucleotide biosynthesis is critical for growth of bacteria in human blood. PLoS Pathog 4:e37. doi: 10.1371/journal.ppat.0040037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537-12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bae T, Banger AK, Wallace A, Glass EM, Åslund F, Schneewind O, Missiakas DM. 2004. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc Natl Acad Sci U S A 101:12312–12317. doi: 10.1073/pnas.0404728101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yee R, Cui P, Shi W, Feng J, Zhang Y. 2015. Genetic screen reveals the role of purine metabolism in Staphylococcus aureus persistence to rifampicin. Antibiotics 4:627–642. doi: 10.3390/antibiotics4040627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bose JL, Daly SM, Hall PR, Bayles KW. 2014. Identification of the Staphylococcus aureus vfrAB operon, a novel virulence factor regulatory locus. Infect Immun 82:1813–1822. doi: 10.1128/IAI.01655-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frydenlund Michelsen C, Hossein Khademi SM, Krogh Johansen H, Ingmer H, Dorrestein PC, Jelsbak L. 2016. Evolution of metabolic divergence in Pseudomonas aeruginosa during long-term infection facilitates a proto-cooperative interspecies interaction. ISME J 10:1323–1336. doi: 10.1038/ismej.2015.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ibberson CB, Jones CL, Singh S, Wise MC, Hart ME, Zurawski DV, Horswill AR. 2014. Staphylococcus aureus hyaluronidase is a CodY-regulated virulence factor. Infect Immun 82:4253–4264. doi: 10.1128/IAI.01710-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prajsnar TK, Cunliffe VT, Foster SJ, Renshaw SA. 2008. A novel vertebrate model of Staphylococcus aureus infection reveals phagocyte-dependent resistance of zebrafish to non-host specialized pathogens. Cell Microbiol 10:2312–2325. doi: 10.1111/j.1462-5822.2008.01213.x. [DOI] [PubMed] [Google Scholar]
- 18.Herbomel P, Thisse B, Thisse C. 1999. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development 126:3735–3745. [DOI] [PubMed] [Google Scholar]
- 19.Lieschke GJ, Oates AC, Crowhurst MO, Ward AC, Layton JE. 2001. Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood 98:3087–3096. doi: 10.1182/blood.V98.10.3087. [DOI] [PubMed] [Google Scholar]
- 20.Kanehisa M, Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA, Holland TA, Keseler IM, Kothari A, Kubo A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Subhraveti P, Weaver DS, Weerasinghe D, Zhang P, Karp PD. 2014. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 42:D459–D471. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McFarland WC, Stocker BA. 1987. Effect of different purine auxotrophic mutations on mouse-virulence of a Vi-positive strain of Salmonella dublin and of two strains of Salmonella typhimurium. Microb Pathog 3:129–141. doi: 10.1016/0882-4010(87)90071-4. [DOI] [PubMed] [Google Scholar]
- 23.Baxter-Gabbard KL, Pattee PA. 1970. Purine biosynthesis in Staphylococcus aureus. Arch Mikrobiol 71:40–48. doi: 10.1007/BF00412233. [DOI] [PubMed] [Google Scholar]
- 24.Wegkamp van Oorschot W, de Vos WM, Smid EJ. 2007. Characterization of the role of para-aminobenzoic acid biosynthesis in folate production by Lactococcus lactis. Appl Environ Microbiol 73:2673–2681. doi: 10.1128/AEM.02174-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Then R, Angehrn P. 1973. Sulphonamide-induced “thymineless death” in Escherichia coli. J Gen Microbiol 76:255–263. doi: 10.1099/00221287-76-2-255. [DOI] [PubMed] [Google Scholar]
- 26.Mathieu LG, de Repentigny J, Turgeon S, Sonea S. 1968. Thymineless death of Staphylococcus aureus and formation of its alpha toxin. Can J Microbiol 14:983–987. doi: 10.1139/m68-163. [DOI] [PubMed] [Google Scholar]
- 27.Kriegeskorte A, Block D, Drescher M, Windmüller N, Mellmann A, Baum C, Neumann C, Lorè NI, Bragonzi A, Liebau E, Hertel P, Seggewiss J, Becker K, Proctor RA, Peters G, Kahl BC. 2014. Inactivation of thyA in Staphylococcus aureus attenuates virulence and has a strong impact on metabolism and virulence gene expression. mBio 5:e01447-14. doi: 10.1128/mBio.01447-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anzaldi LL, Skaar EP. 2010. Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infect Immun 78:4977–4989. doi: 10.1128/IAI.00613-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin H, Everse J. 1987. The cytotoxic activity of hematoheme: evidence for two different mechanisms. Anal Biochem 161:323–331. doi: 10.1016/0003-2697(87)90458-1. [DOI] [PubMed] [Google Scholar]
- 30.Hammer ND, Skaar EP. 2011. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu Rev Microbiol 65:129–147. doi: 10.1146/annurev-micro-090110-102851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Besier S, Zander J, Siegel E, Saum SH, Hunfeld K-P, Ehrhart A, Brade V, Wichelhaus TA. 2008. Thymidine-dependent Staphylococcus aureus small-colony variants: human pathogens that are relevant not only in cases of cystic fibrosis lung disease. J Clin Microbiol 46:3829–3832. doi: 10.1128/JCM.01440-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nunoshiba T, DeRojas-Walker T, Tannenbaum SR, Demple B. 1995. Roles of nitric oxide in inducible resistance of Escherichia coli to activated murine macrophages. Infect Immun 63:794–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wright JA, Nair SP. 2012. The lipoprotein components of the Isd and Hts transport systems are dispensable for acquisition of heme by Staphylococcus aureus. FEMS Microbiol Lett 329:177–185. doi: 10.1111/j.1574-6968.2012.02519.x. [DOI] [PubMed] [Google Scholar]
- 34.Koch AE, Burchall JJ. 1971. Reversal of the antimicrobial activity of trimethoprim by thymidine in commercially prepared media. Appl Microbiol 22:812–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nottebrock H, Then R. 1977. Thymidine concentrations in serum and urine of different animal species and man. Biochem Pharmacol 26:2175–2179. doi: 10.1016/0006-2952(77)90271-4. [DOI] [PubMed] [Google Scholar]
- 36.Chimalapati S, Cohen J, Camberlein E, Durmort C, Baxendale H, de Vogel C, van Belkum A, Brown JS. 2011. Infection with conditionally virulent Streptococcus pneumoniae Δpab strains induces antibody to conserved protein antigens but does not protect against systemic infection with heterologous strains. Infect Immun 79:4965–4976. doi: 10.1128/IAI.05923-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandrini MPB, Clausen AR, Munch-Petersen B, Piskur J. 2006. Thymidine kinase diversity in bacteria. Nucleosides Nucleotides Nucleic Acids 25:1153–1158. doi: 10.1080/15257770600894469. [DOI] [PubMed] [Google Scholar]
- 38.Kehl-Fie TE, Chitayat S, Hood MI, Damo S, Restrepo N, Garcia C, Munro KA, Chazin WJ, Skaar EP. 2011. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe 10:158–164. doi: 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zander J, Besier S, Ackermann H, Wichelhaus TA. 2010. Synergistic antimicrobial activities of folic acid antagonists and nucleoside analogs. Antimicrob Agents Chemother 54:1226–1231. doi: 10.1128/AAC.00705-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zander J, Besier S, Saum SH, Dehghani F, Loitsch S, Brade V, Wichelhaus TA. 2008. Influence of dTMP on the phenotypic appearance and intracellular persistence of Staphylococcus aureus. Infect Immun 76:1333–1339. doi: 10.1128/IAI.01075-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldstein EJC, Proctor RA. 2008. Role of folate antagonists in the treatment of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis 46:584–593. doi: 10.1086/525536. [DOI] [PubMed] [Google Scholar]
- 42.Hussain M, Hastings JG, White PJ. 1991. A chemically defined medium for slime production by coagulase-negative staphylococci. J Med Microbiol 34:143–147. doi: 10.1099/00222615-34-3-143. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 44.Schenk S, Laddaga RA. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett 73:133–138. doi: 10.1111/j.1574-6968.1992.tb05302.x. [DOI] [PubMed] [Google Scholar]
- 45.Novick RP. 1991. Genetic systems in staphylococci. Methods Enzymol 204:587–636. doi: 10.1016/0076-6879(91)04029-N. [DOI] [PubMed] [Google Scholar]
- 46.Bottomley AL, Kabli AF, Hurd AF, Turner RD, Garcia-Lara J, Foster SJ. 2014. Staphylococcus aureus DivIB is a peptidoglycan-binding protein that is required for a morphological checkpoint in cell division. Mol Microbiol 94:1041–1064. doi: 10.1111/mmi.12813. [DOI] [PubMed] [Google Scholar]
- 47.Lee CY, Iandolo JJ. 1986. Integration of staphylococcal phage L54a occurs by site-specific recombination: structural analysis of the attachment sites. Proc Natl Acad Sci U S A 83:5474–5478. doi: 10.1073/pnas.83.15.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bose JL, Fey PD, Bayles KW. 2013. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl Environ Microbiol 79:2218–2224. doi: 10.1128/AEM.00136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rhodes J, Hagen A, Hsu K, Deng M, Liu TX, Look AT, Kanki JP. 2005. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell 8:97–108. doi: 10.1016/j.devcel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 50.McVicker G, Prajsnar TK, Williams A, Wagner NL, Boots M, Renshaw SA, Foster SJ. 2014. Clonal expansion during Staphylococcus aureus infection dynamics reveals the effect of antibiotic intervention. PLoS Pathog 10:e1003959. doi: 10.1371/journal.ppat.1003959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, Foster SJ. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J Bacteriol 184:5457–5467. doi: 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kreiswirth B, Löfdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 53.Bremell T, Lange S, Svensson L, Jennische E, Gröndahl K, Carlsten H, Tarkowski A. 1990. Outbreak of spontaneous staphylococcal arthritis and osteitis in mice. Arthritis Rheum 33:1739–1744. doi: 10.1002/art.1780331120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.