ABSTRACT
Polymicrobial infections often include both fungi and bacteria and can complicate patient treatment and resolution of infection. Cross-kingdom interactions among bacteria, fungi, and/or the immune system during infection can enhance or block virulence mechanisms and influence disease progression. The fungus Candida albicans and the bacterium Pseudomonas aeruginosa are coisolated in the context of polymicrobial infection at a variety of sites throughout the body, including mucosal tissues such as the lung. In vitro, C. albicans and P. aeruginosa have a bidirectional and largely antagonistic relationship. Their interactions in vivo remain poorly understood, specifically regarding host responses in mediating infection. In this study, we examine trikingdom interactions using a transparent juvenile zebrafish to model mucosal lung infection and show that C. albicans and P. aeruginosa are synergistically virulent. We find that high C. albicans burden, fungal epithelial invasion, swimbladder edema, and epithelial extrusion events serve as predictive factors for mortality in our infection model. Longitudinal analyses of fungal, bacterial, and immune dynamics during coinfection suggest that enhanced morbidity is associated with exacerbated C. albicans pathogenesis and elevated inflammation. The P. aeruginosa quorum-sensing-deficient ΔlasR mutant also enhances C. albicans pathogenicity in coinfection and induces extrusion of the swimbladder. Together, these observations suggest that C. albicans-P. aeruginosa cross talk in vivo can benefit both organisms to the detriment of the host.
KEYWORDS: Candida albicans, Pseudomonas aeruginosa, infection, mucosal, polymicrobial, zebrafish
INTRODUCTION
Microorganisms such as fungi and bacteria form complex communities in a variety of niches within the human body in both healthy individuals and in the event of disease (1). During polymicrobial infection, interactions between different microbial species can alter host responses and/or microbial virulence and pathogenesis, often complicating patient treatment and resolution of infection. Cross-kingdom interactions due to bacteria, fungi, the immune system, or any combination of these components can enhance or block virulence mechanisms and influence disease progression.
The fungus Candida albicans and the bacterium Pseudomonas aeruginosa are coisolated at a variety of infection sites, including burn wounds, contaminated catheters, and lung infections, and they influence each other's virulence (1). They are believed to be clinically important copathogens in specific patients, such as those with underlying pulmonary disease (2, 3). C. albicans is among the most commonly isolated fungi in fungal-bacterial coinfections (4, 5). It is a dimorphic opportunistic pathogen with the ability to form invasive hyphal filaments and drug-resistant biofilms, and it produces virulence factors such as secreted aspartyl proteases and the toxin candidalysin (6–8). P. aeruginosa is another opportunistic pathogen with sophisticated virulence mechanisms, including the production of exosomes and toxins and the formation of biofilms (9, 10). The P. aeruginosa transcription factor LasR is the master regulator of the quorum-sensing system that senses cell density and controls virulence factor expression (11, 12).
Cross-kingdom microbial interactions in clinically relevant species such as C. albicans and P. aeruginosa can affect virulence factor production and thereby regulate threats to the host (13, 14). When cocultured in vitro, P. aeruginosa suppresses C. albicans hyphal development through a variety of different pathways that involve quorum-sensing molecules and phenazines (15–17), some of which can be induced in the presence of C. albicans (18). The complexity of interactions increases as one considers observations demonstrating that different genotypes may differ in their interactions (19), that P. aeruginosa interacts differently with fungal cells in different morphologies (16), and that the different species compete for nutrients and, thus, interactions may be influenced by the environment (20). In order to determine how the interactions that occur in vitro relate to the interactions that occur in vivo, additional models for the in vivo dissection of microbe-microbe interactions are needed.
Experiments performed in some murine C. albicans-bacterial coinfection models have indicated synergistic virulence for one or both pathogens, suggesting that the outcome of P. aeruginosa-C. albicans coinfections is not easily predicted from in vitro antagonism (21–26). For example, a murine burn model found that preinfection with P. aeruginosa increased the damage caused by C. albicans (27). However, some studies of C. albicans-P. aeruginosa coinfections in murine and Caenorhabditis elegans models find that combining the two species can negatively regulate overall virulence in the context of coinfection (14, 28). These disparate results suggest that other factors, including the host environment, can play a role in the way bacteria and fungi influence each other.
One important component of the host environment to consider is immune response, because immunopathology can play a driving role in coinfections. Polymicrobial infection can stimulate excessive inflammation that causes morbidity and mortality through neutrophil recruitment, cytokine hyperelicitation, pulmonary edema with alveolar collapse, and severe tissue damage (23, 29–33). Enhanced expression of the proinflammatory cytokine interleukin-6 (IL-6) has been implicated in elevated pathogenesis during polymicrobial infection, including Candida-bacterial coinfection (23, 24, 34–37). Although host responses and immunopathology are critical components of polymicrobial infection outcome, our current understanding of how host immune responses play a role in mediating altered pathogenesis in polymicrobial infection remains limited.
This gap in our knowledge may be bridged by vertebrate infection models that recapitulate mammalian infection but allow simultaneous monitoring of host, bacteria, and fungi. The juvenile zebrafish model offers the power to noninvasively track fungal, bacterial, and immune dynamics simultaneously and at high resolution over the course of a live infection with the ability to longitudinally associate infection factors with mortality. We have recently developed a swimbladder infection route to model mucosal infection with C. albicans in the zebrafish and study how the immune system mediates fungal infection in real time (38–40). The swimbladder is anatomically, morphologically, and transcriptionally similar to the human lung, with an air-epithelial interface that produces epithelial surfactants (41–44). Due to these similarities, polymicrobial infections in the zebrafish swimbladder may share well-conserved aspects of disease that could be applied to understand coinfection in the lung.
Using this model, we find that C. albicans-P. aeruginosa coinfection of the mucosa leads to synergistic virulence and enhanced mortality. Coinfection mortality is associated with higher C. albicans burden and invasive pathogenesis, which serve as strong predictive factors for mortality. Some aspects of the proinflammatory immune response are also enhanced in polymicrobial infection. These data indicate that P. aeruginosa augments C. albicans pathogenesis and host inflammation, which may contribute to a synergistic virulence associated with mucosal C. albicans-P. aeruginosa coinfection.
RESULTS
C. albicans and P. aeruginosa actively contribute to synergistic virulence in vivo.
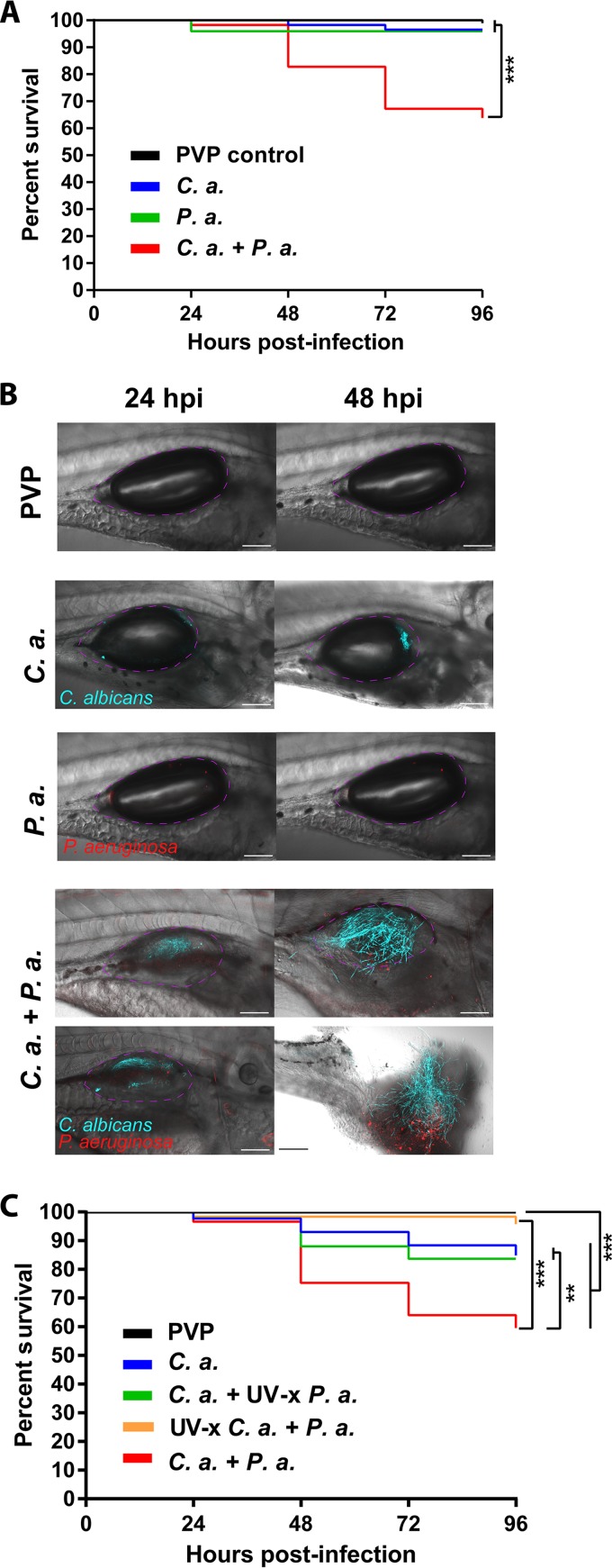
C. albicans and P. aeruginosa are opportunistic pathogens coisolated in mucosal tissues, but their interactions have not yet been well characterized during mucosal infection. To further examine C. albicans-P. aeruginosa interactions in mucosal infection and determine if the host environment influences outcome of coinfection, we adopted our newly described zebrafish swimbladder model (38) to mimic the coinfected lung found in some patients (2). We created a mucosal infection of the swimbladder in immunocompetent 4-day-postfertilization (dpf) zebrafish with C. albicans, P. aeruginosa, and C. albicans plus P. aeruginosa (coinfection). We used a red fluorescent protein-expressing derivative of the P. aeruginosa burn isolate PA14 and a far-red fluorescent protein-expressing derivative of the C. albicans invasive candidiasis isolate SC5314. These are both clinical isolates that have been used in many in vitro coculture experiments and have well-characterized virulence in multiple infection models. We found that infection with each species alone causes little mortality, but C. albicans and P. aeruginosa are synergistically virulent in coinfection. This was the case for both low infection doses (Fig. 1A) and higher doses (see Fig. S1 in the supplemental material). Representative images document the high fungal burden and damage in coinfections (Fig. 1B). This mortality difference was also observed in the enhanced green fluorescence protein (EGFP) neutrophil transgenic Tg(Mpx:EGFP) fish line used in subsequent experiments (Fig. S2). To test if the enhanced virulence associated with coinfection is dependent on live organisms, we established coinfections with UV-inactivated bacteria and fungi and found that microbial synergy depends on the presence of both live C. albicans and live P. aeruginosa (Fig. 1C). Thus, the synergistic virulence of C. albicans-P. aeruginosa mucosal infection requires viable fungi and bacteria to exacerbate mortality.
FIG 1.
C. albicans and P. aeruginosa demonstrate synergistic virulence in mucosal infection of the swimbladder. (A and B) Wild-type zebrafish larvae at 4 days postfertilization were separated into 4 groups and microinjected into the swimbladder with 5 nl of PVP (control), C. albicans (C. a.) at 2.5 × 107 CFU/ml, P. aeruginosa (P. a.) at 2.5 × 108 CFU/ml, or C. albicans and P. aeruginosa (C. a. + P. a.) at 2.5 × 107 CFU/ml and 2.5 × 108 CFU/ml, respectively. Fish were screened immediately postinjection to select for consistent inocula, and mortality was recorded every 24 h out to 96 h postinjection. Confocal images were acquired at ×10 and ×20 magnification, with scale bars at 100 μm and 200 μm. Data are representative of 4 pooled, independent experiments. Pooled numbers of individual fish are the following: n = 98, 58, 49, and 58 for PVP, C. albicans, P. aeruginosa, and C. albicans plus P. aeruginosa, respectively. A Kaplan-Meier survival analysis and log-rank (Mantel-Cox) test with Bonferroni correction demonstrated a significant reduction in survival. (C) Larvae were injected and monitored as described above with the following groups: PVP control, C. albicans (2.5 × 107 CFU/ml) plus UV-inactivated (UV-x) P. aeruginosa (2.5 × 108 CFU/ml), P. aeruginosa (2.5 × 108 CFU/ml) plus UV-x C. albicans (2.5 × 107 CFU/ml), or C. albicans plus P. aeruginosa at 2.5 × 107 CFU/ml and 2.5 × 108 CFU/ml, respectively. Data are representative of six pooled, independent experiments. Pooled numbers of individual fish are the following: n = 142, 86, 92, 33, and 96 for PVP, C. albicans, C. albicans plus UV-x P. aeruginosa, UV-x C. albicans plus P. aeruginosa, and C. albicans plus P. aeruginosa, respectively. A log-rank (Mantel-Cox) test with Bonferroni correction determined significant differences as indicated. Statistical significance was assigned based on GraphPad Prism conventions (not significant [n.s.], P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; adjusted in panels A and C with Bonferroni correction). The swimbladder is outlined in a dotted magenta line for clarity.
Immune responses to polymicrobial infection.
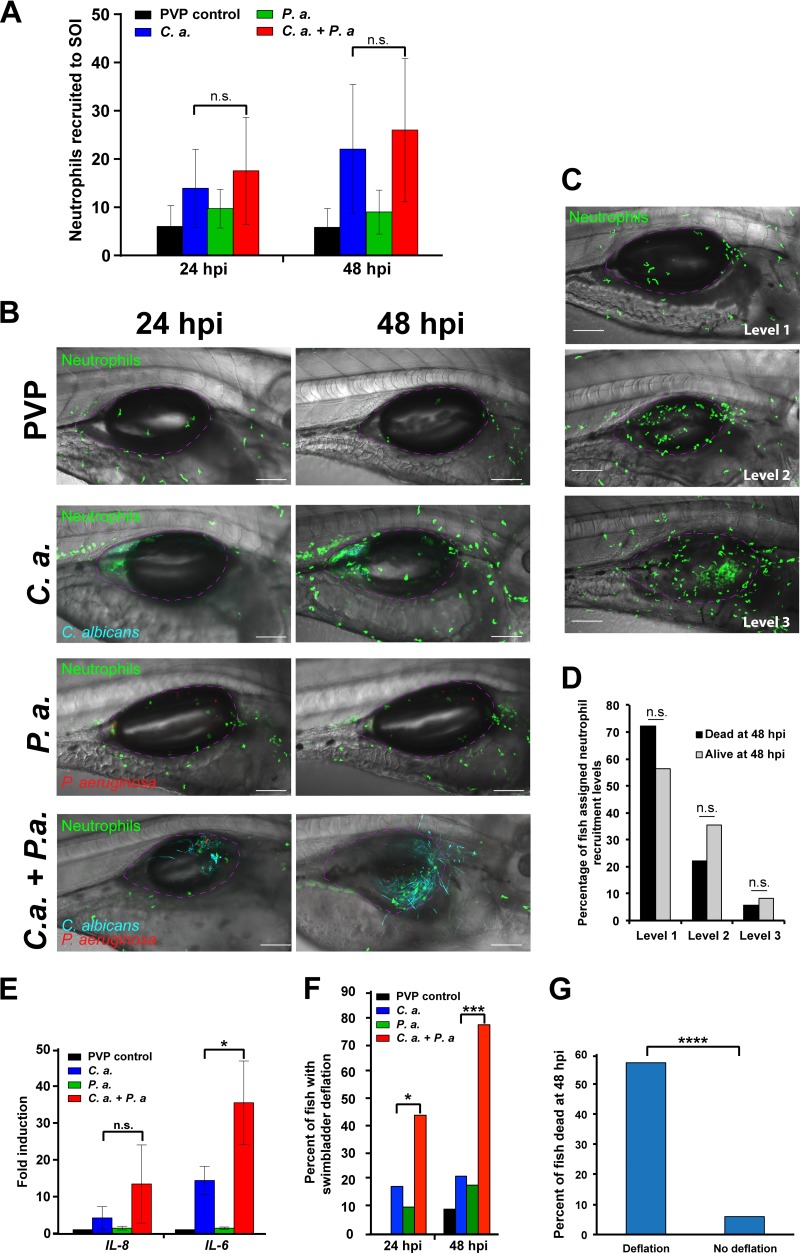
Enhanced virulence in coinfection can be due to changes in bacterial or fungal behavior, altered interactions with the immune system, or any combination of these components that comprise trikingdom interactions. Excessive inflammation, including immune infiltration, cytokine production, and edema, can enhance tissue damage and mortality in the host, so we utilized the Tg(Mpx:EGFP) fish line to determine whether coinfection amplifies neutrophilic recruitment. We quantified neutrophil migration to the site of infection in individual fish at 24 and 48 h postinjection (hpi) using confocal microscopy. In both C. albicans monoinfection and C. albicans plus P. aeruginosa coinfection, neutrophils were highly recruited to the site of infection, with a slight (but not significant [n.s.]) elevation associated with coinfection (Fig. 2A and B). C. albicans infection developed in either the posterior or anterior of the swimbladder, progressing around the air bubble in two-dimensional (2D) space, as detailed in Z-stack animations (see Movies S1 to S4, which are based on Fig. 2B). To determine whether neutrophil recruitment can serve as a predictive factor for mortality, individual fish were first imaged by confocal microscopy at 24 hpi. Z-stacks were blindly scored for neutrophil recruitment as level 1 (low), 2 (medium), or 3 (high), as illustrated in Fig. 2C. Individuals then were monitored for survival until 48 hpi. This analysis found no correlation between the neutrophil recruitment level and survival in individual fish (Fig. 2D). Thus, there is no excessive neutrophil infiltration in coinfection, and neutrophil recruitment is not a predictor for mortality.
FIG 2.
Coinfection stimulates immune infiltration. Tg(Mpx:EGFP) zebrafish larvae at 4 days postfertilization were separated into 4 groups and microinjected into the swimbladder with 5 nl of PVP (control), C. albicans at 2.5 × 107 CFU/ml, P. aeruginosa at 2.5 × 108 CFU/ml, or C. albicans plus P. aeruginosa at 2.5 × 107 CFU/ml and 2.5 × 108 CFU/ml, respectively. Fish were screened immediately postinjection to select for neutrophil fluorescence and consistent inocula. (A and B) Neutrophils at the site of injection (SOI) were qualitatively scored and blinded, and confocal images of representative fish at ×20 magnification were taken at 24 and 48 h postinjection (hpi). Scale bar, 100 μm. Z-stack animations of the 24-hpi and 48-hpi images of a fish infected with C. albicans only are included in the supplemental material as Movies S1 (24 hpi) and S2 (48 hpi), and animations of a fish infected with both C. albicans and P. aeruginosa are included as Movies S3 (24 hpi) and S4 (48 hpi). Data are pooled from 3 independent experiments. Total numbers of individual fish are the following: n = 21, 23, 27, and 18 for PVP, C. albicans, P. aeruginosa, and C. albicans plus P. aeruginosa, respectively. Analysis was conducted using one-way analysis of variance (ANOVA) with Tukey's multiple-comparisons posttest. (C and D) Fish were imaged at 24 hpi and individuals were monitored to 48 hpi to measure survival. (C) Confocal images acquired at 24 hpi were used to stratify fish based on neutrophil recruitment (level 1, low; level 2, medium; level 3, high; as demonstrated by representative confocal images). (D) No significant differences in 24-hpi neutrophil recruitment phenotype were found between fish that survived or died using Fisher's exact test. (E) Representative fish were homogenized at 48 hpi for isolation of total RNA followed by cDNA synthesis for qPCR analysis. Gene expression of IL-6 and IL-8 was normalized to that of gapdh, with PVP control used for the reference (ΔΔCT). Fold induction (2ΔΔCT) is represented. Total RNA was extracted from 3 independent experiments; total numbers were 22, 24, 28, and 21 larvae for PVP, C. albicans, P. aeruginosa, and C. albicans plus P. aeruginosa groups, respectively. A one-way ANOVA with Tukey's multiple-comparisons posttest revealed significantly higher IL-6 expression in the coinfection than the C. albicans monoinfection (P = 0.011). qPCR for both genes was replicated in triplicate. (F) Percentage of fish with swimbladder deflation at 24 and 48 hpi. A Fisher's exact test revealed a significantly higher percentage of fish with swimbladder deflation in the coinfection compared to the C. albicans monoinfection, as indicated; n = 28 fish for C. albicans infection and n = 27 fish for coinfection. (G) Confocal images acquired at 24 hpi were stratified based on the survival of individual fish at 48 hpi relative to swimbladder deflation, with n = 28 with deflation and n = 34 without deflation. According to a Fisher's exact test, fish that died by 48 hpi had significantly higher incidences of swimbladder deflation than those that survived, as indicated. Statistical significance was assigned based on GraphPad Prism convention (n.s., P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001). The swimbladder is outlined in a dotted magenta line for clarity.
Another key cause and indicator of pathogenic inflammation is proinflammatory cytokine and chemokine release. Therefore, we examined a number of well-known proinflammatory cytokines and chemokines associated with polymicrobial infection, including tumor necrosis factor alpha (TNF-α), IL-17, gamma interferon (IFN-γ), IL-8, and IL-6. Of the assessed proinflammatory mediators, we found infection-associated increases in the expression of both IL-8 and IL-6 (Fig. 2E; Fig. S3). The chemokine IL-8 is a potent neutrophil chemoattractant (45), and in coinfection its expression displayed a slight but not statistically significant elevation (Fig. 2E). The proinflammatory cytokine IL-6 is produced by several cell types and has been linked to edema, sepsis, and increased mortality in both monoinfections and C. albicans-S. aureus coinfection (22–24, 46). Interestingly, this proinflammatory cytokine was significantly upregulated in C. albicans-P. aeruginosa coinfection compared to the C. albicans monoinfection (Fig. 2E), suggesting that IL-6 proinflammatory gene expression contributes to increased mortality. However, since quantitative PCR (qPCR) is an endpoint assay, we were not able to determine if higher IL-6 production in individual fish predicted mortality.
Inflammation and IL-6 production are associated with edema, alveolar collapse, and acute injury in the lung, suggesting that loss of the air bubble in the swimbladder (deflation) serves as a noninvasive parallel indicator of inflammation and edema (33, 39, 47, 48). To longitudinally test association of this inflammatory phenotype with mortality, we compared the proportion of fish with swimbladder deflation in single infection and coinfections. At both 24 and 48 hpi, a significantly higher percentage of fish had swimbladder deflation in coinfection compared to monoinfection (Fig. 2F). To further assess the relationship between swimbladder deflation and mortality, individual fish were tracked and the incidence of deflation at 24 hpi was correlated to their survival at 48 hpi. Fish with deflation had a significantly lower survival rate than those with inflated swimbladders (Fig. 2G). Thus, swimbladder deflation is higher in coinfection and serves as an early predictive factor for mortality in the zebrafish. Together, these experiments link coinfection to enhanced inflammatory cytokine responses and suggest that local inflammation contributes to mortality in C. albicans-P. aeruginosa coinfection of the mucosa.
Coinfection morbidity does not result from high P. aeruginosa burden or dissemination.
Given that live P. aeruginosa organisms are required for enhanced mortality in coinfection, we aimed to further characterize the role of the bacteria in mediating virulence in C. albicans-P. aeruginosa infection. We took advantage of the zebrafish larva's small size and transparency to determine if bacteria leave the swimbladder and cause systemic disease. However, within the limits of detection, bacterial dissemination from the swimbladder to other parts of the zebrafish was never observed in more than 30 experiments (more than 300 coinfected fish). Given the strong fluorescence of the red fluorescent P. aeruginosa used and the success of others at quantifying bacterial dissemination in zebrafish larvae (49–51), our observations suggest that bacteremia is not causing mortality.
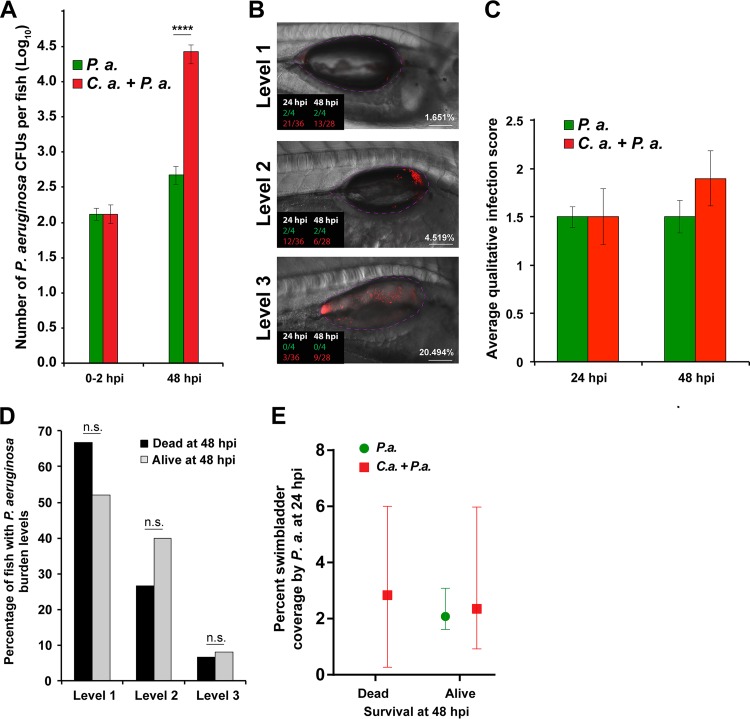
We then measured bacterial burden to determine if C. albicans enhances bacterial growth or retention in the swimbladder. To quantify bacterial burden in the infected fish, we conducted a CFU analysis by homogenizing representative fish from each group at 0 and 48 hpi and plating the homogenate on selective media. The initial P. aeruginosa load was the same for animals with P. aeruginosa alone and with combined P. aeruginosa and C. albicans, but there was a significantly higher P. aeruginosa burden at 48 hpi in coinfected fish (Fig. 3A). To investigate the correlation between bacterial burden and death, P. aeruginosa infection levels were scored in individual fish as high (level 3), medium (level 2), or low (level 1) burden via confocal microscopy at 24 hpi (Fig. 3B and C). Based on these assigned infection levels, there was no statistical difference in bacterial burden between single and coinfections (Fig. 3C). When the infection level at 24 hpi was mapped to survival in individual fish at 48 hpi, no correlation was observed between the level of P. aeruginosa at 24 hpi and survival of the fish at 48 hpi (Fig. 3B and D). P. aeruginosa burden was then quantitatively analyzed by microscopy to determine the percent swimbladder coverage by the bacteria using ImageJ software to calculate bacterial fluorescence in the outlined swimbladder area. Swimbladder coverage was further analyzed relative to fish survival, and we found that there is no significant correlation between bacterial burden as a measure of swimbladder coverage at 24 hpi and probability of death by 48 hpi (Fig. 3E). Taken together, these data indicate that although levels of P. aeruginosa are elevated with the coinfection, within the population of coinfected fish there is no relationship between elevated bacterial burden and mortality.
FIG 3.
P. aeruginosa burden does not directly contribute to mortality. Tg(Mpx:EGFP) zebrafish larvae were infected and screened as previously described. Three to 4 representative fish per group were subsequently homogenized for quantifying CFU of P. aeruginosa by using selective media, with the remaining fish monitored out to 48 hpi. Representative fish were imaged at 24 and 48 hpi via confocal microscopy. (A) Data are representative of 6 pooled, independent experiments. A total of 22 individual fish were homogenized per time point (0 hpi and 48 hpi) for CFU quantification and plotted on a log10 scale. Student's t test demonstrated a significant increase in the number of P. aeruginosa organisms between mono- and coinfection groups at 48 hpi, as indicated. (B, C, and D) Confocal images acquired at 24 hpi were blinded and qualitatively scored based on Pseudomonas burden (level 1, low; level 2, medium; level 3, high), and then individuals were monitored to determine their survival at 48 hpi. Total numbers of fish analyzed were 15 dead and 25 alive at 48 hpi. (B) Representative images of each level of burden. Fractions in the lower left corner indicate the number of fish of a given phenotype that were scored at a given time point postinfection (green, P. aeruginosa monoinfection; red, P. aeruginosa plus C. albicans). Percentage shown at the lower right is from ImageJ quantification of burden, which correlates well with blinded qualitative scoring. (C) No significant differences in average infection level were observed between mono- and coinfected fish based on qualitative scoring of Pseudomonas burden according to Student's t test. (D) No significant differences were found in the percentages of fish with different qualitatively scored levels of P. aeruginosa burden between mono- and coinfected fish, as tested by Fisher's exact test. (E) Percent swimbladder coverage by P. aeruginosa was quantified via ImageJ analysis of microbial fluorescence from confocal images acquired for representative fish at 24 hpi. The calculated percent coverage from this analysis is also shown in panel B. For fish that died at 48 hpi, n = 11 for C. albicans plus P. aeruginosa; for fish that lived at 48 hpi, n = 4 for P. aeruginosa and n = 22 for C. albicans plus P. aeruginosa. No differences were observed using an unpaired Mann-Whitney test. Confocal images were acquired at ×20 magnification. Scale bar, 100 μm. Statistical significance was assigned based on GraphPad Prism convention (n.s., P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001). The swimbladder is outlined in a dotted magenta line for clarity.
Coinfection enhances C. albicans virulence in the swimbladder.
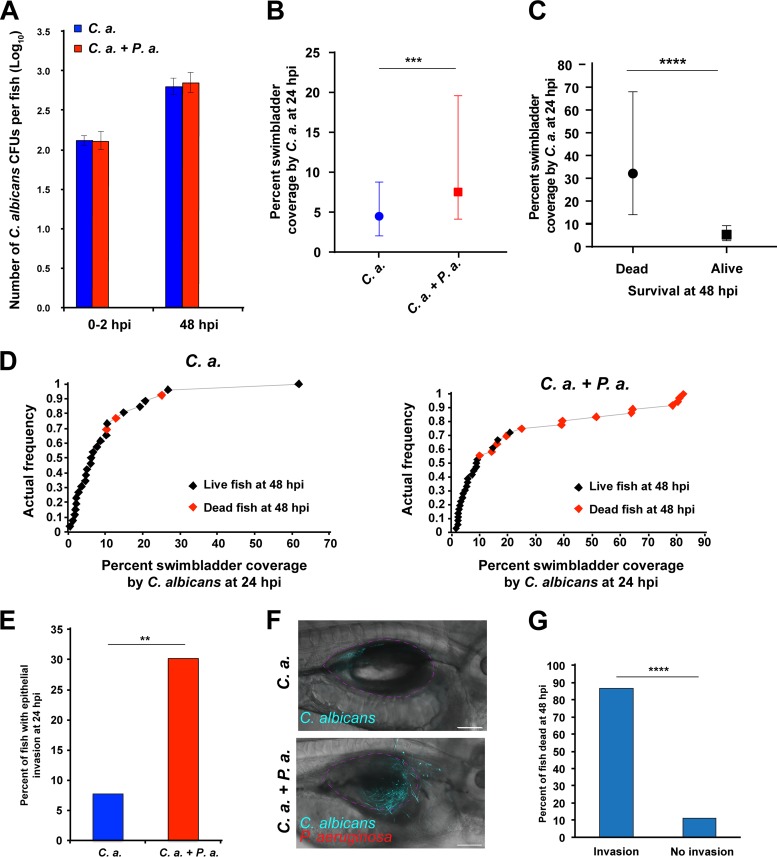
Since bacterial pathogenesis was not directly associated with enhanced mortality, we investigated whether C. albicans plays a more direct role in the synergistic virulence of coinfection. C. albicans burden was quantified by conducting a CFU assay, as previously described for P. aeruginosa. This analysis revealed no differences in C. albicans burden between the single infection and coinfection at 48 hpi (Fig. 4A). However, C. albicans filaments risk being underdetected by CFU assays due to strong adhesion and cell-cell clustering (52, 53). Therefore, fungal burden was also measured as the percent coverage of the swimbladder area by C. albicans using ImageJ software, as described above for P. aeruginosa. At 24 hpi, we observed significantly higher swimbladder coverage by C. albicans in the coinfection compared to that in the C. albicans monoinfection (Fig. 4B). To determine if this was associated with a change in dimorphism or hyphal gene expression, we performed infections with C. albicans expressing a protein fusion to the hypha-specific Hwp1p protein, with or without P. aeruginosa. Quantification of fungal morphology revealed that there was no difference in the relative yeast/hypha ratio, while the moderate increase in median fungal burden is consistent with quantification of live fish (Fig. S4A and B). No differences in Hwp1-EGFP levels were noted between mono- and coinfection conditions, although green autofluorescence prevented quantification of Hwp1-EGFP levels (Fig. S4C).
FIG 4.
Coinfection enhances C. albicans pathogenicity. Tg(Mpx:EGFP) zebrafish larvae infected and screened as previously described and 3 to 4 random fish per group were subsequently homogenized for quantifying CFU of C. albicans by using selective media, with the remaining fish monitored out to 48 hpi. Random fish were imaged at 24 and 48 hpi via confocal microscopy, and another 3 to 4 fish per group at 48 hpi were homogenized to quantify CFU as described above. Data are representative of 6 to 8 pooled, independent experiments. (A) A total of 22 individual fish were homogenized per time point for CFU quantification and plotted on a log10 scale. There was no significant difference in C. albicans CFU between single infection and coinfection according to Student's t test. (B) Median percent coverage of the swimbladder by C. albicans was quantified via ImageJ analysis of confocal images acquired of representative fish at 24 hpi, with n = 55 and 63 for C. albicans and C. albicans plus P. aeruginosa infections, respectively. An unpaired Mann-Whitney test revealed a significant difference between C. albicans single infection and coinfection, as indicated. (C) Confocal images acquired at 24 hpi were stratified based on the survival of individual fish at 48 hpi relative to the median percent swimbladder coverage by C. albicans, with n = 18 dead and n = 44 alive. Scale bar, 100 μm. According to an unpaired Mann-Whitney test, fish that died by 48 hpi had significantly higher swimbladder coverage by C. albicans than those that survived, as indicated. (D) Cumulative frequency distribution plots representing percent swimbladder coverage by C. albicans at 24 hpi. Fish that died at 48 hpi are indicated by red points; for C. albicans, n = 26; for C. albicans plus P. aeruginosa, n = 36. (E) Percentage of fish with epithelial invasion at 24 hpi; for C. albicans, n = 52; for C. albicans plus P. aeruginosa, n = 63. According to a Fisher's exact test, there was a significant difference between C. albicans and C. albicans plus P. aeruginosa infection, as indicated. (F and G) Confocal images acquired at 24 hpi were quantified by ImageJ and stratified based on the survival of individual fish at 48 hpi relative to epithelial invasion by C. albicans, with n = 15 with invasion and n = 45 without invasion. According to a Fisher's exact test, fish that died by 48 hpi had significantly higher incidences of C. albicans invasion than those that survived, as indicated. Statistical significance was assigned based on GraphPad Prism conventions (n.s., P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001). The swimbladder is outlined in a dotted magenta line for clarity.
To further assess the impact of swimbladder coverage by C. albicans on mortality, the fish were categorized based on their survival at 48 hpi (Fig. 4C). Fish that died at 48 hpi had significantly more C. albicans swimbladder coverage at 24 hpi than fish that survived to 48 hpi (Fig. 4C). This difference is consistent with cumulative plots showing that higher swimbladder coverage at 24 hpi by C. albicans correlates to death at 48 hpi (Fig. 4D). Therefore, there is greater swimbladder coverage by C. albicans at 24 hpi in the coinfection and higher early burden is associated with mortality.
The observation that C. albicans burden is more closely associated with mortality than immune recruitment or bacterial burden suggests that fungal pathogenesis underlies exacerbated mortality in coinfection. Given that a well-known virulence factor of C. albicans is hyphal penetration of the host epithelium causing tissue damage (7, 40), we assayed fungal epithelial invasion of the swimbladder. Overall, there were significantly more fish with invasive C. albicans filaments penetrating the epithelium at 24 hpi in the coinfection than in the monoinfection (Fig. 4E and F). Furthermore, fish with epithelial invasion at 24 hpi showed significantly lower survival to 48 hpi (Fig. 4G). Therefore, C. albicans epithelial invasion may serve as an additional early predictive factor for mortality, occurring more frequently during coinfection. Taken together, these data implicate C. albicans pathogenesis as a risk factor in coinfection, as indicated by associations of both elevated C. albicans burden and epithelial invasion with mortality.
Because both swimbladder collapse, indicative of immunopathology, and fungal invasion, indicative of fungal virulence, were independent predictors of mortality, we sought to determine if there is a relationship between these two events. We analyzed the cooccurrence of swimbladder collapse and fungal invasion at 24 and 48 hpi and scored changes in each over this time. Remarkably, we found that invasion only occurred in fish with deflated swimbladders, suggesting that deflation precedes invasion (Fig. S5A). Further, we found that infections in fish with deflation but no invasion progressed to deflation plus invasion between 24 and 48 hpi (Fig. S5B). Taken together, these data indicate that swimbladder deflation precedes fungal invasion, although they do not establish a cause-and-effect relationship.
P. aeruginosa quorum-sensing-deficient ΔlasR mutant enhances mortality and C. albicans pathogenicity in coinfection.
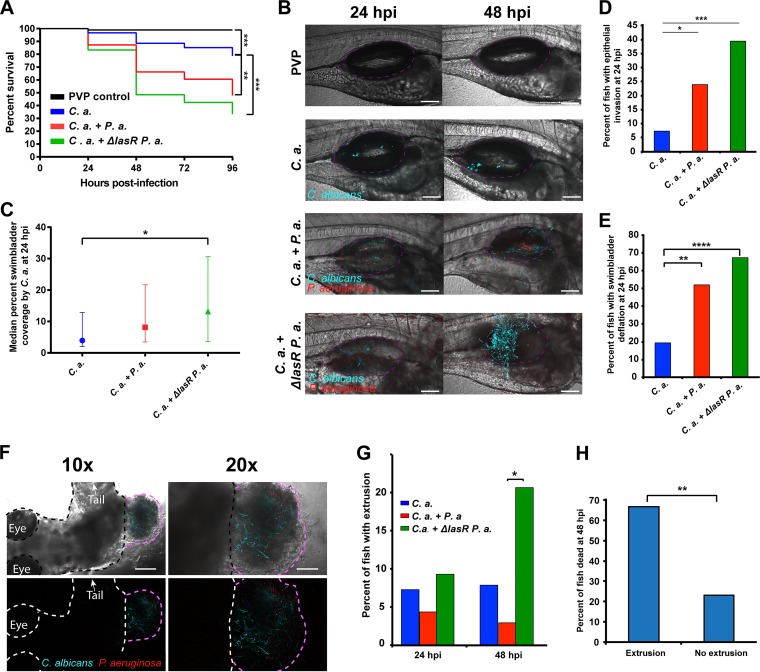
Our data suggest that C. albicans virulence is enhanced when combined with live P. aeruginosa, so we sought to further investigate how P. aeruginosa modulates fungal pathogenesis. In vitro, P. aeruginosa-produced 3OC12HSL can suppress C. albicans virulence in the context of a mixed-species biofilm through the lasR-controlled production of quorum-sensing molecules (18, 54, 55). In contrast, the production of toxic phenazines is strongly stimulated by C. albicans in lasR mutant strains through activation of downstream components of the quorum-sensing pathway (19). Therefore, the role of P. aeruginosa quorum sensing on C. albicans-P. aeruginosa coinfection was assessed using fluorescent derivatives of a lasR-defective (ΔlasR) P. aeruginosa strain (16) and its parental PA14 wild-type strain. First, we analyzed mortality in coinfections of C. albicans and either the ΔlasR mutant or wild-type parental PA14 strain. As previously observed, the combination of C. albicans and wild-type P. aeruginosa exacerbated mortality compared to that of the C. albicans-only infection (Fig. 5A and B). Unexpectedly, the combination of C. albicans and the P. aeruginosa ΔlasR mutant showed mortality similar to and, if anything, slightly greater than that of the coinfection with the wild-type strain (Fig. 5A and B). Overall, there was no statistically significant difference in survival between the two coinfection groups (wild type versus ΔlasR mutant). This suggests that the enhanced virulence in coinfections does not depend on lasR-mediated signaling in P. aeruginosa, but loss of lasR may even further enhance pathogenesis.
FIG 5.
P. aeruginosa quorum-sensing-deficient ΔlasR mutant enhances C. albicans pathogenicity in coinfection. Tg(Mpx:EGFP) zebrafish larvae at 4 days postfertilization were separated into four groups and microinjected into the swimbladder with 5 nl of PVP (control), C. albicans at 2.5 × 107 CFU/ml, C. albicans plus P. aeruginosa at 2.5 × 107 CFU/ml and 2.5 × 108 CFU/ml, respectively, or C. albicans plus ΔlasR mutant P. aeruginosa at 2.5 × 107 CFU/ml and 2.5 × 108 CFU/ml, respectively. Fish were screened immediately postinjection to select for neutrophil fluorescence and consistent inocula. Mortality was recorded every 24 h out to 96 hpi, and representative fish were imaged at 24 and 48 hpi via confocal microscopy. Data are representative of 5 to 8 pooled, independent experiments. (A and B) Kaplan-Meier survival analysis and representative images. Pooled numbers of individual fish are the following: n = 96, 61, 71, and 66 for PVP, C. albicans, C. albicans plus P. aeruginosa, and C. albicans plus ΔlasR mutant P. aeruginosa, respectively. A log-rank (Mantel-Cox) test with Bonferroni correction demonstrated a significant reduction in survival between C. albicans and C. albicans plus P. aeruginosa and between C. albicans and C. albicans plus ΔlasR mutant P. aeruginosa, as indicated. Images were acquired at ×20 magnification; scale bar, 100 μm. (C) Median percent coverage of the swimbladder by C. albicans was quantified via ImageJ analysis of confocal images acquired of representative fish at 24 hpi, with n = 41, 44, and 44 for C. albicans, C. albicans plus P. aeruginosa, and C. albicans plus ΔlasR mutant P. aeruginosa, respectively. According to a Kruskal-Wallis test, there was a significant difference between C. albicans and C. albicans plus ΔlasR mutant P. aeruginosa infections, as indicated. (D) Percentage of fish with epithelial invasion at 24 hpi: C. albicans, n = 41; C. albicans plus P. aeruginosa, n = 46; C. albicans plus ΔlasR mutant P. aeruginosa, n = 43. According to a Fisher's exact test, there was a significant difference between C. albicans and C. albicans plus P. aeruginosa infections and between C. albicans and C. albicans plus ΔlasR mutant P. aeruginosa, as indicated. (E) Percentage of fish with swimbladder deflation at 24 hpi. According to a Fisher's exact test, there was a significantly higher percentage of fish with swimbladder deflation in the coinfection compared to the C. albicans monoinfection and a significant difference between C. albicans and C. albicans plus ΔlasR mutant P. aeruginosa, as indicated. The numbers of fish were 41, 46, and 43 for C. albicans, C. albicans plus P. aeruginosa, and C. albicans plus ΔlasR mutant P. aeruginosa, respectively. (F and G) Extrusion events were quantified in confocal images. (F) Representative low- and high-power images of an extrusion event. The fish is outlined in black/white, and the extruded infected tissue is outlined in pink. Scale bars are 200 μm for the 10× images (left) and 100 μm for the ×20 images (right) (G) According to a Fisher's exact analysis, extrusion events at 48 hpi are of significantly higher frequency in C. albicans plus ΔlasR mutant P. aeruginosa than in C. albicans infection, as indicated; for 24 hpi, n = 41, 46, and 43 for C. albicans, C. albicans plus P. aeruginosa, and C. albicans plus ΔlasR mutant P. aeruginosa, respectively; for 48 hpi, n = 38, 34, and 29 for C. albicans, C. albicans plus P. aeruginosa, and C. albicans plus ΔlasR mutant P. aeruginosa, respectively. (H) Confocal images acquired at 24 hpi were quantified by ImageJ and stratified based on the survival of individual fish at 48 hpi relative to extrusion events; n = 9 with extrusion and n = 121 without extrusion. According to a Fisher's exact test, fish that died by 48 hpi had significantly higher incidence of extrusion events than those that survived, as indicated. Statistical significance was assigned based on GraphPad Prism conventions (n.s., P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; adjusted with Bonferroni correction for panel A). The swimbladder is outlined throughout in a dotted magenta line for clarity.
To determine if disease progression was altered in ΔlasR mutant coinfections, we performed the same longitudinal analyses to evaluate bacterial, fungal, or immune function in the enhanced virulence of ΔlasR mutant coinfections. CFU analysis revealed slightly elevated levels of P. aeruginosa over time in the wild-type coinfection, but there was no increase over time in the mutant coinfection (Fig. S6A), suggesting that bacterial burden is even less likely to contribute to enhanced mortality in the ΔlasR mutant coinfections. Bacterial dissemination was not seen in these coinfections, as was the case with the wild-type PA14-C. albicans coinfections.
CFU analysis of fungal burden revealed that C. albicans levels were slightly elevated over time in all cohorts, but there were no significant differences between groups (Fig. S6B). A higher percent swimbladder coverage by C. albicans was observed at 24 hpi in both coinfection groups and, if anything, a higher degree in coinfections with the ΔlasR mutant strain (Fig. 5C). Furthermore, higher C. albicans swimbladder coverage correlated with mortality (Fig. S6C and D). Therefore, we conclude that lasR-deficient P. aeruginosa can enhance C. albicans burden, which is associated with death. Both C. albicans epithelial invasion and swimbladder deflation were enhanced with the coinfection and, if anything, there was even greater enhancement with the ΔlasR mutant P. aeruginosa (Fig. 5D and E). Furthermore, these events correlated with higher mortality, as previously observed (Fig. S6E and F). Overall, these data suggest that ΔlasR mutant-enhanced virulence in C. albicans coinfection is associated with a set of morbidity traits similar to those already linked to wild-type PA14-enhanced virulence.
A dramatic indicator of enhanced pathogenicity that was pronounced with the combination of C. albicans and ΔlasR mutant P. aeruginosa was the occurrence of extrusion events in which infected tissue containing both C. albicans and P. aeruginosa protruded from the side of the fish. To measure these events, confocal images acquired at 24 and 48 hpi were analyzed in 3D stacks (Fig. 5F). Interestingly, swimbladder extrusion was observed at a significantly higher frequency at 48 hpi with the combination of C. albicans and ΔlasR mutant P. aeruginosa (Fig. 5G), and early extrusion events were linked to higher mortality (Fig. 5H). Altogether, these data suggest that loss of P. aeruginosa lasR functionality further stimulates C. albicans pathogenicity during coinfection, as indicated by severe fungal burden and invasion, swimbladder deflation, and dramatic host epithelial extrusion.
DISCUSSION
Coinfections with C. albicans and P. aeruginosa threaten a variety of patients, including those with mucosal infections that occur in the context of underlying pulmonary disease (2, 3). Using our newly developed zebrafish swimbladder model, we showed that polymicrobial infection of the mucosa with C. albicans and P. aeruginosa results in synergistic virulence that is closely associated with increased C. albicans pathogenesis and enhanced inflammation. Overall, the high-content longitudinal imaging in our study clearly implicates the fungus and host in coinfection-associated mortality while providing arguments against bacterial dissemination and disease. In the context of coinfection, we find that C. albicans burden, fungal epithelial invasion, edema, and IL-6 expression are all associated with the synergistic virulence of C. albicans-P. aeruginosa coinfection, while neutrophil recruitment and P. aeruginosa burden are not quantitative predictors of infection dynamics. Furthermore, we demonstrated how the loss of LasR activity in P. aeruginosa may further stimulate virulence, perhaps due to the absence of 3OC12HSL production, which can suppress hyphal growth (16), combined with enhanced virulence factor production in the presence of C. albicans (19).
The synergistic virulence and enhanced filamentous invasion of C. albicans in coinfections were unexpected based on known antagonistic interactions of the two microbes in vitro (1). However, the enhanced virulence is consistent with most mouse C. albicans-bacterial coinfection models, including organotypic models, which result in enhanced virulence, cytokine production, and/or fungal invasion (22, 23, 26, 27, 56–58). The fact that the same strains of C. albicans and P. aeruginosa were used in these in vivo experiments as have been used in vitro suggests that the switch from antagonism to synergy is due to the host environment. There is high diversity among both C. albicans and P. aeruginosa clinical isolates, and a limitation of the current study is that it does not address the important question of whether there are differences among clinical isolates that regulate synergistic virulence of the two pathogens. Many environmental factors are both different and dynamic between in vitro and in vivo conditions, including nutrient levels, microbe-substrate interactions, cell-cell interactions, secreted products from the host, immune pressure, and biophysical effects of the three-dimensional environment (1).
Our quantitative longitudinal analysis of infection phenotypes revealed that C. albicans burden and epithelial invasion are most closely linked to enhanced morbidity in the coinfected swimbladder. Similar enhancement of fungal burden and invasion is seen in murine C. albicans-bacterial coinfection and epithelial disease models (56–60). This suggests that bacteria stimulate C. albicans virulence directly and/or indirectly through modulation of host activity. Increases in fungal burden and filamentous invasion of the epithelium suggest the former, while enhanced IL-6 levels and swimbladder edema in coinfected fish are consistent with the latter. Given the links between epithelial damage and inflammation, it is possible that epithelial-immune cross talk also plays an important role in the greater pathology of coinfection.
The enhanced proinflammatory cytokine induction and swimbladder deflation we observe in coinfections suggests that immunopathology plays a role in enhancing coinfection mortality. The inflammatory role of IL-6 in immunopathology is well established, including its association with lethal C. albicans-bacterial intraperitoneal infections (22, 23, 61), but it is not known if this cytokine's action is necessary or sufficient to enhance C. albicans pathogenesis. Mucosal polymicrobial infections in humans stimulate immune cell infiltration, pulmonary edema, acute lung injury, and alveolar collapse, all phenotypes that are associated with severe morbidity and mortality (33, 62–65). The parallels between these pathologies and what we have observed in the swimbladder are striking and are consistent with the idea that immunopathology is a factor in enhanced fungal virulence of swimbladder coinfection. In fact, further analyses of our longitudinal data indicate that fungal invasion only occurs in fish with deflated swimbladders. Although this does not establish a causal relationship, this finding is consistent with the idea that immunopathology precedes fungal invasion.
Although P. aeruginosa burden was higher in coinfections with wild-type bacteria, bacterial burden did not correlate with mortality risk and bacterial dissemination was never observed. Furthermore, bacterial burden was not higher in ΔlasR mutant coinfections, despite the strong ability of this mutant to enhance mortality. Increased bacterial burden and hematogenous spread were not seen, although both are hallmarks of mortality in P. aeruginosa monoinfections of mice and humans (66, 67). This disconnect suggests that P. aeruginosa is playing a different, indirect role in enhancing mortality in the context of this coinfection model. Taken together, our results indicate that C. albicans enhances the ability of P. aeruginosa to colonize the site of infection without enhancing bacterial virulence, or that coinfection requires only a low threshold for P. aeruginosa burden to enhance mortality, either of which would be consistent with the LasR-independent ability of P. aeruginosa to promote virulence in our model. Depending on the context, infections with LasR-defective P. aeruginosa can be more virulent in mice and humans (68–72). Interestingly, coculture of ΔlasR mutants with C. albicans restores some bacterial virulence gene expression, suggesting that interactions between these two common human commensals enhance pathogenesis (19).
In summary, this zebrafish mucosal infection model offers high-resolution longitudinal analysis as a powerful new tool to disentangle the contributions of the immune system and coinfecting bacteria and fungi. The power of this technique is illustrated by the ability to demonstrate that P. aeruginosa induces changes that lead to enhanced epithelial invasion by C. albicans and death of individual host animals. The unexpected finding that P. aeruginosa has a positive influence on C. albicans virulence in vivo raises questions about what aspects of infection change this bacterial-fungal cross talk from something antagonistic in vitro into a positive interaction in vivo. With new clinical studies indicating that fungi exacerbate bacterial lung infection, which can be ameliorated with antifungal treatment, the importance of understanding bacterium-fungus-host interactions in vivo is critical (2, 3, 24). Hopefully, new in vivo tools and models such as the one described in this study will result in a more complete picture of these trikingdom interactions in disease.
MATERIALS AND METHODS
Zebrafish care and maintenance.
Adult zebrafish used for breeding embryos were housed in recirculating systems (Aquatic Habitats, Apopka, FL) at the University of Maine Zebrafish Facility. All zebrafish care protocols and experiments were performed in accordance with NIH guidelines under Institutional Animal Care and Use Committee (IACUC) protocol A2015-11-03. Larvae were reared at a density of 150/dish in 150-mm petri dishes containing 150 ml of E3 (5 mM sodium chloride, 0.174 mM potassium chloride, 0.33 mM calcium chloride, 0.332 mM magnesium sulfate, 2 mM HEPES in Nanopure water, pH 7) supplemented with 0.02 mg/ml of 1-phenyl-2-thiourea (PTU) (Sigma-Aldrich, St. Louis, MO) to prevent pigmentation, as well as 0.3 mg/liter methylene blue (VWR, Radnor, PA) for the first 24 h to prevent microbial growth. Larvae were manually dechorionated at 24 h postfertilization, transferred into media containing E3 and PTU, and incubated at 33°C over the course of experiments. This temperature was chosen as the highest safe temperature for zebrafish health and is regularly used for experiments with temperature-sensitive alleles. Experiments were conducted using wild-type (AB) zebrafish and Tg(Mpx:EGFP) (73) transgenic fish expressing enhanced green fluorescent protein in neutrophils.
Strains and growth conditions.
The Caf2-FR C. albicans strain was constructed by transforming the Caf2-1 strain (74) with the pENO1-iRFP-NATr plasmid. The pENO1-iRFP-NATr plasmid contains a codon-optimized version of the iRFP670 gene (75) under the control of the constitutive ENO1 promoter with a nourseothricin resistance marker (NATr). Codon-optimized iRFP670 was ordered from GenScript (see Fig. S7 in the supplemental material) and was digested with NcoI and PacI and cloned into Peno1-dTomato (39). After NotI digestion of the plasmid, C. albicans transformation was performed using the lithium acetate protocol previously published (76) with nourseothricin resistance as a selection marker (100 μg/ml NAT; Werner Bioagents, Jena, Germany). At least 10 colonies were selected and screened for fluorescence via epifluorescence microscopy and flow cytometry (640-nm excitation laser, 655-nm to 685-nm emission filter) on an LSRII cytometer (Becton Dickinson, Franklin Lakes, NJ). Correct integration of the pENO1-iRFP-NATr plasmid was confirmed using primers PENO1 Fw (5′-TCCTTGGCTGGCACTGAACTCG-3′) and iRFP Rv (5′-ATCACATGAAGTCAAATCAACTTTTCTAGC-3′).
The HWP1-EGFP-FR strain was constructed by first transforming HWP1-EGFP (77) with CIp30 (78, 79) to complement the uracil auxotrophy. This integration was confirmed with primers CIp10-IS (5′-GATATCGAATTCACGCGTAG) and RP10-G (5′-GTACATTCCTACTCCGTTCG). The strain next was transformed, screened, and verified for iRFP integration as described above for the CAF2-FR strain.
C. albicans was grown on yeast-peptone-dextrose (YPD) agar (20 g/liter peptone, 10 g/liter yeast extract, 20 g/liter glucose, 2% agar; Difco, Livonia, MI) for 48 h on solid media at 30°C. For infections, liquid cultures of C. albicans were grown by rotating overnight in YPD at 30°C. Overnight cultures were washed twice and resuspended in calcium- and magnesium-free phosphate-buffered saline (PBS; Lonza, Walkersville, MD). Due to the inability of the human eye to detect near-infrared fluorescent protein fluorescence, C. albicans was stained with the amine-reactive green fluorophore fluorescein-5-isothiocyanate (FITC) to enhance inoculum quantification by microscopy (1.67 mg/ml; Molecular Probes, Eugene, OR) in PBS with sodium bicarbonate (0.037 M final concentration, pH 8.2), incubating C. albicans in the dark for 1 h with occasional vortexing. Alternatively, in HWP1-GFP-FR experiments C. albicans was stained for 5 min with calcofluor white (Sigma-Aldrich, St. Louis, MO) at a final concentration of 750 μg/ml and then rinsed once with PBS. This was conducted to visualize the inoculum by eye without using FITC, which would have overlapped with the HWP1-GFP fluorescence. The culture was then washed four times in PBS and resuspended in PBS. Using a hemocytometer, C. albicans concentration was measured and adjusted to 2.5 × 107 CFU/ml and 5 × 107 CFU/ml for mono- and coinfection (varied by experiment), respectively, in PBS with 5%, wt/vol, polyvinylpyrrolidone (PVP) (Sigma-Aldrich, St. Louis, MO) to ensure a consistent small-volume injection dose with large particles, such as yeast.
A red fluorescent protein-expressing derivative of Pseudomonas aeruginosa clinical isolate strain PA14 (PA14-dTom [80]) or the PA14-derived ΔlasR (16), transformed with the same plasmid, was grown on LB media (10 g/liter Bacto tryptone, 5 g/liter Bacto yeast extract, 10 g/liter sodium chloride, 1.2% agar; BD, San Jose, CA) supplemented with 750 μg/ml ampicillin (EMD Millipore, Billerica, MA) overnight at 37°C. For infections, liquid cultures of P. aeruginosa were grown by rotating overnight in LB-Amp (750 μg/ml) at 37°C. Overnight cultures were washed twice and resuspended in PBS. P. aeruginosa concentration was determined using a spectrophotometer to measure the optical density at 600 nm (OD600) of culture and adjusted to 2.5 × 108 CFU/ml and 5 × 108 CFU/ml for mono- and coinfections, respectively, in 5%, wt/vol, PVP.
UV inactivation of cultures.
Caf2-FR C. albicans and PA14-dTom liquid cultures were grown overnight, washed, and resuspended in PBS, and concentrations for C. albicans and P. aeruginosa were calculated as described above. A volume of 2 ml from each overnight culture was taken to UV inactivate 2.5 × 107 CFU/ml of C. albicans and 1 × 109 CFU/ml of P. aeruginosa, placed in uncovered 100-mm by 15-mm polystyrene petri dishes (Fisher Scientific, Waltham, MA), and exposed to 100,000 μJ/cm2 four times using a CL-1000 UV cross-linker (UVP, Vernon Hills, IL), swirling cultures between exposures. After UV inactivation, cultures were stained with FITC as described above and adjusted to 5 × 107 CFU/ml and 5 × 108 CFU/ml for C. albicans and P. aeruginosa, respectively, to be mixed prior to swimbladder injection. UV inactivation was confirmed by plating C. albicans and P. aeruginosa inactivated and live liquid cultures on YPD and LB-Amp (750 μg/ml), respectively (including PBS control), to confirm lack of growth.
Swimbladder infections via microinjection.
At 4 days postfertilization, zebrafish larvae were anesthetized in Tris-buffered tricaine methane sulfonate (160 μg/ml; Tricaine; Western Chemicals, Inc., Ferndale, WA) and selected for swimbladder inflation. Larvae were transferred to E3 containing PTU and 0.033%, vol/vol, dimethyl sulfoxide (DMSO; Fisher Bioreagents, Pittsburgh, PA) to allow future experiments to be conducted using dexamethasone in DMSO vehicle for repetition in an immunocompromised model. Fish were microinjected as previously described (38) with a 5-nl volume of PVP control, C. albicans at 2.5 × 107 CFU/ml, P. aeruginosa at 2.5 ×108 CFU/ml, or a C. albicans-P. aeruginosa mixture at 2.5 × 107 CFU/ml and 2.5 ×108 CFU/ml, respectively. The C. albicans-P. aeruginosa coculture was prepared by combining equal volumes of C. albicans at 5 × 107 CFU/ml and P. aeruginosa at 5 × 108 CFU/ml prior to injection. Within 1 h of injection, larvae were screened and selected to ensure proper inocula and neutrophil fluorescence using a Zeiss Axio Observer Z1 microscope equipped with a Vivatome system (Carl Zeiss Microimaging, Thornwood, NJ). For mortality experiments, fish were kept at 33°C in E3 containing PTU out to 96 h postinjection and then euthanized by Tricaine overdose.
Confocal laser scanning fluorescence microscopy.
At 24 and 48 h postinjection, larvae were anesthetized in Tricaine and immobilized in 0.5% low-melting-point agarose (Lonza, Switzerland) in E3 containing Tricaine in a 96-well glass-bottom imaging dish (Greiner Bio-One, Monroe, NC). Confocal images were acquired using an Olympus IX-81 inverted microscope with an FV-1000 laser scanning confocal system (Olympus, Waltham, MA). The EGFP, dTomato, and Far-Red fluorescent proteins were detected by laser/optical filters with a 10× (numeric aperture [NA], 0.4), 20× (NA, 0.7), or 40× objective (NA, 0.9) for excitation/emission at 488 nm/505 to 525 nm, 543 nm/560 to 620 nm, and 635 nm/655 to 755 nm, respectively. Z-stacks of 10 to 25 slices, with an interslice interval between 7 and 13 μm, were collected and processed using FluoView (Olympus, Waltham, MA), Photoshop (Adobe Systems, Inc., San Jose, CA), and ImageJ (81). For scoring swimbladder deflation, >90% of all fish had full inflation or full deflation. Any with a smaller than usual air bubble were still scored as inflated to avoid any ambiguity. For neutrophil quantification, neutrophils recruited to the swimbladder were quantified as the average between the number counted by eye via epifluorescence and the number counted by blindly scoring z-stacks from the same fish. For burden quantification, acquisition parameters such as laser power, photomultiplier voltage, and dwell time were consistent for all images collected.
Morphology quantification.
Zebrafish infected with either HWP1-GFP-FR C. albicans or HWP1-GFP-FR C. albicans and PA14-dTomato P. aeruginosa were euthanized at 24 hpi, and random fish were mounted onto microscope slides using 0.5% low-melting-point agarose and flattened under a coverslip. Flattened fish were imaged by epifluorescence microscopy, essentially as described previously (82). Briefly, images were acquired, filaments and yeast were each manually outlined in Photoshop, and the areas of each were measured in ImageJ.
RNA isolation and qPCR analysis.
Total RNA was isolated from 6 to 10 whole larvae per group (all survivors at the time point) in three independent experiments, using a combination of TRIzol (Invitrogen, Carlsbad, CA) and Direct-zol RNA miniprep kit (Zymo Research, Irvine, CA). Briefly, the Direct-zol RNA isolation protocol was followed and the TRIzol-ethanol mixture containing RNA was transferred to a Zymo-Spin IIC column, followed by the manufacturer's recommended wash steps. Total RNA was eluted in 25 μl of nuclease-free water and stored at −80°C. cDNA was synthesized from 500 ng of RNA per sample using iScript reverse transcription (RT) supermix for RT-qPCR (Bio-Rad, Hercules, CA). qPCR primers used are shown in Table 1. A CFX96 thermocycler (Bio-Rad, Hercules, CA) was used under the following conditions: 95°C for 35 s, followed by 40 cycles of 95°C for 5 s and 60°C for 20 s; the final step was 95°C for 10 s followed by 65°C for 5 s and included a dissociation curve. Threshold cycles (CT) and dissociation curves were analyzed with Bio-Rad CFX Manager software. Gene expression levels were normalized to zebrafish gapdh (ΔCT) and compared to the noninfected controls (ΔΔCT). Fold induction (2ΔΔCT) is represented.
TABLE 1.
qPCR primer information
| Gene | Sequencea (5′–3′) | Reference or source |
|---|---|---|
| gapdh | Fw, TGGGCCCATGAAAGGAAT | 39 |
| Rv, ACCAGCGTCAAAGATGGATG | ||
| IL-6 | Fw, GGACGTGAAGACACTCAGAGACG | This study |
| Rv, AAGGTTTGAGGAGAGGAGTGCTG | ||
| IL-8 | Fw, TGCATTGAAACAGAAAGCCGACG | This study |
| Rv, ATCTCCTGTCCAGTTGTCATCAAGG | ||
| IL-17a | Fw, CAATCTGAGGACGGAAAGGG | This study |
| Rv, ACTGGGCTTCAAAGATGACC | ||
| IFN-γ | Fw, TGGGCGATCAAGGAAAACGA | This study |
| Rv, TTGATGCTTTAGCCTGCCGT | ||
| TNF-α | Fw, CGCATTTCACAAGCGAATTT | 39 |
| Rv, CTGGTCCTGGTCATCTCTCC |
Fw, forward; Rv, reverse.
CFU assessments.
For CFU quantification, 3 to 4 randomly selected infected larvae were pooled and homogenized at 0 hpi and 48 hpi in 500 to 600 μl of 1× PBS. For plating, 50 μl of homogenate from groups was plated on both YPD agar supplemented with 250 μg/ml penicillin-streptomycin (Lonza), 30 μg/ml gentamicin sulfate (BioWhittaker, Lonza), and 3 μg/ml vancomycin hydrochloride (Amresco, Solon, OH) and on Pseudomonas isolation agar (Sigma-Aldrich) supplemented with 750 μg/ml ampicillin for C. albicans and P. aeruginosa selection, respectively. To achieve a countable number of colonies, homogenate (neat) and 1:2 (homogenate:1× PBS) dilutions were plated at 0 hpi, while 1:5 (homogenate:1× PBS) and 1:50 (homogenate:1× PBS) dilutions were plated for the 48-hpi time point. Plates were incubated overnight at 37°C, colonies were counted the following day, and CFU/fish was calculated.
Statistical and ImageJ analyses.
To calculate the percentage of C. albicans and P. aeruginosa organisms covering the swimbladder, confocal images acquired at 24 and 48 hpi were processed by manually outlining the swimbladder area and calculating the percent covered by C. albicans or P. aeruginosa as a measure of microbial fluorescence in ImageJ.
Statistical analyses were conducted using GraphPad Prism 6 software (GraphPad Software, Inc., La Jolla, CA). All significant differences are indicated in the figures, with *, **, ***, and **** indicating P values of <0.05, <0.01, <0.001, and <0.0001, respectively. Bonferroni correction was used to assess significant differences in Kaplan-Meier survival curves. All statistical results are available in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge John Singer for providing the BW19851(p67T1) strain, Judy Berman for providing the HWP1-EGFP strain, Megan Lenardon and Al Brown for providing the CIp30 plasmid, Remi Gratacap for technical advice, Mark Nilan for superior fish care, and members of the Wheeler laboratory for technical assistance and stimulating discussions.
Footnotes
This article is Maine Agricultural and Forest Experiment Station publication number 3556.
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00475-17.
REFERENCES
- 1.Peleg AY, Hogan DA, Mylonakis E. 2010. Medically important bacterial-fungal interactions. Nat Rev Microbiol 8:340–349. doi: 10.1038/nrmicro2313. [DOI] [PubMed] [Google Scholar]
- 2.Leclair LW, Hogan DA. 2010. Mixed bacterial-fungal infections in the CF respiratory tract. Med Mycol 48:S125–S132. doi: 10.3109/13693786.2010.521522. [DOI] [PubMed] [Google Scholar]
- 3.Schwarz C, Hartl D, Eickmeier O, Hector A, Benden C, Durieu I, Sole A, Gartner S, Milla CE, Barry PJ. 31 July 2017. Progress in definition, prevention and treatment of fungal infections in cystic fibrosis. Mycopathologia doi: 10.1007/s11046-017-0182-0. [DOI] [PubMed] [Google Scholar]
- 4.Kim SH, Clark ST, Surendra A, Copeland JK, Wang PW, Ammar R, Collins C, Tullis DE, Nislow C, Hwang DM, Guttman DS, Cowen LE. 2015. Global analysis of the fungal microbiome in cystic fibrosis patients reveals loss of function of the transcriptional repressor Nrg1 as a mechanism of pathogen adaptation. PLoS Pathog 11:e1005308. doi: 10.1371/journal.ppat.1005308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valenza G, Tappe D, Turnwald D, Frosch M, König C, Hebestreit H, Abele-Horn M. 2008. Prevalence and antimicrobial susceptibility of microorganisms isolated from sputa of patients with cystic fibrosis. J Cystic Fibrosis 7:123–127. doi: 10.1016/j.jcf.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Naglik JR, Challacombe SJ, Hube B. 2003. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev 67:400–428. doi: 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayer FL, Wilson D, Hube B. 2013. Candida albicans pathogenicity mechanisms. Virulence 4:119–128. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moyes DL, Naglik JR. 2011. Mucosal immunity and Candida albicans infection. Clin Dev Immunol 2011:346307. doi: 10.1155/2011/346307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balasubramanian D, Schneper L, Kumari H, Mathee K. 2013. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res 41:1–20. doi: 10.1093/nar/gks1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bomberger JM, MacEachran DP, Coutermarsh BA, Ye S, O'Toole GA, Stanton BA. 2009. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog 5:e1000382. doi: 10.1371/journal.ppat.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rumbaugh KP, Griswold JA, Hamood AN. 2000. The role of quorum sensing in the in vivo virulence of Pseudomonas aeruginosa. Microbes Infect 2:1721–1731. doi: 10.1016/S1286-4579(00)01327-7. [DOI] [PubMed] [Google Scholar]
- 12.Schuster M, Lostroh CP, Ogi T, Greenberg EP. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol 185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mear JB, Kipnis E, Faure E, Dessein R, Schurtz G, Faure K, Guery B. 2013. Candida albicans and Pseudomonas aeruginosa interactions: more than an opportunistic criminal association? Med Mal Infect 43:146–151. doi: 10.1016/j.medmal.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Peleg AY, Tampakakis E, Fuchs BB, Eliopoulos GM, Moellering RC, Mylonakis E. 2008. Prokaryote–eukaryote interactions identified by using Caenorhabditis elegans. Proc Natl Acad Sci U S A 105:14585–14590. doi: 10.1073/pnas.0805048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall RA, Turner KJ, Chaloupka J, Cottier F, De Sordi L, Sanglard D, Levin LR, Buck J, Muhlschlegel FA. 2011. The quorum-sensing molecules farnesol/homoserine lactone and dodecanol operate via distinct modes of action in Candida albicans. Eukaryot Cell 10:1034–1042. doi: 10.1128/EC.05060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogan DA, Vik A, Kolter R. 2004. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol 54:1212–1223. doi: 10.1111/j.1365-2958.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- 17.Morales DK, Grahl N, Okegbe C, Dietrich LEP, Jacobs NJ, Hogan DA. 2013. Control of Candida albicans metabolism and biofilm formation by Pseudomonas aeruginosa phenazines. mBio 4:e00526-12. doi: 10.1128/mBio.00526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibson J, Sood A, Hogan DA. 2009. Pseudomonas aeruginosa-Candida albicans interactions: localization and fungal toxicity of a phenazine derivative. Appl Environ Microbiol 75:504–513. doi: 10.1128/AEM.01037-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cugini C, Morales DK, Hogan DA. 2010. Candida albicans-produced farnesol stimulates Pseudomonas quinolone signal production in LasR-defective Pseudomonas aeruginosa strains. Microbiology 156:3096–3107. doi: 10.1099/mic.0.037911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purschke FG, Hiller E, Trick I, Rupp S. 2012. Flexible survival strategies of Pseudomonas aeruginosa in biofilms result in increased fitness compared with Candida albicans. Mol Cell Proteomics 11:1652–1669. doi: 10.1074/mcp.M112.017673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dongari-Bagtzoglou A, Kashleva H, Dwivedi P, Diaz P, Vasilakos J. 2009. Characterization of mucosal Candida albicans biofilms. PLoS One 4:e7967. doi: 10.1371/journal.pone.0007967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nash EE, Peters BM, Fidel PL, Noverr MC. 2015. Morphology-independent virulence of candida species during polymicrobial intra-abdominal infections with Staphylococcus aureus. Infect Immun 84:90–98. doi: 10.1128/IAI.01059-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nash EE, Peters BM, Palmer GE, Fidel PL, Noverr MC. 2014. Morphogenesis is not required for Candida albicans-Staphylococcus aureus intra-abdominal infection-mediated dissemination and lethal sepsis. Infect Immun 82:3426–3435. doi: 10.1128/IAI.01746-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pendleton KM, Huffnagle GB, Dickson RP. 2017. The significance of Candida in the human respiratory tract: our evolving understanding. Pathog Dis 75:ftx029. doi: 10.1093/femspd/ftx029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlecht LM, Peters BM, Krom BP, Freiberg JA, Hänsch GM, Filler SG, Jabra-Rizk MA, Shirtliff ME. 2015. Systemic Staphylococcus aureus infection mediated by Candida albicans hyphal invasion of mucosal tissue. Microbiology (Reading, Engl) 161:168–181. doi: 10.1099/mic.0.083485-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu H, Sobue T, Thompson A, Xie Z, Poon K, Ricker A, Cervantes J, Diaz PI, Dongari-Bagtzoglou A. 2014. Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell Microbiol 16:214–231. doi: 10.1111/cmi.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neely AN, Law EJ, Holder IA. 1986. Increased susceptibility to lethal Candida infections in burned mice preinfected with Pseudomonas aeruginosa or pretreated with proteolytic enzymes. Infect Immun 52:200–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez-Medina E, Fan D, Coughlin LA, Ho EX, Lamont IL, Reimmann C, Hooper LV, Koh AY. 2015. Candida albicans inhibits Pseudomonas aeruginosa virulence through suppression of pyochelin and pyoverdine biosynthesis. PLoS Pathog 11:e1005129. doi: 10.1371/journal.ppat.1005129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allard JB, Rinaldi L, Wargo M, Allen G, Akira S, Uematsu S, Poynter ME, Hogan DA, Rincon M, Whittaker LA. 2009. Th2 allergic immune response to inhaled fungal antigens is modulated by TLR-4-independent bacterial products. Eur J Immunol 39:776–788. doi: 10.1002/eji.200838932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moldoveanu B, Otmishi P, Jani P, Walker J, Sarmiento X, Guardiola J, Saad M, Yu J. 2009. Inflammatory mechanisms in the lung. J Inflamm Res 2:1–11. [PMC free article] [PubMed] [Google Scholar]
- 31.Nayar G, Gauna A, Chukkapalli S, Velsko I, Kesavalu L, Cha S. 2016. Polymicrobial infection alter inflammatory microRNA in rat salivary glands during periodontal disease. Anaerobe 38:70–75. doi: 10.1016/j.anaerobe.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roux D, Gaudry S, Dreyfuss D, El-Benna J, de Prost N, Denamur E, Saumon G, Ricard J-D. 2009. Candida albicans impairs macrophage function and facilitates Pseudomonas aeruginosa pneumonia in rat. Crit Care Med 37:1062–1067. doi: 10.1097/CCM.0b013e31819629d2. [DOI] [PubMed] [Google Scholar]
- 33.Mizgerd JP. 2008. Acute lower respiratory tract infection. N Engl J Med 358:716–727. doi: 10.1056/NEJMra074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chekabab SM, Silverman RJ, Lafayette SL, Luo Y, Rousseau S, Nguyen D. 2015. Staphylococcus aureus inhibits IL-8 responses induced by Pseudomonas aeruginosa in airway epithelial cells. PLoS One 10:e0137753. doi: 10.1371/journal.pone.0137753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Condotta SA, Khan SH, Rai D, Griffith TS, Badovinac VP. 2015. Poly-microbial sepsis increases susceptibility to chronic viral infection and exacerbates CD8+ T cell exhaustion. J Immunol (Baltimore, Md) 195:116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freitas A, Alves-Filho JC, Victoni T, Secher T, Lemos HP, Sônego F, Cunha FQ, Ryffel B. 2009. IL-17 receptor signaling is required to control polymicrobial sepsis. J Immunol (Baltimore, Md) 182:7846–7854. [DOI] [PubMed] [Google Scholar]
- 37.Nahid MA, Rivera M, Lucas A, Chan EKL, Kesavalu L. 2011. Polymicrobial infection with periodontal pathogens specifically enhances microRNA miR-146a in ApoE-/- mice during experimental periodontal disease. Infect Immun 79:1597–1605. doi: 10.1128/IAI.01062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gratacap RL, Bergeron AC, Wheeler RT. 2014. Modeling mucosal candidiasis in larval zebrafish by swimbladder injection. J Vis Exp 2014. doi: 10.3791/52182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gratacap RL, Rawls JF, Wheeler RT. 2013. Mucosal candidiasis elicits NF-κB activation, proinflammatory gene expression and localized neutrophilia in zebrafish. Dis Models Mech 6:1260–1270. doi: 10.1242/dmm.012039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gratacap RL, Scherer AK, Seman BG, Wheeler RT. 12 June 2017. Control of mucosal candidiasis in the zebrafish swimbladder depends on neutrophils that block filament invasion and drive extracellular trap production. Infect Immun doi: 10.1128/IAI.00276-17. [DOI] [PMC free article] [PubMed]
- 41.Zheng W, Wang Z, Collins JE, Andrews RM, Stemple D, Gong Z. 2011. Comparative transcriptome analyses indicate molecular homology of zebrafish swimbladder and mammalian lung. PLoS One 6:e24019. doi: 10.1371/journal.pone.0024019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cass AN, Servetnick MD, McCune AR. 2013. Expression of a lung developmental cassette in the adult and developing zebrafish swimbladder. Evol Dev 15:119–132. doi: 10.1111/ede.12022. [DOI] [PubMed] [Google Scholar]
- 43.Robertson GN, Croll RP, Smith FM. 2014. The structure of the caudal wall of the zebrafish (Danio rerio) swim bladder: evidence of localized lamellar body secretion and a proximate neural plexus. J Morphol 275:933–948. doi: 10.1002/jmor.20274. [DOI] [PubMed] [Google Scholar]
- 44.Winata CL, Korzh S, Kondrychyn I, Zheng W, Korzh V, Gong Z. 2009. Development of zebrafish swimbladder: the requirement of Hedgehog signaling in specification and organization of the three tissue layers. Dev Biol 331:222–236. doi: 10.1016/j.ydbio.2009.04.035. [DOI] [PubMed] [Google Scholar]
- 45.Hammond ME, Lapointe GR, Feucht PH, Hilt S, Gallegos CA, Gordon CA, Giedlin MA, Mullenbach G, Tekamp-Olson P. 1995. IL-8 induces neutrophil chemotaxis predominantly via type I IL-8 receptors. J Immunol (Baltimore, Md) 155:1428–1433. [PubMed] [Google Scholar]
- 46.Spittler A, Razenberger M, Kupper H, Kaul M, Hackl W, Boltz-Nitulescu G, Függer R, Roth E. 2000. Relationship between interleukin-6 plasma concentration in patients with sepsis, monocyte phenotype, monocyte phagocytic properties, and cytokine production. Clin Infect Dis 31:1338–1342. doi: 10.1086/317499. [DOI] [PubMed] [Google Scholar]
- 47.Bellingan GJ. 2002. The pulmonary physician in critical care. 6. The pathogenesis of ALI/ARDS. Thorax 57:540–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Liu H, Yao J, Huang Y, Qin S, Sun Z, Xu Y, Wan S, Cheng H, Li C, Zhang X, Ke Y. 2016. Manipulating the air-filled zebrafish swim bladder as a neutrophilic inflammation model for acute lung injury. Cell Death Dis 7:e2470. doi: 10.1038/cddis.2016.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brannon MK, Davis JM, Mathias JR, Hall CJ, Emerson JC, Crosier PS, Huttenlocher A, Ramakrishnan L, Moskowitz SM. 2009. Pseudomonas aeruginosa type III secretion system interacts with phagocytes to modulate systemic infection of zebrafish embryos. Cell Microbiol 11:755–768. doi: 10.1111/j.1462-5822.2009.01288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clay H, Davis JM, Beery D, Huttenlocher A, Lyons SE, Ramakrishnan L. 2007. Dichotomous role of the macrophage in early Mycobacterium marinum infection of the zebrafish. Cell Host Microbe 2:29–39. doi: 10.1016/j.chom.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Torraca V, Masud S, Spaink HP, Meijer AH. 2014. Macrophage-pathogen interactions in infectious diseases: new therapeutic insights from the zebrafish host model. Dis Model Mech 7:785–797. doi: 10.1242/dmm.015594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bendel CM, Hess DJ, Garni RM, Henry-Stanley M, Wells CL. 2003. Comparative virulence of Candida albicans yeast and filamentous forms in orally and intravenously inoculated mice. Crit Care Med 31:501–507. doi: 10.1097/01.CCM.0000049954.48239.A1. [DOI] [PubMed] [Google Scholar]
- 53.Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. 2003. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell 2:1053–1060. doi: 10.1128/EC.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davis-Hanna A, Piispanen AE, Stateva LI, Hogan DA. 2008. Farnesol and dodecanol effects on the Candida albicans Ras1-cAMP signalling pathway and the regulation of morphogenesis. Mol Microbiol 67:47–62. doi: 10.1111/j.1365-2958.2007.06013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kerr JR, Taylor GW, Rutman A, Høiby N, Cole PJ, Wilson R. 1999. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J Clin Pathol 52:385–387. doi: 10.1136/jcp.52.5.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bertolini MM, Xu H, Sobue T, Nobile CJ, Del Bel Cury AA, Dongari-Bagtzoglou A. 2015. Candida-streptococcal mucosal biofilms display distinct structural and virulence characteristics depending on growth conditions and hyphal morphotypes. Mol Oral Microbiol 30:307–322. doi: 10.1111/omi.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diaz PI, Xie Z, Sobue T, Thompson A, Biyikoglu B, Ricker A, Ikonomou L, Dongari-Bagtzoglou A. 2012. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect Immun 80:620–632. doi: 10.1128/IAI.05896-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Villar CC, Kashleva H, Mitchell AP, Dongari-Bagtzoglou A. 2005. Invasive phenotype of Candida albicans affects the host proinflammatory response to infection. Infect Immun 73:4588–4595. doi: 10.1128/IAI.73.8.4588-4595.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cavalcanti YW, Morse DJ, Silva WJd Del-Bel-Cury AA, Wei X, Wilson M, Milward P, Lewis M, Bradshaw D, Williams DW. 2015. Virulence and pathogenicity of Candida albicans is enhanced in biofilms containing oral bacteria. Biofouling 31:27–38. doi: 10.1080/08927014.2014.996143. [DOI] [PubMed] [Google Scholar]
- 60.Falsetta ML, Klein MI, Colonne PM, Scott-Anne K, Gregoire S, Pai C-H, Gonzalez-Begne M, Watson G, Krysan DJ, Bowen WH, Koo H. 2014. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect Immun 82:1968–1981. doi: 10.1128/IAI.00087-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peters BM, Noverr MC. 2013. Candida albicans-Staphylococcus aureus polymicrobial peritonitis modulates host innate immunity. Infect Immun 81:2178–2189. doi: 10.1128/IAI.00265-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson JE, Gonzales RA, Olson SJ, Wright PF, Graham BS. 2007. The histopathology of fatal untreated human respiratory syncytial virus infection. Modern Pathol 20:108–119. doi: 10.1038/modpathol.3800725. [DOI] [PubMed] [Google Scholar]
- 63.Lange M, Cox RA, Traber DL, Hamahata A, Nakano Y, Traber LD, Enkhbaatar P. 2013. Time course of early histopathological lung changes in an ovine model of acute lung injury and pulmonary infection. Exp Lung Res 39:201–206. doi: 10.3109/01902148.2013.794254. [DOI] [PubMed] [Google Scholar]
- 64.Murakami K, Bjertnaes LJ, Schmalstieg FC, McGuire R, Cox RA, Hawkins HK, Herndon DN, Traber LD, Traber DL. 2002. A novel animal model of sepsis after acute lung injury in sheep. Crit Care Med 30:2083–2090. doi: 10.1097/00003246-200209000-00022. [DOI] [PubMed] [Google Scholar]
- 65.Nitta K, Kobayashi T. 1994. Impairment of surfactant activity and ventilation by proteins in lung edema fluid. Resp Physiol 95:43–51. doi: 10.1016/0034-5687(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 66.Juan C, Pena C, Oliver A. 2017. Host and Pathogen Biomarkers for Severe Pseudomonas aeruginosa Infections. J Infect Dis 215:S44–S51. doi: 10.1093/infdis/jiw299. [DOI] [PubMed] [Google Scholar]
- 67.Lorenz A, Pawar V, Haussler S, Weiss S. 2016. Insights into host-pathogen interactions from state-of-the-art animal models of respiratory Pseudomonas aeruginosa infections. FEBS Lett 590:3941–3959. doi: 10.1002/1873-3468.12454. [DOI] [PubMed] [Google Scholar]
- 68.D'Argenio DA, Wu M, Hoffman LR, Kulasekara HD, Déziel E, Smith EE, Nguyen H, Ernst RK, Larson Freeman TJ, Spencer DH, Brittnacher M, Hayden HS, Selgrade S, Klausen M, Goodlett DR, Burns JL, Ramsey BW, Miller SI. 2007. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol 64:512–533. doi: 10.1111/j.1365-2958.2007.05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hammond JH, Hebert WP, Naimie A, Ray K, Van Gelder RD, DiGiandomenico A, Lalitha P, Srinivasan M, Acharya NR, Lietman T, Hogan DA, Zegans ME. 2016. Environmentally endemic Pseudomonas aeruginosa strains with mutations in lasR are associated with increased disease severity in corneal ulcers. mSphere 1:e00140-16. doi: 10.1128/mSphere.00140-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hoffman LR, Kulasekara HD, Emerson J, Houston LS, Burns JL, Ramsey BW, Miller SI. 2009. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J Cystic Fibrosis 8:66–70. doi: 10.1016/j.jcf.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hoffman LR, Richardson AR, Houston LS, Kulasekara HD, Martens-Habbena W, Klausen M, Burns JL, Stahl DA, Hassett DJ, Fang FC, Miller SI. 2010. Nutrient availability as a mechanism for selection of antibiotic tolerant Pseudomonas aeruginosa within the CF airway. PLoS Pathog 6:e1000712. doi: 10.1371/journal.ppat.1000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.LaFayette SL, Houle D, Beaudoin T, Wojewodka G, Radzioch D, Hoffman LR, Burns JL, Dandekar AA, Smalley NE, Chandler JR, Zlosnik JE, Speert DP, Bernier J, Matouk E, Brochiero E, Rousseau S, Nguyen D. 2015. Cystic fibrosis-adapted Pseudomonas aeruginosa quorum sensing lasR mutants cause hyperinflammatory responses. Sci Adv 1:e1500199. doi: 10.1126/sciadv.1500199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Renshaw SA, Loynes CA, Trushell DMI, Elworthy S, Ingham PW, Whyte MKB. 2006. A transgenic zebrafish model of neutrophilic inflammation. Blood 108:3976–3978. doi: 10.1182/blood-2006-05-024075. [DOI] [PubMed] [Google Scholar]
- 74.Fonzi WA, Irwin MY. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shcherbakova DM, Verkhusha VV. 2013. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat Methods 10:751–754. doi: 10.1038/nmeth.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gietz RD, Schiestl RH, Willems AR, Woods RA. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 77.Gerami-Nejad M, Dulmage K, Berman J. 2009. Additional cassettes for epitope and fluorescent fusion proteins in Candida albicans. Yeast 26:399–406. doi: 10.1002/yea.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dennison PM, Ramsdale M, Manson CL, Brown AJ. 2005. Gene disruption in Candida albicans using a synthetic, codon-optimised Cre-loxP system. Fungal Genet Biol 42:737–748. doi: 10.1016/j.fgb.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 79.Murad AM, Lee PR, Broadbent ID, Barelle CJ, Brown AJ. 2000. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16:325–327. [DOI] [PubMed] [Google Scholar]
- 80.Singer JT, Phennicie RT, Sullivan MJ, Porter LA, Shaffer VJ, Kim CH. 2010. Broad-host-range plasmids for red fluorescent protein labeling of Gram-negative bacteria for use in the zebrafish model system. Appl Environ Microbiol 76:3467–3474. doi: 10.1128/AEM.01679-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brothers KM, Newman ZR, Wheeler RT. 2011. Live imaging of disseminated candidiasis in zebrafish reveals role of phagocyte oxidase in limiting filamentous growth. Eukaryot Cell 10:932–944. doi: 10.1128/EC.05005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.